Abstract
The gene doublesex of Anastrepha obliqua is composed of four instead of the usual six exons. It is transcribed in both sexes and its primary transcript undergoes sex-specific splicing, producing female DsxF and male DsxM proteins, which have in common the amino-terminal region but which differ at the carboxyl-terminal region.
THE gene doublesex (dsx) of Drosophila is the last gene in the genetic cascade that controls sex determination. It is transcribed in both sexes but gives rise to female (DsxF) and male (DsxM) proteins through sex-specific splicing of its primary transcript (reviewed in Sánchez et al. 2005).
In recent years, molecular mechanisms regulating sex determination have received special attention due to their potential use in sterile insect technique (SIT) programs for the control and eradication of insect pests. The genes of the sex determination cascade, like dsx, have been proposed as candidates for such purposes (Pannuti et al. 2000; Saccone et al. 2002). Tephritids have a serious detrimental economic impact on agriculture. Among fruit flies, the gene dsx has been characterized in Bactrocera tryoni (Queensland fruit fly) (Shearman and Frommer 1998), B. oleae (olive tree fly) (Lagos et al. 2005), and Ceratitis capitata (medfly) (Saccone et al. 2002 cited in Pane et al. 2002). As in Drosophila, dsx in these species encodes male- and female-specific proteins, which are produced by sex-specific splicing of its primary transcript.
Although the biological investigation of some tephritids (Ceratitis and Bactrocera) is already well under way, other species, like the genus Anastrepha, are less extensively analyzed. Considering the potential use of the dsx gene in control programs of fruit flies, our objective was to isolate and characterize this gene in Anastrepha obliqua (fruit fly), a species of great economic importance on the American continent.
A first step in the isolation of the A. obliqua dsx gene (Aodsx) was a PCR reaction on A. obliqua genomic DNA to amplify the well-conserved cysteine-rich DNA binding domain Dsx-DM domain. The sequence of the amplified fragment corresponded to this putative domain. Next we used the amplified fragment as probe to screen a genomic library from A. obliqua that we constructed. A positive phage, Dsx7.1A, was isolated. A total of 4757 bp of its genomic insert were sequenced in the 5′ and 3′ directions, and its conceptual translation was compared to the Dsx protein of B. oleae, demonstrating that we isolated the amino-terminal region enclosing the DM domain of the putative AoDsx protein.
A specific primer was synthesized from the sequence corresponding to the beginning of the putative AoDsx protein. This primer and an oligo(dT) were used in PCR with cDNA of A. obliqua male and female adults separately. A band of ∼1.6 kb in males and a band of ∼1.5 kb in females were amplified, cloned, and sequenced. Their translation and their comparison with the sequence of the genomic insert of phage Dsx7.1A indicated that we had amplified the genomic sequences corresponding to the amino-terminal region of male and female AoDsx proteins. None of the other isolated genomic phages carried the 3′ region of the Aodsx gene. To determine the molecular organization of this gene, we used the following plan.
First, specific primers from the putative A. obliqua exons were synthesized after comparison of the A. obliqua and B. oleae male and female dsx-cDNAs, respectively. These primers were then used to amplify genomic DNA of A. obliqua. Amplified fragments were obtained only with primers from the putative exon 3 and exon 4 and with primers from putative exon 5 and exon 6. The sequences of these fragments were compared with the male and female cDNA sequences mentioned above. The first amplified genomic fragment contained an intron of 126 bp, and the second one contained only exon sequences, indicating that exons 5 and 6 form a unique exon in the Aodsx gene. The Genome Walker kit methodology was next applied to determine the exon/intron junctions through genomic walking on A. obliqua genomic DNA. The sequences of the genomic fragments that were generated were compared with the A. obliqua male and female cDNA sequences found previously. In this way, the exon/intron junctions were unambiguously determined.
Second, to determine the beginning of the Aodsx transcription unit, 5′-RACE analysis was performed. The 5′-RACE product ran as a clear band on the gel, indicating that most likely there are no further exons 5′ of exon 1. Thus, the Aodsx gene lacks the intron located in the 5′-UTR region of the gene dsx of Drosophila and Bactrocera. In addition, it indicated that the transcription start site would lie 638 bp upstream of the beginning of the ORF. Therefore, the sizes of the complete male and female cDNAs are 2265 and 2229 bp, respectively.
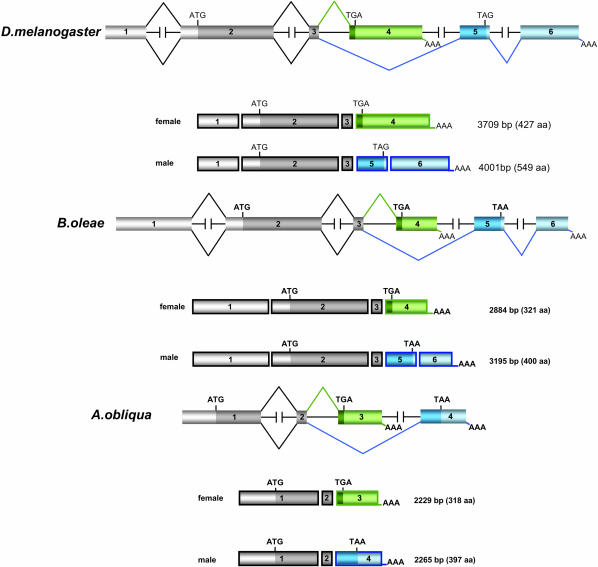
The molecular organization of Aodsx gene and its comparison with that of the dsx gene of Drosophila melanogaster and of B. oleae is shown in Figure 1. The Aodsx gene is composed of four instead of six exons: the first two are common to both sexes, whereas exon 3 is female specific and exon 4 is male specific. Exon 1 of Aodsx corresponds to the fusion of exons 1 and 2, and exon 4 of Aodsx corresponds to the fusion of male-specific exons 5 and 6.
Figure 1.
The molecular organization of the A. obliqua dsx gene and its comparison with dsx of D. melanogaster and B. oleae. Exons (boxes) and introns (lines that when broken indicate that the length of the intron remains unknown) are not drawn to scale. The numbers inside the boxes indicate the number of the exon. The beginning and the end of the ORF are indicated by ATG and TGA or TAA, respectively. The longest cDNA variant is shown. We constructed the genomic library from A. obliqua using the λ-DASH II/EcoRI vector kit (Stratagene, La Jolla, CA) following the manufacturer's instructions. The screening of the A. obliqua genomic library was performed using the PCR fragment amplified from genomic DNA, using degenerated primers for the DM region found in Dsx proteins. Hybridization and identification of positive clones were performed using the protocols described by Maniatis et al. (1982). The 5′-RACE was performed using the Marathon cDNA amplification kit (CLONTECH, Palo Alto, CA) following the manufacturer's instructions. To determine the exon/intron junctions of Aodsx, the BD Genome Walker universal kit (BD Biosience) was used following the manufacturer's instructions. The 5′-RACE product and all PCR and RT-PCR products were subcloned in TOPO-TA cloning vector (Stratagene). Sequencing was performed using an automatic 377 DNA sequencer (Applied Biosystems, Foster City, CA). The accession numbers for the ORFs of DsxF and DsxM are AY948420 and AY948421, respectively. For the sequences of primers, see supplemental data at http://www.genetics.org/supplemental/.
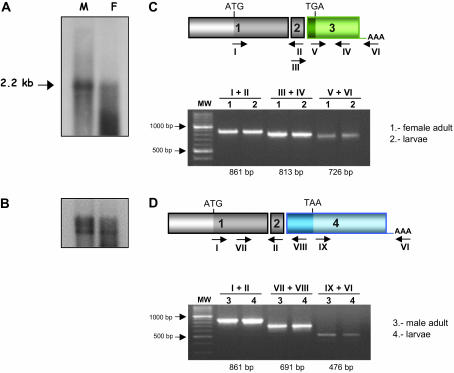
Northern blots (Figure 2, A and B) and overlapping RT-PCRs (Figure 2, C and D) of male and female A. obliqua adults and of a mixture of male plus female larvae of different developmental stages indicated that a single dsx transcript of ∼2.2 kb is present in males and in females, in agreement with the size of the male and female cDNAs determined previously, and confirmed that the gene dsx is transcribed in both sexes, producing two different spliced mRNAs, one in each sex, during larval development and in adult life. The RT-PCR analyses revealed also that females have four dsx cDNA variants, differing in the length of their 3′-UTR and corresponding to four different polyadenylation sites in the female-specific exon (see below). These variants are 2229 bp (found in female larvae and adults), 1860 bp (found in adult females), and 1759 and 1701 bp (found in female larvae). Only the biggest variant contains the 13-nucleotide repeats (dsxRE) and the purine-rich element (PRE) element involved in splicing regulation (see below). Two variants of 2265 and 2223 bp, differing at their 3′-UTR, were found in males. The former is present in larvae and adults, and the latter is present in adults (data not shown).
Figure 2.
Expression of the gene dsx of A. obliqua. (A) Northern blots of total RNA from males (M) and females (F) adults. The Northerns were hybridized with a riboprobe produced from the Aodsx male cDNA. Hybridization with the D. melanogaster rDNA probe (pDm238) (Roiha et al. 1981) was used as a loading control (B). Overlapping RT-PCR analyses from total RNA of female (C) and male (D) adults or a mixture of male and female larvae. (C and D, top) The molecular organization of the corresponding cDNAs is shown. The locations of the primers are shown by arrows and identified by Roman numerals (for the sequences of primers see supplemental data at http://www.genetics.org/supplemental/). Ten micrograms of total RNA from larvae (a mixture of males and females) and adults (males and females separately) were reverse transcribed with the Superscript II RNase H− reverse transcriptase (Invitrogen, San Diego) using primer VI and following the manufacturer's instructions. Ten percent of the synthesized cDNA was amplified by PCR. RT-PCR products were analyzed by electrophoresis in agarose gels, and the amplified fragments were subcloned using the TOPO TA-cloning kit (Invitrogen) following the manufacturer's instructions. These were then sequenced using an automatic 377 DNA sequencer (Applied Biosystems) from the universal forward and reverse primers.
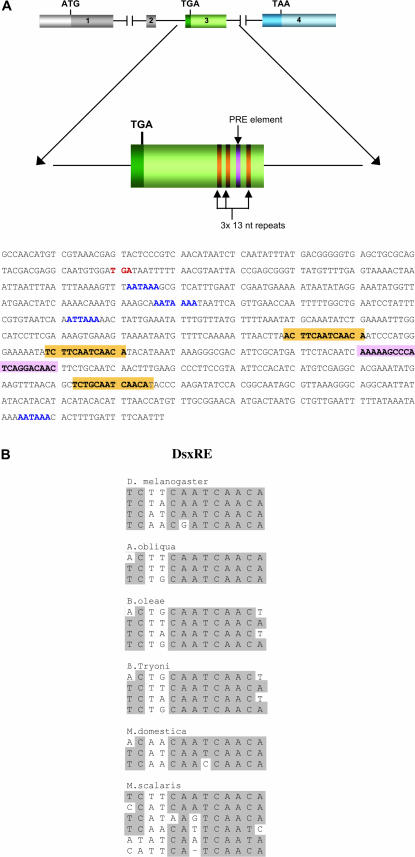
The comparison of Aodsx mRNA molecular organization between males and females suggests that in A. obliqua the male-splicing pathway represents the default mode. First, the putative female-specific amino acid region is skipped over in males. And second, the female-specific exon 3 contains three putative dsxRE targets for the Tra-Tra2 complex as well as for the PRE inserted between dsxRE targets 2 and 3 (Figure 3). The dsxRE elements are highly conserved in the different species (Figure 3). In addition, the female-specific exon contains four polyadenylation sequences, whose function determines the four dsx-mRNAs variants found in A. obliqua females.
Figure 3.
The female-specific exon of A. obliqua dsx. (A) Distribution of the 13 nucleotide repeats and the PRE, which are marked in orange and pink, respectively, in the sequence. Polyadenylation signals are in blue and the translational stop codon (TGA) is in red. (B) Comparison of the dsxRE elements present in the female-specific exon in different species. The shading indicates identical amino acids.
The conceptual translation of the male and female Aodsx mRNAs shows that they encode two polypeptides of 397 and 319 amino acids, respectively. Their comparison with the Dsx proteins of the other insects is presented in Figure 4 and Table 1. The number of amino acids for the non-sex-specific and male-specific regions varies among the species, whereas the female-specific region shows conservation, except for Bombyx mori, which is composed of more amino acids. The degree of similarity is higher for the female-specific than for the non-sex-specific and the male-specific regions. The similarity is higher between the dipteran species than between the dipteran species and the lepidopteran B. mori. Among the dipterans, it is higher among the tephritids A. obliqua, B. oleae, and B. tryoni. The number of amino acids and the similarity of the OD1 and OD2 domains are very high among all species, as expected, since these domains endow the Dsx proteins with the capacity to interact with other proteins and with DNA (An et al. 1996; Cho and Wensink 1997).
Figure 4.
Comparison of the Dsx predicted polypeptides in D. melanogaster (Dm) (Burtis and Baker 1989), A. obliqua (Ao) (this work), B. oleae (Bo) (Lagos et al. 2005), B. tryoni (Bt) (Shearman and Frommer 1998), Musca domestica (housefly) (Md) (Hediger et al. 2004), Megaselia scalaris (phorid fly) (Ms) (Sievert et al. 1997), and B. mori (silkworm) (Bm) (Ohbayashi et al. 2001; Suzuki et al. 2001). (A) Sequence common to both sexes; (B) female-specific sequence; and (C) male-specific sequence. The DNA-binding domain OD1 and the oligomerization domain OD2 are shaded. Gaps were introduced in the alignments to maximize similarity. The comparison of protein sequences was performed using ClustalW (1.82).
TABLE 1.
Comparative analysis of Dsx proteins
| No. of amino acids (% similarity)
|
|||||
|---|---|---|---|---|---|
| Species | Non-sex-specific region | Female-specific region | Male-specific region | OD1 domain | OD2 domain |
| D. melanogaster | 397 | 30 | 152 | 63 | 64 |
| A. obliqua | 287 (65.2) | 30 (100) | 109 (53.9) | 63 (100) | 64 (100) |
| B. oleae | 291 (67) | 30 (100) | 109 (53.2) | 63 (100) | 64 (100) |
| B. tryoni | 289 (66.5) | 30 (100) | 109 (54.6) | 63 (100) | 64 (100) |
| M. domestica | 367 (71.5) | 30 (93) | 160 (52.6) | 63 (93.6) | 64 (96.8) |
| M. scalaris | 280 (56.4) | 30 (96.6) | 293 (57.9) | 63 (96.8) | 63 (89) |
| B. mori | 215 (44) | 49 (90) | 53 (18.4) | 63 (93.6) | 64 (89) |
To allow for a more accurate comparison of the Dsx proteins, they were divided into three regions: non-sex-specific, female-specific, and male-specific regions. In addition, the OD1 and OD2 domains were also compared. The percentage of similarity refers to the identical plus conservative amino acids. The Dsx proteins of D. melanogaster were used as reference.
In summary, the gene dsx of A. obliqua is transcribed during development and in adult life in both sexes but its primary transcript undergoes sex-specific splicing, producing the female DsxF and male DsxM proteins. This sex-specific regulation makes dsx a good candidate to be used in the future for the development of molecular tools that can improve the SIT technique to control the Anastrepha pests.
Acknowledgments
This work was financed by grant BMC2002-02858 awarded to L. Sánchez by the Dirección General de Investigación Cienfica y Técnica, by grants from a Joint Programme of Consejo Superior de Invesigaciones Cientifícas (20004BR0005 to L. Sánchez, Madrid) and Conseilho Nacional do Desenvolvimento Cientifico e Tecnologico (690088/02-7 to A. P. L. Perondini, Sao Paulo, Brazil), and by a Fundaçao de Amparo a Pesquisas do Estado de Sao Paulo grant (03/02698-7 to D. Selivon, Sao Paulo, Brazil).
References
- An, W., S. Cho, H. Ishii and P. C. Wensin, 1996. Sex-specific and non-sex-specific oligomerization domains in both of the Doublesex transcription factors from Drosophila melanogaster. Mol. Cell. Biol. 16: 3106–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtis, K. C., and B. S. Baker, 1989. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56: 997–1010. [DOI] [PubMed] [Google Scholar]
- Cho, S., and P. C. Wensink, 1997. DNA binding by the male and female doublesex proteins of Drosophila melanogaster. J. Biol. Chem. 272: 3185–3189. [DOI] [PubMed] [Google Scholar]
- Hediger, M., G. Burghardt, C. Siegenthaler, N. Buser, D. Hilfiker-Kleiner et al., 2004. Sex determination in Drosophila melanogaster and Musca domestica converges at the level of the terminal regulator doublesex. Dev. Genes Evol. 214: 29–42. [DOI] [PubMed] [Google Scholar]
- Lagos, D., M. F. Ruiz, L. Sánchez and K. Komitopoulou, 2005. Isolation and characterization of the Bactrocera oleae genes orthologous to the sex determination Sex-lethal and doublesex genes of Drosophila melanogaster. Gene 348: 111–121. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., E. F. Fritsch and J. Sambrook, 1982. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Ohbayashi, F., M. G. Suzuki, K. Mita, K. Okano and T. Shimada, 2001. A homologue of the Drosophila doublesex gene is transcribed into sex-specific mRNA isoforms in silkworm, Bombyx mori. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 128: 145–158. [DOI] [PubMed] [Google Scholar]
- Pane, A., M. Salvemini, P. D. Bovi, C. Polito and G. Saccone, 2002. The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development 129: 3715–3725. [DOI] [PubMed] [Google Scholar]
- Pannuti, A., T. Kocacitak and J. C. Lucchesi, 2000. Drosophila as a model for the study of sex determination in anopheline and aedine mosquitoes, pp. 263–270 in Area-wide Control of Fruit Flies and Other Insect Pests, edited by Keng-Hong Tan. Palau Pinang, Penang, Malaysia.
- Roiha, H., J. R. Miller, L. C. Woods and D. M. Glover, 1981. Arrangements and rearrengements of sequences flanking the two types of rDNA insertion in D. melanogaster. Nature 290: 49–53. [DOI] [PubMed] [Google Scholar]
- Saccone, G., A. Pane and L. C. Polito, 2002. Sex determination in flies, fruit flies and butterflies. Genetica 116: 15–23. [DOI] [PubMed] [Google Scholar]
- Sánchez, L., N. Gorfinkiel and I. Guerrero, 2005. Sex determination and the development of the genital disc, pp. 1–38 in Comprehensive Molecular Insect Science, Vol. 1, edited by L. I. Gilbert, K. Iatrou and S. S. Gill. Elsevier Pergamon, Oxford.
- Shearman, D. C. A., and M. Frommer, 1998. The Bactrocera tryoni homologue of the Drosophila melanogaster sex-determination gene doublesex. Insect Mol. Biol. 7: 1–12. [DOI] [PubMed] [Google Scholar]
- Sievert, V., S. Kuhn and W. Traut, 1997. Expression of the sex determination cascade genes Sex-lethal and doublesex in the phorid fly Megaselia scalaris. Genome 40: 211–214. [DOI] [PubMed] [Google Scholar]
- Suzuki, M. G., F. Ohbayashi, K. Mita and T. Shimada, 2001. The mechanism of sex-specific splicing at the doublesex gene is different between Drosophila melanogaster and Bombyx mori. Insect Biochem. Mol. Biol. 31: 1201–1211. [DOI] [PubMed] [Google Scholar]