Abstract
Heliconius melpomene is a mimetic butterfly that exhibits great geographic variation in color pattern. We present here a genetic linkage map based on analysis of genetic markers in 73 individuals from a single F2 family, offspring of a cross between H. m. cythera from western Ecuador and H. m. melpomene from French Guiana. A novel “three-step method” is described for the analysis of dominant markers in an F2 cross, using outbred parental strains and taking advantage of the lack of crossing over in female Lepidoptera. This method is likely to prove useful for future mapping studies in outbred species with crossing over restricted to one sex, such as the Lepidoptera and Drosophila. The resulting linkage map has 21 linkage groups corresponding to the 21 chromosomes of H. melpomene and includes 219 AFLP markers, 23 microsatellites, 19 single-copy nuclear genes, and the color pattern switch genes Yb and Sb. The marker density is high, averaging >1/7 cM. The total map length is 1616 cM and the average chromosome length is 77 cM. The genome size of H. melpomene was estimated to be 292 Mb, giving a relationship of physical-to-map distance of 180 kb/cM. This map forms the basis for future comparative linkage analysis of color pattern evolution in Heliconius.
BUTTERFLIES in the genus Heliconius have long been models for the study of ecology, natural selection, and speciation (Brown 1981). This is largely due to their bright wing-color patterns and Müllerian mimicry among species. Field experiments have shown strong purifying selection on wing pattern, thought to result from predators learning to avoid the color patterns of these distasteful insects (Benson 1972; Mallet and Barton 1989; Kapan 2001). This strong selection explains mimicry, the striking convergence of pattern between distantly related species that benefit from a shared signal to predators, but also acts to maintain diversity of pattern by stabilizing hybrid zones between intraspecific populations with distinct color patterns (Mallet and Barton 1989). Thus, many species are divided into distinct color pattern races across their range.
Color pattern differentiation contributes to both pre- and postmating isolation and therefore plays an important role in speciation. Heliconius melpomene and its close relative, H. cydno, use their mimetic color patterns as cues in mate recognition (Jiggins et al. 2001b). In addition, hybrids have intermediate, nonmimetic color patterns and are likely selected against. Across the genus as a whole, the evolution of mimicry seems to be associated with increased rates of species diversification (Turner 1976; Gilbert 1991). Thus, the vivid wing patterns of Heliconius provide one of the clearest examples of a morphological trait under strong natural selection with a key role in adaptive radiation.
The Mendelian genetics of mimetic color patterns in Heliconius have been well studied using crosses between races and species (Sheppard et al. 1985; Mallet 1989; Jiggins and McMillan 1997; Linares 1997; Gilbert 2003; Naisbit et al. 2003). Studies of two species in particular, Heliconius erato and H. melpomene, have resulted in a detailed description of the genetic basis of intraspecific color pattern diversity (Sheppard et al. 1985 and references therein). In both species, dramatically divergent color pattern races differ at just a handful of loci, each with very major effects on color pattern. These loci segregate in predicted Mendelian ratios and often affect many aspects of the wing pattern. For example, a single region in H. melpomene determines the presence of a yellow hind-wing bar, a submarginal white wing margin, and a yellow forewing band. These three phenotypic elements are likely controlled by distinct but tightly linked loci, known as Yb, Sb, and N, respectively (Emsley 1964; Naisbit et al. 2003). There is also strong epistasis between genes, with particular alleles having very different effects depending on genetic background. Despite this classical genetics background, to date nothing is known of the molecular basis of these regulatory loci.
The fact that Heliconius butterflies with distinct color patterns can be readily crossed in the laboratory allows the generation of genetic linkage maps and will ultimately facilitate identification of the molecular basis of the color pattern switch genes. Such linkage maps can be used to (1) identify markers linked to color pattern genes, (2) map candidate loci that are thought to be involved in color pattern development, and (3) enable a comparative approach to understanding convergent evolution between mimetic species such as H. erato and H. melpomene.
Candidate loci that might be involved in wing development can be identified from studies of pattern formation and pigment synthesis in other butterfly species. First, studies of eyespots in Bicyclus and Precis butterflies have shown that expression patterns of several genes, which are most likely upstream regulators of pigment formation, correlate with wing pattern phenotypes (Keys et al. 1999; Brunetti et al. 2001). We can hypothesize that in Heliconius the upstream regulators of pattern formation are similarly signaling proteins and transcription factors, such as those that signal embryo segmentation and wing differentiation in Drosophila (Carroll et al. 1994; Wilkins 2002). Wing patterns consist of regions of colored scales containing ommochrome (yellow, orange, and red) and melanin (black) pigments. The gene pathways involved in the formation of both of these pigment classes are well characterized (Linzen 1974; Summers et al. 1982; Gilbert et al. 1988; Koch 1993; Koch et al. 1998; Wittkopp et al. 2002). Pigment synthesis and signaling pathways therefore constitute likely candidates for genes involved in pattern evolution. Alleles that segregate in crossing experiments must reflect changes in coding sequence or cis-regulatory elements of genes involved in some aspect of pattern formation (Stern 2000). Hence, identifying linkage relationships between color pattern switch genes and the conserved protein-coding regions of candidate loci is the first step in determining whether they are one and the same.
In addition to specific candidate genes, large numbers of anonymous markers are necessary to ensure coverage of all chromosomes (n = 21 in H. melpomene). Here we use the AFLP technique that generates many segregating markers from a single PCR reaction. These markers increase the probability that loci will be identified linked to gene regions involved in color pattern evolution. A novel three-step method for analysis of AFLP markers that takes advantage of crosses carried out between outbred strains, such that loci of all possible segregation types are found in the same brood, is presented. In combination with the lack of crossing over during meiosis in female Lepidoptera, this greatly facilitates the assignment of loci to linkage groups and the production of an integrated linkage map from a relatively small number of loci and individuals. This technique is therefore likely to be of use in future mapping studies based on wild populations of Lepidoptera. Here we present the first linkage map of H. melpomene that incorporates AFLP, microsatellite, and single-copy nuclear genes.
MATERIALS AND METHODS
Collection and crossing experiments:
H. melpomene cythera were collected in western Ecuador around Mindo (0.065° S, 78.789° W), and H. melpomene melpomene were collected near Cayenne in French Guiana (4.913° N, 52.360° W). Male H. m. cythera were crossed to female H. m. melpomene (the reverse cross produces sterile F1 females; cf. Jiggins et al. 2001a) in insectaries in Gamboa, Panama. The F1 offspring were then crossed together and females maintained for as long as possible to obtain large F2 broods. Adults were kept with ample pollen from fresh Psiguria warcsewiczii flowers to maximize female egg laying and male mating enthusiasm. Single females were kept in 1 × 1 × 2-m cages and all eggs were collected daily. A single brood female (brood 33) produced a family of 152 adult offspring and was chosen for the analysis presented here.
AFLP analysis:
DNA was extracted using the QIAGEN (Chatsworth, CA) DNeasy tissue kit following recommended protocols. Final elution of DNA was carried out in 250 μl of AE buffer. AFLP analysis was carried out on 88 offspring, two parents, and four grandparents of brood 33 on a 96-well PCR plate. The restriction and digestion steps of the AFLP technique were carried out using the GIBCO BRL (Gaithersburg, MD) AFLP plant mapping kit, using half-volume reactions. After the restriction-ligation steps, the DNA was diluted 1:10 and 5 μl was used for the preselective PCR, using Eco-N and Mse-N primers. The 50-μl reaction contained 1× buffer, 0.8 mm dNTPs, 0.3 mm of each primer, and 0.02 units/μl QIAGEN Taq polymerase and was run with a cycling profile of 94° for 2 min, followed by 20 cycles of 94° for 30 sec, 56° for 1 min, and 72° for 1 min. A 1:50 dilution of the product was used for selective PCR. The 10-μl selective PCR mix contained 2.5 μl of diluted preselective DNA, 1× buffer, 0.8 mm dNTPs, 0.375 mm of fluorescent 6-FAM-labeled Eco-NN primer, 0.375 mm of Mse-NNN primer, and 0.025 units/μl QIAGEN Taq polymerase and was run using a cycling profile of 94° for 2 min followed by 12 cycles of 94° for 30 sec, 65° for 30 sec, reduced by −0.7° per cycle to 56.6° and 72° for 1 min, and then 23 cycles of 94° for 30 sec, 56° for 30 sec, and 72° for 1 min. The selective PCR reaction was then cleaned through 700 μl of G-50 sephadex in Whatman Unifilter 96-well plates. The cleaned product was diluted 1:10 and 1 μl was loaded on a 20-cm acrylamide gel on an MJ Research (Watertown, MA) base station sequencing machine with Genescan Rox 500 size standard (Applied Biosystems, Foster City, CA) in each lane. A Hex-labeled microsatellite was used as a lane marker in lanes 24, 48, 72, and 96 to assist in lane tracking.
There was considerable variation in the number of bands produced with different primer combinations, so combinations were initially tested with either two or six samples. Those giving the largest number of bands were selected and run for all samples. Gel images were analyzed in Cartographer (MJ Research) by defining allele ranges using a combined chromatogram of all samples from a given primer combination. Data were then exported and all monomorphic bands were removed from further analysis.
Microsatellites:
Locus identification:
A previous study identified eight loci for H. melpomene from a CA-repeat-enriched microsatellite library (Flanagan et al. 2002). Of those, two did not give consistent amplification results in our study (Hel12 and Hel15) and one locus was duplicated (Hel11 is the same locus as Hel8). The remaining loci, Hel02, Hel04, Hel05, Hel08, and Hel14, along with one other locus, Hel17, developed from the same library, were scored here. Three loci, Hm17, Hm20, and Hm21, were obtained by Michael J. Blum using a partial (400- to 800-bp) genomic DNA library of H. melpomene (Blum 2002). An additional 18 loci (Hm1–16, Hm18, and Hm19) were obtained from a CA-enriched microsatellite library of H. melpomene (J. Mavarez and M. Gonzalez, unpublished results).
Genotyping:
Twenty-five polymorphic microsatellite loci showing clear unambiguous length polymorphisms were identified for mapping. The 10-μl reactions contained 1× buffer (QIAGEN), 1.5 mm MgCl2, 0.2 mm dNTPs, 0.025 units/μl Taq polymerase (QIAGEN), and 10–40 ng genomic DNA. Loci were scored using one of two alternative methods: (a) 0.1 μm dye-labeled (6-Fam, Hex, or Tet) forward primers and 0.5 μm reverse primers or (b) 0.01 μm m13-tailed forward primer, 0.4 μm dye-labeled (6-Fam, Hex, or Tet) m13 primer (5′-CACGACGTTGTAAAACGAC-3′), and 0.4 μm reverse primer. The PCR reaction was performed in a PTC-100 or PTC-200 thermal cycler (MJ Research) using a first incubation of 94° for 2 min, followed by 30 cycles of 92° for 20 sec, 55° for 30 sec, and 72° for 30 sec. A final extension of 15 min at 72° was added. Reaction products were diluted (usually 1:20, 1:30, and 1:40 for Hex, Tet, and 6-Fam labeled products, respectively) and resolved in 6% Long Ranger (BioWhittaker Molecular Applications) denaturing polyacrylamide gels on a MJ base station sequencing machine (MJ Research) following the manufacturer's protocols. Allele sizes were determined using Cartographer software (MJ Research) with Genescan Rox-400 or -500 (Applied Biosystems) as size standards.
Single-copy nuclear genes:
A number of single-copy nuclear loci (SCNL) were amplified using either previously published or novel primers (Table 1). For the ribosomal proteins shown in Table 1, sequences from a cDNA library derived from wing tissue of H. erato were used (W. O. McMillan, unpublished results; EST sequences deposited in GenBank with accession nos. CO377782–90). These were aligned with homologs from Bombyx mori, available from SilkBase (http://www.ab.a.u-tokyo.ac.jp/silkbase/). Primers were then designed on the basis of the H. erato sequence, but situated in regions of high sequence conservation between Bombyx and Heliconius. Primers for Vermilion were designed from a cDNA sequence obtained from Robert Reed (GenBank accession no. AY691422). Primers for Apterous and Patched were developed by A. Tobler (personal communication). Loci used for mapping were first amplified in the two parents of the brood and sequenced. A standard amplification protocol was used with a 20-μl reaction containing 1× NH4 buffer, 2.0 mm MgCl2, 0.8 mm dNTPs, 0.4 mm of each primer, and 0.02 units/μl BioTaq polymerase (Bioline, London) and was run with a cycling profile of 94° for 2 min, followed by 30 cycles of 94° for 20 sec, 55° for 40 sec, and 72° for 1 min. Several distinct strategies were employed to search for variation for mapping. First, 5 μl of this product was run on a 1.5% agarose gel stained with ethidium bromide to search for length variation between parental alleles. If no length variation was evident, the PCR products were sequenced. In some cases PCR products were cloned using the pGEM-T Easy Vector System (Promega, Madison, WI) and five to six clones were sequenced for each parent. Cloning and sequencing protocols are as described previously (Beltrán et al. 2002). Alternatively, PCR products were sequenced directly and the sequence chromatograms were searched for double peaks indicative of heterozygous sites. Once polymorphic sites were identified, either as double peaks in directly sequenced chromatograms or as variability between clone sequences, the sequences were searched to identify diagnostic restriction enzymes at variable sites.
TABLE 1.
Anchor loci and primers
| Gene name | Abbreviation | Primer sequences | Intron(s)? |
|---|---|---|---|
| Dopa-decarboxylase | Ddc | Ddc-fo: CAGAGGGTCAAGGAACAGCAC | Yes |
| Ddc-ri: CGAAAGCGCAAGACGATGTAG | |||
| Decapentaplegic | Dpp | Dpp-f34: AGAGAACGTGGCGAGACACTG | No |
| Dpp-r327: GAGGAAAGTTGCGTAGGAACG | |||
| Patched | Ptc | Ptc-F(7): CTCCGAAGAAGGTCTGCCGCAAG | Yes |
| Ptc-R(364): AATTCGTGCTCGTCGTATTTTC | |||
| Cubitus interruptus | Ci | Ci-fi: ATGCGGAGACATACTGGTGAA | Yes |
| Ci-ro: TGTATCTTTTAGTGCAACCCG | |||
| Apterous | Ap | Ap-f35: TGAATCCTGAATACCTGGAGA | No |
| Ap-r286: CTTTTCCGCCATTTTGTTCTC | |||
| Vermilion | V | Ve-220f: CGCTCGACCTTATGGATTTC | Yes |
| Ve-465r: GAGTGTTAAGTCCCGGCGTA | |||
| Ribosomal protein L3 | RpL3 | RpL3-F59: CCGTCATCGTGGTAAAGTGA | Yes |
| RpL3-R537: TCTCCATGATATGGGCCTTC | |||
| Ribosomal protein L5 | RpL5 | RpL5-F23: CACCCAAGTACCGTTTGATTG | Yes |
| RpL5-R441: CATATCCTGGGAATCTCTTGATG | |||
| Ribosomal protein L10a | RpL10 | RpL10-F68: AATGCGGTCCTTCAATCATC | Yes |
| RpL10-R665: GTCCCATGGTGGACTTCATA | |||
| Ribosomal protein L11 | RpL11 | RpL11-F138: CTGTGTCGGTGAATCTGGTG | Yes |
| RpL11-R571: CTGTTGGAACCACTTCATGG | |||
| Ribosomal protein L19 | RpL19 | RpL19-F131: CCCGACAGAACATCCGTAAG | Yes |
| RpL19-R529: CGGGCTTCCTTCACTCTGT | |||
| Ribosomal protein P0 | RpP0 | RpP0-F142: ACCCAAAATGTTTCATCGTG | Yes |
| RpP0-R565: TCACCAGGCTTCAAGATGTG | |||
| Ribosomal protein S5 | RpS5 | RpS5-F31: GGTTGAGGAAAACTGGAACG | No |
| RpS5-R535: CAACAGCCAGATAGCCTGGT | |||
| Ribosomal protein S8 | RpS8 | RpS8-F56: GCCCATTCGTAAGAAGAGGAA | Yes |
| RpS8-R624: CTTTGCCCTCTTGGATTTGA | |||
| Ribosomal protein S9 | RpS9 | RpS9-F12: GCCATCATGGTGAACAACAG | Yes |
| RpS9-R601: TCTTCCTCCTCATCATTGGT | |||
| Elongation factor 1α | Ef1a | EF1H-F: GAGAAGGAAGCCCAGGAAAT | No |
| EF1H-R: CCTTGACRGACACGTTCTTT | |||
| Long-wavelength opsin | OPS1 | 5′-RACER: TGGTTACAATAGGGCTGA | Yes |
| RLWFD4: ACAAAAATCAATGGGAGAGTA | |||
| Triose-phosphate isomerase | Tpi | Tpi-1: GGTCACTCTGAAAGGAGAACCATCTT | Yes |
| Tpi-2: CACAACATTTGCCCAGTTGTTGCCAA | |||
| Mannose-phosphate isomerase | Mpi | Mpi-F: ATTCAAGCTCATCCAACTAAGG | Yes |
| Mpi-R: TTATGAAGTTGTTCTGCATGGT |
Full gene names, abbreviations, and primer sequences are shown. Primers are shown 5′–3′ with the forward primer listed first. “Introns (?)” indicates whether the fragment amplified contains one or more introns. OPS1, Tpi, Mpi, ci, dpp, and Ddc primers were published previously (Hsu et al. 2001; Beltrán et al. 2002; Tobler et al. 2004).
All individuals for the mapping family were amplified using the standard amplification protocol described above. If length variation occurred between parental genotypes, PCR products were visualized in 1.5% agarose gels stained with ethidium bromide. Alternatively, enzyme digests were carried out using 10 μl of the PCR reaction, following manufacturer's protocols, prior to visualization of the product.
Genome size estimation:
The size of the H. melpomene genome was determined using flow cytometery after Bennett et al. (2003). The brain of a single adult H. melpomene and the head of a single adult Drosophila melanogaster Iso-1 female standard were added to 1 ml of cold Galbraith buffer (Galbraith et al. 1983) in a 1.5-ml Kontes dounce tissue grinder, stroked 15 times with an A pestle, and filtered through a 20-μm nylon filter. Propidium iodide was added to a final concentration of 50 ppm, and the mixture costained in the dark at 4° for 30–40 min. The mean fluorescence of costained nuclei in five replicate samples of each sex was quantified using a Coulter Epics Elite (Coulter Electronics, Hialeah, FL) with a laser tuned at 488 nm and 45 mW. Fluorescence was detected by a photomultiplier screened by a long-pass filter. DNA content was determined by comparing the ratio of the 2C mean of the sample with the 2C mean for Drosophila (1C = 175 Mb).
Linkage analysis:
Dominant markers such as AFLPs typically segregate in a 3:1 ratio in F2 mapping families resulting from crosses between inbred lines. In a single family, markers are inherited in one of two linkage phases, depending on whether the band-present genotypes are present in parental strain A or B. In contrast, the loci analyzed here were derived from two outbred populations with considerable genetic variation present in parental individuals. As a result, in a large number of backcross markers (BC) the AFLP band was present in one F1 parent only, resulting in a 1:1 segregation ratio in the F2 progeny. When these backcross markers were present only in the F1 female, they were termed female informative, and when present only in the F1 male, they were termed male informative. Linkage mapping in Lepidoptera is also unusual because female meiosis is achiasmatic; i.e., recombination within chromosomal linkage groups is restricted to males (Suomalainen et al. 1973; Heckel et al. 1999). These peculiarities facilitate the generation of an integrated linkage map from dominant markers, using the following three-step method.
Step 1—identification of the chromosome print:
Female-informative BC markers on the same chromosome are inherited in complete linkage and can be used to determine for each individual offspring whether the nonrecombinant chromosome is derived from the maternal grandmother or grandfather, a pattern known as the “chromosome print” (Yasukochi 1998). In contrast to the analysis of Yasukochi (1998), we can use the female-informative BC markers to observe the chromosome print directly, instead of inferring it from the segregation patterns of F2 markers. AFLP loci were first sorted according to parental genotypes. All female-informative markers were grouped into linkage groups with a LOD score of >8 using the program JoinMap 3.0. Of course, we actually expect complete linkage and a maximal LOD score, but in reality a percentage of genotyping errors need to be accounted for in the analysis. These linkage groups were then inspected by eye and a single chromosome print was determined for each linkage group by assuming that apparent recombinant genotypes observed at a single locus were genotyping errors.
Step 2—grouping of F2 markers into linkage groups:
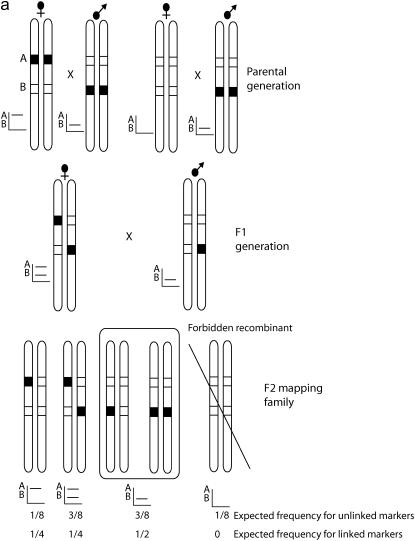
This chromosome print was then used to group F2 markers (i.e., loci with a band-present genotype in both F1 parents) into linkage groups (Figure 1) on the basis of the method of forbidden recombinants (Shi et al. 1995a; Heckel et al. 1999). When a chromosome print is compared with an F2 marker, there is one class of genotypes that is impossible if the two are found on the same chromosome (Figure 1). We carried out this analysis using Joinmap and identified linkage groups that were supported with LOD scores of between 3 and 4 and included F2 markers with a chromosome print. The expected pattern was then confirmed manually and any markers with more than three forbidden recombinants in all linkage group comparisons were excluded from further analysis. For the 73 offspring in our mapping family, we expect on average 9.1 (73/8) forbidden recombinants for unlinked loci. The binomial probability of getting only three forbidden recombinants for unlinked loci, given 73 offspring, is 0.011. Hence three forbidden recombinants was chosen as an arbitrary cutoff, indicating that the markers were linked but taking into account some genotyping errors.
Figure 1.
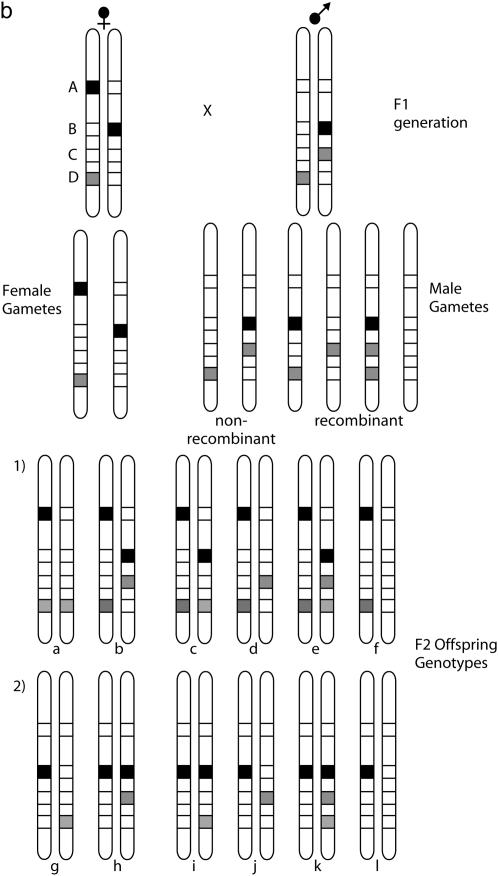
Segregation of markers in an F2 mapping family. (a) The segregation of two markers demonstrating assignment of an F2 locus to a linkage group by the forbidden recombinant method. Two loci are shown: A, inherited as a female-informative locus and corresponding to the chromosome print for this linkage group, and B, a locus that shows a fixed difference between the two parental strains and is inherited as an F2 marker in a 3:1 ratio of band present:band absent in the mapping family. The AFLP banding pattern for the two markers alongside each diploid genotype is shown. Occurrence of forbidden recombinant genotypes, in which both bands are absent, would indicate that the two markers are not syntenic. These genotypes would be expected to occur in one-eighth of the F2 family for unlinked markers. The offspring shown in the box are those that are rescored as missing data for analysis of the recombination map. (b) In more detail, the problem of linkage analysis between dominant markers inherited in repulsion, demonstrated here by loci B and D. Locus C is a male-informative marker that can be used to link B and D into a single map. The first group of progeny, a–f, are informative with respect to linkage of B and C, with c and d being the recombinant genotypes. The second group, g–l, are informative with respect to linkage of C and D. Note that because of dominance, a–f are uninformative with respect to locus D, and g–l are uninformative with respect to locus B.
Step 3—calculation of a backcross recombination map:
The F2 markers were then converted into BC markers for analysis of recombination distances as follows. For each linkage group, the F2 markers were separated into two groups depending on linkage phase, i.e., whether the band-present genotype was derived from H. m. cythera or from H. m. melpomene (depicted as markers B and D in Figure 1b). All individuals that have inherited the band-present allele on the maternal nonrecombining chromosome must show the band-present genotype and therefore the genotype of the paternal recombinant chromosome cannot be determined for half the F2 individuals (Figure 1b). These individuals are thus excluded from the analysis and are scored as missing data. The remaining 50% of the offspring have the band-absent allele on their nonrecombinant chromosome, and thus the genotype of the recombinant chromosome can be determined with certainty. For each chromosome, this analysis thus produces two nonoverlapping sets of genotypes for the F2 markers inherited in repulsion; i.e., for markers derived from a band-present genotype in H. m. cythera, one-half of the F2 offspring have genotypes for recombinant analysis, but this same group of individuals has been excluded from the analysis for all markers derived from a H. m. melpomene band-present genotype (F2 genotype groups 1 and 2 in Figure 1b).
The F2 markers inherited in repulsion to one another cannot be combined directly into a recombinational linkage map. However, here the male-informative BC markers that have recombinant genotypes for all offspring can be used to produce an integrated map for markers derived from both populations (Figure 1b). To carry this out, the complete data matrix of F2 and male-informative BC markers was run in Joinmap and linkage groups were identified.
The Z chromosome was analyzed separately by first searching for AFLP markers that were present in both parents and present in all male offspring, but segregated in a 1:1 ratio in female offspring—i.e., those markers in which the band-present allele was present as a single copy in both parents. Male offspring were then rescored as missing data and the remaining female offspring genotypes were analyzed with the male-informative data set to identify markers linked to these loci (i.e., those markers present only in the father that segregate in a 1:1 ratio in all offspring). These loci were then treated as discussed below for calculation of the Z chromosome recombination linkage map using Mapmaker.
Female- and male-informative data from codominant markers were also analyzed separately. Female informative alleles were first used to determine linkage group assignments by comparing the chromosome prints using Joinmap. As with the AFLP markers, LOD scores of >8 indicated that markers were syntenic. Male-informative alleles were scored separately to carry out recombination mapping for each chromosome.
Recombination linkage maps were initially assembled using JoinMap 3.0. Each linkage group was tested for improbable genotypes, i.e., genotypes indicative of a double recombination event on either side of the locus in question. When a single locus is responsible for many such genotypes, this can indicate genotyping errors. AFLP chromatograms for problem loci were carefully checked and genotypes were rescored where necessary. Several problematic loci were removed from the analysis at this stage, usually when it seemed possible that two or more loci were so similar in size that accurate scoring was impossible. In addition, we identified several loci in which two bands scored on the AFLP gel as distinct loci were in fact alternate alleles at the same locus, as indicated by very tight linkage. Such cases were rescored as a single locus.
Final recombination linkage maps were produced using a likelihood analysis implemented in Mapmaker 3.0 (Lander et al. 1987). Data were rescored in F2 backcross format by manual estimation of linkage phase and rescoring of all loci in the same linkage phase. Linkage orders for small linkage groups (fewer than eight loci) were determined by comparison of the likelihoods of all possible linkage orders using the “compare” command. For larger linkage groups, a subset of markers was analyzed in a similar manner, and then remaining markers were added singly using the “try” command. Final linkage orders for all chromosomes were tested using the “ripple” command, which compares the likelihoods of alternative linkage orders among groups of adjacent markers. The final “best” linkage order chosen was then used to estimate recombination distances using the “map” command. The male-informative genotype data used in this analysis are available as supplementary material at http://www.genetics.org/supplemental/. Finally, the most likely position of the color pattern locus was investigated first by assigning it to a linkage group using the method of forbidden recombinants and then by calculating the likelihood of placing the locus in each interval along that linkage group using the “try” command in Mapmaker.
RESULTS
Color pattern genetics:
A total of 152 offspring were obtained for brood 33, although some of these could not be scored for color pattern due to failure at emergence. This total does not include brood contaminants that were identified during genotyping (see below). Among these offspring, four color pattern loci that segregated in Mendelian ratios were identified. The Yb locus controls the underside hind-wing yellow bar of H. m. cythera (Figure 2). This locus was originally identified in crosses carried out using the same race, H. m. cythera, as studied here (Emsley 1964), but was more fully described in later crosses involving races from other geographic areas, such as Brazil (Sheppard et al. 1985) and Peru (Mallet 1989). We denote here the allele specific to H. m. cythera as ybc. The phenotypic effect of ybc is distinct from that of yb, as the yellow band is expressed only on the underside of the hind wing in H. m. cythera as opposed to the upper and underside expression associated with yb in races from other areas. Heterozygotes in all cases can be identified by a “shadow” bar of reflective melanic scales (note that this phenotype is not shown in Figure 2), giving an observed monohybrid segregation ratio of 43:72:37 for no bar:shadow:yellow bar expression (YbYb:Ybybc:ybcybc).
Figure 2.
Parental and hybrid phenotypes. (A) H. melpomene cythera (ventral), (B) H. melpomene melpomene (dorsal), and the four major hybrid phenotypic classes. The parental genotypes are ybc ybc sbsb AcAc KK for H. m. cythera and YbYb SbSb acac kk for H. m. melpomene. Note that the phenotypes generated by the loci K and Ac are not expressed in either parental strain.
The Yb locus is tightly linked to Sb, which controls the marginal white band on the hind wing of H. m. cythera. In our cross, the two phenotypes were inherited in complete association, except for a single recombinant genotype that expressed the yellow HW bar but not the white margin, a phenotype that we interpret as being ybybSb−. As recombinants would have been detected in only 50% of the F2 family (i.e., in individuals that inherit the recessive ybsb chromosome from their mother), this is equivalent to a recombination rate of 1.3% (1/76). Previous H. cydno × H. melpomene and H. cydno interracial crosses have revealed similar recombinant phenotypes, suggesting that these actually are two distinct loci and are most probably homologous in the two species (Linares 1996, 1997; Naisbit et al. 2003). Other color pattern characters seen segregating in this brood could not be used for mapping due to the small number of informative offspring (the loci K and Ac; Figure 1).
Linkage analysis:
A total of 24 AFLP primer combinations were scored for a subset of the mapping family, consisting of 94 individuals including the parents and grandparents of brood 33. The primer combinations used are shown in Figure 3 and in supplementary material at http://www.genetics.org/supplemental/. A total of 649 polymorphic bands were scored, excluding bands that were extremely close together (<2 bp) and could not be easily separated. Of those, a total of 502 polymorphic loci showed meaningful segregation patterns, given the parental genotypes, and did not show significantly skewed segregation ratios (G-test at P < 0.01). These consisted of 194 F2 loci (i.e., loci with the band present in both parents and a 3:1 segregation of presence:absence in the family), 167 male-informative, and 141 female-informative backcross loci (i.e., loci with the band present in either the father or the mother, respectively, and a 1:1 segregation ratio in the offspring). Among the excluded loci there were no well-supported linkage groups that might have indicated that genomic regions showing significant segregation distortion were being excluded from the analysis. During analysis, we identified eight individuals that had been misidentified and were not members of this brood (these individuals were also excluded from the phenotypic analysis described above), and another seven individuals failed to produce consistent results in the AFLP gels. This is a commonly observed, if rarely reported, result in AFLP analysis and may be due to incomplete digestion of genomic DNA (D. Heckel, personal communication). The final analysis was therefore carried out with 73 offspring.
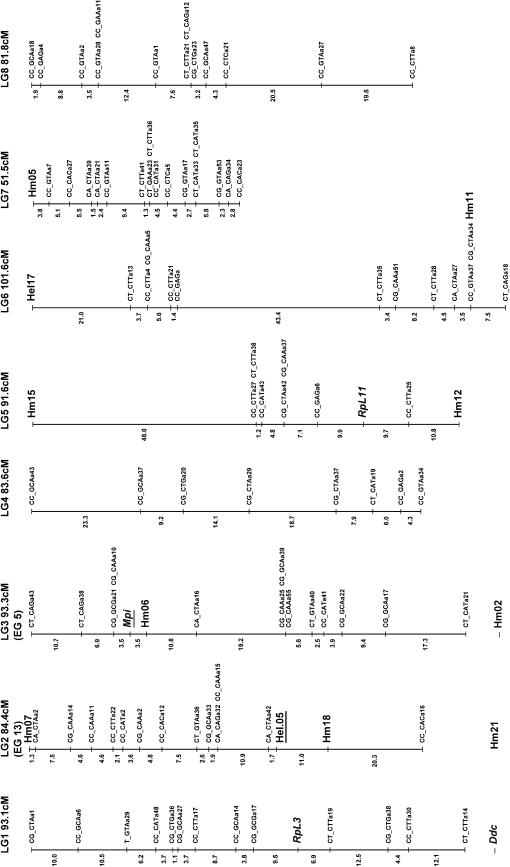
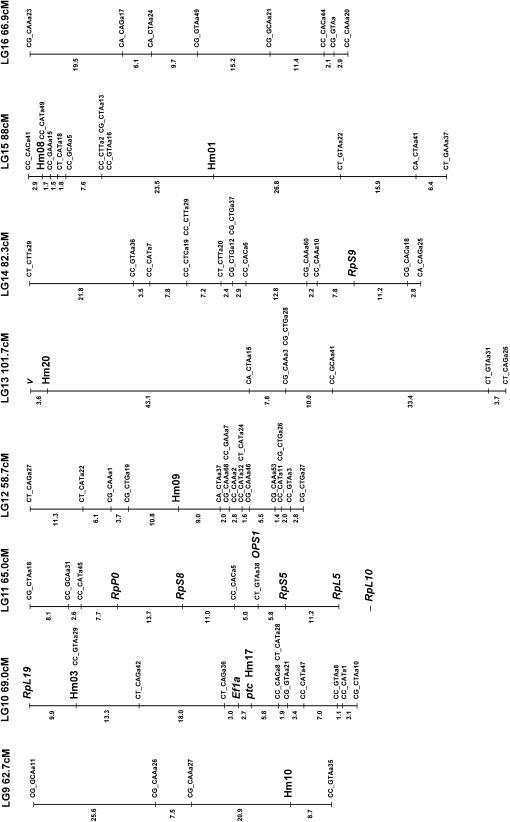
Figure 3.
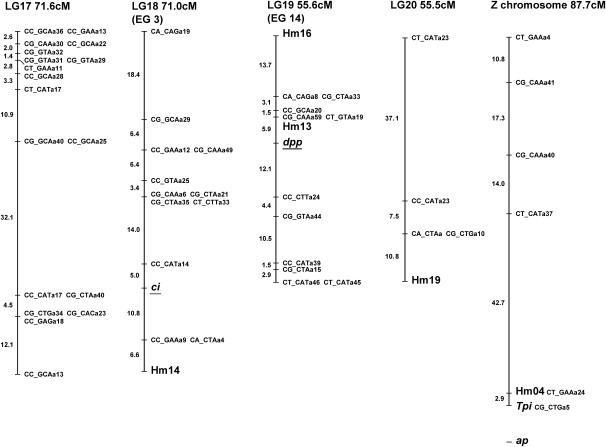
Linkage map of H. melpomene. Microsatellite and SCNP markers are shown in larger type, using standard gene nomenclature for the latter. All remaining markers are AFLPs and are coded with the two selective EcoRI nucleotides, followed by the three selective MseI nucleotides, and finally an identifier code; e.g., CT_CAGa38 is band a38 scored from the EcoRI-CT MseI-CAG primer combination. Markers assigned to linkage groups for which there is no information on map position are shown separately below the linkage group. Possible homology with linkage groups in H. erato is shown in parentheses below each linkage group number with the H. erato linkage group number; e.g., linkage group 2 (LG2) corresponds to linkage group 13 in H. erato (EG13; see Tobler et al. 2004) on the basis of the shared Hel.05 marker. These comparisons are based on a single marker in each case so chromosomal homology is tentative. The marker used to establish homology is underlined in each case.
Of the 141 female-informative AFLP loci, 121 fell into 21 linkage groups (corresponding to the 21 chromosomes of H. melpomene) with LOD scores >8.0, the remaining 20 loci failing to form well-supported groups. Three linkage groups showed some evidence for segregation distortion, although this was not significant in any case (LG1 28:43, G1 = 1.59, NS; LG6 27:41, G1 = 1.45, NS; LG11 28:43, G1 = 1.59, NS). The lack of strong segregation distortion in these linkage groups suggests that marked genomic incompatibilities are not expressed in this brood. This further justifies the exclusion of loci that deviated strongly from expected Mendelian ratios, as these most likely were subject to genotyping errors.
The F2 loci were then analyzed in Joinmap along with the chromosome print of the 20 autosomes. Of the 194 F2 loci included in this analysis, 134 grouped into the 20 autosomal linkage groups with LOD scores >3.0. Of the 134 markers, 23 showed between one and three forbidden recombinant genotypes, indicating genotyping errors. The small size of the mapping family compounded by genotyping errors probably explains why the 60 remaining loci failed to group into chromosomal linkage groups. These F2 loci were then converted into male informative loci as described in materials and methods. These converted F2 markers were then analyzed in Joinmap with the 1:1 male-informative markers. A total of 138 male-informative markers grouped into linkage groups with the converted F2 markers. Separate data files were then assembled for each linkage group and recombination maps were constructed for each chromosome. In three cases, linked AFLP markers generated from the same primer combination were identified as most likely alternate alleles at the same locus. In one case, two dominant loci could be rescored as a single codominant locus.
The microsatellite loci all gave readily interpretable segregating variation, except for three loci (Hel.04, Hel14, and Hel02) that were monomorphic and another that gave inconsistent banding patterns (Hel08). Where available, female-informative alleles were identified from the maternal genotype, scored separately, and analyzed to test for the chromosomal assignment of each locus. Male-informative alleles were then scored and included in the recombination map for the relevant linkage group.
A total of 19 SCNL loci were successfully amplified and their identity confirmed by translated BLAST (tBLASTx) searches of publicly available databases. All loci showed either length variation or segregating sites that could be scored with diagnostic restriction enzymes and allowed genotyping of offspring. Only the wingless gene, sequenced using primers described previously (Brower and Egan 1997), showed single-nucleotide polymorphisms in the parents, but no diagnostic restriction digests were found. Furthermore, in a few cases we were able to obtain only female-informative linkage information, allowing assignment of markers to chromosomes but not recombination mapping (Figure 3). Linkage analysis was carried out as for microsatellite loci.
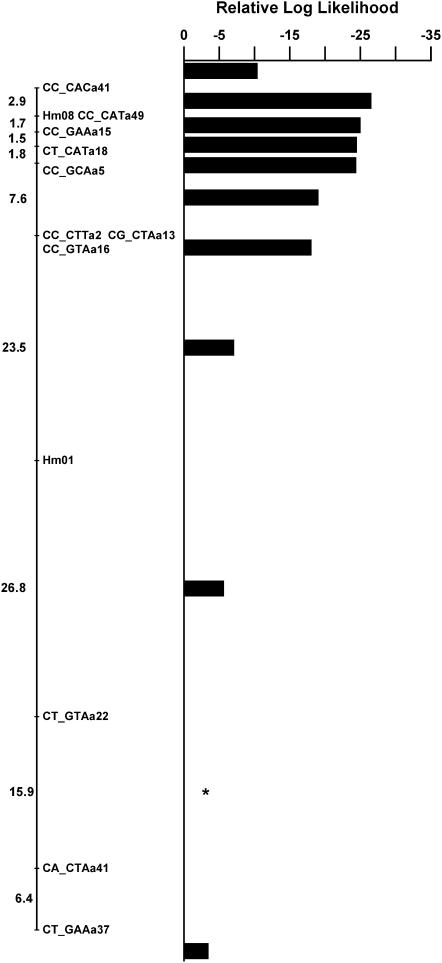
The color pattern locus Yb was unambiguously assigned to linkage group 15 by comparing the chromosome prints of all linkage groups. The likelihood of placing this gene into all intervals along this linkage group was then calculated. Marker density in the region of the color pattern locus was fairly low, but it was nonetheless possible to isolate the locus to a region 22 cM in length (Figure 4). A single AFLP band was extremely close to the Yb locus with no recombinants found in the 73 offspring genotyped.
Figure 4.
Placement of the color pattern locus Yb on linkage group 15. The log likelihood is shown for placement of the color pattern locus in each mapping interval along the linkage group, relative to the most likely location that is set at zero and is indicated by a star. Values <−2 indicate a significantly worse fit to the data at ∼95%, so placement of the locus in all intervals except for 2 can be ruled out. The Yb locus therefore must be located in the 22-cM interval between the markers CT_GTAa22 and CT_GAAa37.
Genome size estimation:
Genome sizes of the male and female H. melpomene were nearly identical and two-thirds larger than the genome of D. melanogaster. The mean and standard error of the genome size in megabase pairs were 1C = 292 ± 1.8 Mb and 1C = 292.2 ± 2.4 Mb, for the female and male, respectively. This is 74% as large as the genome of the closely related species H. erato (1C = 395 and 396 for the H. erato male and female, respectively).
DISCUSSION
The detailed linkage map presented here lays the foundation for future studies of the development and evolution of mimetic color patterns in Heliconius butterflies. The map is derived from a single mapping family derived from outbred strains of H. melpomene, includes 219 AFLP loci, 23 microsatellite loci, and 19 SCNL, with at least 5 loci on each of the 21 linkage groups. This map is therefore a tool that can be used to identify and study the evolutionary history of markers linked to regions controlling adaptive variation in H. melpomene. As a first step toward this goal, one color pattern locus is included in the map described here. Furthermore, we have identified a single AFLP band that is located within a centimorgan of the color pattern locus. In the future, the markers and techniques described here can be used to map all the known H. melpomene color pattern loci onto a single linkage map, to map other candidate genes, and to generate a comparative analysis of the genes underlying the convergent patterns found in the co-mimic species H. erato.
The total map length of the 21 H. melpomene chromosomes is 1616 cM and the estimated genome size is 1C = 292 Mb. Compared to a recent estimate of map length in H. erato of 2400 cM (Tobler et al. 2004) and a genome size estimate of 395 Mb, our genome size estimate for H. melpomene is 74% of the genome size of H. erato and the map length is 67% of that estimated for H. erato. The discrepancy in genome size and map length is surprisingly similar, although map length estimates for both species are likely to be revised as more detailed maps are generated. Given the 292-Mb genome size of H. melpomene, the relationship between physical and recombination distance is ∼180 kb/cM, which is similar to that estimated for B. mori (Yasukochi 1998). The average chromosome length is 77 cM with surprising consistency in length among chromosomes (Figure 3).
The three-step method described here shows how it is possible to construct an integrated linkage map from dominant markers with a relatively small sample size of loci and individuals. This is likely to prove useful in studies of Lepidoptera and other insects (including Drosophila species) in which one sex is achiasmatic and crosses are carried out between outbred populations. In our final analysis using Mapmaker, the brood is considered as if it were a backcross, so the likelihood estimates of recombination distances assume that all segregating variation is derived from the male parent (Lander et al. 1987). Hence, the linkage map is not directly affected by the lack of recombination in females. Nonetheless, it is hoped that this and future studies will stimulate the development of linkage analysis software that is specifically designed to accommodate the lack of recombination and complicated segregation patterns shown in data sets such as this (Shi et al. 1995b; Heckel et al. 1999).
One major goal of this work is to generate integrated linkage maps in which all the loci that control geographic variation in H. melpomene can be compared with homologous regions in H. erato. The various color pattern genes that have been identified in both species are found in different allelic combinations in different geographic races (Sheppard et al. 1985), so only a few of the many loci segregate in any one mapping family. Therefore, to generate an integrated map for a single species, anchor loci that can be scored reliably in different crosses between races of the same species are needed. For this goal the best markers are likely to be microsatellites, given the facility with which segregating variation can be scored among members of the same species. Only 3 of the loci scored here were monomorphic and therefore impossible to map giving a success rate of almost 90% among the loci scored for this mapping family. Our map now contains 23 microsatellite loci distributed across 14 chromosomes (Figure 2).
Nonetheless, a striking result was that more than half of the microsatellite loci used showed patterns of segregation that were best interpreted by hypothesizing the presence of null alleles (Table 2). In some cases nulls were “recovered” by redesigning primers from the original clone sequence to amplify a larger PCR product. However, poor primer design alone probably cannot account for the high frequency of nulls. One possibility might be that priming sites were too close to repeat regions, but the average distance between priming sites and repeat regions in our loci is comparable to other insect microsatellites (J. Mavarez and M. Gonzalez, unpublished results). It has been suggested previously that microsatellite loci are particularly problematic in the Lepidoptera and often show a higher-than-expected rate of null alleles. However, the reasons for this remain unclear (Meglecz and Solignac 1998; Daly et al. 2004). In general, we were able to map such loci, as segregation patterns were readily interpretable. However, in the few cases where null/null genotypes occurred in the brood, this required careful confirmation of genotypes by rechecking that the lack of amplification products genuinely represented a null genotype.
TABLE 2.
Microsatellite loci and primer sequences
| Locus | Primer sequence | Cross type | Allele sizes (bp) |
|---|---|---|---|
| Hm01 | CGCGGTAGAAATAGCACAAG | AB × C0 | 159, 168, 176, null |
| CGAGAAGCCCTACAAGTGTG | |||
| Hm02 | TATTTGCACGATGGAAACCC | AB × A0 | 170, 178, null |
| GCGAGGTGGAGACAAAAGAC | |||
| Hm03 | GACGTCACAGCGGGGAAC | AB × CD | 298, 324, 376, 444 |
| AGAGGGGAACGGAGTGTCAT | |||
| Hm04 | CCTGGCTTATCTACGACGACA | AB × A | 378, 392 |
| ATGCAGCTTACTCGCTGGTT | |||
| Hm05 | GCGGTAAGGTAAAACCGTGA | A0 × B0 | 150, 161, null |
| CAGAAGAAAATGGTTGGATGG | |||
| Hm06 | AAATAGTGTGCGGCGGAATA | AB × AA | 219, 221 |
| TGGAGTAGAAATGCGGGTTTA | |||
| Hm07 | GCAGAGGGAACCTCGTGTTA | A0 × B0 | 264, 266, null |
| CGCAGTTTGTGCGAATTACA | |||
| Hm08 | AAAGCCTGAGTGCCGTATTG | AB × CD | 285, 293, 295, 299 |
| GCAATGTCAGCATCGAATGT | |||
| Hm09 | m13-CAACTGCAATGACCCATCAC | A0 × B0 | 200, 218, null |
| AATGTCGTGCTCCCATGAAG | |||
| Hm10 | m13-GGCCGCTTTGTAAGAATGTC | AB × CD | 220, 242, 246, 268 |
| TGTGTAAATGAAATCCATAATTGGTC | |||
| Hm11 | m13-TTCTGGTGTCTAGCGGTTATG | A0 × B0 | 404, 416, null |
| AATAGCGACCATGCTGAGAG | |||
| Hm12 | m13-TGTCTTATCATTGGCGTTGC | AB × CD | 255, 263, 264, 281 |
| CAACCGTCGTTCCAGACG | |||
| Hm13 | m13-TCACTAGTTTTCGGCTTATCG | A0 × B0 | 185, 195, null |
| AAGGCTAAATGATGCCTAAAG | |||
| Hm14 | m13-ATGCTGTAACCCGCATAGC | A0 × B0 | 157, 195, null |
| TGCATTTATGATGTAAAAGTTCG | |||
| Hm15 | m13-TTTCGCCACCATAATCTTTC | AB × 00 | 234, 241, null |
| CACATCGCAGGTATTCCATC | |||
| Hm16 | m13-CGGATAGACATTTGTTAAAGTGTG | AB × BC | 266, 282, 291 |
| ACGAGGATGCGGACTACG | |||
| Hm17 | CAACGAGATATTCCAGAGAGTGTT | A0 × B0 | 355, 375, null |
| TGGAACGTATAGAAAGCGCA | |||
| Hm18 | AATTACATATCGTTTCATTA | AB × C0 | 218, 224, 254, null |
| CTACGACAAGACCCTCCTGA | |||
| Hm19 | m13-CGCTAATTCAAAGGAAAGAGGA | AB × C0 | 170, 174, 198, null |
| AGTGCTGTCATGGCTAACGA | |||
| Hm20 | GGGATCGATGAAAAAGAGCA | AB × B0 | — |
| AGAGCCTTCATTTACCCCGT | |||
| Hm21 | GAACTCCAGAAGGTTACCCCA | AB × BB | — |
| GCCGGTCTTTGTCTATTGGA | |||
| Hel.05 | TGCTGTCCATACCCAACTCA | AB × BC | 280, 294, 330 |
| CGAACTCACAACCATCAGTCA | |||
| Hel.17 | GGCGCACAGACGAGACTAC | A0 × B0 | 201, 207, null |
| ACTGCCGCACGAATAATAAC |
Primers are shown 5′–3′ with the forward primer listed first. The m13 sequence used in amplifications with m13-tailed primers is 5′-CACGACGTTGTAAAACGAC-3′ (see text for details). Segregation patterns and allele sizes are also shown. Hm04 is sex linked. Null alleles are shown as 0 in the segregation pattern column. Hm20 and Hm21 were scored on agarose gels so exact allele sizes are not shown.
For the goal of comparative linkage mapping between H. erato and H. melpomene, the utility of microsatellites is limited because only a relatively small proportion of loci cross amplify in such distantly related species. For example, in a recent study of microsatellites in H. erato only half of the described loci worked in H. melpomene (Flanagan et al. 2002). In contrast, all the SCNL primers that have been tested so far amplify equally well in both species, suggesting that these loci are likely to be the most useful for comparative mapping. The map presented here contains 19 SCNL loci distributed across eight chromosomes. Indeed, we can already identify possible homology with the H. erato map for markers on four different chromosomes, albeit with only one marker per chromosome (three SCNL and one microsatellite; see Figure 2).
Another major reason to use SCNL markers is to map candidate genes with the goal of determining the identity of the switch gene loci. Six loci included on the present map were included as candidates: Deca-pentaplegic, Patched, Cubitus Interruptus, Apterous, and the pigment pathway enzymes, Vermilion and Dopa-Decarboxylase. None of these mapped to the same linkage group as the single color pattern locus Yb. Additional SCNL loci will be useful as anchor loci for comparative mapping both in Heliconius and across the Lepidoptera (Heckel 1993). In particular, the ribosomal proteins are useful as they show extremely high sequence conservation between Heliconius and the moth B. mori and are also distributed widely across the genome. We have identified a cluster of five ribosomal proteins on a single linkage group (LG11) that was not predicted from linkage relationships of the homologous loci in Drosophila. Linkage groups of conserved loci such as these will prove useful in estimating the degree of synteny across the Lepidoptera, as these loci can now be readily mapped in other species.
This linkage map represents the first step toward identifying genomic regions involved in color pattern evolution in H. melpomene. Indeed, here we have identified an AFLP marker that is within a centimorgan of the color pattern locus Yb. If this marker can be cloned and converted into a codominant marker, it will prove useful as a probe for identification of BAC clones covering the region of interest. Our map will also allow an estimate of the degree of conservation of synteny between H. melpomene and other Heliconius species where mapping projects are underway, notably H. numata and H. erato. Once conserved linkage relationships are established for regions of interest, this will allow a test of whether genes for mimicry are homologous among distantly related Heliconius species. This is of particular interest in the case of H. melpomene and H. erato where convergence of pattern due to mimicry is so precise and where it has been hypothesized that the same switch genes might be involved in both cases (Nijhout 1991).
Acknowledgments
We thank Robert Reed, James Mallet, Durrell Kapan, and Mathieu Joron for discussion and sharing of sequences. We thank the Smithsonian Tropical Research Institute and the Biotechnology and Biological Sciences Research Council for financial support during this work. C.D.J. is funded by the Royal Society.
References
- Beltrán, M., C. D. Jiggins, V. Bull, W. O. McMillan, E. Bermingham et al., 2002. Phylogenetic discordance at the species boundary: gene genealogies in Heliconius butterflies. Mol. Biol. Evol. 19: 2176–2190. [DOI] [PubMed] [Google Scholar]
- Bennett, M. D., I. J. Leitch, H. J. Price and J. S. Johnston, 2003. Comparisons with Caenorhabditis (∼100 Mb) and Drosophila (∼175 Mb) using flow cytometry show genome size in Arabidopsis to be ∼157 Mb and thus ∼25% larger than the Arabidopsis initiative estimate of ∼125 Mb. Ann. Bot. 91: 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, W. W., 1972. Natural selection for Müllerian mimicry in Heliconius erato in Costa Rica. Science 176: 936–939. [DOI] [PubMed] [Google Scholar]
- Blum, M. J., 2002. Neotropical hybrid zone stability and formation. Ph.D. Thesis, Duke University, Durham, NC.
- Brower, A. V. Z., and E. G. Egan, 1997. Cladistic analysis of Heliconius butterflies and relatives (Nymphalidae: Heliconiiti): a revised phylogenetic position for Eueides based on sequences from mtDNA and a nuclear gene. Proc. R. Soc. Lond. Ser. B 264: 969–977. [Google Scholar]
- Brown, K. S., 1981. The biology of Heliconius and related genera. Annu. Rev. Entomol. 26: 427–456. [Google Scholar]
- Brunetti, C. R., J. Selegue, A. Monteiro, V. French, P. M. Brakefield et al., 2001. The generation and diversification of butterfly eyespot color patterns. Curr. Biol. 11: 1578–1585. [DOI] [PubMed] [Google Scholar]
- Carroll, S. B., J. Gates, D. N. Keys, S. W. Paddock, G. E. F. Panganiban et al., 1994. Pattern formation and eyespot determination in butterfly wings. Science 265: 109–114. [DOI] [PubMed] [Google Scholar]
- Daly, D., K. Walthan, J. Mulley, P. C. Watts, A. Rosin et al., 2004. Trinucleotide microsatellite loci for the peppered moth (Biston betularia). Mol. Ecol. Notes 4: 179–181. [Google Scholar]
- Emsley, M. G., 1964. The geographical distribution of the color-pattern components of Heliconius erato and Heliconius melpomene with genetical evidence for the systematic relationship between the two species. Zoologica 49: 245–286. [Google Scholar]
- Flanagan, N. S., A. Davison, M. Alamo, R. Albarran, K. Faulhaber et al., 2002. Characterisation of microsatellite loci in neotropical Heliconius butterflies. Mol. Ecol. Notes 2: 398–401. [Google Scholar]
- Galbraith, D. W., K. R. Harkins, J. M. Maddox, N. M. Ayres, D. P. Sharma et al., 1983. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051. [DOI] [PubMed] [Google Scholar]
- Gilbert, L. E., 1991. Biodiversity of a Central American Heliconius community: pattern, process, and problems, pp. 403–427 in Plant-Animal Interactions: Evolutionary Ecology in Tropical and Temperate Regions, edited by P. W. Price, T. M. Lewinsohn, T. W. Fernandes and W. W. Benson. John Wiley & Sons, New York.
- Gilbert, L. E., 2003. Adaptive novelty through introgression in Heliconius wing patterns: evidence for shared genetic ‘tool box’ from synthetic hybrid zones and a theory of diversification, pp. 281–318 in Ecology and Evolution Taking Flight: Butterflies as Model Systems, edited by C. L. Boggs, W. B. Watt and P. R. Ehrlich. University of Chicago Press, Chicago.
- Gilbert, L. E., H. S. Forrest, T. D. Schultz and D. J. Harvey, 1988. Correlations of ultrastructure and pigmentation suggest how genes control development of wing scales of Heliconius butterflies. J. Res. Lepidoptera 26: 141–160. [Google Scholar]
- Heckel, D. A., L. J. Gahan, Y. Liu and B. E. Tabashnik, 1999. Genetic mapping of resistance to Bacillus thuringiensis toxins in diamondback moth using biphasic linkage analysis. Proc. Natl. Acad. Sci. USA 96: 8373–8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckel, D. G., 1993. Comparative linkage mapping in insects. Annu. Rev. Entomol. 38: 381–408. [Google Scholar]
- Hsu, R., A. D. Briscoe, B. S. W. Chang and N. E. Pierce, 2001. Molecular evolution of a long wavelength-sensitive opsin in mimetic Heliconius butterflies (Lepidoptera: Nymphalidae). Biol. J. Linn. Soc. 72: 435–449. [Google Scholar]
- Jiggins, C. D., and W. O. McMillan, 1997. The genetic basis of an adaptive radiation: mimicry in two Heliconius sibling species. Proc. R. Soc. Lond. Ser. B 264: 1167–1175. [Google Scholar]
- Jiggins, C. D., M. Linares, R. E. Nasbit, C. Salazar, Z. H. Yang et al., 2001. a Sex-linked hybrid sterility in a butterfly. Evolution 55: 1631–1638. [DOI] [PubMed] [Google Scholar]
- Jiggins, C. D., R. E. Naisbit, R. L. Coe and J. Mallet, 2001. b Reproductive isolation caused by colour pattern mimicry. Nature 411: 302–305. [DOI] [PubMed] [Google Scholar]
- Kapan, D. D., 2001. Three-butterfly system provides a field test of Müllerian mimicry. Nature 409: 338–340. [DOI] [PubMed] [Google Scholar]
- Keys, D. N., D. L. Lewis, J. E. Selegue, B. J. Pearson, L. V. Goodrich et al., 1999. Recruitment of a hedgehog regulatory circuit in butterfly eyespot evolution. Science 283: 532–534. [DOI] [PubMed] [Google Scholar]
- Koch, P. B., 1993. Production of C-14 labeled 3-hydroxy-L-kynurenine in a butterfly, Heliconius charitonia L. (Heliconidae), and precursor studies in butterfly wing ommatins. Pigment Cell Res. 6: 85–90. [DOI] [PubMed] [Google Scholar]
- Koch, P. B., D. N. Keys, T. Rocheleau, K. Aronstein, M. Blackburn et al., 1998. Regulation of dopa decarboxylase expression during colour pattern formation in wild-type and melanic tiger swallowtail butterflies. Development 125: 2303–2313. [DOI] [PubMed] [Google Scholar]
- Lander, E., J. Abrahamson, A. Barlow, M. Daly, S. Lincoln et al., 1987. Mapmaker a computer package for constructing genetic-linkage maps. Cytogenet. Cell Genet. 46: 642. [DOI] [PubMed] [Google Scholar]
- Linares, M., 1996. The genetics of mimetic coloration in the butterfly Heliconius cydno weymeri. J. Hered. 87: 142–149. [Google Scholar]
- Linares, M., 1997. The ghost of mimicry past: laboratory reconstruction of an extinct butterfly ‘race’. Heredity 78: 628–635. [Google Scholar]
- Linzen, B., 1974. The tryptophan → ommochrome pathway in insects. Adv. Insect Physiol. 10: 117–246. [Google Scholar]
- Mallet, J., 1989. The genetics of warning colour in Peruvian hybrid zones of Heliconius erato and H. melpomene. Proc. R. Soc. Lond. Ser. B 236: 163–185. [Google Scholar]
- Mallet, J., and N. H. Barton, 1989. Strong natural selection in a warning color hybrid zone. Evolution 43: 421–431. [DOI] [PubMed] [Google Scholar]
- Meglecz, E., and M. Solignac, 1998. Microsatellite loci for Parnassius mnemosyne (Lepidoptera). Hereditas 128: 179–180. [Google Scholar]
- Naisbit, R. E., C. D. Jiggins and J. Mallet, 2003. Mimicry: developmental genes that contribute to speciation. Evol. Dev. 5: 269–280. [DOI] [PubMed] [Google Scholar]
- Nijhout, H. F., 1991. The Development and Evolution of Butterfly Wing Patterns. Smithsonian Institution Press, Washington, DC.
- Sheppard, P. M., J. R. G. Turner, K. S. Brown, W. W. Benson and M. C. Singer, 1985. Genetics and the evolution of Muellerian mimicry in Heliconius butterflies. Philos. Trans. R. Soc. Lond. Ser. B 308: 433–613. [Google Scholar]
- Shi, J., D. G. Heckel and M. R. Goldsmith, 1995. a A genetic linkage map for the domesticated silkworm, Bombyx mori, based on restriction fragment length polymorphisms. Genet. Res. 66: 109–126. [Google Scholar]
- Shi, J. R., D. G. Heckel and M. R. Goldsmith, 1995. b A genetic-linkage map for the domesticated silkworm, Bombyx mori, based on restriction-fragment-length-polymorphisms. Genet. Res. 66: 109–126. [Google Scholar]
- Stern, D. L., 2000. Perspective: evolutionary developmental biology and the problem of variation. Evolution 54: 1079–1091. [DOI] [PubMed] [Google Scholar]
- Summers, K. M., A. J. Howells and N. A. Pyliotis, 1982. Biology of eye pigmentation in insects. Adv. Insect Physiol. 16: 119–166. [Google Scholar]
- Suomalainen, E., L. M. Cook and J. R. G. Turner, 1973. Achiasmatic oogenesis in the heliconiine butterflies. Hereditas 74: 302–304. [Google Scholar]
- Tobler, A., D. D. Kapan, N. S. Flanagan, C. Gonzalez, E. Peterson et al., 2005. First generation linkage map of the warningly-colored butterfly Heliconius erato. Heredity 94: 408–417. [DOI] [PubMed] [Google Scholar]
- Turner, J. R. G., 1976. Adaptive radiation and convergence in subdivisions of the butterfly genus Heliconius (Lepidoptera: Nymphalidae). Zool. J. Linn. Soc. 58: 297–308. [Google Scholar]
- Wilkins, A. S., 2002. The Evolution of Developmental Pathways. Sinauer Associates, Sunderland, MA.
- Wittkopp, P. J., J. R. True and S. B. Carroll, 2002. Reciprocal functions of the Drosophila Yellow and Ebony proteins in the development and evolution of pigment patterns. Development 129: 1849–1858. [DOI] [PubMed] [Google Scholar]
- Yasukochi, Y., 1998. A dense genetic map of the silkworm, Bombyx mori, covering all chromosomes based on 1018 molecular markers. Genetics 150: 1513–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]