Abstract
Over 1000 genetically linked RFLP loci in Brassica napus were mapped to homologous positions in the Arabidopsis genome on the basis of sequence similarity. Blocks of genetically linked loci in B. napus frequently corresponded to physically linked markers in Arabidopsis. This comparative analysis allowed the identification of a minimum of 21 conserved genomic units within the Arabidopsis genome, which can be duplicated and rearranged to generate the present-day B. napus genome. The conserved regions extended over lengths as great as 50 cM in the B. napus genetic map, equivalent to ∼9 Mb of contiguous sequence in the Arabidopsis genome. There was also evidence for conservation of chromosome landmarks, particularly centromeric regions, between the two species. The observed segmental structure of the Brassica genome strongly suggests that the extant Brassica diploid species evolved from a hexaploid ancestor. The comparative map assists in exploiting the Arabidopsis genomic sequence for marker and candidate gene identification within the larger, intractable genomes of the Brassica polyploids.
ARABIDOPSIS thaliana (hereafter referred to as Arabidopsis) is one of almost 3500 species that make up the monophyletic family of the Brassicaceae (Price et al. 1994). Arabidopsis thus shares recent common ancestry with a large number of species of significant economic importance, including a diverse range of vegetable and oil producing crops, the majority of which are Brassica species. Arabidopsis is an excellent model system for the Brassicaceae, with a small and relatively simple genome, efficient transformation system, diverse range of genetic and genomic resources, and a completed genome sequence (Arabidopsis Genome Initiative 2000).
Over the past 10 years, plant comparative mapping has taken prominence as a powerful tool, first, for uncovering the processes and rate of genome evolution and, second, for allowing the transfer of genetic resources between species. Comparative mapping has been most extensively applied to the grasses where the genetic maps of 11 species, including the model monocot rice, have been aligned. These include 11 diverse species varying dramatically in haploid chromosome number, genome size, and phenotype (reviewed in Devos and Gale 2000). Perhaps the most striking observation from the cereal studies was the extensive genome conservation observed between species that diverged millions of years ago. Using rice as the basal genome, <30 conserved blocks were identified, which could be rearranged and/or duplicated to form each of the other grass genomes. Comparative mapping studies among members of the Brassicaceae have been more ambiguous in their conclusions, leading to ongoing discussions about the level of genome duplication prevalent in modern Brassica cultivars and the extent of the genome rearrangements that have occurred in the evolution of these cultivars from a common ancestor (Lagercrantz 1998; Lan et al. 2000; Lukens et al. 2003).
This study focuses on the genome of the oilseed crop Brassica napus, which is an amphidiploid species formed from multiple independent fusion events between ancestors of the diploids B. rapa (A genome donor) and B. oleracea (C genome donor) (U 1935; Palmer et al. 1983; Parkin et al. 1995). Polyploidy is a prevalent evolutionary mechanism within angiosperms since it has been estimated that 30–70% of modern plant species have evolved through a polyploid ancestor (reviewed in Wendel 2000). Polyploidy can occur through either the duplication of whole-chromosome complements or the fusion of related chromosome complements, and stabilization of the newly expanded karyotype must then take place to ensure normal diploid inheritance. Diploidization of the novel polyploid can occur through chromosomal restructuring or genetic control of illegitimate recombination events or a combination of both mechanisms. It is widely accepted that the progenitor diploid genomes of B. napus are ancient polyploids and that large-scale chromosome rearrangements have occurred since their evolution from a lower-chromosome-number progenitor (Schmidt et al. 2001). What is more contentious is whether the diploids evolved through a hexaploid ancestor or whether they were formed via segmental duplication of one or two ancestral genomes (Lukens et al. 2004). B. napus, a relatively young amphidiploid, is somewhat of an anomaly since it has been established that no major chromosomal rearrangements have occurred since the fusion of the progenitor A and C genomes, but homeologous recombination events between these two related genomes are common in newly resynthesized B. napus lines and have been observed at low levels in established canola cultivars (Parkin et al. 1995; Sharpe et al. 1995; Udall et al. 2004). It has yet to be established if B. napus has evolved or inherited a locus controlling homologous pairing similar to the Ph1 locus in hexaploid wheat (Jenczewski et al. 2003).
Comparative mapping between B. napus and Arabidopsis has thus far targeted small regions of the Arabidopsis genome, generally identifying three colinear segments in each of the diploid genomes for every region of Arabidopsis studied, thereby promoting the idea that the diploid Brassica species may have evolved through a hexaploid ancestor (Osborn et al. 1997; Cavell et al. 1998; Parkin et al. 2002). However, at the same time regions suggesting a more complex relationship between the two species were also identified (Osborn et al. 1997; Parkin et al. 2002). In the earliest published global comparison between one of the diploid Brassicas, B. nigra (black mustard), and Arabidopsis, an extensive number of rearrangements were invoked to explain how the two extant diploid genomes evolved from a common hexaploid ancestor (Lagercrantz 1998). There have been four global comparisons of the genomes of B. oleracea and Arabidopsis. Although all have been limited by a low density of common loci, three identified extensive synteny between the two genomes but were inconclusive in assessing the level of duplication of the colinear segments (Lan et al. 2000; Babula et al. 2003; Lukens et al. 2003). A more recent comparison of the B. oleracea and Arabidopsis genomes refuted the possibility of a hexaploid ancestor, citing evidence of colinear blocks ranging in copy number from 1 to 7 (Li et al. 2003).
This study describes a comprehensive comparison of a Brassica genome with that of Arabidopsis. Sequences of 359 probes derived from Brassica and Arabidopsis that detect 1232 genetically mapped loci in B. napus were used to query the Arabidopsis genome, revealing 550 homologous sequences and their inferred chromosomal positions. The data provide strong evidence to support the hypothesis that the Brassica diploid genomes evolved through a hexaploid ancestor and suggest conservation of some centromeric regions between the two species. The postulated ancestor appears to have been formed from duplication events that occurred subsequent to the putative global duplication events that took place between 65 and 90 million years ago (MYA) during the evolution of Arabidopsis (Lynch and Conery 2000; Simillion et al. 2002; Raes et al. 2003). The resultant genetic and physical comparative map can be used not only to infer genome rearrangements during the evolution of the Brassica species but also to identify regions of the Arabidopsis genome that may harbor genes of interest and should potentiate the exploitation of Arabidopsis genomic tools in Brassica research.
MATERIALS AND METHODS
Genetic linkage analysis:
Genetic linkage analysis in B. napus was carried out as described previously except hybridizations with Arabidopsis clones were washed only at low stringency (2× SSC, 0.1% SDS) (Sharpe et al. 1995). The B. napus population consisted of 60 doubled haploid lines derived from crosses between a winter B. napus breeding line (CPB87/5) and a newly resynthesized B. napus line (SYN1) as described in Parkin et al. (1995). The genetic map also includes loci positioned through previously described map alignments with a second linkage map of B. napus and one of B. oleracea (Bohuon et al. 1996; Parkin and Lydiate 1997). Briefly, common parental genotypes allowed corresponding loci to be identified between the maps through the inheritance of identical restriction fragment length polymorphism (RFLP) alleles. Loci mapped in only one population that cosegregated with such common loci were positioned at that locus in the combined map. Loci mapped in only one population positioned between common loci were placed in the corresponding interval in the combined map on the basis of their relative position in the map of origin. The RFLP probes consisted of 213 Brassica genomic clones (pN, pO, pR, pW: Sharpe et al. 1995), 61 Brassica cDNA clones (CA, es), 88 Arabidopsis cDNA clones (I, N, R, Z: Sillito et al. 2000), and 6 cloned Brassica or Arabidopsis genes (ACYL, CONSTANS, FCA, HS1, oleosin: pC2, Δ9 desaturase: pC3). The genetic linkage map was constructed using Mapmaker v3 with a LOD score of 4.0 (Lander et al. 1987) and the linkage groups were drawn using Mapchart (Voorrips 2002). Irregularities in meiotic pairing in the resynthesized B. napus parental line of the doubled haploid population used for the initial and the additional mapping caused a nondisjunction event that prevented the accurate mapping of further loci to linkage group N16 (Parkin et al. 1995). A limited map of N16 derived from the alignment of N16 from B. napus, described in Sharpe et al. (1995) and O6 from B. oleracea, described in Bohuon et al. (1996), has been used in the present analysis. A similar alignment of N16 and O6 was discussed in Ryder et al. (2001).
Sequence analysis:
Brassica genomic or cDNA clones were sequenced from each end using the BigDye v2 Terminator cycle sequencing kit according to the instructions of the manufacturer and subsequently the reactions were run out on an automated ABI377 DNA Sequencer (Applied Biosystems, Foster City, CA). The Brassica sequences were analyzed using Sequencher (Gene Codes, Ann Arbor, MI) to trim vector sequence, identify overlaps, and generate contigs. Brassica and Arabidopsis sequences were analyzed for homology to the The Institute for Genomic Research Arabidopsis pseudochromosome genomic sequence version 5.0 (ftp://ftp.tigr.org/pub/data/a_thaliana/) using the BLAST programs of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) housed on a Linux server. Low-complexity sequences were filtered in the BLAST analysis, and default values for cost (mismatch cost, −3.0), reward (match reward, 1.0), and word size (11 bp) were selected. The default gap opening penalty (5.0) and the gap extension penalty (2.0) were also selected. Perl script was used to extract the base-pair position in the Arabidopsis genomic sequence of each high-scoring segment pair (HSP), identified with BLASTN, for each clone where the primary HSP had an E-value of ≤1E-07 (supplemental Table S1 at http://www.genetics.org/supplemental/).
RESULTS
Comparative map of B. napus and Arabidopsis:
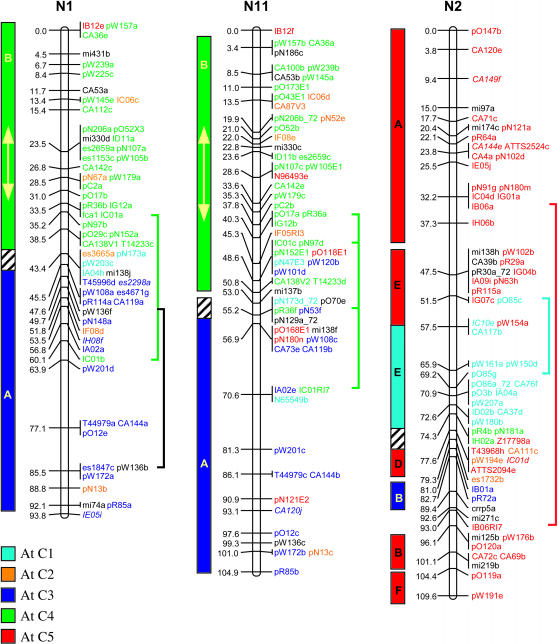
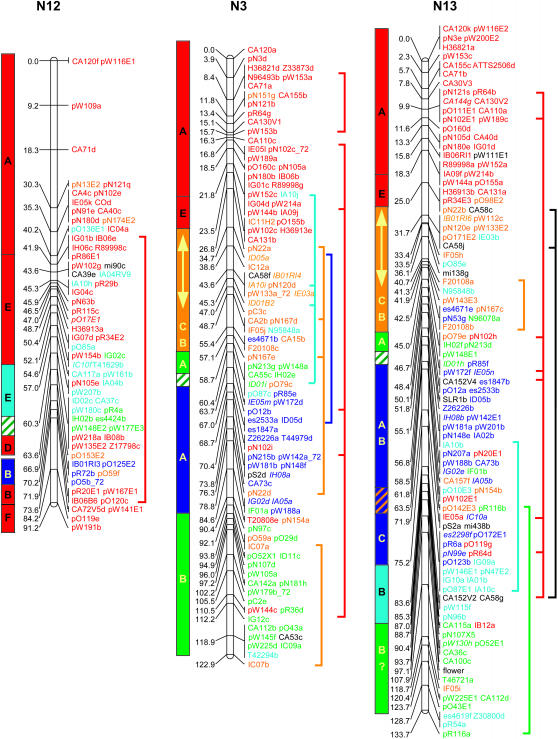
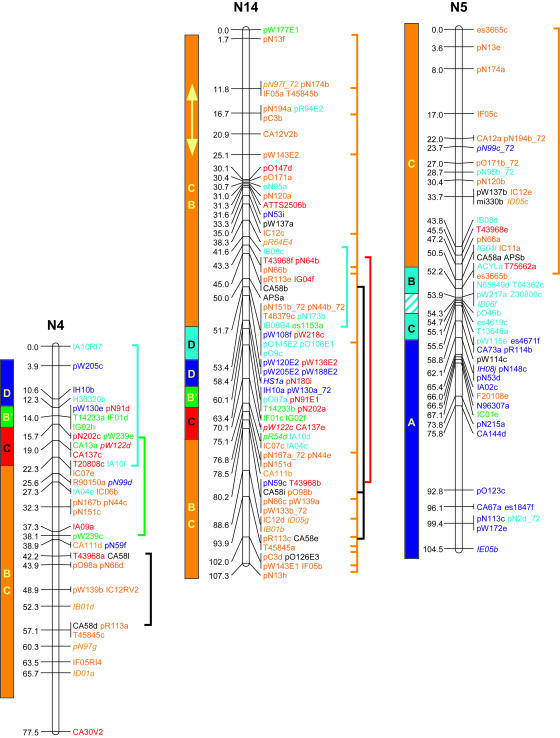
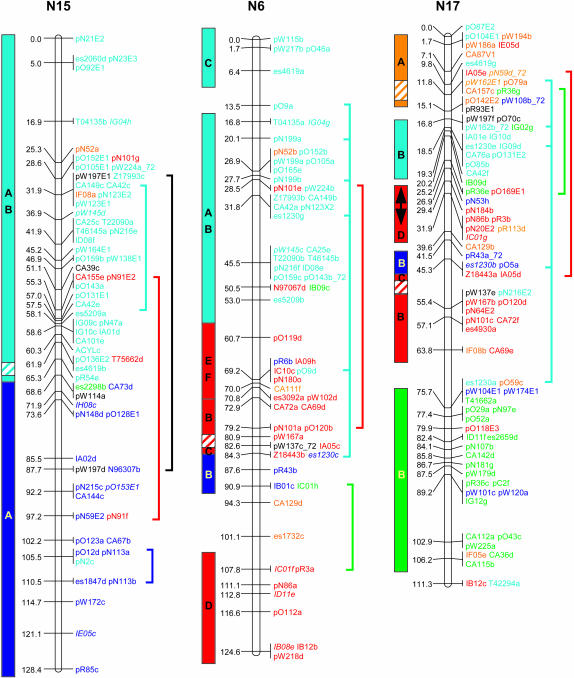
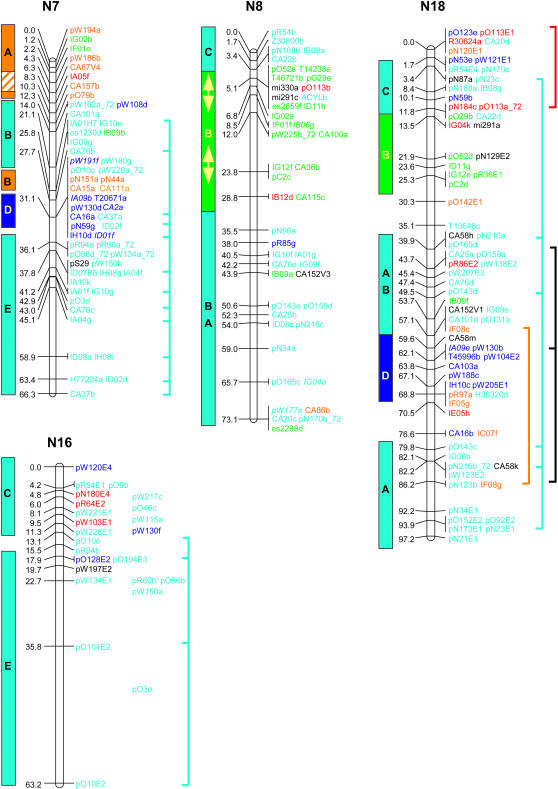
Genetic linkage mapping of RFLPs identified with 183 Brassica and Arabidopsis cDNA clones added another 646 loci to the published aligned map of B. napus (Bohuon et al. 1996; Parkin and Lydiate 1997). The complete B. napus linkage map is presented in Figure 1 and consists of 1317 genetic loci distributed over 19 linkage groups with a combined map length of 1968 cM.
Figure 1.
Genetic linkage map of B. napus. Linkage groups are arranged according to the regions of primary homology between the A (N1–N10) and C (N11–N19) genomes (Parkin et al. 2003), with centimorgan distances indicated to the left of each group. Each genetic locus is colored according to the presumed Arabidopsis homolog: light blue, chromosome 1; orange, chromosome 2; dark blue, chromosome 3; green, chromosome 4; and red, chromosome 5. Loci in italics were found within conserved blocks on the basis of secondary or tertiary hits within the Arabidopsis genome. Loci duplicated within a B. napus linkage group are indicated by vertical lines to the right of the group. Identified genome blocks showing conservation of marker content and marker order between the Arabidopsis and B. napus genomes are shown to the left of each linkage group. Each block is colored and labeled according to the identified homologous region in Arabidopsis (see Figure 2). Inversions identified in Brassica relative to Arabidopsis are indicated by arrows. Regions of the B. napus genome that have been tentatively aligned with Arabidopsis centromeric regions are indicated by hatched blocks.
The genetic linkage map was generated from segregating loci detected by 368 DNA clones, 274 of which were derived from anonymous Brassica genomic or complementary DNA; 5 were Brassica homologs of known genes and the remaining 89 clones were derived from Arabidopsis cDNA. Sequence data were obtained for 267 of the anonymous Brassica clones and BLASTN analysis was used to identify homologous loci within the Arabidopsis genome for each clone. A fairly low expect value (E-value) was used as the exclusion cutoff (1E-07) (supplemental Table S1 at http://www.genetics.org/supplemental/). The low E-value was adopted to maximize the number of markers positioned, since the majority of the probes were derived from genomic, potentially intergenic, DNA. A total of 258 of the Brassica clones displayed homology to 404 regions within the Arabidopsis genome, with an average sequence identity of 86% over all aligned HSPs. The majority of these hits were to genic regions and the most similar Arabidopsis gene was identified for each clone (supplemental Table S1 at http://www.genetics.org/supplemental/). A less stringent E-value can lead to the identification of a large number of small nonspecific regions of homology (Lukens et al. 2003). Of the 258 clones, 58 identified regions of similarity with bit scores <82, a value suggested as a cutoff for identifying orthologous sequences within the Arabidopsis genome for Brassica markers (Lukens et al. 2003). For 11 clones, these lower-scoring hits represented their only or primary region of homology within the Arabidopsis genome; the data for these clones were included in the comparative analysis described below. The remainder of the low-scoring hits represented secondary or tertiary regions of homology, which generally fell within predefined duplicated regions within the Arabidopsis genome (Arabidopsis Genome Initiative 2000), and these data did not impact on the comparative analysis. Ten of the Brassica genomic clones showed no significant homology to the Arabidopsis genomic sequence at an E-value of 1E-07; 1 clone, pR113, mapped to the Arabidopsis genome over multiple adjacent HSPs, but with an E-value of 1E-06. Subsequent BLASTX analysis of the remaining 9 clones identified related sequence for 2 clones, pR30 and pN87, which showed significant (1E-44 and 1E-94, respectively) homology to an annotated retroelement pol polyprotein sequence (At3g29156). Perhaps not surprisingly, neither of these clones mapped to colinear regions between the two genomes (see below).
To position the Arabidopsis clones accurately relative to the Brassica sequences, all the clones were compared to the Arabidopsis pseudochromosome sequence using BLASTN analysis. In total, 550 loci were physically positioned within the Arabidopsis genome on the basis of sequence identity (an average of one comparative marker every 214 kb). These same clones identified 1232 RFLP loci on the genetic linkage map of B. napus (an average of one comparative marker every 1.6 cM). In Figure 1 each of the B. napus genetic loci has been color coded according to the most significant BLASTN hit for the probe that detected that locus. Forty-two percent of the RFLP clone probes showed sequence similarity to more than one region of the Arabidopsis genome. Some of the mapped homologous loci in B. napus may represent orthologs of these secondary hits within the model genome. Brassica loci whose position within a conserved block in Arabidopsis was dependent upon such secondary hits are color coded according to the appropriate duplicate hit and are identified by italics in Figure 1.
All of the B. napus linkage groups were composed of loci identified by probes related to sequence from each of the five Arabidopsis chromosomes (Table 1 and Figure 1). If the Brassica genomes evolved through simple polyploidy from a lower chromosome ancestor similar to Arabidopsis, it might be expected that the comparative loci mapped within the B. napus genome would be equally represented across the Arabidopsis genome. However, the number of loci originating from each Arabidopsis chromosome was not evenly distributed, with significantly fewer loci than expected detected by probes showing homology to Arabidopsis chromosomes 2 and 3 and significantly more loci than expected detected by probes with homology to Arabidopsis chromosome 5 (P < 0.001 for a goodness-of-fit test) (Table 1). This nonrandom distribution could be a function of a reduction in chromosome number in the Arabidopsis lineage and/or a function of gene loss occurring after genome duplication events within Arabidopsis.
TABLE 1.
Number of loci originating from each Arabidopsis chromosome, on the basis of sequence homology, for each B. napus linkage group
| Linkage group | C1 | C2 | C3 | C4 | C5 | ND | Total |
|---|---|---|---|---|---|---|---|
| N1 | 4 | 5 | 14 | 29 | 2 | 6 | 60 |
| N11 | 3 | 6 | 13 | 27 | 7 | 7 | 63 |
| N2 | 14 | 3 | 3 | 4 | 34 | 9 | 67 |
| N12 | 12 | 4 | 4 | 7 | 41 | 2 | 70 |
| N3 | 9 | 19 | 17 | 24 | 36 | 2 | 107 |
| N13 | 19 | 16 | 25 | 17 | 42 | 11 | 130 |
| N4 | 6 | 14 | 6 | 7 | 7 | 2 | 42 |
| N14 | 13 | 33 | 10 | 6 | 13 | 6 | 81 |
| N5 | 13 | 13 | 16 | 1 | 5 | 5 | 53 |
| N15 | 36 | 3 | 16 | 2 | 7 | 4 | 68 |
| N6 | 29 | 4 | 3 | 5 | 21 | 1 | 63 |
| N17 | 17 | 11 | 9 | 23 | 18 | 4 | 82 |
| N7 | 32 | 9 | 7 | 3 | 3 | 1 | 55 |
| N16 | 17 | 0 | 3 | 0 | 3 | 1 | 24 |
| N8 | 20 | 1 | 1 | 17 | 4 | 3 | 46 |
| N18 | 31 | 7 | 12 | 7 | 8 | 7 | 72 |
| N9 | 27 | 10 | 9 | 12 | 17 | 1 | 76 |
| N19 | 10 | 4 | 5 | 8 | 57 | 7 | 91 |
| N10 | 14 | 2 | 3 | 2 | 40 | 6 | 67 |
| Total (%) | 326 (26) | 164 (13) | 176 (13) | 201 (16) | 365 (30) | 85 | 1317 |
| Expecteda | 313 | 209 | 240 | 187 | 281 |
ND, not determined.
Expected number of loci to originate from each Arabidopsis chromosome on the basis of random distribution of loci across five chromosomes with the following approximate sizes: C1, 30 Mb; C2, 20 Mb (not including the NOR region); C3, 23 Mb; C4, 18 Mb (not including the NOR region); and C5, 27 Mb.
Identification of conserved blocks between Arabidopsis and B. napus:
For each B. napus linkage group it was possible to identify blocks of conserved synteny between B. napus and Arabidopsis, which represent chromosomal segments that have been maintained since the divergence of Arabidopsis and Brassica from a common ancestor (Figures 1 and 2).
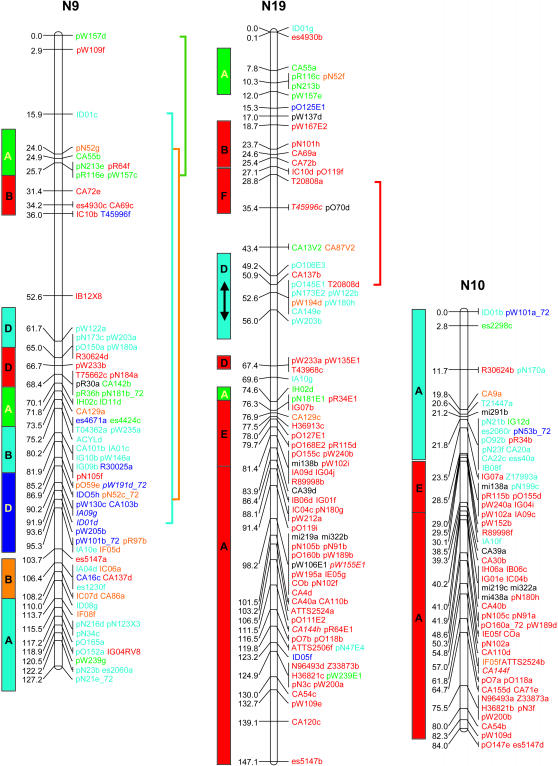
Figure 2.
A representation of the Arabidopsis genome based on the primary location of each sequenced B. napus RFLP marker on the Arabidopsis pseudochromosomes (megabase distances are indicated to the right of the chromosomes). Duplicate marker locations are indicated in parentheses. Blocks of markers found to be genetically linked in B. napus are indicated by shading and capital letters (A–F). In the majority of cases, C4B is conserved as a complete block, but in two instances, on N4 and N14, a small section of the block was observed and is represented by C4B′.
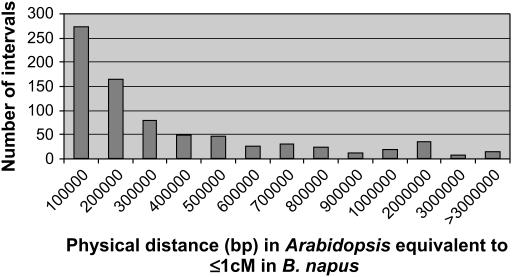
A conserved block is defined as a region that contains several closely linked homologous loci in both the Arabidopsis and Brassica genomes. Each block has a minimum of four mapped loci with at least one shared locus every 5 cM in B. napus and at least one shared locus every 1 Mb in Arabidopsis. Using these criteria, each conserved block contained on average 7.8 shared loci and had an average length of 14.8 cM in B. napus and 4.8 Mb in Arabidopsis. Together, the blocks covered almost 90% of the mapped length of the B. napus genome. The average physical distance covered in the Arabidopsis genome per 1 cM of genetic distance in the B. napus genome was calculated for every pair of comparative markers identified within the conserved blocks (Figure 3). The distribution was skewed, with 35% of the intervals tested giving a ratio of 1 cM of the B. napus genetic map to ≤100,000 bp of Arabidopsis sequence, with a median ratio of 1 cM to 160,767 bp.
Figure 3.
Distribution of the physical distance in Arabidopsis compared to the genetic distance in B. napus for each pair of linked comparative markers found with the conserved blocks.
On the basis of the conserved blocks, 21 segments were identified within the Arabidopsis genome, which could be duplicated and rearranged to form the skeleton of the B. napus genome (Figures 1 and 2). Although coverage of the two genomes is extensive, there are areas where marker density is limited, specifically the regions spanning the Arabidopsis centromeres (Figure 2). The low-copy-number sequences utilized in the Brassica mapping would be expected to have lower levels of similarity to centromeres, since the centromeres tend to be located within gene-poor transposable-element-rich regions (Arabidopsis Genome Initiative 2000).
Comparative genome organization:
The organization of the B. napus genome in comparison to the Arabidopsis genome as depicted in Figures 1 and 2 has been summarized for each of the linkage groups. Due to the close homology between the A (N1–N10) and C (N11–N19) genomes of B. napus, the primary homeologs in B. napus (described in Parkin et al. 2003) are indicated in the comparison.
N1/N11:
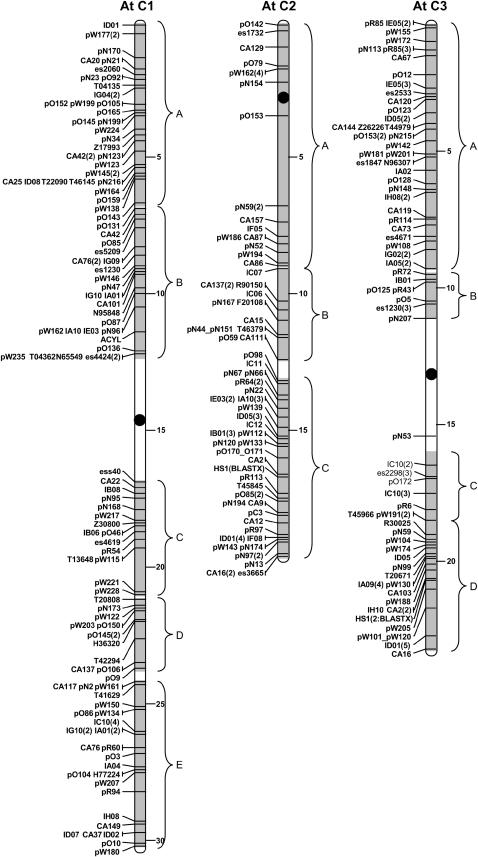
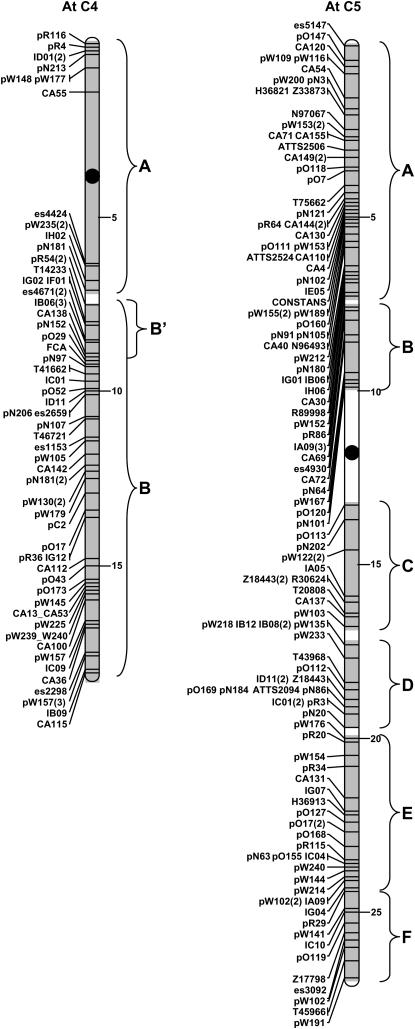
These two B. napus linkage groups are homologous along their entire length. The top half of each linkage group shows significant homology to the long arm of Arabidopsis chromosome 4 (block C4B) with one inversion, previously noted in Cavell et al. (1998), disrupting the colinearity between the two genomes. The inversion appears to be specific to N1/N11 and is not present in the homologous regions of linkage groups N3/N17 and N8/N18 where copies of block C4B were found. The lower half of N1/N11 is homologous to the top arm of Arabidopsis chromosome 3 (block C3A). This block is also strongly conserved in N5/N15 and N3/N13. In each case, the distal end of the Arabidopsis chromosome corresponds with the terminal end of the linkage groups. At the breakpoint between the two large stretches of colinearity, there are three markers that span the centromere on Arabidopsis C3 and additional markers that do not identify a conserved region. One gross chromosomal rearrangement would be sufficient to generate N1/N11 from the blocks defined in Figure 2.
N2/N12:
These two linkage groups are homologous along their mapped length. Parkin et al. (2002) previously described the relationship between N2/N12 and Arabidopsis C5, where the upper region of N2/N12 is homologous to the top 8 Mb of Arabidopsis C5 (block C5A) and an inversion on Arabidopsis C5 has moved block C5E to lie adjacent to block C5A. This pattern of C5A-C5E is conserved on linkage groups N3/N13 and N10/N19. The same inversion moved blocks C5B and C5D to the bottom of N2/N12. N2/N12 share a region of homology with Arabidopsis C1, block C1E, adjacent to which are five markers that flank the centromere on Arabidopsis C4. Two more small conserved regions were identified on N2/N12, C3B, and C5F. One inversion on Arabidopsis C5 and three insertion/deletion/translocation events represent the least number of rearrangements, which could generate the present organization of N2/N12.
N3(N17)/N13:
The homology of N3/N13 to C5 is described above, below which N3/N13 share homology with Arabidopsis C2 (block C2BC). Block C2BC on N3/N13 was defined by a lower density of comparative markers, which were further rearranged by an inversion, compared to the duplicated copies of C2BC found on N4/N14 and N5. The lower end of C2BC on N3/N13, which borders the centromere on C2, lies adjacent to a conserved block originating from the centromeric region of Arabidopsis C4 (block C4A). Below C4A, N3/N13 share homology with block C3A as described above. At the junction of C3A, which lies proximal to the centromere on C3, N3 is no longer homologous to N13 but instead shares homology with linkage group N17 and Arabidopsis C4 as described above. The remainder of linkage group N13 has no clear region of homeology in the B. napus A genome. However, in relation to Arabidopsis, this region of N13 shares homology with the blocks flanking the centromere of C3 (C3B-C3C), block C1B, and block C4B. In the area that would be homologous to the centromeric region of C3, there are eight markers with homology to different Arabidopsis chromosomes, three of which flank the centromere on C2. At least three gross chromosomal rearrangements and two inversions are necessary to generate N3 from the identified conserved blocks; assuming that C3ABC has been essentially conserved, one additional translocation/insertion would be necessary to generate N13.
N4/N14/N5:
The majority of N4 and N14 (65 and 75% of the mapped length, respectively) and the upper half of N5 share homology with Arabidopsis C2. The organization of N14 suggests that of an isocentric chromosome with the upper and lower arms sharing numerous common markers mapped in inverse orientation with respect to each other. The top of N4 and the homeologous central section of N14 show small blocks of colinearity with Arabidopsis C3, C4, and C5; N14 has one additional block from C1. Three gross chromosomal rearrangements are sufficient to describe the organization of N4 and one additional inversion and two translocation/insertions would describe N14.
N5/N15/N6:
The lower half of N5 and N15 as described above (for N1/N11) are colinear with the long arm of Arabidopsis C3. At the center of N5/N15, the markers originate from Arabidopsis C1, with comparative markers flanking the centromere on C1. This central region on N15 is part of a larger block, which is colinear with the upper arm of Arabidopsis C1 and the homeologous region of B. napus N6. One and two large chromosomal rearrangements would generate the present organization of N15 and N5, respectively.
N6/N17:
The lower half of N6 shows homology to sections of Arabidopsis C5 and C3. The region from block C5B to the bottom of N6 is homeologous but inverted with respect to N17. Two markers on N6/N17 (CA129 and es1732) identify sequences on the short arm of Arabidopsis C2. There were insufficient marker data from this region to identify a conserved block; however, fine mapping of a dwarf gene in B. rapa has subsequently aligned this region of N6 with the short arm of Arabidopsis C2 (Muangprom and Osborn 2004). It is to be expected that for regions such as these, which flank the Arabidopsis centromeres where there is a dearth of comparative markers, further conserved blocks will be identified. The comparison of N6/N17 to Arabidopsis is complex relative to other B. napus linkage groups and at least five and six chromosomal rearrangements need to be invoked to generate N6 and N17, respectively.
N7(N16)/N17:
The top of N7/N17 is homologous to the short arm of Arabidopsis C2, including comparative markers that flank the centromere on C2. Homeology between N7 and N17 breaks down after block C1B, where the lower half of N7 is homologous with N16 and Arabidopsis C1. Due to the constraints of the mapping population (see materials and methods), there are limited markers mapped to N16, making the number of rearrangements difficult to interpret. The data suggest that at least three translocations/deletions/insertions of conserved blocks have taken place to give N7, and at least one chromosomal rearrangement gave rise to N16.
N8/N18/N9:
The whole of N8 appears to be homeologous with N18 and is syntenous with Arabidopsis C1C, C4B, and C1AB; however, block C1AB is inverted on N18 with respect to N8. The remainder of N18 is homeologous to the lower portion of N9 and is syntenous with Arabidopsis C3D, C2B, a fraction of C1B, and C1A. The latter block forms part of an internal duplication on N18. One insertion of block C4B into the centromeric region lying between C1AB and C1C and two inversions (in C4B) could describe N8. The same insertion of C4B found on N8, duplication of C1A, and translocation/insertion of C3D would generate N18.
N9/N19/N10:
N10 and N19 share a region that is syntenous with Arabidopsis C5 as described above (for N2/N12). The end of C5E, which coincides with the break in homology between N10 and N19, separates a region of apparent conservation between the two species from one that is fragmented. The tops of N9 and N19 share loci from comparative markers, which are assigned to a number of blocks, running from the top of N9/N19 in the order C4A-C5B-C5F-C1D-C5D-C4A. There is no clear region of homology in the B. napus C genome for the top of N10, which is syntenous with Arabidopsis C1. N9 has the most complex segmental pattern of all the linkage groups, necessitating at least nine chromosomal rearrangements to generate the mapped group. One inversion on C5 (as described for N2/N12) and one translocation would explain N10, and one inversion and six other rearrangements would explain N19.
At least 74 translocations, fusions, deletions, or inversions of the 21 conserved segments found within the Arabidopsis genome are necessary to generate the present-day B. napus genome. However, 28 of these rearrangements are common to both the A and C genomes of B. napus, suggesting that they occurred prior to their divergence from a common ancestor. As described above, a number of the breakpoints between conserved segments correspond to previously defined translocation end points, which separate the A and C genomes of B. napus (Parkin et al. 2003). In a number of instances, the junctions of conserved blocks coincide with telomeric or centromeric regions of Arabidopsis, suggesting that centric fission and fusion have played a role in the chromosomal restructuring.
Duplication within the Brassica genome:
Counting the number of times that a single Arabidopsis region is found within the B. napus genome provides an estimate of the level of genome duplication within Brassica compared to the model genome. Each conserved chromosomal segment was represented between four and seven times within the B. napus genome (Table 2). However, the organization of the different duplicated copies of each block varied with respect to each other, either by the presence of additional rearrangements (see description for N1/N11 above) or by the number of comparative markers [see description for N3(N17)/N13 above]. In Arabidopsis, 81% of the comparative loci positioned on the genome mapped to conserved regions present in at least six copies within the B. napus genome (Table 2). Eighty-six percent of the mapped length of the B. napus genome, which was arranged in conserved blocks, was found in at least six copies (Table 2). These results corroborate previous suggestions based on more limited data that the Brassica diploid genomes have evolved through a hexaploid ancestor. However, the presence of seven copies of some Arabidopsis regions within the B. napus genome suggests that further segmental duplication events may have occurred subsequent to any polyploidy event(s).
TABLE 2.
Description of each conserved block found in Arabidopsis
| Conserved blocka | No. of times replicated | No. of comparative markers in Arabidopsis (nonsyntenous)b | No. of comparative loci in B. napusc | Total cM coverage in B. napus | Length of conserved block (Mb) in Arabidopsisd |
|---|---|---|---|---|---|
| C1A | 7 | 31 (1) | 88 | 192.1 | 6.68 |
| C1B | 7 | 22 (2) | 54 | 64.7 | 5.32 |
| C1C | 7 | 14 (1) | 28 | 38.1 | 4.12 |
| C1D | 4 | 10 (1) | 18 | 24.1 | 2.49 |
| C1E | 4 | 23 (2) | 42 | 106.6 | 6.07 |
| C2A | 5 | 12 (2) | 19 | 42.1 | 8.71e |
| C2B | 6 | 11 (1) | 24 | 34.15 | 3.42 |
| C2C | 6 | 31 (3) | 65 | 186 | 6.35 |
| C3A | 6 | 31 (1) | 88 | 244.6 | 9.27 |
| C3Bf | 5–8 | 7 | 13 | 24.75 | 1.66 |
| C3Cf | 1–4 | 6 | 8 | 4.35 | 3.07 |
| C3D | 6 | 18 (1) | 34 | 36.85 | 4.7 |
| C4A | 6 | 11 | 31 | 25.65 | 7.09e |
| C4B′ | 6 | 8 | 18 | 5.85 | 1.45 |
| C4B | 6 | 35 (2) | 106 | 244.85 | 8.96 |
| C5A | 6 | 44 (3) | 142 | 245.7 | 7.55 |
| C5B | 6 | 7 | 25 | 55 | 1.98 |
| C5C | 4 | 7 (2) | 11 | 27.55 | 3.54 |
| C5D | 6 | 16 (1) | 26 | 45.05 | 2.42 |
| C5E | 6 | 18 (3) | 52 | 45.35 | 4.32 |
| C5F | 5 | 6 (1) | 14 | 13.15 | 1.99 |
| Total(%) | 115–121 | 368 (27) | 906 | 1706.55 (86.7%) | 101.16 (85.3%) |
Conserved blocks are indicated in Figures 1 and 2. Those blocks that are present in at least three copies in each of the diploid Brassica genomes are indicated by italics.
Number of comparative loci originating from the conserved block in Arabidopsis, which are mapped in a conserved region within the A and/or C genomes of B. napus. Loci within the conserved block that have not been mapped to a colinear position in B. napus are indicated in parentheses.
Total number of mapped loci within B. napus that originate from the conserved block and are found in a colinear position.
Physical length of the designated conserved block in Arabidopsis as shown in Figure 2. The complete block may not be represented in each of the duplicate copies.
Blocks that include centromeric regions in Arabidopsis.
Limited marker data in the region flanking the centromere on Arabidopsis chromosome 3 make it difficult to accurately identify these regions within the B. napus genome.
Consequences of duplication within the Arabidopsis genome:
The majority of the conserved Arabidopsis blocks, including those known to be part of duplicated regions within Arabidopsis, are each found between five and seven times within the B. napus genome. Effectively, this means that the duplicated regions of the Arabidopsis genome are found between 10 and 14 times within the B. napus genome; similarly, recent physical mapping carried out in B. napus identified 12 regions within the B. napus genome homologous to a small duplicated region of the Arabidopsis genome (Rana et al. 2004). These data suggest that the large segmental genomic duplications found within Arabidopsis occurred in the common ancestor of the two lineages prior to the formation of a Brassica hexaploid ancestor. These data are also consistent with the fact that the last round of genome duplication is believed to have occurred in Arabidopsis between 65 and 90 million years ago (Lynch and Conery 2000; Simillion et al. 2002; Raes et al. 2003) whereas the separation of the Arabidopsis and Brassica lineages is dated somewhere between 12 and 24 million years ago (Koch et al. 2000).
Since the divergence of these two species one would expect the independent loss of redundant duplicate genes from both species. Several such losses from the Arabidopsis genome were observed. For example, on N1 and N11, the upper parts of the linkage groups are colinear with the long arm of Arabidopsis chromosome 4 (Figure 1). Nonetheless, a number of Brassica loci were identified by probes (IC06, CA87, pN52, pN67) originating from Arabidopsis chromosome 2. Although these probes were found in regions identified as being duplicated between chromosomes 2 and 4 of Arabidopsis (http://www.tigr.org/tdb/e2k1/ath1/Arabidopsis_genome_duplication.shtml), they showed no homology to the Arabidopsis chromosome 4 sequence. Thus, Brassica has maintained duplicate copies of these sequences within the region equivalent to chromosome 4, whereas Arabidopsis has lost them.
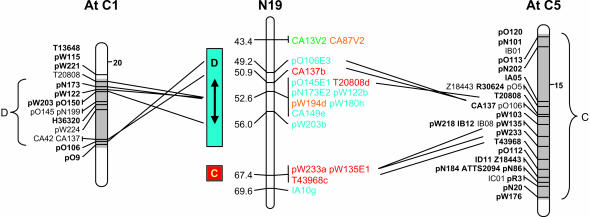
In some instances the duplications evident within the Arabidopsis genome have made it difficult to identify the most similar region shared between the two species. For example, loci on B. napus linkage group N19 show strong homology to both chromosome 5 block C and the duplicated region on Arabidopsis chromosome 1 block D (Figure 4).
Figure 4.
Alignment of segment of B. napus linkage group N19 with both Arabidopsis chromosome 1 and chromosome 5, highlighting the difficulty in identifying the most related Arabidopsis region where there are ancient duplications in the model genome.
Conservation of chromosome landmarks between the two species:
The position of each Brassica centromere has yet to be accurately determined relative to the genetic linkage maps. However, RFLP mapping of artifactual telocentric chromosomes in Brassica aneuploids placed the centromere of linkage group N12 between markers pW177E3 and pO5b, the centromere of group N13 between pW181a and pN96b, and the centromere of group N14 between markers pN151b and pW130a (Kelly 1996). Additionally, integration of the cytogenetic and genetic linkage maps of B. oleracea positioned the centromere of linkage group O1 (equivalent to N11) between markers pN152E1 and pO168E1 (Howell et al. 2002).
In the proposed centromeric region of N12, four coincident markers were mapped with homology to Arabidopsis sequences that span the centromere on chromosome 4, suggesting conservation of chromosome position between the species. It is possible that with sufficient marker data the Arabidopsis centromeric positions could be used to predict functional and ancestral centromeric regions in Brassica chromosomes. The latter would arise since a hexaploid derived from a lower chromosome progenitor, which likely had between 5 and 8 chromosomes, originally would have had between 15 and 24 functional centromeres, which were then reduced to 10 and 9 in the Brassica A and C genomes, respectively. As in the case of N12, there were a number of instances where the density of markers across the Arabidopsis centromere was insufficient to identify a conserved block in B. napus. However, the loci identified by these same markers were tightly linked in B. napus, and in the case of N11, N12, and N13 there was further cytological evidence suggesting the centromere location. Each of these putative centromeric regions is indicated in Figure 1. As evidenced by numerous small segments of colinearity flanking these provisional centromeric regions on N11, N12, and N14, it appears that the neighboring regions are prone to rearrangements and evolve rapidly compared to more distal regions.
The karyotype of B. oleracea indicates that linkage group O7 (equivalent to N17) is an acrocentric chromosome and has a strongly hybridizing 45S locus at the terminus of the short arm (Howell et al. 2002). This region of N17 shows homology to the short arm of Arabidopsis chromosome 2 and coincidently one of the two nucleolar organizer regions (NORs) of Arabidopsis also maps to the terminus of the short arm of chromosome 2 (Franz et al. 1998).
DISCUSSION
In this study, by allowing minor disruptions in conserved regions it was possible to identify 21 conserved blocks within Arabidopsis, which could be replicated and rearranged to cover almost 90% of the mapped length of B. napus. A minimum number of 74 gross rearrangements, with 38 in the A genome and 36 in the C genome, can be estimated to have separated the two lineages since their divergence 14–24 MYA (Koch et al. 2000). This lies between two previously published figures derived from Brassica/Arabidopsis comparative mapping: 19 chromosomal rearrangements separating B. oleracea from Arabidopsis (Lan et al. 2000) and 90 separating B. nigra from Arabidopsis (Lagercrantz 1998). Detecting rearrangements is influenced by a number of variables including the number and type of available comparative markers, the level of polymorphism within a mapping population, and the method of determining colinearity between species. For Lan et al. (2000) the lower figure was probably due to a low density of comparative markers and for Lagercrantz et al. (1998) the much higher figure was due in part to the approach used to identify syntenous regions, with no allowance made for minor disruptions of colinearity, and was exacerbated by the inclusion of markers thought to be single copy in Arabidopsis but now known to be multi-copy. Comparing estimates of the level of rearrangements in lineages is problematic because of the inherent difficulties in comparing data sets and due to variation in the estimated divergence times. With that proviso, considering the data presented here, the level of rearrangement observed in the Brassiceae tribe, as represented by the A and C genomes of B. napus, is relatively high when compared with related species from the Brassicaceae family. Recently, the genetic maps of Capsella rubella (Lepideae tribe) and Arabidopsis lyrata (Sisymbrieae tribe) have been compared to the sequence map of A. thaliana (Boivin et al. 2004; Kuittinen et al. 2004). On the basis of the comparison to the A. thaliana genome, analysis of the two maps indicates equivalent linkage group organization, with the eight chromosomes of C. rubella, A–H, aligning with the A. lyrata chromosomes, AL1–AL8, respectively. This demonstrates that both species evolved from a common ancestor. A. lyrata and C. rubella are estimated to have diverged from Arabidopsis 5 and 10 MYA, respectively (Boivin et al. 2004; Kuittinen et al. 2004). A limited number of major chromosomal rearrangements, ∼6–13, separate these two species from A. thaliana. In addition, no major rearrangements have separated A. lyrata from C. rubella. Although it is not possible to align all the conserved blocks identified in this study with the C. rubella and A. lyrata genomes, the junctions of a number of the rearrangements identified between these two species and A. thaliana correspond to the ends of conserved blocks identified in this study. However, none of the chromosomal rearrangements that separate A. lyrata and C. rubella from A. thaliana appear to be common to the Brassiceae lineage.
The fact that the majority of the identified conserved segments are found in at least six copies in B. napus and that 81% of the comparative loci, which define the conserved blocks in Arabidopsis, are mapped to these triplicated regions, is consistent with a proposed hexaploid ancestor for the diploid Brassica progenitor. However, it could still be argued that the observed pattern of duplicated segments is the result of several smaller independent segmental duplications following a single whole-genome duplication event, a mode of evolution that would require a significant number of independent duplication events. Polyploidy has been a prevalent mechanism of evolution within the angiosperms and it has been estimated that 30–70% of species have undergone at least one round of chromosome doubling during their evolutionary development (reviewed in Wendel 2000). There is also well-documented evidence for extensive chromosomal rearrangements in newly resynthesized Brassica polyploids (Parkin et al. 1995; Song et al. 1995). Thus genome triplication followed by a small number of insertions/deletions/translocations would provide the simplest explanation for the present structure of the Brassica diploid genome.
In this study, the overall picture is one of conservation of gene content and gene order between the genomes of Arabidopsis and B. napus. The average length of the conserved blocks identified between the two species was 14.8 cM in B. napus and 4.8 Mb in Arabidopsis. However, for at least seven B. napus linkage groups, half their mapped length was equivalent to one conserved region of the Arabidopsis genome. Undoubtedly, the Brassica genomes have undergone restructuring during their evolution from a common ancestor of Arabidopsis, but this has not prevented the maintenance of large stretches of similarity, in some cases equivalent to 9 Mb of contiguous Arabidopsis genomic sequence. In a number of instances, the comparative mapping provisionally suggests correspondence of centromere positions between the two species. The large conserved regions found across the different genomes, punctuated by numerous smaller blocks of similarity, suggest that there are preferential regions for chromosome breakage and subsequent rearrangements.
The publication of the genome sequence of Arabidopsis has opened up many avenues of research with the expectation that these endeavors would have applications in the study of the more complex genomes of crop plants (Arabidopsis Genome Initiative 2000). The complete sequence allowed the resolution of the exact physical positions for ∼30,000 genes, 50% of which have no known function and any of which could hold the key to understanding a number of important agronomic traits. The comparative map suggests that the model genome of Arabidopsis can be widely exploited to infer the genetic basis of traits in its economically valuable Brassica crop relatives. In the identified conserved regions, the Arabidopsis genomic sequence should be an excellent resource for identifying useful markers, targeting the genic regions, since they show on average 86% sequence identity. Accurately mapping the genes controlling target phenotypes in large segregating Brassica populations should allow candidate genes to be inferred from the Arabidopsis sequence. However, due to the duplicated nature of the Brassica genomes it will be difficult to predict whether any particular Arabidopsis gene will have been maintained in all the duplicate copies. Comparative genomic sequencing in other plant species suggest that there almost certainly will have been numerous rearrangements at the level of microsynteny (Bennetzen and Ramakrishna 2002). Limited physical mapping in B. oleracea identified only one potential inversion and one gene in a nonsyntenic position; however, there was obvious interspersed gene loss from the different triplicated regions (O'Neill and Bancroft 2000). In addition, recent physical mapping in the B. napus genome uncovered a similarly small number of disruptions in the microsynteny but evidence of changes in gene content between the homologous Brassica segments compared to the homologous Arabidopsis regions (Rana et al. 2004). Genomic sequence data of such regions from Brassica species will allow the extent to which the duplicate copies have been conserved to be determined, provide insights into the mechanism underlying the rearrangements differentiating the different copies, and allow an estimate of the relative age of the different duplication events.
Acknowledgments
We thank Cambridge Plant Breeders, Twyfords, and Advanta who supported the development of the Brassica RFLP probes (pN, pO, pR) and the construction of the genetic linkage maps of B. napus described in Parkin et al. and in Sharpe et al. We thank Christopher Lewis from the Agriculture and Agri-Food Canada Saskatoon Research Centre, who assisted with the Blast analysis and provided Perl scripts for handling the data. We also thank Stephen Robinson and Steve Barnes for critical reading of the manuscript. This research was funded by the Saskatchewan Agri-Food Innovation Fund.
References
- Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Babula, D., M. Kaczmarek, A. Barakat, M. Delseny, C. F. Quiros et al., 2003. Chromosomal mapping of Brassica oleracea based on ESTs from Arabidopsis thaliana: complexity of the comparative map. Mol. Genet. Genomics 268: 656–665. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J. L., and W. Ramakrishna, 2002. Numerous small rearrangements of gene content, order and orientation differentiate grass genomes. Plant Mol. Biol. 48: 821–827. [DOI] [PubMed] [Google Scholar]
- Bohuon, E. J. R., D. J. Keith, I. A. P. Parkin, A. G. Sharpe and D. J. Lydiate, 1996. Alignment of the conserved C genomes of Brassica oleracea and Brassica napus. Theor. Appl. Genet. 93: 833–839. [DOI] [PubMed] [Google Scholar]
- Boivin, K., A. Acarkan, R. S. Mbulu, O. Clarenz and R. Schmidt, 2004. The Arabidopsis genome sequence as a tool for genome analysis in Brassicaceae: a comparison of the Arabidopsis and Capsella rubella genomes. Plant Physiol. 135: 735–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavell, A. C., D. J. Lydiate, I. A. P. Parkin, C. Dean and M. Trick, 1998. Collinearity between a 30-centimorgan segment of Arabidopsis thaliana chromosome 4 and duplicated regions within the Brassica napus genome. Genome 41: 62–69. [PubMed] [Google Scholar]
- Devos, K. M., and M. D. Gale, 2000. Genome relationships: the grass model in current research. Plant Cell 12: 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz, P., S. Armstrong, C. Alonso-Blanco, T. C. Fischer, R. A. Torres-Rulz et al., 1998. Cytogenetics of the model system Arabidopsis thaliana. Plant J. 13: 867–876. [DOI] [PubMed] [Google Scholar]
- Howell, E. C., G. C. Barker, G. H. Jones, M. J. Kearsey, G. J. King et al., 2002. Integration of the cytogenetic and genetic linkage maps of Brassica oleracea. Genetics 161: 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenczewski, E., F. Eber, A. Grimaud, S. Huet, M. O. Lucas et al., 2003. PrBn, a major gene controlling homeologous pairing in oilseed rape (Brassica napus) haploids. Genetics 164: 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, A., 1996. The genetic basis of petal number and pod orientation in oilseed rape (B. napus). Ph.D. Thesis, University of Newcastle, Newcastle, UK.
- Koch, M. A., B. Haubold and T. Mitchell-Olds, 2000. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17: 1483–1498. [DOI] [PubMed] [Google Scholar]
- Kuittinen, H., A. A. De Haan, C. Vogl, S. Oikarinen, J. Leppala et al., 2004. Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics 168: 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz, U., 1998. Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics 150: 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, T. H., T. A. Delmonte, K. P. Reischmann, J. Hyman, S. P. Kowalski et al., 2000. An EST-enriched comparative map of Brassica oleracea and Arabidopsis thaliana. Genome Res. 10: 776–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., J. Abrahamson, A. Barlow, M. Daley, S. Lincoln et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Li, G., M. Gao, B. Yang and C. F. Quiros, 2003. Gene for gene alignment between the Brassica and Arabidopsis genomes by direct transcriptome mapping. Theor. Appl. Genet. 107: 168–180. [DOI] [PubMed] [Google Scholar]
- Lukens, L., F. Zou, D. Lydiate, I. Parkin and T. Osborn, 2003. Comparison of a Brassica oleracea genetic map with the genome of Arabidopsis thaliana. Genetics 164: 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens, L. N., P. A. Quijada, J. Udall, J. C. Pires, M. E. Schranz et al., 2004. Genome redundancy and plasticity within ancient and recent Brassica crop species. Biol. J. Linn. Soc. 82: 665–674. [Google Scholar]
- Lynch, M., and J. S. Conery, 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Muangprom, A., and T. C. Osborn, 2004. Characterization of a dwarf gene in Brassica rapa, including the identification of a candidate gene. Theor. Appl. Genet. 108: 1378–1384. [DOI] [PubMed] [Google Scholar]
- O'Neill, C., and I. Bancroft, 2000. Comparative physical mapping of segments of the genome of Brassica oleracea var. alboglabra that are homoeologous to sequenced regions of chromosomes 4 and 5 of Arabidopsis thaliana. Plant J. 23: 233–243. [DOI] [PubMed] [Google Scholar]
- Osborn, T. C., C. Kale, I. A. P. Parkin, A. G. Sharpe, M. Kuiper et al., 1997. Comparison of flowering time genes in Brassica rapa, B. napus and Arabidopsis thaliana. Genetics 146: 1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, J. D., C. R. Shields, D. B. Cohen and T. J. Orton, 1983. Chloroplast DNA evolution and the origin of amphidiploid Brassica species. Theor. Appl. Genet. 65: 181–189. [DOI] [PubMed] [Google Scholar]
- Parkin, I. A. P., and D. J. Lydiate, 1997. Conserved patterns of chromosome pairing and recombination in Brassica napus crosses. Genome 40: 496–504. [DOI] [PubMed] [Google Scholar]
- Parkin, I. A. P., A. G. Sharpe, D. J. Keith and D. J. Lydiate, 1995. Identification of the A and C genomes of the amphidiploid Brassica napus (oilseed rape). Genome 38: 1122–1131. [DOI] [PubMed] [Google Scholar]
- Parkin, I. A. P., D. J. Lydiate and M. Trick, 2002. Assessing the level of collinearity between Arabidopsis thaliana and Brassica napus for A. thaliana chromosome 5. Genome 45: 356–366. [DOI] [PubMed] [Google Scholar]
- Parkin, I. A., A. G. Sharpe and D. J. Lydiate, 2003. Patterns of genome duplication within the Brassica napus genome. Genome 46: 291–303. [DOI] [PubMed] [Google Scholar]
- Price, R. A., J. D. Palmer and I. A. Al-Shehbaz, 1994. Sytematic relationships of Arabidopsis: a molecular and orphological perspective, pp. 7–19 in Arabidopsis, edited by E. M. Meyerowitz and C. R. Somerville. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Raes, J., K. Vandepoele, C. Simillion, Y. Saeys and Y. Van De Peer, 2003. Investigating ancient duplication events in the Arabidopsis genome. J. Struct. Funct. Genomics 3: 117–129. [PubMed] [Google Scholar]
- Rana, D., T. Van Den Boogaart, C. M. O'Neill, L. Hynes, E. Bent et al., 2004. Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J. 40: 725–733. [DOI] [PubMed] [Google Scholar]
- Ryder, C. D., L. B. Smith, G. R. Teakle and G. J. King, 2001. Contrasting genome organisation: two regions of the Brassica oleracea genome compared with collinear regions of the Arabidopsis thaliana genome. Genome 44: 808–817. [PubMed] [Google Scholar]
- Schmidt, R., A. Acarkan and K. Boivin, 2001. Comparative structural genomics in the Brassicaceae family. Plant Physiol. Biochem. 39: 253–262. [Google Scholar]
- Sharpe, A. G., 1997. Marker assisted breeding in oilseed rape (Brassica napus). Ph.D. Thesis, University of East Anglia, Norwich, UK.
- Sharpe, A. G., I. A. P. Parkin, D. J. Keith and D. J. Lydiate, 1995. Frequent non-reciprocal translocations in the amphidiploid genome of oilseed rape. Genome 38: 1112–1121. [DOI] [PubMed] [Google Scholar]
- Sillito, D., I. A. Parkin, R. Mayerhofer, D. J. Lydiate and A. G. Good, 2000. Arabidopsis thaliana: a source of candidate disease-resistance genes for Brassica napus. Genome 43: 452–460. [DOI] [PubMed] [Google Scholar]
- Simillion, C., K. Vandepoele, M. C. Van Montagu, M. Zabeau and Y. Van De Peer, 2002. The hidden duplication past of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 99: 13627–13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, K., P. Lu, K. Tang and T. C. Osborn, 1995. Rapid changes in synthetic polyploids of Brassica and its implications for polyploidy evolution. Proc. Natl. Acad. Sci. USA 92: 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U, N., 1935. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilisation. Jpn. J. Bot. 7: 389–452. [Google Scholar]
- Udall, J., P. Quijada and T. C. Osborn, 2004. Detection of chromosomal rearrangements derived from homologous recombination in four mapping populations of Brassica napus L. Genetics 169: 967–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips, R. E., 2002. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93: 77–78. [DOI] [PubMed] [Google Scholar]
- Wendel, J. F., 2000. Genome evolution in polyploids. Plant Mol. Biol. 42: 225–249. [PubMed] [Google Scholar]