Abstract
The simple cellular composition and array of distally pointing hairs has made the Drosophila wing a favored system for studying planar polarity and the coordination of cellular and tissue level morphogenesis. We carried out a gene expression screen to identify candidate genes that functioned in wing and wing hair morphogenesis. Pupal wing RNA was isolated from tissue prior to, during, and after hair growth and used to probe Affymetrix Drosophila gene chips. We identified 435 genes whose expression changed at least fivefold during this period and 1335 whose expression changed at least twofold. As a functional validation we chose 10 genes where genetic reagents existed but where there was little or no evidence for a wing phenotype. New phenotypes were found for 9 of these genes, providing functional validation for the collection of identified genes. Among the phenotypes seen were a delay in hair initiation, defects in hair maturation, defects in cuticle formation and pigmentation, and abnormal wing hair polarity. The collection of identified genes should be a valuable data set for future studies on hair and bristle morphogenesis, cuticle synthesis, and planar polarity.
MORPHOGENESIS at the interface between the cellular and tissue levels is poorly understood but of substantial interest. One of the prime model systems for studying this is the Drosophila wing. The wing is the largest Drosophila appendage and a great deal has been learned about the genetic basis for wing patterning and the regulation of wing cell proliferation (e.g., Serrano and O'Farrell 1997; Teleman and Cohen 2000; Irvine and Rauskolb 2001; De Celis 2003; Martin et al. 2004). The flat simple structure of both the pupal and adult cuticular wing has also made it a favored system for studies on cell and tissue level morphogenesis and planar polarity (Eaton 1997, 2003; Adler 2002; Baum 2002). The vast majority of wing blade cells differentiate a single distally pointing cuticular hair. The cellular extension that forms the hair contains both actin filaments and microtubules and the function of both cytoskeletons is required for normal differentiation (Eaton et al. 1996; Turner and Adler 1998). The distal polarity of hairs is under the control of the frizzled (fz) tissue polarity pathway (Wong and Adler 1993). The timing of hair initiation is at least indirectly under the control of the ecdysone pathway, but relatively little is known about how temporal aspects of wing cell differentiation are controlled (Wong and Adler 1993; Thummel 2001). Among the genes previously implicated as having a role in regulating the time of hair initiation are grainy head (Lee and Adler 2004) and kojak (He and Adler 2002).
In the prepupa the wing everts and adopts a shape that appears to be a miniature version of the adult wing (Turner and Adler 1995). A pupal cuticle is secreted and the cells remain attached to the pupal cuticle for several hours. Apolysis occurs first over the wing blade, but is delayed along the wing margin. Cell division ends by ∼24 hr after white prepupae (awp) and terminal differentiation of the wing cells begins. The first sign of wing planar polarity is the accumulation of protein complexes along the distal [Fz, Dishevelled (Dsh), and Starry night (Stan)/Flamingo (Fmi)] and proximal [Prickle (Pk), Van Gogh (Vang)/Strabismus (Stbm), Stan/Fmi, and Inturned (In)] (Usui et al. 1999; Axelrod 2001; Feiguin et al. 2001; Shimada et al. 2001; Strutt 2001; Tree et al. 2002; Bastock et al. 2003; Adler et al. 2004) sides of the cells. This is seen by 24 hr awp and is retained for some time after hair initiation at 32 hr. The accumulation of these protein complexes is thought to provide a cortical mark that organizes planar cell polarity. Hair extension proceeds rapidly and is largely complete by about 38 hr awp. Once hair elongation is largely complete the hair moves to the center of the apical surface of the cell, where it is transiently located on a pedestal (Mitchell et al. 1990; Fristrom et al. 1993). At this time the wing cells begin to flatten, resulting in an increase in wing surface area. As part of this expansion the wing becomes folded inside of the pupal cuticle sac. The wing subsequently straightens after eclosion.
The regulation of gene expression is fundamental to much of biology but it remains unclear to what extent morphogenesis will be regulated by changing the expression of genes vs. regulation at the level of protein activity and localization. Mutations in a number of genes that encode transcription factors implicate these proteins and their modulation of gene expression as as being important for the terminal differentiation of the wing. Among the most interesting are ovo/svb, which is required for development of a hair (Delon et al. 2003), and grh, which regulates the expression of the planar polarity genes starry night and inturned and hence planar polarity (Adler et al. 2004; Lee and Adler 2004). Clones mutant for grh often show delayed hair formation, suggesting that it also regulates the expression of one or more genes involved in hair initiation. We report here the characterization of gene expression in the pupal wing, using Affymetrix Drosophila genome chips. The time points analyzed were prior to the start of hair initiation, early in hair development, and at the end of hair extension. We identified 1335 genes whose expression changed at least twofold between two time points and 435 whose expression changed at least fivefold. More than 40 of these were previously described as having a role in wing development.
We undertook the gene chip analysis with the goal of identifying candidate genes that play an important role in wing hair morphogenesis. On the basis of what was known about wing development we expected that genes involved in cell division would have a low level of expression at 32 and 40 hr and might be identified as genes whose expression fell during the period covered. It seemed likely that at least some genes that played a key role in the elaboration of the hair would show increased expression between 24 and 32 hr. Similarly we predicted that genes likely to play a role in the movement of the hair and flattening of wing cells would be likely to have their highest level of expression at 40 hr. Given the complexity of cuticle synthesis and modification we thought that genes involved in these processes would likely have modulated expression, although a number of patterns seemed possible. Genes that fit all of these expectations were found. As a functional test of the collection of identified genes we examined 10 genes where mutations existed but where there was no indication of a substantial wing phenotype. We found a new wing phenotype for 9 of these 10 genes, suggesting that many of the genes identified in our collection are important for morphogenesis.
MATERIALS AND METHODS
Fly stocks:
Alleles of knk, kkv, brat, kst, Koj, SelD, kermit, baz, ddc, ken, Hmgs, dy, m,Uch-L3, FRT/FLP, GFP-expressing and Deficency-carrying chromosomes were obtained from the Drosophila Stock Center in Bloomington, Indiana. Mutations in HR46 and flies carrying hs-HR46 transgenes were kindly provided by Carl Thummel. Flies carrying a mutation in EIP78DC were kindly provided by Adelaide Carpenter. Flies carrying the not1 allele were kindly provided by Iris Salecker.
Clonal analysis:
Somatic clones were generated using the FRT/FLP system (Xu and Rubin 1993). Pupal wing clones were marked by the loss of GFP. Unmarked clones were detected by mutant phenotypes.
Cytological techniques:
White prepupae were collected and aged until dissection. Immunostaining was done by standard techniques. Fluorescent secondary antibodies and fluorescent phalloidin for staining the actin cytoskeleton were obtained from Molecular Probes (Eugene, OR). Confocal images were obtained on an ATTO CARV confocal unit attached to a Nikon microscope. In situ hybridization on pupal wings was done as described previously (Geng et al. 2000), using digoxygenin-labeled probes.
RNA isolation:
Wings were dissected from timed pupae in cold PBS and then frozen until homogenized in TRIzol reagent (GIBCO BRL, Life Technologies, Gaithersburg, MD) and RNA was isolated. In these experiments we routinely isolated RNA from 100 to 300 pupal wings. Due to the difficulty of dissecting unfixed pupal wings we did not worry about contamination with small amounts of muscle, fat body, and thoracic epidermis. Consistent with this policy a number of flight muscle genes were detected as being expressed and having their expression level change. We also detected the expression of genes thought to be expressed in fat body. We used total RNA in making probes. The amount of RNA in a single probe was equivalent to that isolated from 30 pupal wings. In control experiments we isolated RNA from whole pupae.
Gene chip experiments:
Affymetrix Drosophila genome chips were probed using standard Affymetrix protocols at the University of Virginia Nucleic Acids Center.
Data were analyzed initially using Affymetrix software and then using the Dchip program. Experiments were done in duplicate and a comparison of duplicates showed good agreement (Figure 1). Pairs of samples from different time points were compared using Dchip. The parameters chosen have a substantial influence on the set of genes returned. We used many different parameter sets but in this article we present the results from a single pair of analyses. The data were normalized by Dchip on the basis of the sample with the median level of signal. The data were analyzed using model-based expression (the PM-MM difference method). We used the Dchip t-test function to identify genes whose expression differed significantly (P < 0.05) and we then filtered these for those that showed a fivefold or twofold or greater change. As one would expect there is not a simple relationship between fold changes, the P-value for expression being different at the two time points being considered. We used Dchip to estimate the empirical false detection rate (FDR) by permutation. In all of the analyses reported the median FDR was <2% and for most conditions it was <1%. The data from these experiments are available in the public access database of the gene expression open source system (GEOSS) at https://genes.med.virginia.edu/public_data/index.cgi.
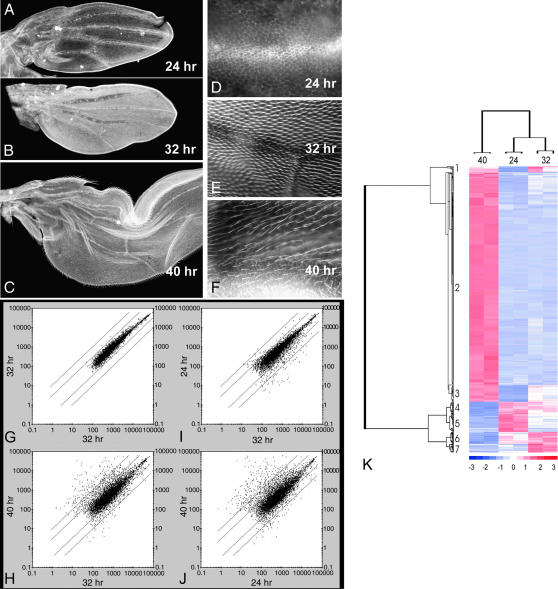
Figure 1.
Gene expression during wing differentiation. (A–C) Low-magnification images of 24-, 32-, and 40-hr pupal wings stained with a fluorescent phalloidin. All images are shown at the same magnification. The flattening of the wing cells results in the increased size of the wing at 40 hr vs. 32 hr. (D–F) Higher-magnification images of the same wings. Note that at 24 hr no hairs have started to form, at 32 hr short bright staining hairs are visible, and at 40 hr the hairs are longer, thinner, and do not stain quite as brightly as at 32 hr. (G–J) Scatter plots from Affymetrix gene chip experiments. Only genes/RNAs scored as present are plotted. The diagonal lines represent 3- and 10-fold differences in expression. (G) Replicate experiments for 32-hr wing RNA. Note the good agreement in expression levels in the 32-hr replicates. H–J show 24 hr vs. 32 hr, 40 hr vs. 32 hr, and 40 hr vs. 24 hr. As might be expected the greatest differences are seen in the 40-hr vs. 24-hr plot. Note that more genes show highly increased levels of expression at 40 hr than at 24 hr. (K) The clustering analysis (from Dchip) for the replicate 24-, 32-, and 40-hr samples. Only genes whose expression changed fivefold (P = 0.05) are included. Note that more than two-thirds of the genes that show substantially higher expression at 40 hr than at 24 hr or 32 hr fall into group 2. Note also that the 24- and 32-hr expression patterns cluster together compared to the 40-hr pattern.
RESULTS
Wing development from 24 to 40 hr:
Development of hairs is not synchronous across the wing. Differentiation starts distally and moves proximally in a patchy manner (Wong and Adler 1993). Thus any time point will contain cells of somewhat different developmental stages. We chose three time points for analyzing the pupal wing transcriptome. Twenty-four-hour pupal wings (Figure 1, A and D) do not show any signs of hair differentiation but the differentiation of the marginal row bristles has begun. At 32 hr cells over all except the most proximal regions have begun to elaborate hairs (Figure 1, B and E). The hairs at this time point stain strongly for F-actin. At 40 hr the wings have expanded and thinned (Figure 1, C and F). The hairs are long and do not stain as strongly for F-actin. In vivo the 40-hr wing is folded in a sac of pupal cuticle; however, such wings tend to spread out during the processing of the tissue. This property of asynchronous differentiation across a tissue is not unusual and is seen in other body regions such as the legs. In addition, there are substantial temporal differences between tissues. For example, the differentiation of the abdominal epidermis is substantially later than that of the wing.
Transcriptome analysis:
RNA was isolated from pupal wings and used to probe Affymetrix Drosophila gene chips. In replicate experiments we found good reproducibility for each time point (Figure 1G). RNA for ∼7500 genes was scored as present in each of the samples. A total of 9394 Drosophila genes (actually probe sets) were expressed in at least one time point and only 4576 were scored as being absent in all samples. We used the Dchip program to identify genes whose expression differed significantly between time points (P < 0.05). We found 2152 genes whose expression differed between at least two time points. Of these, 1335 genes had an expression change of twofold or greater and 435 had an expression change of fivefold or greater (Table 1). A total of 8059 genes (actually probe sets) were expressed at least at one time point whose expression did not differ significantly twofold or greater between any time points. The largest changes were seen when we compared the 24- and 40-hr samples (351 genes more than fivefold) with a much larger number of genes changing expression from 32 to 40 hr (270 genes more than fivefold) than from 24 to 32 hr (21 genes more than fivefold). There were also many more genes whose expression increased rather than decreased during each time period (Figure 1, H–J). For example, from 32 to 40 hr 228 genes had a fivefold or greater increase in expression while only 42 had a fivefold or greater decrease. This likely reflects the need for greatly increased expression of many genes for the elaboration of the adult cuticle, hairs, and bristles.
TABLE 1.
Comparison of gene expression at different times
| Compare times (hr) | No. ≥ ±2-fold | No. ≥ +2-fold | No. ≤ −2-fold | No. unique to time period | Also in pupae (2-fold) | No. ≥ ±5-fold | No. ≥ +5-fold | No. ≤ −5-fold | No. unique to time period | Also in pupae (5-fold) |
|---|---|---|---|---|---|---|---|---|---|---|
| 24 vs. 32 | 119 | 68 | 51 | 37 | 20 | 21 | 19 | 2 | 8 | 1 |
| 24 vs. 40 | 1048 | 729 | 319 | 235 | 421 | 351 | 306 | 45 | 149 | 115 |
| 32 vs. 40 | 711 | 538 | 173 | 495 | 253 | 270 | 228 | 42 | 74 | 67 |
A total of 1335 individual genes changed twofold or greater. A total of 436 individual genes changed fivefold or greater.
We used the Dchip program (Li and Hung Wong 2001) to cluster the 435 genes whose expression changed at least 5-fold and they almost all fell into seven expression groups (Table 2). By far the largest of these was expression group 2, which contained 321 of the 435 genes. This group was characterized by a modest increase in gene expression between 24 and 32 hr (median change of 1.3-fold) followed by a large increase from 32 to 40 hr (median change of 7.7-fold). Many of these changes appear to be related to the beginning of cuticle deposition during this time interval. Selected groups of genes whose expression changed from 24 to 32 hr (Table 3), from 32 to 40 hr (Table 4), and from 24 to 40 hr (Table 5) are presented and a full listing of the genes whose expression changed ≥2-fold can be found in supplementary Tables S1, S2, and S3, respectively, at http://www.genetics.org/supplemental/. In examining the lists of genes whose expression changed we found that many of the most interesting genes from a biological context showed changes between 2- and 5-fold so we did not limit our consideration to those genes that showed the greatest changes.
TABLE 2.
Clustering of genes whose expression changed fivefold or greater
| Expression group
|
No. of genes
|
Median FC
|
||
|---|---|---|---|---|
| 32/24 | 40/24 | 40/32 | ||
| 1 | 9 | 9.3 | 8.9 | −1.1 |
| 2 | 321 | 1.3 | 10.3 | 7.7 |
| 3 | 26 | 5.7 | 10.3 | 1.9 |
| 4 | 19 | −1.2 | −6.3 | −5.3 |
| 5 | 27 | −2.4 | −7.7 | −3.0 |
| 6 | 18 | 1.5 | −7.7 | −10.4 |
| 7 | 13 | 4.5 | −2.64 | −11.2 |
FC, fold change (ratio of expression at the two indicated times).
TABLE 3.
Examples of genes whose expression changed from 24 to 32 hr
| Fold change
|
Experimental group
|
In pupae
|
||||
|---|---|---|---|---|---|---|
| P-value | Twofold | Fivefold | Gene | Comment | ||
| −7.31 | 0.05 | 5 | No | No | Cyclin-dependent kinase (Cks) | Regulator of cell division |
| −3.84 | 0.01 | No | No | CG9307 | Chitinase | |
| −3.13 | 0.03 | No | No | mutagen-sensitive 29 (mus209) | PCNA, DNA repair | |
| −2.91 | 0.05 | No | No | gluon | Condensin family, chromosome mechanics | |
| −2.85 | 0.02 | No | No | Klp61F | Kinesin family | |
| −2.22 | 0.02 | 5 | No | No | Twin of m4 (Tom) | Thoracic bristle phenotype |
| −2.19 | 0.02 | 5 | No | No | scute (sc) | Transcription factor, bristle sense organ |
| −2.15 | 0.04 | No | No | string (stg) | Protein tyrosine phosphatase, cell cycle | |
| −2.14 | 0.03 | 5 | No | No | Domina (Dom) | Required for imaginal disc growth |
| 4.49 | 0.01 | 7 | No | No | Inscuteable (insc) | Asymmetric cell division in PNS lineage |
| 4.67 | 0.03 | 3 | No | No | Dopamine N-acetyltransferase (Dat) | Catecholamine metabolism |
| 4.98 | 0.04 | Yes | No | Punch (Pu) | GTP cyclohydrolase I | |
| 5.30 | 0.05 | 2 | No | No | yellow-d2 | Related to yellow |
| 6.17 | 0.01 | 2 | No | No | osi7 | Member of osirus family |
| 12.62 | 0.01 | 2 | No | No | ectodermal (ect) | |
TABLE 4.
Examples of genes whose expression changed from 32 to 40 hr
| Fold change
|
Experimental group
|
In pupae
|
||||
|---|---|---|---|---|---|---|
| P-value | Twofold | Fivefold | Gene | Comment | ||
| −6.79 | 0.01 | 7 | Yes | No | Inscuteable (insc) | Asymmetric cell division in PNS lineage |
| −5.20 | 0.02 | 5 | Yes | No | scute (sc) | Transcription factor, bristle sense organ |
| −3.17 | 0.03 | No | No | non-stop (not) | Ubiquitin hydrolase, eye planar polarity | |
| −3.08 | 0.05 | No | No | TNF-receptor-associated factor 1 (Traf1) | Upstream of Jnk pathway and msn | |
| −2.98 | 0.01 | Yes | No | sugarless (sgl) | UDP-glucose 6-dehydrogenase, signaling | |
| −2.91 | 0.04 | No | No | Cyclin A (CycA) | Cell division | |
| −2.53 | 0.02 | No | No | Beadex (Bx) | Transcription factor, wing scalloping | |
| −2.52 | 0.01 | No | No | twine | Protein tyrosine phosphatase, cell division | |
| −2.35 | 0.01 | No | No | wingless (wg) | Wnt ligand, wing growth | |
| −2.30 | 0.04 | No | No | Ubiquitin C-terminal hydrolase (Uch-L3) | Ubiquitin hydrolase | |
| −2.27 | 0.03 | Yes | No | Cyclin B (CycB) | Cell division | |
| −2.04 | 0.03 | No | No | inturned (in) | Planar polarity | |
| 2.37 | 0.03 | 3 | No | No | quail (qua) | Gelsolin/villin, actin bundling, female sterile |
| 3.10 | 0.00 | Yes | No | CG11546 | Drosophila Kermit, planar polarity | |
| 3.12 | 0.00 | No | No | Rala | Ras family GTPase, hairs and bristles | |
| 3.95 | 0.03 | Yes | No | forked (f) | Actin bundling, twisted bristles and hairs | |
| 3.95 | 0.04 | Yes | No | Ecdysone receptor (EcR) | Nuclear receptor, wing phenotype | |
| 4.94 | 0.02 | Yes | No | krotzkopf verkehrt (kkv) | Blimp embryonic cuticle phenotype | |
| 5.25 | 0.01 | 2 | No | No | Ecdysone-induced protein 71CD | |
| 5.55 | 0.00 | 2 | Yes | No | karst (kst) | Heavy β-spectrin, wing phenotype |
| 6.24 | 0.01 | 2 | No | No | furrowed (fw) | Bristles, notum, and wing phenotypes |
| 6.43 | 0.01 | 2 | No | No | jbug | Filamen |
| 6.51 | 0.00 | 2 | No | No | yellow-d2 | Related to yellow |
| 6.79 | 0.01 | 2 | No | No | ken and barbie (ken) | Transcription factor, loss of genitalia |
| 6.92 | 0.03 | 2 | Yes | No | knickkopf (knk) | Embryonic blimp phenotype |
| 9.89 | 0.01 | 2 | No | No | vrille (vri) | Wing hair, vein, marginal row phenotypes |
| 12.91 | 0.03 | 2 | Yes | Yes | CG7214 | Cuticle protein |
| 13.35 | 0.03 | 2 | Yes | No | CG12045 | Cuticle protein |
| 13.45 | 0.00 | 2 | Yes | No | CG9295 | Cuticle protein |
| 13.50 | 0.00 | 2 | No | No | Buffy | Regulator of apoptosis |
| 16.66 | 0.05 | 2 | Yes | Yes | dusky (dy) | Small dark wing phenotype, related to m |
| 19.33 | 0.03 | 2 | Yes | Yes | CG6458 | Structural constituent of larval cuticle |
| 25.65 | 0.01 | 2 | No | No | osi11 | Member of osirus family |
| 34.39 | 0.04 | 2 | Yes | Yes | Myosin heavy chain (Mhc) | Muscle protein |
| 41.52 | 0.00 | 2 | Yes | Yes | osi3 | Member of osirus family |
| 41.95 | 0.02 | 2 | Yes | Yes | CG1869 | Chitinase |
| 57.63 | 0.00 | 2 | Yes | Yes | Drip | Aquaporin-like |
| 68.85 | 0.02 | 2 | Yes | Yes | CG17355 | Protease inhibitor |
| 117.91 | 0.00 | 2 | Yes | Yes | CG15013 | Related to dy and m |
| 118.20 | 0.00 | 2 | Yes | Yes | osi12 | Member of osirus family |
TABLE 5.
Examples of genes whose expression changed from 24 to 40 hr
| Fold change
|
Experimental group
|
In pupae
|
||||
|---|---|---|---|---|---|---|
| P-value | Twofold | Fivefold | Gene | Comment | ||
| −11.38 | 0.01 | 5 | Yes | No | scute (sc) | Transcription factor, bristle development |
| −7.65 | 0.01 | 5 | No | No | Domina (Dom) | Required for imaginal disc growth. |
| −5.35 | 0.00 | 5 | Yes | No | Twin of m4 (Tom) | Thoracic bristle phenotype |
| −3.91 | 0.04 | No | No | non-stop (not) | Ubiquitin hydrolase, eye planar polarity | |
| −3.58 | 0.04 | No | No | Daughters against dpp (Dad) | dpp pathway | |
| −3.55 | 0.01 | Yes | No | Cyclin B (CycB) | Cell division | |
| −2.90 | 0.02 | Yes | No | Ubiquitin C-terminal hydrolase (UchL3) | Ubiquitin hydrolase | |
| −2.63 | 0.02 | Yes | No | wingless (wg) | Secreted factor, wing growth and margin | |
| −2.42 | 0.01 | No | No | schnurri (shn) | Transcription factor, dpp signaling | |
| −2.27 | 0.02 | No | No | Apc2 | Wnt signaling | |
| −2.06 | 0.04 | No | No | brinker (brk) | dpp signal transduction, wing growth | |
| 2.01 | 0.05 | No | No | puckered (puc) | JUN kinase phosphatase | |
| 2.21 | 0.01 | No | No | singed (sn) | Fascin, actin bundling, hair and bristle | |
| 2.57 | 0.05 | Yes | No | ebony (e) | Mutations result in dark body | |
| 2.59 | 0.04 | No | No | Moesin (Moe) | Disc cell morphology, cytoskeleton | |
| 2.65 | 0.05 | No | No | Tyramine beta hydroxylase (Tbh) | Catecholamine metabolism | |
| 2.76 | 0.05 | Yes | No | Stubble (Sb) | Endopeptidase, short stout bristles | |
| 3.02 | 0.03 | No | No | krotzkopf verkehrt (kkv) | Blimp embryonic cuticle phenotype | |
| 3.13 | 0.01 | No | No | Dopa decarboxylase (Ddc) | Cuticle crosslinking and pigmentation | |
| 3.47 | 0.00 | No | No | Rala | Ras family GTPase, hairs and bristles | |
| 3.87 | 0.00 | Yes | No | CG11546 | Drosophila kermit, planar polarity | |
| 4.18 | 0.01 | 2 | Yes | No | karst (kst) | Heavy β-spectrin, wing |
| 5.58 | 0.00 | 2 | No | No | ken and barbie (ken) | Transcription factor, loss of genitalia |
| 5.67 | 0.04 | 2 | No | No | knickkopf (knk) | Embryonic blimp phenotype |
| 6.02 | 0.00 | 3 | Yes | No | cheerio (cher) | Filamen protein, actin binding, female sterile |
| 6.46 | 0.00 | 2 | Yes | No | pawn (pwn) | Mutations affect hairs and bristles |
| 7.47 | 0.01 | 3 | Yes | No | Dopamine N-acetyltransferase (Dat) | Catecholamine metabolism |
| 8.11 | 0.04 | 3 | No | No | quail (qua) | Gelsolin/villin, actin bundling, female sterile |
| 11.84 | 0.01 | 1 | Yes | Yes | CG13209 | kojak gene (N. Ren and P. N. Adler, unpublished data) |
| 34.51 | 0.00 | 2 | Yes | Yes | yellow-d2 | Related to yellow |
| 37.22 | 0.00 | 2 | Yes | No | Buffy | Regulator of apoptosis |
| 39.77 | 0.03 | 2 | Yes | No | CG12045 | Cuticle protein |
| 42.53 | 0.03 | 2 | Yes | Yes | CG1869 | Chitinase |
| 58.05 | 0.02 | 3 | Yes | Yes | miniature (m) | Related to dy, small wing phenotype |
| 59.33 | 0.01 | 7 | No | No | Hormone receptor-like in 46 (Hr46) | Nuclear receptor; wing, bristle phenotypes |
| 99.26 | 0.00 | 2 | Yes | Yes | Drip | Aquaporin-like |
| 329.89 | 0.05 | 2 | No | No | dy (dusky) | Small dark wing, related to m |
| 398.96 | 0.00 | 2 | Yes | Yes | CG15013 | Related to dy |
| 467.26 | 0.05 | 2 | Yes | Yes | Myosin heavy chain (Mhc) | Muscle protein |
| 4589.75 | 0.02 | 2 | Yes | Yes | osi1 | Member of osirus family |
As a first step in validating the array data we selected seven genes and characterized the expression of these genes by real time PCR (Table 6). Six of the seven genes showed increased expression at the later time points on the basis of the chip data. The RT-PCR data showed similar changes, giving us confidence in the data set. Only one of the seven genes (forked) showed much-reduced changes in expression when assayed by RT-PCR. The reason for this is unclear. Several genes showed increased changes when assayed by RT-PCR. The largest differences in expression ratios were for dy and cg8213 and involved 24-hr samples with a very low absolute level of expression. These values are likely subject to greater errors and a small absolute error here would translate into a big difference in the fold change ratio. For one of the selected genes, CG13209, we used three different sets of primers to examine different parts of the mRNA by real time PCR. Good agreement was obtained for the results from the different primers.
TABLE 6.
Comparison of gene expression changes by RT-PCR and gene chips
| Gene | 24 | 32/24 Aa | 32/24 RT-PCR | 40/32 Aa | 40/32 RT-PCR | 40/24 Aa | 40/24 RT-PCR |
|---|---|---|---|---|---|---|---|
| CG13209A | 1 | 10.2 | 12.3 | 1.2 | 0.55 | 11.8 | 6.87 |
| CG13209B | 1 | 10.2 | 11.79 | 1.2 | 0.58 | 11.8 | 6.89 |
| CG13209C | 1 | 10.2 | 18.25 | 1.2 | 0.53 | 11.8 | 9.71 |
| CG1869 | 1 | 1.01 | 3.5 | 42.8 | 15.98 | 43.4 | 56.1 |
| CG8213 | 1 | 15.6 | 102.5 | 8.4 | 3.83 | 130.2 | 393.4 |
| Fkbp13 | 1 | 1.9 | 2.28 | 4.7 | 2.32 | 9.1 | 5.31 |
| dy | 1 | 19.8 | 21.71 | 16.7 | 35.01 | 329.9 | 760.1 |
| f | 1 | 8.4 | 0.9 | 3.9 | 1.02 | 33.4 | 1.72 |
| stg | 1 | 0.47 | 2.68 | 0.017 | 0.07 | 0.08 | 1.57 |
Affymetrix gene chip assay.
Metamorphosis is under hormonal control and all cell types in the pupae are likely to be responding in some way. We had isolated RNA after the laborious manual dissection of unfixed pupal wings because of our interest in the morphogenesis of that tissue and the likelihood that since this tissue represents a small fraction of the mass of the pupae it would not be informative to examine whole pupal RNA. To determine if this was correct we also used RNA isolated from whole pupae to probe Affymetrix chips. We found that only a minority of the genes identified in our pupal wing experiments were similarly identified in the whole pupae experiments. The greatest difference was seen for genes whose pupal wing RNA levels changed fivefold or more from 24 to 32 hr (Table 1). For this condition only, only 1 of 21 genes had a fivefold or greater change in whole pupal RNA. A greater overlap was seen for genes whose expression changed fivefold from 24 to 40 hr in pupal wings. For this condition 115 of 351 genes had a fivefold or greater change in whole pupal RNA.
Literature analysis of identified genes:
The literature provided additional validation of the gene chip data. Among the genes identified are >40 where a mutant phenotype is known in the wing. Some of these are suggestive of a specific role for the gene in the differentiation of the pupal wing during the time covered by our experiment. Among the most striking are miniature and dusky, which encode related proteins (DiBartolomeis et al. 2002; Roch et al. 2003). Mutations in both of these genes result in small dark wings. In miniature mutants the cell outlines are still visible in the adult wing, suggesting a defect in late cellular mophogenesis or cuticle deposition (Roch et al. 2003). The expression of both of these genes increased substantially (58-fold for miniature and 330-fold for dusky) from 24 to 40 hr. The expression of a third related gene, CG15013, increased 399-fold during this time period. No mutations have been described for CG15013. The biochemical function of these proteins is unclear, but all three proteins are predicted to contain a short C-terminal cytoplasmic tail and a large extracellular N-terminal region containing a ZP domain. The proteins are localized apically in pupal wing cells (Roch et al. 2003). It has been suggested that they mediate interactions between the cytoskeleton, membrane, and forming cuticle. The mutant phenotype is consistent with these genes functioning in wing expansion as they are expressed most highly at 40 hr, and under our conditions expression starts at ∼38 hr and continues for several hours.
Mutations in three genes whose expression increased from 24 to 32 hr [forked (33.9-fold), singed (2.2-fold), and pawn (6.5-fold)] have dramatic wing hair and bristle morphology phenotypes and indeed mutations in these genes have long been used as cuticle markers in genetic mosaic experiments. Mutations in ebony (increased 2.6-fold from 24 to 40 hr) result in darkly pigmented wings and mutations in Sb (increased 2.8-fold from 24 to 32 hr) result in short fat bristles (this is seen in the marginal row bristles on the wing). Mutations (or overexpression) of many other identified genes result in abnormal wing disc development. These include boule, Ecdysone receptor, karst, furrowed, Rala, inturned, vrille, sugarless, hephaestus, HR46, Dopa decarboxylase, ovo, APC2, inscuteable, twins, Beadex, Hairless, Suppressor of Hairless, brinker, Dichaete, wingless, scute, scalloped, Domina, Moesin, schnurri, and minidiscs. Descriptions of all of these genes can be found in FlyBase.
The development of wing hairs is known to involve the actin and microtubule cytoskeletons (Eaton et al. 1996; Turner and Adler 1998) and we detected changes in the expression of a number of proteins that encode cytoskeleton interacting proteins. These include forked, singed (fascin), cheerio (filamen), quail (villin), karst (BH-spectrin), jitterbug (filamen), Moesin, paxillin, scraps (anillin), an uncoventional myosin XV, mapmodulin, lamin C, Klp3A, enabled, beta-tubulin97EF, betaTub60D, and three septins (CG9699, CG16953, and CG2916). The apolysis of the pupal cuticle and the beginning of deposition of adult cuticle take place during the time period examined and, not surprisingly, among the genes whose expression was strongly modulated we identified 15 genes thought to encode cuticle proteins (CG9077, CG8515, CG15013, CG7076, CG6458, CG6469, CG9295, CG12045, CG7214, CG4818, CG13214, CG9036, CG2555, CG15884, and Lcp65Ag2), 3 genes that encode chitinases (CG2989, CG1869, and CG9307), a chitin synthetase (kkv), and 3 genes that encode chitin-binding proteins (Gasp, peritrophin-like, and CG3426). Also among the genes whose expression was sharply increased were a number of genes known to be important for cuticle pigmentation and sclerotinization (e, Ddc, and amd). Two genes whose expression increased during the time period were kkv and knk, which share an unusual embryonic cuticular phenotype (Ostrowski et al. 2002).
Cell division ends in the wing around the time of our earliest time point. Hence it is not surprising that we found a decrease in the expression of many genes known to be important for growth of the wing (and other) discs. These include string, sugarless, cyclin dependent kinase, fizzy, fizzy related, Bub1, wingless, schnurri, twine, gluon, cdc2c, abnormal spindle, disc proliferation abnormal, Domina, Dad, Mad, Cyclin B, Cyclin A, twins, mus209 (PCNA), RfC3 (DNA replication factor complex 3), and brinker. Another prominent group of genes whose expression was altered were genes involved in ecdysone action or response. These included the Ecdysone receptor, ImpL1, Eip71CD, Eip74EF, Eip63F1, Edg84A, Hr39, Eig71Eb, ImpE3, and HR46. Ten members of the osiris gene family were in group 2. Six of these showed a >50-fold increase in expression from 24 to 32 hr (see Table 5 and supplementary Table S3 at http://www.genetics.org/supplemental/). The biochemical function of this gene family is obscure, but it has been suggested that it corresponds to the triplo lethal gene region (Dorer et al. 2003). Consistent with our observation that our samples were contaminated with muscle we found that a number of muscle genes had increased expression (e.g., Mhc, Mlc1, Mlc2, and Act88F) from 24 to 40 hr awp.
Five of the genes identified in our chip experiment (inturned, l(2)02045, non-stop, rala, and traf1) are notable for having a link to planar polarity. The inturned (in) gene functions downstream of frizzled and the In protein has been found to localize to the proximal side of pupal wing cells under the instruction of the upstream frizzled pathway genes (Lee and Adler 2002; Adler et al. 2004). Mutations in in result in abnormal wing hair polarity and the formation of many multiple hair cells. The level of in mRNA fell 2-fold from 32 to 40 hr (Table 4). The l(2)02045 gene (also known as CG11546) is the Drosophila homolog of the Xenopus kermit gene (Tan et al. 2001) and we refer to it as kermit. The amount of kermit mRNA increased 3.1-fold from 32 to 40 hr (Table 4). This gene was originally identified by an EP insertion that resulted in abnormal hair polarity and multiple hair cells when overexpressed using several GAL4 drivers (Toba et al. 1999). The Xenopus homolog of this gene was identified in a two-hybrid screen by virtue of its binding to the carboxy-terminal tail of Fz (Rasmussen et al. 2001; Tan et al. 2001). Members of this family of proteins have been found to be resident in the Golgi and to interact with G-coupled receptors (Katoh 2002). The expression of traf1 decreased 3.1-fold between 32 and 40 hr (Table 4). This gene is thought to function upstream of misshapen and the JNK pathway (Liu et al. 1999), which have been implicated in planar polarity (Boutros et al. 1998; Paricio et al. 1999). The expression of the small GTPase Rala increased 3.5-fold between 24 and 40 hr (Table 5). This gene has been suggested to act upstream of the JNK pathway in flies and the expression of a dominant negative form of this protein results in a multiple hair cell phenotype (Sawamoto et al. 1999; Mirey et al. 2003). Finally, the expression of the non-stop gene decreased ∼3.9 fold between 24 and 40 hr (Table 5) and mutations in this gene have been found to display a weak planar polarity phenotype in the eye (Martin et al. 1995).
We detected temporal changes in the mean expression levels of a number of genes known to be important for wing and wing hair development that were not significant at the 0.05 level. On the basis of the known biology we expect that some of these are likely to be real and interesting changes and it suggests a limitation of our experiment or analysis. For example, the ovo/svb complex gene encodes a set of transcription factors. It is known to be important for hair formation as mutations result in a loss of hair development (Delon et al. 2003). ovo was picked up as a gene whose expression decreased 5.6-fold from 32 to 40 hr, but the difference was not significant (P = 0.13). A second interesting example is SelD, which encodes a selenophosphate synthetase. Loss-of-function mutations in SelD result in multiple hair cells and abnormal polarity (Alsina et al. 1998; Morey et al. 2001). We found SelD expression fell 3.1-fold between 24 and 40 hr, but once again this was not significant (P = 0.11). A third example is CG13209. The 11.8-fold increase in the expression of this gene between 24 and 40 hr was significant (P = 0.01), but the 10.2-fold change between 24 and 32 hr was not significant (P = 0.16). This gene was one that we validated by RT-PCR using three different sets of primers and we found an average 14.1-fold increase in expression between 24 and 32 hr (Table 6).
Testing of candidate genes:
Our primary goal in characterizing pupal wing gene expression was to identify genes that play an important role in pupal wing morphogenesis. As a test of the approach we selected 10 genes where mutant stocks were available but where a wing phenotype had not been described in any detail. Our prediction was that mutations in at least several would produce a previously unappreciated wing phenotype. As described below this turned out to be the case.
ken and barbie (ken) encodes a DNA-binding transcription factor that contains an N-terminal BTB/POZ domain and three C2H2 zinc fingers (Lukacsovich et al. 2003). It was a member of gene expression cluster 2 and its expression increased 6.8-fold from 32 to 40 hr. Loss-of-function mutations in ken are semilethal. Escaper adults have unpigmented aristae and often lack external genitalia (hence the gene name) (Lukacsovich et al. 2003). We examined wings from ken mutant escapers and also in genetic mosaics. We saw that the triple row bristles on the wing margin were lightly pigmented, reminiscent of the arista phenotype. This was most obvious in mosaics where the lightly pigmented bristles stood out from their wild-type neighbors (Figure 2K). We did not see a hair phenotype, but a subtle hair pigmentation phenotype would be difficult to see.
Figure 2.
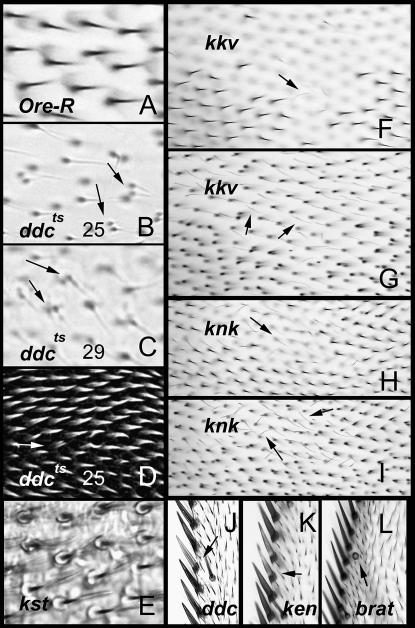
Phenotypes in adult wings. (A–C) Micrographs of adult wings from Oregon R, Ddcts raised at 25° and Ddcts raised at 29°. The arrows point to double hair cells on the Ddcts wings. Note how light and fine the Ddcts wings are. (D) A Ddcts pupal wing stained with a fluorescent phalloidin. Note that the hairs are of normal thickness (compare to Ore-R from Figure 1E). An arrow marks a double hair cell. (E) A micrograph of an adult wing from a kst escaper fly. The wing is resting on a glass microscope slide but is not in mounting media. Note the circular pedestals around the hairs. (F and G) Wings that contain unmarked kkv clones. F and G are of different focal planes. F shows a region with normal hairs and a region (presumably a clone) with little or no sign of hairs (an arrow points to one faint hair). The hairs appear to be missing because they are faint and lay flat on the wing surface unlike wild-type hairs, which are elevated. At a lower focal plane the hairs can be seen (G). (H and I) An equivalent pair for a wing that contains unmarked knk clones. (J–L) The wing margin from three mutants. J and K show wings that contain unmarked Ddc and ken clones, respectively. The lightly pigmented bristles are presumably part of mutant clones. L shows a region from a bratts mutant that is missing a bristle shaft. This phenotype was common in such wings.
The HMGS gene encodes the Drosophila HMG Coenzyme A synthase, a key enzyme in steroid and isoprenoid metabolism (Dobrosotskaya et al. 2002). It was also a member of gene expression cluster 2 and we found its expression increased 8.4-fold from 32 to 40 hr. Individuals homozygous for a P-insertion allele die as pharate adults or pupae. The pharate adults are notable for a melanotic liquid that accumulates principally near the ventral head. Mutations that result in weak cuticle often show such melanotic leakage, suggesting that HMGS may be required for normal cuticle elaboration. The reason for the phenotype being seen primarily in the ventral head is unclear. We did not see evidence for a specific wing phenotype.
The karst gene, which encodes the Drosophila BHeavy-spectrin (Thomas et al. 1998), was also a member of cluster 2. We found its expression increased 5.5-fold from 32 to 40 hr. Spectrin typically contains four chains, two a and two b, that are known to link the actin cytoskeleton to the plasma membrane. Somewhat surprisingly kst mutants are viable (at reduced levels) and female sterile due to defects in the follicular epithelium (Thomas et al. 1998). Adult kst mutants have rough eyes and their wings often are cupped downward. We examined kst wings and found an additional mutant phenotype that is nicely correlated with its expression profile. kst wing cells produce normal-looking hairs but the hairs are often found on a small pedestal (Figure 2E). The wing cell surface (that is not hair) is rough and at times remnants of cell outlines are visible. This phenotype can also be seen in mosaic clones. The clones can be recognized under the stereo microscope as they are often associated with a dimpling of the wing surface.
Both the kkv and knk genes were identified in a screen for having an unusual defect in embryonic cuticle—the blimp phenotype (Ostrowski et al. 2002). Mutant embryo cuticles were seen to expand in cuticle preparations. The kkv gene encodes a chitin synthase implicating it in cuticle synthesis and its expression increased 4.9-fold from 32 to 40 hr. The knk gene encodes a novel gene that is well conserved only in the ecdysozoa, suggesting a role in cuticle metabolism. The amino acid sequence shows homology to what is thought to be a dopamine-binding domain, suggesting knk might be involved in crosslinking of cuticle. The expression of knk increased 7-fold between 32 and 40 hr and it was a member of expression group 2. Mutations in both of these genes are embryonic lethals so we examined mosaic clones of cells carrying mutations in either of these genes. The phenotypes seen in the adult cuticle were quite similar to one another. Most notably wing mutant wing hairs displayed a lack of pigmentation and were thinner and flimsier than normal (Figure 2, F–I). This phenotype is dramatic and at low magnification it often appears as if hairs were not formed by mutant cells. The hairs appeared normal in size and shape when clones were examined in pupal wings (data not shown), arguing that the mutations affect a process after hair outgrowth (e.g., cuticle synthesis or maturation). Clones in other body regions such as the abdomen and thorax also showed a dramatic loss of pigmentation. In all of these cases the borders between pigmented and unpigmented were relatively sharp. Consistent with these mutations resulting in weak cuticle we often saw locations where internal tissues and hemolymph appeared to be erupting from the animal. This was usually seen on the dorsal abdomen, particularly in the region of the intersegmental membrane. The eruptions could be related to the blimp phenotype seen in embryos.
The expression of brain tumor (brat) decreased 5.5-fold from 24 to 40 hr. This gene has been studied primarily due to the neural tumor phenotype seen in loss-of-function mutants (Arama et al. 2000; Sonoda and Wharton 2001). We examined the wings of bratts/Df brat flies raised under semipermissive conditions. We did not see a hair phenotype but we did see the occasional loss of sensory bristle shaft cells (principally distally along the anterior margin) and occasional duplicated bristle cells (principally in the costa; Figure 2L). These phenotypes are suggestive of a role for brat in specifying cell fate or in Notch-mediated lateral inhibition.
The expression of dopa decarboxylase (Ddc) increased 6-fold from 24 to 32 hr and then decreased 1.9-fold from 32 to 40 hr. This well-characterized gene is known to function in the epidermis for the crosslinking of cuticle and in the formation of melanin (Hirsh and Davidson 1981; Konrad and Marsh 1987; Wright 1996). Loss of Ddc function results in fragile and pale cuticle with thin bristles. No detailed description of the wing phenotype has been reported previously. Ddc null alleles are recessive embryonic lethals so we first examined adults that contained clones mutant for Ddc. On the abdomen (and some other parts of the body) we could see clones where there were lightly pigmented cuticle and bristles. We did not see any wing phenotype other than apparent clones resulting in lighter triple-row bristles (Figure 2J). The abdominal clone boundaries were not sharp as we had seen for grh, knk, or kkv, which also give rise to lightly pigmented cuticle, suggesting that the Ddc cells might be rescued by the diffusion of dopamine from neighboring cells. We therefore examined adults homozygous for a temperature-sensitive Ddc allele. Animals raised at 25° showed a much stronger phenotype in general than what we saw in clones, suggesting that Ddc acts nonautonously in the wing. The phenotype was even stronger in animals raised at 29°. The wings of Ddc mutants were characterized by very thin wispy hairs, occasional multiple hair cells, and an overall faint appearance (Figure 2, A–C). When we examined Ddcts pupal wings the early hairs appeared normal in morphology. Thus, the wispy appearance of the adult wing hairs is presumably due to a late defect. We suggest that Ddc-dependent crosslinking of the cuticle is essential for maintaining the structure of the hair and in the absence of this crosslinking the hair collapses after the actin cytoskeleton is disassembled. Occasional multiple hair cells were seen in the Ddcts pupal wings (Figure 2D); thus that defect is likely due to a different process also being affected in the mutant. The formation of multiple hair cells has previously been associated with planar polarity defects (Wong and Adler 1993; Adler 2002) or due to disruptions of the cytoskeleton (Eaton et al. 1996; Turner and Adler 1998; Adler 2002).
The HR46 gene (also known as DHR3) encodes a nuclear receptor and is an essential gene known to be important for the ecdysone cascade (Lam et al. 1999; Thummel 2001). Large clones of loss-of-function alleles result in wing (folded and curved) and notum defects (rough short bristles and pale pigmentation). The expression of this gene increased 250-fold from 24 to 32 hr and then decreased 4.3-fold from 32 to 40 hr. We first examined moderate-sized wing clones of cells lacking HR46, but we did not see a clear cut phenotype. In pupal wing clones examined a couple of hours after hair formation mutant hairs appeared somewhat thicker but this alteration was transient (Figure 3H). The Eip78CD gene encodes a related nuclear receptor. The expression of this nonessential gene increased 3-fold from 24 to 32 hr followed by a 3-fold drop from 32 to 40 hr (but the differences were not significant; P = 0.22 and P = 0.23, respectively), suggesting it might be functionally redundant with HR46 (Russell et al. 1996). To test this hypothesis we examined Eip78CD wings that also contained HR46 mutant clones. We did not see any mutant phenotype in the clones, suggesting either that there is an alternative redundant gene or that HR46 is not essential for hair morphogenesis. Since the level of HR46 expression fell dramatically between 32 and 40 hr it seemed possible that declining HR46 expression could be important for hair development. To test this we induced the overexpression of HR46 from a transgene containing a hs promoter. This resulted in a dramatic loss of hair formation, leading to wings with extensive bald regions (Figure 3, G, I, and J). The strongest phenotype was seen when the transgene was induced by heat-shocking 6–8 hr prior to the time of hair initiation. The phenotype was dose sensitive and directly related to the number of transgenes and length and temperature of transgene induction (data not shown).
Figure 3.
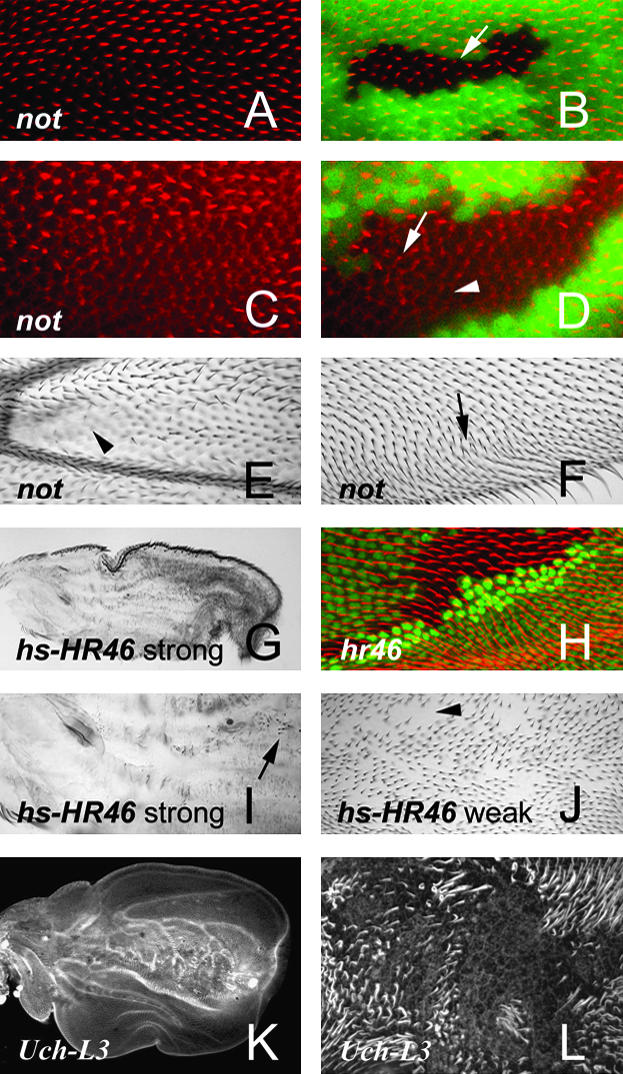
Phenotypes in pupal and adult wings. (A–D) not1 clones marked by the loss of GFP. The pupal wings were stained with a fluorescent phalloidin to stain the actin cytoskeleton. Note that in A and B the hairs in the clones are smaller than neighboring wild-type hairs. In C and D many of the clone cells have not yet started to elaborate hairs. The arrows point to clone cells with small hairs and the arrowhead to a cell that has not yet started to form a hair. E and F show micrographs of adult wings bearing unmarked not1 clones. The arrowhead points to a region without hairs and the arrow in F points to double hair cells in a region of polarity disruption. (G–J) Experiments with HR-46. (G) A low-magnification image of a wing where the overexpression of HR-46 produced a strong hair loss phenotype. (I) A higher magnification micrograph from such a wing. (J) A wing with a weaker phenotype. The arrowhead points to a region lacking hairs in the weak phenotype wing and the arrow points to a hair in the strong phenotype wing. (H) An HR-46 loss-of-function clone in a pupal wing marked by the loss of GFP. The wing was stained with a fluorescent phalloidin to show the actin cytoskeleton. Note that hairs inside the clone are slightly stouter than their neighbors. (K and L) Pupal wings from Uch-L3P mutants stained with a fluorescent phalloidin. In the low-magnification image (K) note the abnormal shape of the wing. It is shorter and fatter than normal (compare to pupal wings in Figure 1B). (L) A higher-magnification micrograph shows a region with cells that failed to form hairs. Hair polarity appears abnormal but this is not a routinely seen phenotype in Uch-L3 mutants and could possibly be a mounting artifact. Further experiments will be required to determine the significance of this observation.
The expression of the non-stop (not) gene decreased 3.9-fold from 24 to 40 hr. Mutations in not result in photoreceptor neurons projecting through the lamina instead of terminating there (Martin et al. 1995). The mutations also result in ∼20% of ommatidia being misoriented—a planar polarity phenotype. Strong alleles of not die as prepupae so we examined not clones in both adult and pupal wings. In our initial experiments we examined wings from vg-gal4 UAS-flp; not1 FRT80/ubi-GFP FRT80 flies. These wings contain large numbers of clones (marked in pupal wings but unmarked in the adult) due to driving expression of flp with vg-Gal4. Perhaps 25% of wing cells are found in clones. All adult wings of this genotype had regions where cells failed to form hairs or had very small hairs (Figure 3E). These were found only in proximal medial regions on the ventral wing surface. All such wings also had subtle polarity abnormalities—small groups of hairs with slightly abnormal polarity in all regions of the wing. Similar defects can be produced as mounting artifacts but consistently finding such defects leads us to conclude that these were due to not clones. Of 47 such wings examined 27 also contained multiple hair cells and a further 10 contained regions with planar polarity defects reminiscent of genes such as fz and dsh (Figure 3F). When we examined marked not clones in pupal wings we found that most, but not all, showed cells where hair differentiation was delayed or absent (Figure 3, A–D). Such clones were seen in all wing regions. We suggest that all not clones have delayed hair formation. When the clones are located in wing regions where hairs normally form first (distal or peripheral regions) the hairs form later than normal but still have enough time to reach a relatively normal length. In contrast when clones are located in regions where hair formation is normally late (proximal and medial regions on the ventral wing surface) not enough time remains prior to cuticle deposition to produce a normal hair. The not gene encodes a ubiquitin carboxyterminal hydrolase likely to function in the removal of ubiquitin from proteins during protein degradation.
The Uch-L3 gene also encodes a ubiquitin carboxy hydrolase and its expression decreased 2.9-fold between 24 and 40 hr. A P-insertion mutation in this gene is semilethal and escapers have an abnormal eye (Spradling et al. 1999). We did not find any homozygous Uch-L3J2b8 flies that eclosed but we were able to examine animals that died as pharate adults. These animals displayed several morphological defects such as loss of tarsal leg joints, shorter and fatter leg segments, the loss of a discrete antennal segment 4, and a fatter arista that could be due to defects in cell shape or movement. We examined pupal wings from such animals and found wings that were wider and shorter than normal and regions with a loss of hairs (Figure 3, K and L). All of the phenotypes seen in Uch-L3 pupae and pharate adults showed variable expressivity.
DISCUSSION
Wing differentiation genes:
The 1335 genes identified in our analysis of the transcriptome of differentiating wing cells have already proven to be a valuable resource in analyzing wing and wing hair development. As a test of the usefulness of the data set we selected a number of genes where genetic mutants were available and where a wing phenotype had not been described. This was not a random set as the existence of mutations favored genes that were not redundant and we chose genes where either the structure of the encoded protein or the phenotype in other developmental contexts suggested that the gene might be important for wing differentiation. Nonetheless it is notable that 9 of 10 genes selected had a wing phenotype. For several of these the phenotype was dramatic while in others it was modest. Projects to systematically recover insertion mutations in all or most fly genes (Bellen et al. 2004) will aid the further analysis of the many genes identified in our experiments. These stocks are limited by the failure of many insertions to inactivate a gene, although imprecise excision of P elements is straightforward and should lead to loss-of-function mutations. The development of an RNAi approach for Drosophila pupae would be very helpful. The injection of dsRNA into many insects results in a systemic RNAi response (Klingler 2004; Tomoyasu and Denell 2004); however, we have had only limited success with such injections in Drosophila. Perhaps this is due to Drosophila lacking one or several genes required for supporting systemic RNAi. If so the expression of exotic genes that encode such factors could circumvent this limitation. An alternative approach would be the development of collections of transgenic flies that can be induced to express double-stranded RNA for desired genes.
Ecdysone and the program of wing differentiation:
Metamorphosis in Drosophila, as in other insects, is under hormonal control (Riddiford 1993). Pupariation is marked and caused by a peak in ecdysone levels. Levels quickly fall and there is then a major peak at ∼30 hr that promotes adult development (Handler 1982; Riddiford 1993). Thus, our 24-hr samples are at a time of low ecdysone, the 32-hr sample is during a period of rapid increase in ecdysone levels, and the 40-hr sample is from tissues that have been exposed to high ecdysone levels for some hours and where they may have begun to fall. The ecdysone receptor is a member of the steroid receptor superfamily and its activation leads to a transcriptional cascade that is thought to control adult differentiation in a tissue-specific manner (Thummel 1996). Many of the early genes encode transcription factors. The different responses of specific tissues make the analysis of whole-animal gene expression data problematic. Indeed, in our experiments the analysis of whole pupal RNA did not provide insights equivalent to those from examining changes in wing gene expression. When RNA profiling was done on five different tissues at the start of metamorphosis similar numbers of genes had increases and decreases in expression (Li and White 2003). In our experiments we found substantially more genes with increased as opposed to decreased expression. We suggest the difference between these two studies is that we were looking later in development at cells beginning their terminal differentiation.
Seventy-four percent of the genes whose expression changed fivefold or greater clustered into expression group 2. There were substantially higher RNA levels at 40 hr than at 24 or 32 hr for these genes. Given that ecdysone levels are thought to reach their peak around 30 hr (Handler 1982; Riddiford 1993) we think the expression of cluster 2 genes is likely an indirect response to the increase in ecdysone levels that promotes adult development. The induction in cluster 2 expression could be dependent on the induction of one or more genes directly by the increase in ecdysone levels. We think genes in groups 1, 3, or 7 are more likely to be primary responders as these genes show sharply increased expression at 32 hr. One gene in cluster 7 was HR46, which is known to be important for the ecdysone response and is thought to be a direct target of ecdysone at the onset of metamorphosis although its induction is delayed due to a need for protein synthesis (Lam et al. 1999; Thummel 2001). This gene encodes an orphan receptor and it is a candidate for being involved in the induction of the group 2 genes or the repression of the group 4 and/or 6 genes whose expression falls sharply between 32 and 40 hr (Table 2).
Cuticle synthesis:
Many of the genes picked out due to dramatic changes in expression appear to be involved in the synthesis of cuticle. This included genes that encode components of the cuticle (e.g., Cuticle proteins), genes that encode enzymes that are involved in the synthesis of cuticle components (e.g., Chitin synthase), genes that degrade cuticle components (e.g., Chitinases), genes involved in the crosslinking of cuticle (e.g., Dopa decarboxylase), and genes involved in the pigmentation of cuticle (e.g., Ebony). These results are not surprising given that the terminal differentiation of wing cells involves the formation of cuticle and that cuticle deposition begins during the time period examined. Cuticle is a feature of insects and many other invertebrates, but it is not found in vertebrates and hence is a potential target for agents that specifically target invertebrates. The collection of genes uncovered in our chip experiments should lead to the identification of additional potential targets for insecticides. Of particular interest are genes that are well conserved only in insects. The kkv and knk genes were found previously to have an embryonic cuticle phenotype and we have found that they also produce dramatic pupal cuticle mutant phenotypes (Ostrowski et al. 2002). These genes lack close vertebrate homologs, suggesting that they might be good targets for insecticides.
Ubiquitination and hair formation:
not and Ubh-L3 were two of the genes whose expression we found to be strongly modulated during wing development. These genes share the property of encoding ubiquitin carboxy hydrolases that are involved in the removal of ubiquitin during proteosomal-mediated protein degradation. The expression of both of these genes decreased during the time period covered by our experiments. One of the phenotypes seen in both mutants was delayed hair formation, which in some cases led to cells not forming a hair. This phenotype is not only similar to kojak but also similar to that seen in guftagu (gft) mutant cells (Mistry et al. 2004). This gene encodes a Cullin 3 protein, which is part of the SCF ubiquitin ligase complex, which is an upstream component of the ubiquitin-mediated proteolysis pathway. These observations suggest that the regulated degradation of one or more proteins by this pathway will play an important role in regulating hair initiation.
Acknowledgments
We thank the University of Virginia Molecular Biology core facility for help in carrying out the gene chip experiments. We particularly thank Carl Thummel, Adelaide Carpenter, Daniel St. Johnson, and Iris Salecker for providing fly stocks. This work was supported by grants from the National Institute of General Medical Science to P.N.A. The University of Virginia Cancer Center supported core facilities used in carrying out this research.
References
- Adler, P. N., 2002. Planar signaling and morphogenesis in Drosophila. Dev. Cell 2: 525–535. [DOI] [PubMed] [Google Scholar]
- Adler, P. N., C. Zhu and D. Stone, 2004. Inturned localizes to the proximal side of wing cells under the instruction of upstream planar polarity proteins. Curr. Biol. 14: 2046–2051. [DOI] [PubMed] [Google Scholar]
- Alsina, B., F. Serras, J. Baguna and M. Corominas, 1998. patufet, the gene encoding the Drosophila melanogaster homologue of selenophosphate synthetase, is involved in imaginal disc morphogenesis. Mol. Gen. Genet. 257: 113–123. [DOI] [PubMed] [Google Scholar]
- Arama, E., D. Dickman, Z. Kimchie, A. Shearn and Z. Lev, 2000. Mutations in the beta-propeller domain of the Drosophila brain tumor (brat) protein induce neoplasm in the larval brain. Oncogene 19: 3706–3716. [DOI] [PubMed] [Google Scholar]
- Axelrod, J., 2001. Unipolar membrane association of Dishevelled mediates Frizzled planar cell polarity signaling. Genes Dev. 15: 1182–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastock, R., H. Strutt and D. Strutt, 2003. Strabismus is asymmetrically localised and binds to Prickle and Dishevelled during Drosophila planar polarity patterning. Development 130: 3007–3014. [DOI] [PubMed] [Google Scholar]
- Baum, B., 2002. Winging it—actin on the fly. Dev. Cell 2: 125–126. [DOI] [PubMed] [Google Scholar]
- Bellen, H. J., R. W. Levis, G. Liao, Y. He, J. W. Carlson et al., 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros, M., N. Paricio, D. I. Strutt and M. Mlodzik, 1998. Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell 94: 109–118. [DOI] [PubMed] [Google Scholar]
- De Celis, J. F., 2003. Pattern formation in the Drosophila wing: the development of the veins. BioEssays 25: 443–451. [DOI] [PubMed] [Google Scholar]
- Delon, I., H. Chanut-Delalande and F. Payre, 2003. The Ovo/Shavenbaby transcription factor specifies actin remodelling during epidermal differentiation in Drosophila. Mech. Dev. 120: 747–758. [DOI] [PubMed] [Google Scholar]
- DiBartolomeis, S. M., B. Akten, G. Genova, M. A. Roberts and F. R. Jackson, 2002. Molecular analysis of the Drosophila miniature-dusky (m-dy) gene complex: m-dy mRNAs encode transmembrane proteins with similarity to C. elegans cuticulin. Mol. Genet. Genomics 267: 564–576. [DOI] [PubMed] [Google Scholar]
- Dobrosotskaya, I. Y., A. C. Seegmiller, M. S. Brown, J. L. Goldstein and R. B. Rawson, 2002. Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila. Science 296: 879–883. [DOI] [PubMed] [Google Scholar]
- Dorer, D. R., J. A. Rudnick, E. N. Moriyama and A. C. Christensen, 2003. A family of genes clustered at the Triplo-lethal locus of Drosophila melanogaster has an unusual evolutionary history and significant synteny with Anopheles gambiae. Genetics 165: 613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton, S., 1997. Planar polarization of Drosophila and vertebrate epithelia. Curr. Opin. Cell Biol. 9: 860–866. [DOI] [PubMed] [Google Scholar]
- Eaton, S., 2003. Cell biology of planar polarity transmission in the Drosophila wing. Mech. Dev. 120: 1257–1264. [DOI] [PubMed] [Google Scholar]
- Eaton, S., R. Wepf and K. Simons, 1996. Roles for Rac1 and Cdc42 in planar polarization and hair outgrowth in the wing of Drosophila. J. Cell Biol. 135: 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiguin, F., M. Hannus, M. Mlodzik and S. Eaton, 2001. The ankyrin repeat protein Diego mediates Frizzled-dependent planar polarization. Dev. Cell 1: 93–101. [DOI] [PubMed] [Google Scholar]
- Fristrom, D., M. Wilcox and J. Fristrom, 1993. The distribution of PS integrins, laminin A and F-actin during key stages in Drosophila wing development. Development 117: 509–523. [DOI] [PubMed] [Google Scholar]
- Geng, W., B. He, M. Wang and P. N. Adler, 2000. The tricornered gene, which encodes the Drosophila NDR kinase is required to maintain the integrity of cellular extensions. Genetics 156: 1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler, A. M., 1982. Ecdysteroid titers during pupal and adult development in Drosophila melanogaster. Dev. Biol. 93: 73–82. [DOI] [PubMed] [Google Scholar]
- He, B., and P. N. Adler, 2002. The genetic control of arista lateral morphogenesis in Drosophila. Dev. Genes Evol. 212: 218–229. [DOI] [PubMed] [Google Scholar]
- Hirsh, J., and N. Davidson, 1981. Isolation and characterization of the dopa decarboxylase gene of Drosophila melanogaster. Mol. Cell. Biol. 1: 475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine, K. D., and C. Rauskolb, 2001. Boundaries in development: formation and function. Annu. Rev. Cell Dev. Biol. 17: 189–214. [DOI] [PubMed] [Google Scholar]
- Katoh, M., 2002. GIPC gene family. Int. J. Mol. Med. 9: 585–589. [PubMed] [Google Scholar]
- Klingler, M., 2004. Tribolium. Curr. Biol. 14: R639–R640. [DOI] [PubMed] [Google Scholar]
- Konrad, K. D., and J. L. Marsh, 1987. Developmental expression and spatial distribution of dopa decarboxylase in Drosophila. Dev. Biol. 122: 172–185. [DOI] [PubMed] [Google Scholar]
- Lam, G., B. L. Hall, M. Bender and C. S. Thummel, 1999. DHR3 is required for the prepupal-pupal transition and differentiation of adult structures during Drosophila metamorphosis. Dev. Biol. 212: 204–216. [DOI] [PubMed] [Google Scholar]
- Lee, H., and P. N. Adler, 2002. The function of the frizzled pathway in the Drosophila wing is dependent on inturned and fuzzy. Genetics 160: 1535–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H., and P. N. Adler, 2004. The grainy head transcription factor is essential for the function of the frizzled pathway in the Drosophila wing. Mech. Dev. 121: 37–49. [DOI] [PubMed] [Google Scholar]
- Li, C., and W. Hung Wong, 2001. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2: RESEARCH0032. [DOI] [PMC free article] [PubMed]
- Li, T. R., and K. P. White, 2003. Tissue specific gene expression and ecdysone-regulated genomic networks in Drosophila. Dev. Cell 5: 59–72. [DOI] [PubMed] [Google Scholar]
- Liu, H., Y. C. Su, E. Becker, J. Treisman and E. Y. Skolnik, 1999. A Drosophila TNF-receptor-associated factor (TRAF) binds the Ste20 kinase Misshapen and activates Jun kinase. Curr. Biol. 9: 101–104. [DOI] [PubMed] [Google Scholar]
- Lukacsovich, T., K. Yuge, W. Awano, Z. Asztalos, S. Kondo et al., 2003. The ken and barbie gene encoding a putative transcription factor with a BTB domain and three zinc finger motifs functions in terminalia development of Drosophila. Arch. Insect Biochem. Physiol. 54: 77–94. [DOI] [PubMed] [Google Scholar]
- Martin, F. A., A. Perez-Garijo, E. Moreno and G. Morata, 2004. The brinker gradient controls wing growth in Drosophila. Development 131: 4921–4930. [DOI] [PubMed] [Google Scholar]
- Martin, K. A., B. Poeck, H. Roth, A. J. Ebens, L. C. Ballard et al., 1995. Mutations disrupting neuronal connectivity in the Drosophila visual system. Neuron 14: 229–240. [DOI] [PubMed] [Google Scholar]
- Mirey, G., M. Balakireva, S. L'Hoste, C. Rosse, S. Voegeling et al., 2003. A Ral guanine exchange factor-Ral pathway is conserved in Drosophila melanogaster and sheds new light on the connectivity of the Ral, Ras, and Rap pathways. Mol. Cell. Biol. 23: 1112–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry, H., B. A. Wilson, I. J. Roberts, J. C. O'Kane and J. B. Skeath, 2004. Cullin-3 regulates pattern formation, external sensory organ development and cell survival during Drosophila development. Mech. Dev. 121: 1495–1507. [DOI] [PubMed] [Google Scholar]
- Mitchell, H. K., J. Edens and N. S. Petersen, 1990. Stages of cell hair construction in Drosophila. Dev. Genet. 11: 133–140. [DOI] [PubMed] [Google Scholar]
- Morey, M., F. Serras, J. Baguna, E. Hafen and M. Corominas, 2001. Modulation of the Ras/MAPK signalling pathway by the redox function of selenoproteins in Drosophila melanogaster. Dev. Biol. 238: 145–156. [DOI] [PubMed] [Google Scholar]
- Ostrowski, S., H. A. Dierick and A. Bejsovec, 2002. Genetic control of cuticle formation during embryonic development of Drosophila melanogaster. Genetics 161: 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paricio, N., F. Feiguin, M. Boutros, S. Eaton and M. Mlodzik, 1999. The Drosophila STE20-like kinase misshapen is required downstream of the Frizzled receptor in planar polarity signaling. EMBO J. 18: 4669–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, J. T., M. A. Deardorff, C. Tan, M. S. Rao, P. S. Klein et al., 2001. Regulation of eye development by frizzled signaling in Xenopus. Proc. Natl. Acad. Sci. USA 98: 3861–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddiford, L. M., 1993. Hormones and Drosophila development, pp. 899–939 in The Development of Drosophila melanogaster, Vol. II, edited by M. Bate and A. Martinez-Arias. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Roch, F., C. R. Alonso and M. Akam, 2003. Drosophila miniature and dusky encode ZP proteins required for cytoskeletal reorganisation during wing morphogenesis. J. Cell Sci. 116: 1199–1207. [DOI] [PubMed] [Google Scholar]
- Russell, S. R. H., G. Heimbeck, C. M. Goddard, A. T. C. Carpenter and M. Ashburner, 1996. The Drosophila Eip78C gene is not vital but has a role in regulating chromosome puffs. Genetics 144: 159–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamoto, K., P. Winge, S. Koyama, Y. Hirota, C. Yamada et al., 1999. The Drosophila Ral GTPase regulates developmental cell shape changes through the Jun NH(2)-terminal kinase pathway. J. Cell Biol. 146: 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano, N., and P. H. O'Farrell, 1997. Limb morphogenesis: connections between patterning and growth. Curr. Biol. 7: R186–R195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, Y., T. Usui, S. Yanagawa, M. Takeichi and T. Uemura, 2001. Asymmetric colocalization of Flamingo, a seven-pass transmembrane cadherin, and Dishevelled in planar cell polarization. Curr. Biol. 11: 859–863. [DOI] [PubMed] [Google Scholar]
- Sonoda, J., and R. P. Wharton, 2001. Drosophila Brain Tumor is a translational repressor. Genes Dev. 15: 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling, A. C., D. Stern, A. Beaton, E. J. Rhem, T. Laverty et al., 1999. The Berkeley Drosophila genome project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153: 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt, D., 2001. Asymmetric localization of frizzled and the establishment of cell polarity in the Drosophila wing. Mol. Cell 7: 367–375. [DOI] [PubMed] [Google Scholar]
- Tan, C., M. A. Deardorff, J. P. Saint-Jeannet, J. Yang, A. Arzoumanian et al., 2001. Kermit, a frizzled interacting protein, regulates frizzled 3 signaling in neural crest development. Development 128: 3665–3674. [DOI] [PubMed] [Google Scholar]
- Teleman, A. A., and S. M. Cohen, 2000. Dpp gradient formation in the Drosophila wing imaginal disc. Cell 103: 971–980. [DOI] [PubMed] [Google Scholar]
- Thomas, G. H., D. C. Zarnescu, A. E. Juedes, M. A. Bales, A. Londergan et al., 1998. Drosophila beta heavy-spectrin is essential for development and contributes to specific cell fates in the eye. Development 125: 2125–2134. [DOI] [PubMed] [Google Scholar]
- Thummel, C. S., 1996. Flies on steriods—Drosophila metamorphosis and the mechanisms of steroid hormone action. Trends Genet. 12: 306–310. [DOI] [PubMed] [Google Scholar]
- Thummel, C. S., 2001. Molecular mechanisms of developmental timing in C. elegans and Drosophila. Dev. Cell 1: 453–465. [DOI] [PubMed] [Google Scholar]
- Toba, G., T. Ohsako, N. Miyata, T. Ohtsuka, K. H. Seong et al., 1999. The gene search system. A method for efficient detection and rapid molecular identification of genes in Drosophila melanogaster. Genetics 151: 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu, Y., and R. E. Denell, 2004. Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev. Genes Evol. 214: 575–578. [DOI] [PubMed] [Google Scholar]
- Tree, D. R. P., J. M. Shulman, R. Rousset, M. P. Scott, D. Gubb et al., 2002. Prickle mediates feedback amplification to generate asymmetric planar cell polarity signaling. Cell 109: 1–11. [DOI] [PubMed] [Google Scholar]
- Turner, C. M., and P. N. Adler, 1995. Morphogenesis of Drosophila pupal wings in vitro. Mech. Dev. 52: 247–255. [DOI] [PubMed] [Google Scholar]
- Turner, C. M., and P. N. Adler, 1998. Distinct roles for the actin and microtubule cytoskeletons in the morphogenesis of epidermal hairs during wing development in Drosophila. Mech. Dev. 70: 181–192. [DOI] [PubMed] [Google Scholar]
- Usui, T., Y. Shima, Y. Shimada, S. Hirano, R. W. Burgess et al., 1999. Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell 98: 585–595. [DOI] [PubMed] [Google Scholar]
- Wong, L. L., and P. N. Adler, 1993. Tissue polarity genes of Drosophila regulate the subcellular location for prehair initiation in pupal wing cells. J. Cell Biol. 123: 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, T. R. F., 1996. The Wilhelmine E. Key 1992 invitational lecture. Phenotypic analysis of the Dopa decarboxylase gene cluster mutants in Drosophila melanogaster. J. Hered. 87: 175–190. [DOI] [PubMed] [Google Scholar]
- Xu, T., and G. M. Rubin, 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117: 1223–1237. [DOI] [PubMed] [Google Scholar]