Abstract
The med-1 and med-2 genes encode a pair of essentially identical GATA factor-related transcription factors that have been proposed to be necessary for specification of the C. elegans endoderm (intestine or E lineage) as well as part of the C. elegans mesoderm. med-1 and med-2 are proposed to be the direct downstream targets and the principal effectors of the maternally provided SKN-1 transcription factor; med-1 and med-2 would thus occupy the pivotal interface between maternal and zygotic control of gene expression. The conclusion that med-1 and med-2 are necessary for C. elegans endoderm specification was based on a partially penetrant (∼50%) loss of endoderm markers produced by RNA-mediated interference (RNAi). To determine whether this partial penetrance reflects: (i) inefficient RNAi against early zygotic transcripts, (ii) experimental uncertainty in the expected level of endoderm loss in skn-1 nulls, or (iii) additional redundancy in the pathway of endoderm specification, we constructed worm strains that segregate embryos lacking both the med-1 gene (because of a gene-specific deletion) and the med-2 gene (using either of two chromosomal deficiencies). Contrary to expectations, we observe that only ∼3–20% of med-2(−); med-1(−) embryos do not express markers of endoderm differentiation. Furthermore, we found no evidence for a maternal contribution of the med genes to endoderm specification. We conclude that the major pathway(s) for endoderm specification in C. elegans must be independent of the med-1 and med-2 genes.
THE Caenorhabditis elegans endoderm (intestine or E lineage) is clonally derived from a single cell, the E cell, in the eight-cell embryo (Sulston et al. 1983). The early endoderm is one of the few C. elegans lineages for which a plausible specification pathway has been proposed in molecular detail, beginning with maternally provided transcription factors, progressing through several waves of zygotically produced transcription factors, and ending with gene products that function in the terminally differentiated intestine (see review by Maduro and Rothman 2002; see also Baugh et al. 2003, 2005; Robertson et al. 2004).
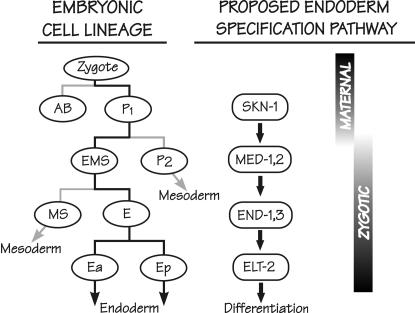
Figure 1 summarizes the regulatory cascade proposed for specification of the C. elegans endoderm. The maternally provided b-ZIP-like transcription factor SKN-1 is essential for correct specification of the fate of the EMS blastomere of the four-cell embryo (Bowerman et al. 1992, 1993). Within the EMS cell, SKN-1 is proposed to directly activate the zygotic expression of two genes called med-1 and med-2, which encode zinc-finger proteins related to GATA-type transcription factors but with atypical binding sites (Maduro et al. 2001; Maduro and Rothman 2002; Broitman-Maduro et al. 2005). These two small intronless genes are 98% identical and, for convenience, are often referred to simply as the med genes (mesendoderm-determining); they are the principal subjects of this article. The med genes are proposed to specify both the C. elegans endoderm and that portion of the C. elegans mesoderm deriving from the MS blastomere (Figure 1). To specify the endoderm, the MED-1 and MED-2 factors are proposed to directly activate the zygotic expression of a redundant pair of genes called end-1 and end-3, which also encode GATA-type factors (Zhu et al. 1997, 1998; Maduro et al. 2001; Maduro and Rothman 2002; Broitman-Maduro et al. 2005). This endoderm specification step takes place in the E cell, the clonal progenitor of the intestine, within a permissive environment associated with lowered nuclear levels of the HMG protein POP-1 (Lin et al. 1995, 1998; Rocheleau et al. 1997; Thorpe et al. 1997; Lo et al. 2004). The END-1/END-3 pair of GATA factors is proposed to directly activate expression of the elt-2 gene, which encodes a GATA factor that may be the principal transcription factor directing subsequent intestinal differentiation (Hawkins and McGhee 1995; Fukushige et al. 1998, 2005).
Figure 1.
Cell lineage of the early C. elegans embryo (left), aligned with the proposed transcription factor cascade that leads to specification of the C. elegans endoderm (right). Lineages that lead to the intestine are solid; other lineages are shaded. Only transcription factors that are on the proposed endoderm specification pathway are shown; in particular, roles for SKN-1 and MED-1,2 in specification of the MS lineage are not shown. The proposed activation by SKN-1 of the med-1 and med-2 genes marks the transition from maternal to zygotic control of gene expression. This figure was redrawn from Figure 4 of Maduro and Rothman (2002).
The properties of the med genes have generated substantial interest for at least two reasons: (i) they are proposed to occupy the important interface between maternal and zygotic control of gene expression (Figure 1), and (ii) their proposed involvement in specifying both MS mesoderm and E endoderm has been used as evidence for an ancient “mesendoderm” region of the embryo, specified by a transcription factor network conserved in all bilateral metazoons (Maduro et al. 2001; Rodaway and Patient 2001; Maduro and Rothman 2002; Broitman-Maduro et al. 2005). Maduro et al. (2001, p. 481) have proposed that “the meds are activated by, and function downstream of, SKN-1 in the EMS lineage and are essential to specify E and MS fates in any context.” In this article, we test only part of this assertion, that is, whether the med genes are indeed necessary to specify the C. elegans endoderm. To be clear about the expectations of our experiments, it is important to realize that loss of endoderm caused by loss of maternal skn-1 function is not fully penetrant (Bowerman et al. 1992). Furthermore, the penetrance of this endoderm loss is temperature dependent, such that at 15° and 25°, respectively, 54–81% of embryos produced by skn-1(zu67) (strong loss-of-function allele) mothers do not produce endoderm (Bowerman et al. 1992). Maduro et al. (2001) performed RNA-mediated interference (RNAi) to target the med genes and found that, among the fraction of embryos that arrested and that were collected from 7 to 9 hr following injection, 52% (75/143) failed to express endoderm markers; this proportion is lower than but possibly within experimental error of the 60–80% of nonexpressing embryos expected to be produced by a skn-1 null under their experimental conditions. The necessity of the med genes for endoderm specification can now be tested, without relying on RNAi, by combining a recently available gene-specific deletion of the med-1 gene (see below) with a chromosomal deficiency that removes the med-2 gene. As described in this study, the unexpected result is that only a minority (3–20%), not the expected majority, of embryos lacking both the med-1 and the med-2 gene do not express markers diagnostic for endoderm, suggesting that there must be a signficant pathway for endoderm specification that does not require the med-1 and med-2 genes.
MATERIALS AND METHODS
Strains and alleles:
C. elegans was maintained and manipulated by standard procedures (Brenner 1974; Wood 1988). The strain RB930 containing the med-1(ok804) allele was obtained from the C. elegans Genome Knockout Consortium and outcrossed three times to wild type, following the med-1(ok804) allele by PCR (see below) and choosing a strain of high viability; for convenience, we retain the original strain designation RB930 [med-1(ok804)]. The deletion was validated by both Southern blotting and PCR. Southern blotting was by standard methods (Sambrook and Russell 2001), using a 1-kb probe amplified and cloned from the med-2(+) gene (see Figure 2B); final washing was at 55° and Na+ concentration of 0.2 m to allow for simultaneous detection of both the med-2 gene and the highly similar med-1 gene. The hybridization pattern was consistent with the complete removal of the med-1 sequence, as depicted in Figure 2B; specifically, there was no evidence of an “extra” band, suggesting an unsuspected duplication of the med-1 gene elsewhere in the genome (that could possibly have occurred during the mutagenesis producing the ok804 deletion). The absence of an extra med-1 copy elsewhere in the genome was also verified by PCR using several different sets of primers. One particular PCR search for an extra med-1 sequence used two primers (oJM306, TCTTTGACAATACCCAGCAAC, and oJM307, CACCATTCGTTTCCTGTACC, which anneal near the 5′- and 3′-ends, respectively, of the coding regions of both med-1 and med-2) to amplify a 474-bp band, using template DNA purified from either wild-type N2 worms or strain RB930 [med-1(ok804)]. The two amplified fragments were gel purified and sequenced on both strands. Sequence traces obtained with the fragment amplified from wild-type DNA clearly showed the multiple sequence ambiguities expected for a mixture of med-1 and med-2 DNA (Maduro et al. 2001); in contrast, sequence traces obtained with the fragment amplified from RB930 DNA showed only a single sequence, corresponding to med-2(+) (data not shown). The most definitive (but still negative) PCR search for an extra copy of med-1 followed this same strategy but used two primers that hybridize within the DNA-binding domain of both med-1 and med-2 (oJM308, TGAAACAATTCGTTGGAGGA; oJM309, TGTTAACGGCAGTG-ACTGGA). The expected med-1/med-2 polymorphism within the zinc-finger sequence (Maduro et al. 2001) could be detected in the sequence traces when N2 DNA was used as template but only the med-2 sequence could be detected when RB930 [med-1(ok804)] DNA was used as template. We conclude that any extra copy of the med-1 gene can neither be intact nor have a DNA-binding domain.
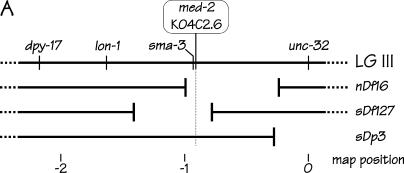
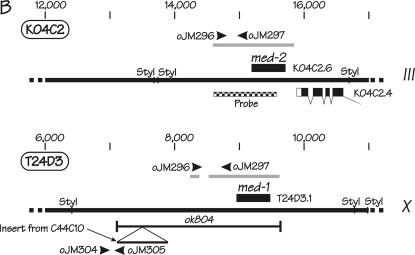
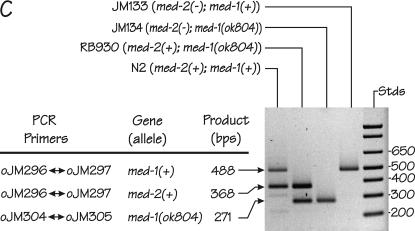
Figure 2.
Genetic positions and molecular characteristics of the med-1 and med-2 genes. (A) The genetic map of the middle of the C. elegans chromosome III. The med-2 gene is located on cosmid K04C2, which is removed by the chromosomal deficiencies sDf127 and nDf16. As described in the text, the free duplication sDp3 is used to balance the deficiency sDf127; a chromosome marked with lon-1 unc-32 is used to balance the deficiency nDf16. (B) Expanded views of the chromosomal regions surrounding the med-2 gene (top) and the med-1 gene (bottom). Coordinates correspond to the cosmid sequences K04C2 and T24D3 for med-2 and med-1, respectively. The intronless ORFs corresponding to med-1 and med-2 are indicated by the solid boxes. The shaded bars above the med-2 and med-1 ORFs indicate the chromosomal regions that show high sequence similarity between the two genes (see Maduro et al. 2001 for details). Because of this region of high sequence similarity, the presence of med-1 and med-2 genes can be assayed simultaneously and independently by the same set of primers, oJM296 and oJM297. The extent of the med-1 deletion allele ok804 is indicated. We note that an alternative structure of the med-1 gene (containing a 3′-intron) has previously appeared in the C. elegans sequence annotation: the ok804 allele removes the predicted med-1 zinc-finger DNA-binding domain and would be predicted to produce a genetic null for either gene model. The ok804 deletion is associated with a 790-bp insertion of a (non-ORF-containing) sequence from cosmid C44C10, allowing the deletion to be detected by PCR primers oJM304 and oJM305, as indicated. The cross-hatched region beneath the med-2 gene was amplified by PCR, cloned, and used as a probe to detect both med-2 and med-1 sequences by Southern blotting on genomic DNA digested with the restriction enzyme StyI. (C) PCR detection of med-1(+), med-1(ok804), and med-2(+) alleles in individual arrested embryos produced by strains N2 (wild type), RB930 [med-1(ok804)], JM134 med-1(−), and JM133 med-1(+). The deduced status of the med genes in the individual embryos are indicated beside the strain names. Locations of PCR primers within the med-1 and med-2 genes are shown in B.
To produce a strain in which the med-2 gene was removed by the chromosomal deficiency sDf127, RB930 [med-1(ok804)] males were crossed to hermaphrodites of strain BC4638 [dpy-17(e164) sDf127(s2428) unc-32(e189) III; sDp3 (III,f)] and Unc Non-Dpy F2 hermaphrodites that segregated dead eggs were selected. The status of the med-1 gene in populations segregating from these hermaphrodites was determined by PCR to produce the two final strains: JM133 [dpy-17 sDf127 unc-32 III; sDp3 (III,f); med-1(+) X] and JM134 [dpy-17 sDf127 unc-32 III; sDp3 (III,f); med-1(ok804) X]. The presence of the sDf127 deficiency was confirmed by crossing JM133 and JM134 hermaphrodites to sma-3(e491)/+ males and observing Sma males in the progeny; the sma-3 gene is 20–25 kb upstream of med-2. The strains were also validated by PCR, as described in the text.
To produce a strain in which the med-2 gene was removed by the chromosomal deficiency nDf16, RB930 [med-1(ok804)] males were crossed to hermaphrodites from strain CB1918 [lon-1(e185) unc-32(e189) III]; wild-type F1 hermaphrodites were picked and allowed to self and a progeny strain that was homozygous lon-1 unc-32 and also homozygous med-1(ok804) was identified by PCR. Hermaphrodites from this intermediate strain were crossed to N2 males, F1 males were crossed to hermaphrodites from strain CX2914 [nDf16/dpy-17(e164) unc-32(e189) III] and a wild-type F1 hermaphrodite [nDf16/lon-1 unc-32 III; med-1(ok804)/+ X] that segregates dead eggs, Lon Unc worms, and (slow-growing) wild types was identified. Individual populations derived from single hermaphrodites were then screened by PCR to identify two strains: JM135 [nDf16/lon-1 unc-32 III; med-1(+) X] and JM136 [nDf16/lon-1 unc-32 III; med-1(ok804) X]. The presence of nDf16 was verified by crossing JM135 and JM136 hermaphrodites to sma-3(e491)/+ males and observing Sma males in the F1 progeny.
Individual arrested embryos were picked (from plates incubated in parallel to the slides used for the birefringence assays, using a fresh capillary for each embryo) and digested with proteinase K, essentially as described (Williams et al. 1992). The following primers (see Figure 2, B and C) were used to detect med-1 and med-2 alleles in individual arrested embryos: oJM296, AGGTATGAAGCAGGCGTAGGC; oJM297, ACAGAGGTGCAAGGTGGTCC; oJM304, GTTTCATCACTTTTTGCTGTGG; and oJM305, CAAAATAGGCTTGCTTTTACGG. Amplification conditions were as follows: initial denaturation at 94° for 2 min, followed by 30 cycles of 94° for 30 sec, 62° for 30 sec, and 72° for 30 sec, ending with a final extension at 72° for 7 min. Identities of the amplified fragments were verified by restriction enzyme digestions (data not shown). Hermaphrodite mothers removed from the assay slides were frequently allowed to produce a further set of embryos, which were then genotyped by PCR to verify the hermaphrodite genotype and to ensure that deficiency chromosomes remained balanced.
Double-stranded RNAi:
Double-stranded RNAi corresponding to the genomic med-1, glp-1, skn-1, and let-413 loci (Kamath et al. 2003) as well as to the coding sequence of green fluorescent protein (GFP) were produced, purified, and dissolved at a concentration of 1 mg/ml, as previously described (Fukushige et al. 2005). Double-stranded RNA was also produced corresponding to the med-1 coding region (using as template plasmid pMM239 and primer sequences T7MED1A, taatacgactcactatagggaggCCTACCCTTACCCCG, and T7MED1C, taatacgactcactatagggaggAAATCTTGAGTTATGAT, kindly provided by M. Maduro; lowercase letters correspond to the added sequence of the T7 promoter) and, as an additional control, to a portion of the coding sequence of lacZ (using as template plasmid pPD16.43 and primer sequences oJM333, taatacgactcactatagggaggTAATCACGACGCGCTGTATC, and oJM334, taatacgactcactatagggaggCGGATAAACGGAACTGGAAA); the latter two double-stranded RNAs (dsRNAs) were purified and dissolved at a final concentration of 2 mg/ml.
In our standard protocol, RNA was injected once into a gonad arm and once into the intestine cytoplasm/body cavity of young wild-type hermaphrodites; if worms are judged to be unhealthy or fragile (e.g., the deficiency-containing strains), hermaphrodites were injected only once into the intestine/body cavity. Injected worms were allowed to recover for 8–18 hr and then transferred to the birefringence assay slides (see below) at regular intervals over the next 2–3 days. All incubations were at 20 ± 0.1°. RNA injections were also performed according to a protocol provided by M. Maduro: young adult hermaphrodites (usually zero to five embryos in the uterus) were injected in both gonad arms, such that the (nondehydrated) gonad volume was estimated to increase by 20–50%. At least 80% of the worms were successfully injected in both gonads; the remaining 20% of the worms were injected in at least one gonad arm but when the second gonad injection proved difficult or ambiguous, the intestine/body cavity was also injected. Injected worms were incubated at 20 ± 0.1° for 7 hr and then transferred to the birefringence assay slides (see below) for 2 hr for embryo collection; injected hermaphrodites were often transferred to fresh slides to collect later embryos. To ensure maximal effectiveness of the RNAi, we aimed to produce ∼20% lethality in the injected hermaphrodites. We routinely verified, using gel electrophoresis, that the dsRNA aliquot used for the injection remained completely undegraded; in half of our injection series, excess uninjected dsRNA was recovered from the injection needle and verified to be intact. In all med RNAi experiments, control injections (lacZ or GFP dsRNA) were performed in exact parallel and were also performed “blind”; i.e., the person injecting and scoring the phenotype did not know the identity of the RNA. At the conclusion of the injections, wild-type worms were verified to indeed be RNAi sensitive by injection of skn-1 dsRNA.
Assays for endoderm differentiation:
To assay for gut-granule expression with minimal embryo manipulation, 25–50 gravid hermaphrodites of the various strains were placed on thin (∼1 mm) NGM-agar (Wood 1988) pads poured directly onto microscope slides and lightly seeded with Escherichia coli strain OP50. After 7 ± 2 hr of egg laying, adults were removed and the slides were incubated in a tightly sealed humidified chamber; all incubations were at 20 ± 0.1° for times ranging from 17 to 46 hr (to demonstrate that the results are essentially independent of incubation time and are not influenced by any slow-developing embryos). Following incubation, a small volume (50–100 μl) of egg salts (Edgar and McGhee 1986) was added to the agar pad, which was then overlaid with a 22 × 50-mm coverslip, prior to viewing arrested embryos with birefringence optics. Images were obtained using a Zeiss Axioplan 2i microscope and a Hamamatsu (Bridgewater, NJ) OrcaER digital camera, using identical exposure settings for comparisons of the med-1(−) and med-1(+) strains; images were processed using only the default “best-fit” (γ = 1) setting of the Axiovision software.
The histochemical assay for GES-1 activity was performed essentially as described (Edgar and McGhee 1986); a detailed protocol is available upon request. Immunohistochemical staining of arrested embryos was performed essentially as described (Bossinger et al. 2004), using monoclonal antibody MH33 (1/50–1/100 dilution of a hybridoma supernatant). Labeled secondary antibodies were obtained from Molecular Probes (Eugene, OR) and used at a dilution of 1/500.
RESULTS
Background and expected results:
As explained in the Introduction, loss of maternal skn-1 function produces an impenetrant loss of endoderm, somewhere in the range of 50–80% on the basis of the original description of the skn-1(zu67) strong loss-of-function mutant and depending on the incubation temperature (Bowerman et al. 1992). To be more precise about experimental expectations, we injected dsRNA corresponding to the skn-1 gene into wild-type worms, incubated them under the same experimental conditions used for other endoderm assays performed in this study (20 ± 0.1° with hermaphrodites brooded on seeded microscope slides), and observed that 70% of the arrested embryos do not express gut granules (136/216, 416/630, and 366/472 gut-granule-negative embryos/total embryos counted at the peak of the RNAi effect following three independent injection series with a total of 36 injected hermaphrodites). Thus the expectation is that, if the zygotic med genes are the sole downstream effectors of SKN-1 in the pathway of endoderm specification as shown in Figure 1, ∼70% of med-2(−); med-1(−) embryos should not produce endoderm.
Our attempts to perform RNAi by standard protocols that are effective against both zygotic and maternal genes (Montgomery et al. 1998; Shi and Mello 1998; see materials and methods) failed to detect significant differences among embryos produced by mothers injected with med-1 dsRNA or with control dsRNA corresponding to GFP. We also note that no med-1 or med-2 RNAi effects have been reported by others when the dsRNA is administered by either feeding (Kamath et al. 2003) or injection (Sonnichsen et al. 2005). We also collected embryos in the time window from 7 to 9 hr post-injection, as described by Maduro et al. (2001). From a total of 75 injected hermaphrodites in six independent injection series, an average of 31 ± 20% (all reported uncertainties are standard deviations) embryos produced by med RNA-injected mothers arrested development and, of these arrested embryos, 24 ± 21% did not express gut granules; for comparision, 12 ± 9% of embryos whose mothers had been injected with either dsGFP or dslacZ RNA arrested and, of these, 25 ± 26% were gut-granule negative. Among the arrested embryos in two sets of injections, we observed a subset of partially elongated embryos that resembled the phenotype described by Maduro et al. (2001) and, just as they reported, these partially elongated embryos could be either gut-granule positive or gut-granule negative. However, in our judgment, the most important overall conclusion that should be drawn from the above injections is the small size of the experimental signal: hermaphrodites injected with dsmed RNA produce an average of 0.4 gut-granule-negative embryos per injected hermaphrodite; injections with dsGFP or dslacZ RNA produce an average of 0.2 gut-granule-negative embryos per injected hermaphrodite. We think that this experimental signal is so small that the specificity of such an effect would be difficult to establish unambiguously.
Maduro et al. (2001) also reported that endoderm markers were ablated when med RNAi was performed by heat-shock overexpression of sense and antisense cDNAs from a transgenic array, according to the method of Tavernarakis et al. (2000). This heat-shock-induced med RNAi could provide an alternative method for producing large numbers of med RNAi-arrested embryos, avoiding any limitations of our injection technique. However, when we performed this protocol on N2 control worms (collecting embryos 11–14 hr at 20° after a heat shock of 35° for 4 hr), heat-shocked L4 worms (as in the protocol) were completely sterile; heat-shocked adults produced an average of 0.6 embryo per hermaphrodite, roughly half of which were gut-granule negative because they arrested early in embryogenesis (data not shown). Overall, we believe that it is worthwhile to investigate whether the med genes are necessary for endoderm specification using a technique other than RNAi.
RNAi-independent strategy to test whether zygotic expression of the med-1 and med-2 genes is necessary for endoderm specification:
Figure 2A shows the location of the med-2 gene near the center of chromosome III (Maduro et al. 2001), together with the other genetic markers and chromosomal rearrangements used in this study. The med-1 gene is located on the right arm of the X chromosome (Maduro et al. 2001). The central experiment of this study was to investigate endoderm formation in embryos segregating from a strain homozygous for a gene-specific med-1 knockout allele (see below) and heterozygous for a chromosomal deficiency that removes the med-2 gene. As shown in Figure 2A, med-2 can be removed by either of two overlapping chromosomal deficiencies, sDf127 (Janke et al. 1997) or nDf16 (Thomas et al. 1990), together with ∼100 and ∼300 neighboring genes, respectively, with several dozen genes in addition to the meds being in common. Those embryos lacking both copies of both med-1 and med-2 will arrest development because the chromosomal deficiency removes essential genes in the vicinity of med-2. Nonetheless, such arrested embryos remain alive for days and can easily be assayed for endoderm-specific markers. Qualifications and justifications of the use of chromosomal deletions are discussed in another section.
Gene-specific deletion of med-1:
A med-1 gene-specific deletion (ok804) was provided by the C. elegans Genome Knockout Consortium and, as shown in Figure 2B, removes the entire med-1 coding region. Southern blotting and extensive PCR validated this deletion; in particular, no unsuspected med-1 gene duplication associated with the mutagenesis could be detected elsewhere in the genome (see materials and methods). After outcrossing three times to wild type in an attempt to remove extraneous mutations, the strain RB930 containing the med-1(ok804) deletion appears quite healthy (occasional protruding vulvae) and fertile (brood size was 269 ± 53 with five total broods counted) but harbors a low level of embryonic lethality (percentage hatching was 92.5 ± 3.2). We do not know whether this low level of lethality is caused by med-1(ok804) or by a mutation in an unrelated gene within the strain; in any case, >98% of the arrested RB930 embryos still produce endoderm (data not shown).
Removal of the med-2 gene by the chromosomal deficiency sDf127:
Strain characterization:
The strain JM134 [dpy-17 sDf127 unc-32 III; sDp3(III,f); med-1(ok804) X] was constructed and validated as described in materials and methods. sDp3 is a (single-copy) free duplication that covers most of the left half of chromosome III, including all of sDf127 (Rosenbluth et al. 1985). The genetic properties of sDp3 have been well studied (Hedgecock and Herman 1995) and it is expected that ∼40% of JM134 offspring will not receive the free duplication and will thus arrest because they are homozygous for the sDf127 deficiency; this number will be verified below. As we will also show below, animals that receive two (or more) copies of the duplication must be rare; in any case, embryos that retain sDp3 can easily be identified by PCR. The control strain JM133 [dpy-17 sDf127 unc-32; sDp3; med-1(+)] was isolated as a med-1(+) segregant from the same cross that generated the med-1(−) strain JM134. For convenience, these two strains will be referred to as JM133 med-1(+) and JM134 med-1(−).
There are few obvious differences between strains JM133 med-1(+) and JM134 med-1(−); i.e., they seem to survive equally well with, on average, three copies (JM133) or only one copy (JM134) of a med gene. As shown in Table 1, brood sizes and hatching rates are comparable. The arrested embryos produced by both strains look similar, have >400 nuclei (Table 1), and appear typical of aneuploid “monsters” (see below). The large majority (>98%) of the arrested embryos of both genotypes continue to exclude the vital dye Trypan Blue after 4 days incubation at 20° (data not shown).
TABLE 1.
Comparison of properties of strains JM133 med-1(+) and JM134 med-1(−)
| JM133 dpy-17 sDf127 unc-32; sDp3; med-1(+)
|
JM134 dpy-17 sDf127 unc-32; sDp3; med-1(ok804)
|
|||||
|---|---|---|---|---|---|---|
| Average | SD | n | Average | SD | n | |
| Brood size | 125 | 31 | 8 broods | 122 | 41 | 7 broods |
| % arrest | 45.2 | 6.4 | 8 broods | 49.4 | 2.0 | 7 broods |
| No. nuclei | 444 | 50 | 9 embryos | 421 | 48 | 10 embryos |
Measuring the proportion of arrested embryos that lack both med-1 and med-2 genes:
Before inspecting the arrested embryos for gut marker expression, we first demonstrate that the majority (∼80%) of the arrested embryos produced by strain JM134 med-1(−) are indeed deficiency homozygotes lacking sDp3 and hence lacking both the med-1 and med-2 genes. The two pairs of PCR primers shown in Figure 2B allow detection of the status of both med-2 and med-1 genes in individual arrested embryos (Figure 2C) and have been designed to produce at least one positive band per reaction to verify successful PCR. Using this set of primers, 83.5% (76/91) of the arrested embryos produced by JM134 med-1(−) were found to have lost the sDp3 balancer and are thus med-2(−); med-1(−) homozygotes. The remaining 16.5% (15/91) contain a med-2(+) allele and must arrest for some other reason, perhaps because of haplo-insufficiency or because they have received multiple copies of sDp3.
An independent estimate of the proportion of deficiency homozygotes among the arrested JM134 med-1(−) embryos can be made from progeny phenotypes. The average proportion of arrested embryos produced by strain JM134 med-1(−) is 49.1% [the mean of 49.4 ± 2.0% estimated using individual total broods (Table 1) and 48.8% calculated from 262 unhatched embryos counted among 537 embryos produced in several hours by 50 adult JM134 hermaphrodites]. From the fraction of Dpy offspring produced by a dpy-17 unc-32 III; sDp3 control strain (five total broods), we estimate that 38.1 ± 4.9% of JM134 “med-1(−)” F1 progeny have lost the sDp3 duplication. In other words, 38.1/49.1 = 77.6% of arrested JM134 “med-1(−)” embryos should be med-2(−); med-1(−), in acceptable agreement with the 83.5% estimated by PCR. We note that the estimated frequency of sDp3 loss from the dpy-17 unc-32; sDp3 control strain (38 ± 5%) agrees well with the loss frequency (∼40%) estimated previously by Hedgecock and Herman (1995) using a different strain. We also note that the dpy-17 unc-32; sDp3 strain produces 4.2% unhatched embryos, thereby providing an upper limit to the frequency of arrested embryos receiving multiple copies of sDp3.
Gut marker expression in embryos lacking both med-1 and med-2 genes:
The arrested embryos produced by JM133 med-1(+) and JM134 med-1(−) were assayed for three biochemically independent markers of endoderm differentiation: (i) gut granules, (ii) histochemical staining for the gut-specific GES-1 esterase, and (iii) immunochemical staining using the monoclonal antibody MH33 to detect the intestine-specific IFB-2 intermediate filament protein. All assays in this study were performed “blind”; i.e., the person manipulating the embryos and/or scoring the differentiation marker did not know the strain identity; experiments were repeated at least five times for each genotype, as described in the legends for Figures 3 and 4:
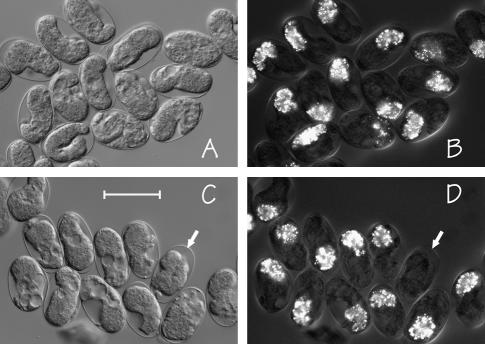
The most convenient (and foolproof) assay for gut differentiation is birefringent gut granules (Chitwood and Chitwood 1974; Laufer et al. 1980), which can first be detected when the embryonic intestine has approximately eight cells and which depend on zygotic transcription soon after endoderm specification (Edgar and McGhee 1988). To eliminate the possibility of selective loss of any particular class of embryos during manipulation, gravid hermaphrodites were placed on NGM-agar pads poured directly onto microscope slides and lightly seeded with bacterial food; after 7 ± 2 hr of egg laying, adults were removed and the slide incubated at 20° to allow viable embryos to hatch. Embryos that failed to hatch (after a wide range of incubation times; see materials and methods) were then inspected with polarized light. Examples of arrested embryos produced by strains JM133 med-1(+) and JM134 med-1(−) are shown in Figure 3, A and B, and Figure 3, C and D, respectively. It is evident that the majority of arrested embryos produced by either strain still express gut granules. However, there is a small fraction of arrested embryos produced by JM134 med-1(−) in which gut granules cannot be detected (see arrows in Figure 3, C and D). Overall, 13.4 ± 3.7% of the arrested embryos produced by strain JM134 med-1(−) do not express gut granules, compared to 0.2% of the arrested embryos produced by the control strain JM133 med-1(+). Correcting for the proportion (measured in the previous section) of arrested JM134 med-1(−) embryos that are not deficiency homozygotes (and assuming that all embryos in this class are marker positive), we estimate that 16.0–17.3% of med-2(−); med-1(−) embryos still express gut granules. On the basis of the penetrance of gut-granule-negative embryos produced by skn-1(null) mothers (measured above), if the meds were solely responsible for skn-1-specified endoderm, we would have expected 70% of the arrested embryos to be gut-granule-negative, rather than our observed 16–18%.
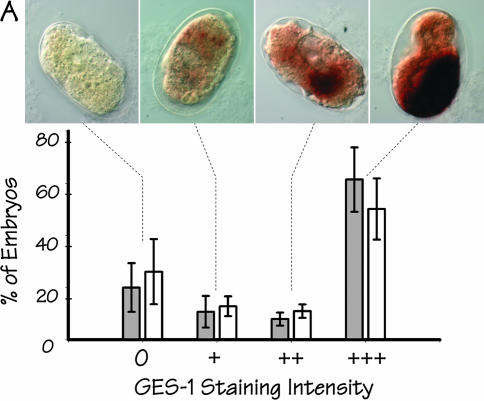
The arrested embryos were stained histochemically for activity of the gut-specific GES-1 esterase (Edgar and McGhee 1986). Because the assay for GES-1 activity is more technically demanding than the simple birefringence assay for gut granules (in particular, the pressure permeabilization-fixation step in GES-1 staining is prone to variability), the intensity of GES-1 staining was scored blind and semiquantitatively as 0, +, ++, or +++. Figure 4A shows a histogram summarizing the staining results, together with typical images of arrested embryos [from strain JM134 med-1(−)] representing each of the four classes of staining intensity. The majority of arrested embryos from both strains stain intensely for GES-1 activity (scored as +++); nonstaining JM134 med-1(−) embryos are only slightly more prevalent (∼6%) than nonstaining JM133 med-1(+) control embryos. Indeed, it is doubtful that any difference in GES-1 staining can be reliably detected between the two sets of embryos (see Figure 4 legend).
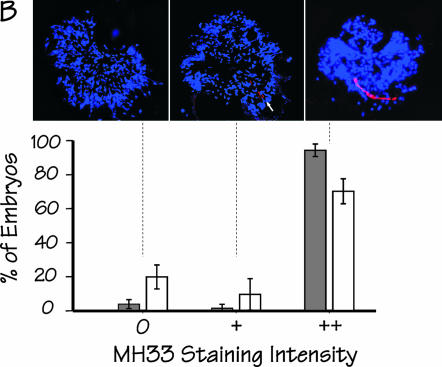
Arrested embryos were stained immunochemically using the monoclonal antibody MH33 (Francis and Waterston 1985), which reacts with IFB-2, an intestine-specific intermediate filament protein (Karabinos et al. 2001; Bossinger et al. 2004); the intensity of staining was scored blind and semiquantitatively as 0, +, or ++. As shown in Figure 4B, the majority of arrested embryos from either JM133 med-1(+) or JM134 med-1(−) stain intensely with MH33 (scored as ++) and exhibit an obvious “endotube” [the sublumenal structure containing IFB-2 (Bossinger et al. 2004)]. After correcting for nonstaining JM133 med-1(+) control embryos, we estimate that ∼16% of all arrested embryos produced by strain JM134 med-1(−) or ∼20% of the med-2(−); med-1(−) embryos do not express the endoderm-specific ifb-2 gene.
Figure 3.
Microscopic images of typical arrested embryos produced by control strain JM133 med-1(+) (A and B) and by strain JM134 med-1(−) (C and D), as seen by differential interference contrast (A and C) or by birefringence optics (B and D). As described in the text, the majority of the arrested embryos produced by either strain still express birefringent gut granules. The arrows in C and D indicate one arrested embryo produced by strain JM134 med-1(−) that does not express gut granules. Bar, 50 μm. This experiment was repeated six times for each genotype; the total number of scored embryos was 671 and 690 for strains JM133 med-1(+) and JM134 med-1(−), respectively.
Figure 4.
Expression of additional markers of endoderm differentiation in arrested embryos produced by strains JM133 med-1(+) and JM134 med-1(−). (A) Histogram showing the distribution of GES-1 staining intensity (classified as 0, +, ++, or +++) within arrested embryos produced by strain JM134 med-1(−) (open bars) or by control strain JM133 med-1(+) (shaded bars). Typical images of each class of stained embryo [from strain JM134 med-1(−)] are shown above the corresponding histogram bar. The experiment was repeated a total of five times for each genotype, scoring a total of 830 and 872 individual arrested embryos produced by strains JM133 med-1(+) and JM134 med-1(−), respectively; error bars are standard deviations. The apparent difference between the JM133 med-1(+) and JM134 med-1(−) histograms is due entirely to one particular pair of slides (corresponding to the longest incubation time) in which some embryos appeared to have degenerated and the staining appeared erratic. If this set of data is excluded from the analysis, the behaviors of JM133 med-1(+) and JM134 med-1(−) are essentially indistinguishable (data not shown). (B) Histogram showing the distribution of MH33 staining intensity (classified as 0, +, ++) within arrested embryos produced by strain JM134 med-1(−) (open bars) or control strain JM133 med-1(+) (shaded bars). Images [from strain JM134 med-1(−)] represent Z-projections of deconvolved image stacks taken for each embryo; blue, DAPI staining; red, MH33 immunofluorescence. A well-formed endotube is obvious in embryos classified as “++”; a rudimentary endotube can be detected in embryos classified as “+” (see arrow). This experiment was repeated five times, scoring a total of 305 and 386 arrested embryos for strains JM133 med-1(+) and JM134 med-1(−), respectively; error bars represent standard deviations. DAPI-stained images from these experiments were used to estimate the total number of nuclei in the arrested embryos (Table 1).
Removal of the med-2 gene by the chromosomal deficiency nDf16:
In an attempt to rule out possible confounding influences associated with the particular chromosomal deficiency (for example, if sDf127 were to remove a repressor of an alternative endoderm-specification pathway), the analysis was repeated using the overlapping deficiency nDf16 (see Figure 2A); nDf16 is a larger deficiency than sDf127 and removes ∼300 genes surrounding med-2; several dozen or so of these genes are also removed by sDf127. We constructed two new strains: JM136 [nDf16/lon-1 unc-32 III; med-1(ok804) X] and the control strain JM135 [nDf16/lon-1 unc-32 III; med-1(+) X]; again, for convenience, these strains will be referred to as JM135 med-1(+) and JM136 med-1(−).
For both strains JM135 med-1(+) and JM136 med-1(−), it is expected that 25% of the F1 progeny should arrest because they are homozygous for nDf16. The control strain JM135 med-1(+) shows a slight degree of excess lethality above the expected 25%: 31% (62/200) of F1 progeny arrest. Thus, ∼6% of the total F1 progeny or 19% of the arrested JM135 med-1(+) embryos fail to hatch for reasons other than nDf16 homozygosity, e.g., haplo-insufficiency. In contrast, strain JM136 med-1(−) shows a substantial degree of excess lethality: ∼66% (428/648) of F1 progeny produced by strain JM136 med-1(−) failed to hatch; that is, only 25/66 = 38% of JM136 arrested embryos are expected to be nDf16 homozygotes and hence med-2(−); med-1(−). In acceptable agreement with this estimate, the direct PCR assay found that 41.5% (39/94) of arrested JM136 embryos are indeed med-2(−); med-1(−) (data not shown). Although this excess lethality is intriguing, it is irrelevant for interpreting the current experiment because 98.8% (2136/2162) of the arrested embryos produced by strain JM136 med-1(−) strongly express birefringent gut granules. For comparison, 97.3% (733/753) of arrested embryos produced by the control strain JM135 med-1(+) are also gut-granule positive. Typical birefringent images of arrested embryos produced by strains JM135 med-1(+) and JM136 med-1(−)” are shown in Figure 5, A and B, respectively. Correcting for the fraction of JM136 med-1(−) arrested embryos that are not homozygous for nDf16, we estimate that only 3% of med-2(−); med-1(−) embryos produced by JM136 med-1(−) do not express gut granules, essentially the same low proportion found in the med-2(−); med-1(+) control embryos.
Figure 5.
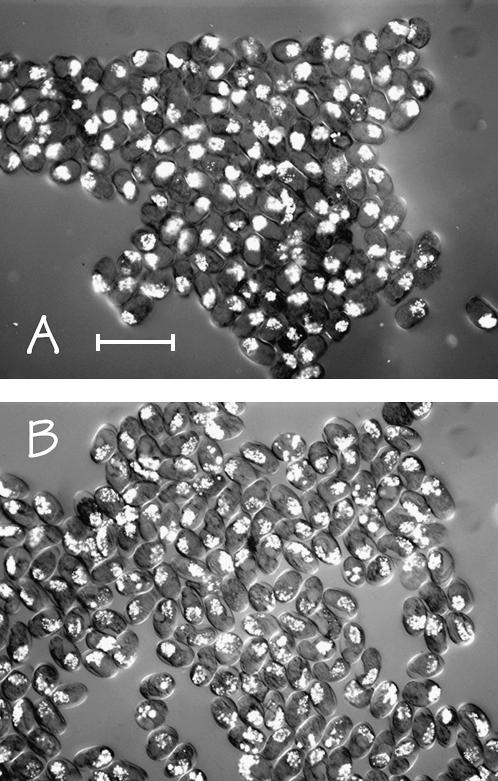
Microscopic images (birefringence optics) of typical arrested embryos produced by (A) strain JM135 med-1(+) and (B) strain JM136 med-1(−). This experiment was repeated a total of eight times for each genotype, scoring a total of 753 and 2162 arrested embryos for strain JM135 med-1(+) and strain JM136 med-1(−), respectively. As described in the text, >97% of arrested embryos produced by either strain still express birefringent gut granules. Bar, 100 μm.
Do med-1 and med-2 genes exhibit a maternal effect?
We consider whether the differences between our results and those of Maduro et al. (2001) can be explained by the med genes specifying endoderm by both a maternal function and a zygotic function. In particular, we test the simplest “maternal med” model: if a particular embryo does not express endoderm markers, it must lack both maternal and zygotic contributions from the med genes; i.e., either maternal or zygotic med function is sufficient to specify endoderm.
A first test of the “maternal med” model is based on the argument that, if a putative med-2 maternal product is necessary to specify embryonic endoderm in the absence of zygotic med genes, then the 15–20% of arrested JM134 med-2(−); med-1(−) embryos that do not express endoderm markers must have been produced by individual mothers who had lost the sDp3 in their germline. Although the rarity of such animals (Hedgecock and Herman 1995) would argue against this possibility, we nonetheless inspected arrested embryos produced by 20 individual JM134 med-1(−) mothers; all 20 broods contained roughly the expected fraction of arrested embryos that do not express gut granules, arguing that the gut-granule-negative embryos do not arise from infrequent mothers lacking sDp3 in their germlines.
As a second test of the “maternal med” model, double-stranded RNA corresponding to the med-1 gene was injected into the sensitized JM134 med-1(−) mothers and the population of arrested F1 embryos was inspected for birefringence. As described in the materials and methods, these injections were performed by standard protocols found to be effective against maternal transcripts (Fire et al. 1998; Shi and Mello 1998), injecting into the intestine/body cavity and monitoring F1 embryos over the next few days. The proportion of med-1 [and therefore, because of high sequence similarity (Maduro et al. 2001), also med-2] RNAi JM134 med-1(−)-arrested embryos that do not express gut granules was only marginally greater (4.1 ± 5.2%) than that found in arrested JM134 med-1(−) embryos whose mothers had been injected with double-stranded RNA corresponding to GFP (experiment repeated three times, injecting 10–20 hermaphrodites per repeat). In addition, RNAi to med-1 did not obviously enhance the level of embryonic arrest. As a control on technique and on the RNAi sensitivity of strain JM134, the same injection protocol using double-stranded skn-1 RNA caused 100% arrest of embryos produced by JM134 med-1(−) mothers. In summary, these two different experimental approaches provide no evidence for a model in which either zygotic or maternal expression of the med genes is necessary (or sufficient) to specify endoderm.
DISCUSSION
In this study, we have tested whether the med-1 and med-2 genes are necessary for specification of the C. elegans endoderm. If the med-1 and med-2 genes are the major effectors downstream of the maternal SKN-1 factor and are responsible for specifying the C. elegans endoderm (Maduro et al. 2001), then 70% of med-2(−); med-1(−) embryos should not produce endoderm. Our results do not agree with this expectation; instead, we find that only ∼3% (∼0% if corrected for the background observed with control embryos) or 15–20% of med-2(−); med-1(−) embryos do not express gut markers, depending on which chromosomal deficiency (nDf16 or sDf127, respectively) was used to remove the med-2 gene. Either result indicates that the C. elegans endoderm can be specified with high efficiency in the complete zygotic absence of the med genes. We emphasize that our experiments have addressed only necessary roles of med-1 and med-2 in endoderm specification and have not addressed other proposed roles for the med genes, such as sufficiency for endoderm marker expression, necessity and sufficiency for specification of the MS lineage, or restriction of the fate of the neighboring C blastomere (Maduro et al. 2001; Maduro and Rothman 2002; Broitman-Maduro et al. 2005).
Two different experimental approaches failed to provide evidence for a maternal contribution of the med genes to endoderm specification: (i) an unsuccessful attempt to identify individual JM134 broods showing a penetrant loss of endoderm in the arrested embryos and (ii) an unsuccessful attempt to perform med RNAi in hermaphrodites that segregate the med-2(−); med-1(−) embryos, using a standard injection protocol that has generally been found to be effective against maternal transcripts (Fire et al. 1998; Shi and Mello 1998). However, it would be difficult to verify that the med-1 RNAi has indeed been effective because, using either oocyte or early embryo RNA, any putative maternal med transcripts are below the detection level of Affymetrix microarrays (Baugh et al. 2003, 2005). Finally, we point out that any maternally provided med contribution (proteins or transcripts) that can rescue the loss of zygotic med function would be inconsistent with the endoderm loss caused by loss of maternal skn-1 (Bowerman et al. 1992).
A secondary issue is to resolve why our estimates of possible med-dependent endoderm vary between the two deficiencies: 15–20% using sDf127 and ∼3% using nDf16. If the sDf127 results are valid, one possibility to explain the nDf16 results is that the 1–2% of the genome included in nDf16 but not in sDf127 contains a repressor of the end-1 or end-3 genes or perhaps a repressor of an alternative pathway to specify endoderm. Alternatively, if the nDf16 results are valid, then the sDf127 results could possibly be explained by increased embryo fragility, a slightly earlier stage of embryo arrest and/or nonspecific loss of endoderm marker expression, all of which could be exacerbated by the fact that sDf127 homozygotes are also homozygous unc-32; null mutations in unc-32 are known to cause embryonic arrest with vacuolated intestines (Pujol et al. 2001) and we found that unc-32 RNAi greatly decreases gut-granule intensity in the arrested embryos, such that ∼20% would be classified as negative by our scoring criteria (data not shown). However, we really have no reason for choosing between these two possibilities and the important point remains that the results obtained with either deficiency are incompatible with the model being tested.
A potential weakness of the present analysis is our use of multigene chromosomal deficiencies to remove the med-2 gene, raising the theoretical possibility of generating an “endoderm permissive environment” within the deficiency embryos. Although such a possibility is difficult to rule out, several considerations suggest that it is unlikely. The perfect candidate for a gene whose removal would generate an “endoderm permissive environment” is the pop-1 gene (Lin et al. 1995, 1998; Rocheleau et al. 1997, 1999) but pop-1 is not removed by either sDf127 or nDf16. Other known endoderm-specifying genes (e.g., skn-1, lit-1, mom-1,2,4,5, etc.) are not removed by either deficiency and, in any case, loss of function of these genes inhibits endoderm formation. Furthermore, genes involved in endoderm specification (up to the med genes) are expected to be maternally provided and thus the homozygous deficiency embryos receive at least a haploid dose of maternal product. As a final defense of the use of chromosomal deficiencies, we suggest that poor embryonic health caused by aneuploidy is more likely to reduce than enhance endoderm gene expression. If only for this final reason, we suggest that the current experimental system should, if anything, overestimate the importance of the med-1 and med-2 genes in endoderm specification.
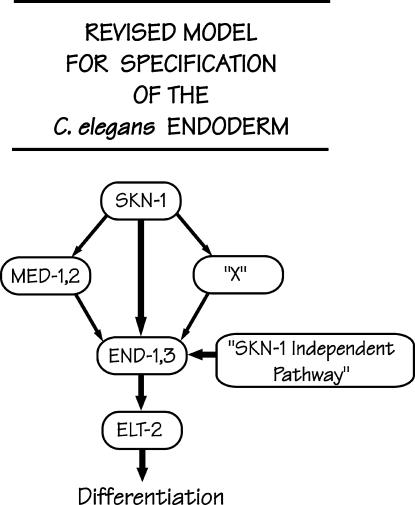
If the med-1 and med-2 genes are not required for (most) C. elegans embryos to produce endoderm, then what is the major endoderm activator? We suggest that, as has been previously proposed and as depicted in Figure 6, the maternal SKN-1 factor may still be the best candidate for initiating the endoderm pathway, for example, by direct activation of the end-1 and end-3 genes within the endoderm precursor E cell (Zhu et al. 1997), apparently in direct conjunction with transcriptional cofactors such as CBP-1 (Shi and Mello 1998; Walker et al. 2000) and in the permissive environment provided by low nuclear concentrations of the POP-1 protein (Lin et al. 1995, 1998; Rocheleau et al. 1997; Thorpe et al. 1997; Lo et al. 2004). It has recently been reported that ablation of MED-binding sites from the 5′-flanking region of the end-1 genes abolishes expression of an end-1 reporter (Broitman-Maduro et al. 2005). However, this alteration was performed in the context of a short promoter fragment (<250 bp) and it had previously been pointed out that six potential SKN-1-binding sites lie within the next 1 kb upstream (Zhu et al. 1997). There is, of course, the possibility that SKN-1 specifies endoderm by acting through a factor other than MED-1/2, and this unknown factor is designated by the traditional X shown in Figure 6. Finally, Figure 6 draws attention to the significant SKN-1 independent pathway for endoderm specification.
Figure 6.
Revised proposal for the transcription factor cascade responsible for specifying the C. elegans endoderm. As described in the text, we suggest that the SKN-1-activated med-1 and med-2 genes may play a relatively minor role in specifying the C. elegans endoderm. We suggest that the major pathway for endoderm specification could involve the SKN-1 transcription factor itself directly activating the end-1 and end-3 genes, as originally proposed by Zhu et al. (1997) or acting through an unknown factor, depicted as X. Finally, there must be a significant SKN-1 independent pathway to specify endoderm (∼30% under the current experimental conditions).
Finally, we point out a limitation in the interpretation of our results that is a common feature of studies of early C. elegans development, namely the analysis of phenotypes in terms of penetrance rather than in terms of expressivity. It is possible that the MED factors could be participating in many acts of transcription that lead to endoderm specification; the present experiments would have detected this participation only if the consequences of med removal were changes in penetrance or substantial changes in expressivity. That is, a 20% lower penetrance of endoderm in the population of arrested embryos was obvious but it is doubtful that a 20% lowering of the intensity of marker expression in the med-2(−); med-1(−) embryos would have been detected. Likewise, the med genes could be intimately involved in endoderm specification but be redundant with some other set of endoderm-specifying genes. These two possible characteristics could, in principle, explain the reported sufficiency of med-1 and med-2 for endoderm marker expression (Maduro et al. 2001; Maduro and Rothman 2002; Broitman-Maduro et al. 2005).
Acknowledgments
We thank the C. elegans Gene Knockout Consortium for providing the med-1(ok804) deletion allele and the C. elegans Genetics Center (funded by the National Center for Research Resources) for providing strains. We also thank Paul Mains, Jeb Gaudet, Dave Hansen, and William Brook (University of Calgary) and Tetsunari Fukushige (National Institutes of Health) for advice and criticism. We especially thank Joel Rothman (University of California, Santa Barbara) and Morris Maduro (University of California, Riverside) for providing detailed protocols and reagents and for frank and constructive discussions. This work was supported by an operating grant from the Canadian Institutes of Health Research. J.D.M. is a Medical Scientist of the Alberta Heritage Foundation for Medical Research and a Canada Research Chair.
Note added in proof: Laurent Segalat (Université Lyon Claude Bernard) has recently identified a Mos insertion within the med-2 gene, thereby producing a likely med-2 null allele (cxP9744). Our preliminary analysis of gut-granule production by med-2 (cxP9744); med-1(ok804) embryos confirms the results obtained with chromosomal deficiencies as described in this article: 16% (18/112) of med-2 (cxP9744); med-1(ok804) embryos do not express gut granules. A more complete analysis of endoderm formation in these embryos will be reported once the strains have been sufficiently outcrossed.
References
- Baugh, L. R., A. A. Hill, D. K. Slonim, E. L. Brown and C. P. Hunter, 2003. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 130: 889–900. [DOI] [PubMed] [Google Scholar]
- Baugh, L. R., A. A. Hill, J. M. Claggett, K. Hill-Harfe, J. C. Wen et al., 2005. The homeodomain protein PAL-1 specifies a lineage-specific regulatory network in the C. elegans embryo. Development 132: 1843–1854. [DOI] [PubMed] [Google Scholar]
- Bossinger, O., T. Fukushige, M. Claeys, G. Borgonie and J. D. McGhee, 2004. The apical disposition of the Caenorhabditis elegans intestinal terminal web is maintained by LET-413. Dev. Biol. 268: 448–456. [DOI] [PubMed] [Google Scholar]
- Bowerman, B., B. A. Eaton and J. R. Priess, 1992. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell 68: 1061–1075. [DOI] [PubMed] [Google Scholar]
- Bowerman, B., B. W. Draper, C. C. Mello and J. R. Priess, 1993. The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell 74: 443–452. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broitman-Maduro, G., M. F. Maduro and J. H. Rothman, 2005. The noncanonical binding site of the MED-1 GATA factor defines differentially regulated target genes in the C. elegans mesendoderm. Dev. Cell 8: 427–433. [DOI] [PubMed] [Google Scholar]
- Chitwood, B., and M. Chitwood, 1974. Introduction to Nematology. University Park Press, Baltimore.
- Edgar, L. G., and J. D. McGhee, 1986. Embryonic expression of a gut-specific esterase in Caenorhabditis elegans. Dev. Biol. 114: 109–118. [DOI] [PubMed] [Google Scholar]
- Edgar, L. G., and J. D. McGhee, 1988. DNA synthesis and the control of embryonic gene expression in C. elegans. Cell 53: 589–599. [DOI] [PubMed] [Google Scholar]
- Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver et al., 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Francis, G. R., and R. H. Waterston, 1985. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J. Cell Biol. 101: 1532–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige, T., M. G. Hawkins and J. D. McGhee, 1998. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev. Biol. 198: 286–302. [PubMed] [Google Scholar]
- Fukushige, T., B. Goszczynski, J. Yan and J. D. McGhee, 2005. Transcriptional control and patterning of the pho-1 gene, an essential acid phosphatase expressed in the C. elegans intestine. Dev. Biol. 279: 446–461. [DOI] [PubMed] [Google Scholar]
- Hawkins, M. G., and J. D. McGhee, 1995. elt-2, a second Gata factor from the nematode Caenorhabditis elegans. J. Biol. Chem. 270: 14666–14671. [DOI] [PubMed] [Google Scholar]
- Hedgecock, E. M., and R. K. Herman, 1995. The ncl-1 gene and genetic mosaics of Caenorhabditis elegans. Genetics 141: 989–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, D. L., J. E. Schein, T. Ha, N. W. Franz, N. J. O'Neil et al., 1997. Interpreting a sequenced genome: toward a cosmid transgenic library of Caenorhabditis elegans. Genome Res. 7: 974–985. [DOI] [PubMed] [Google Scholar]
- Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin et al., 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Karabinos, A., H. Schmidt, J. Harborth, R. Schnabel and K. Weber, 2001. Essential roles for four cytoplasmic intermediate filament proteins in Caenorhabditis elegans development. Proc. Natl. Acad. Sci. USA 98: 7863–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer, J. S., P. Bazzicalupo and W. B. Wood, 1980. Segregation of developmental potential in early embryos of Caenorhabditis elegans. Cell 19: 569–577. [DOI] [PubMed] [Google Scholar]
- Lin, R., S. Thompson and J. R. Priess, 1995. pop-1 encodes an Hmg box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell 83: 599–609. [DOI] [PubMed] [Google Scholar]
- Lin, R., R. J. Hill and J. R. Priess, 1998. POP-1 and anterior-posterior fate decisions in C. elegans embryos. Cell 92: 229–239. [DOI] [PubMed] [Google Scholar]
- Lo, M. C., F. Gay, R. Odom, Y. Shi and R. Lin, 2004. Phosphorylation by the beta-catenin/MAPK complex promotes 14–3-3-mediated nuclear export of TCF/POP-1 in signal-responsive cells in C. elegans. Cell 117: 95–106. [DOI] [PubMed] [Google Scholar]
- Maduro, M. F., and J. H. Rothman, 2002. Making worm guts: the gene regulatory network of the Caenorhabditis elegans endoderm. Dev. Biol. 246: 68–85. [DOI] [PubMed] [Google Scholar]
- Maduro, M. F., M. D. Meneghini, B. Bowerman, G. Broitman-Maduro and J. H. Rothman, 2001. Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3beta homolog is mediated by MED-1 and -2 in C. elegans. Mol. Cell 7: 475–485. [DOI] [PubMed] [Google Scholar]
- Montgomery, M. K., S. Xu and A. Fire, 1998. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 95: 15502–15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol, N., C. Bonnerot, J. J. Ewbank, Y. Kohara and D. Thierry-Mieg, 2001. The Caenorhabditis elegans unc-32 gene encodes alternative forms of a vacuolar ATPase a subunit. J. Biol. Chem. 276: 11913–11921. [DOI] [PubMed] [Google Scholar]
- Robertson, S. M., P. Shetty and R. Lin, 2004. Identification of lineage-specific zygotic transcripts in early Caenorhabditis elegans embryos. Dev. Biol. 276: 493–507. [DOI] [PubMed] [Google Scholar]
- Rocheleau, C. E., W. D. Downs, R. Lin, C. Wittmann, Y. Bei et al., 1997. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell 90: 707–716. [DOI] [PubMed] [Google Scholar]
- Rocheleau, C. E., J. Yasuda, T. H. Shin, R. Lin, H. Sawa et al., 1999. WRM-1 activates the LIT-1 protein kinase to transduce anterior/posterior polarity signals in C. elegans. Cell 97: 717–726. [DOI] [PubMed] [Google Scholar]
- Rodaway, A., and R. K. Patient, 2001. Mesendoderm: An ancient germ layer? Cell 105: 169–172. [DOI] [PubMed] [Google Scholar]
- Rosenbluth, R. E., C. Cuddeford and D. L. Baillie, 1985. Mutagenesis in Caenorhabditis elegans. II. A spectrum of mutational events induced with 1500 r of γ-radiation. Genetics 109: 493–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and D. W. Russell, 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shi, Y., and C. Mello, 1998. A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes Dev. 12: 943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen, B., L. B. Koski, A. Walsh, P. Marschall, B. Neumann et al., 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434: 462–469. [DOI] [PubMed] [Google Scholar]
- Sulston, J. E., E. Schierenberg, J. G. White and J. N. Thomson, 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. [DOI] [PubMed] [Google Scholar]
- Tavernarakis, N., S. L. Wang, M. Dorovkov, A. Ryazanov and M. Driscoll, 2000. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat. Genet. 24: 180–183. [DOI] [PubMed] [Google Scholar]
- Thomas, J. H., M. J. Stern and H. R. Horvitz, 1990. Cell interactions coordinate the development of the C. elegans egg-laying system. Cell 62: 1041–1052. [DOI] [PubMed] [Google Scholar]
- Thorpe, C. J., A. Schlesinger, J. C. Carter and B. Bowerman, 1997. Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell 90: 695–705. [DOI] [PubMed] [Google Scholar]
- Walker, A. K., R. See, C. Batchelder, T. Kophengnavong, J. T. Gronniger et al., 2000. A conserved transcription motif suggesting functional parallels between Caenorhabditis elegans SKN-1 and Cap‘n’Collar-related basic leucine zipper proteins. J. Biol. Chem. 275: 22166–22171. [DOI] [PubMed] [Google Scholar]
- Williams, B. D., B. Schrank, C. Huynh, R. Shownkeen and R. H. Waterston, 1992. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics 131: 609–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, W. B., 1988. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Zhu, J., R. J. Hill, P. J. Heid, M. Fukuyama, A. Sugimoto et al., 1997. end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev. 11: 2883–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., T. Fukushige, J. D. McGhee and J. H. Rothman, 1998. Reprogramming of early embryonic blastomeres into endodermal progenitors by a Caenorhabditis elegans GATA factor. Genes Dev. 12: 3809–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]