Abstract
The role of epistasis as a source of trait variation is well established, but its role as a source of covariation among traits (i.e., as a source of “epistatic pleiotropy”) is rarely considered. In this study we examine the relative importance of epistatic pleiotropy in producing covariation within early and late-developing skull trait complexes in a population of mice derived from an intercross of the Large and Small inbred strains. Significant epistasis was found for several pairwise combinations of the 21 quantitative trait loci (QTL) affecting early developing traits and among the 20 QTL affecting late-developing traits. The majority of the epistatic effects were restricted to single traits but epistatic pleiotropy still contributed significantly to covariances. Because of their proportionally larger effects on variances than on covariances, epistatic effects tended to reduce within-group correlations of traits and reduce their overall degree of integration. The expected contributions of single-locus and two-locus epistatic pleiotropic QTL effects to the genetic covariance between traits were analyzed using a two-locus population genetic model. The model demonstrates that, for single-locus or epistatic pleiotropy to contribute to trait covariances in the study population, both traits must show the same pattern of single-locus or epistatic effects. As a result, a large number of the cases where loci show pleiotropic effects do not contribute to the covariance between traits in this population because the loci show a different pattern of effect on the different traits. In general, covariance patterns produced by single-locus and epistatic pleiotropy predicted by the model agreed well with actual values calculated from the QTL analysis. Nearly all single-locus and epistatic pleiotropic effects contributed positive components to covariances between traits, suggesting that genetic integration in the skull is achieved by a complex combination of pleiotropic effects.
THE burgeoning data on genetic architecture (GA) from quantitative trait locus (QTL) analyses are making it evident that variation in most traits of interest has a complex multifactorial genetic basis (Mackay 2001). The recognition of such complexity has emerged as the QTL paradigm has embraced increasingly complex models of the architecture of the genotype-phenotype relationship (Cheverud 2000; Mackay 2001). However, while advances in QTL analyses have provided valuable insights into the architecture of genetic effects underlying trait variation, few studies have examined the architecture of pleiotropy underlying trait covariation, and those that have generally assume a simple GA of pleiotropy (Cheverud et al. 1997 but see Cheverud 2001, 2004; Cheverud et al. 2004). Achieving an understanding of the GA of pleiotropy and genetic covariation is important for evolutionary quantitative genetics, where genetic covariances play a central role in determining the dynamics of multivariate evolution (see Roff 1997) and form the foundation of theories of morphological integration (Cheverud 1982). Genetic covariances also are becoming increasingly recognized as important for the molecular dissection of complex traits, where there is a current push toward analyses focused on the expression of trait suites rather than on single traits in isolation (Sing et al. 2003).
Nearly all existing studies of the GA of pleiotropic effects have focused on cases where single loci show additive or dominance effects on multiple traits (Cheverud 2001, but see Cheverud et al. 2004). Analyses of the GA of variation in single traits have indicated that epistatic interactions between loci can be an important component of GA of trait variation and can significantly complicate evolutionary dynamics of traits (see Wolf et al. 2000). For multiple traits there is the possibility that epistatic interactions between loci contribute to the GA of pleiotropic effects underlying trait covariation (Cheverud et al. 2004). Although conceptually straightforward, the contribution of “epistatic pleiotropy” to the GA of trait covariation has not been previously investigated (but see Mode and Robinson 1959 for a theoretical analysis of variance components). Although the importance of epistatic pleiotropy is not known, evolutionary studies of genetic correlations appear to indicate that a simple genetic basis of genetic covariation is unlikely (Gromko et al. 1991; Phillips et al. 2001; Whitlock et al. 2002), suggesting a possible role for epistasis in the GA of covariation. The presence of epistatic pleiotropy may be particularly important for the evolution of covariance structure and trait integration because it can allow for the evolution of pleiotropic effects of loci (Cheverud 2004).
There are multiple ways to conceptualize the phenomenon of epistatic pleiotropy. It can be viewed as a form of genetic effect, where the interaction between loci affects the coexpression of multiple traits. In this case, one would interpret pleiotropy as the property of a multilocus system. Alternatively, one can take the viewpoint that it is individual loci that are pleiotropic, but that the pleiotropic effects of individual loci are dependent upon the alleles present at other loci (Cheverud 2004). For epistatic effects on single traits, the relationship between the physiological effects of loci (e.g., physiological epistasis sensu Cheverud and Routman 1995) and genetic variances has been modeled for single-locus and two-locus epistatic effects (see Cheverud 2000). However, the way in which physiological epistatic pleiotropic effects of loci contribute to correlations between traits remains mostly unknown (but see Mode and Robinson 1959 for a treatment of variance components). Thus, to understand the contribution of epistatic pleiotropy to patterns of covariation between traits, we present a simple model of genetic covariances herein.
Leamy et al. (1999) previously analyzed early and late-developing skull trait groups in mice and showed that pleiotropic effects of QTL were mostly restricted to the traits in one, but not both, of these integrated groups. Given the large number of QTL identified for these traits and the demonstrably higher phenotypic integration (correlation) of traits within groups compared to that between groups (Leamy et al. 1999), these data appeared to be ideally suited for an analysis of epistatic pleiotropy. We therefore estimated the strength and patterns of physiological epistatic pleiotropy brought about by interactions among these QTL affecting traits within these two skull groups and compared the results to those for single-locus pleiotropic effects. These results are combined with the two-locus population genetic model of epistatic pleiotropy to examine how various patterns of two-locus genetic effects contribute to covariation between traits and corresponding patterns of integration. As will be seen, the theoretical extent of epistatic pleiotropy predicted by this model for trait pairs matches quite well with actual values calculated from the QTL analysis.
MODEL
To understand how single-locus and epistatic genetic effects contribute to the genetic covariance between the skull traits, we develop a two-locus population genetic model. We extend the Cockerham (1954) single-trait model of epistasis to analyze the covariance between a pair of traits (see Kao and Zeng 2002 for a complete description of the single-trait model).
Genotypic values:
With two loci (A and B) and two alleles at each locus (alleles A1, A2 at locus A and B1, B2 at locus B) there are nine genotypes. With two traits (X and Y) there are, correspondingly, nine genotypic values for each trait. We designate the genotypic values of traits X and Y as Xi and Yi, respectively, where the subscript indicates the two-locus genotype (A1A1B1B1 = 1, A1A1B1B2 = 2, A1A1B2B2 = 3, A1A2B1B1 = 4, A1A2B1B2 = 5, A1A2B2B2 = 6, A2A2B1B1 = 7, A2A2B1B2 = 8, and A2A2B2B2 = 9). The genotypic values are arrayed in the vectors X and Y.
The nine genotypic values for each of the two traits can be completely described by a constant (C) and a set of eight genetic parameters. Four single-locus effects correspond to the additive (a) and dominance (d) effects of each of the two loci and four interaction effects correspond to pairwise interactions between the four single-locus effects. We refer to the first four as “single-locus” effects to indicate that they correspond to marginal effects of the two loci. For the single-locus effects, two subscripts are used to indicate the locus (A or B) and the trait (X or Y) that the effect corresponds to. For example, aAX is the additive effect of locus A on trait X. The four interaction effects are additive-by-additive (aa), additive-by-dominance (ad), dominance-by-additive (da), and dominance-by-dominance (dd). Interaction effects have a single subscript indicating the trait that they are associated with. The constant and the eight genetic parameters define a pair of vectors (GX and GY) describing the genetic effects of the two-locus system for each trait:
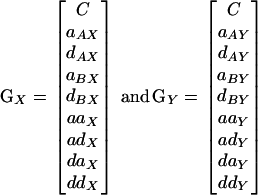 |
(1) |
A matrix of index values (V) defines the contribution of the eight genetic effects and the constant to the nine genotypic values. The Cockerham model of genetic effects used in the QTL analysis corresponds to the following matrix of index values:
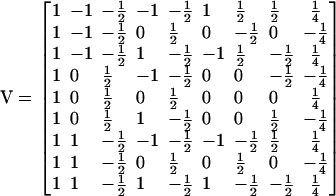 |
(2) |
Thus, the vectors of genotypic values for the two traits can be described by the equations
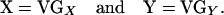 |
Because both are based on the same matrix of index values, the eight regression coefficients estimated in the epistasis analysis (see below) correspond to the eight parameters in the model of genotypic values.
Variances and covariances:
The frequencies of the nine two-locus genotypes are designated Fi, where the subscripting follows that of the genotypic values, and are given in Table 1. These frequencies are arrayed in the vector F.
TABLE 1.
The frequencies of the nine two-locus genotypes
|
A locus genotype
|
|||
|---|---|---|---|
| B locus genotype | A1A1 | A1A2 | A2A2 |
| B1B1 |
 (0.0625) (0.0625) |
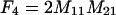 (0.125) (0.125) |
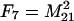 (0.0625) (0.0625) |
| B1B2 | F2 = 2M11M12 (0.125) | F5 = 2(M11M22 + M12M21) (0.25) | F8 = 2M21M22 (0.125) |
| B2B2 |
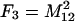 (0.0625) (0.0625) |
F6 = 2M12M22 (0.125) |
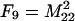 (0.0625) (0.0625) |
Each genotype frequency (Fi) is calculated from the frequencies of the four haplotypes (A1B1, A1B2, A2B1, and A2B2), which are defined as M11 = p1q1 + d, M12 = p1q2 − d, M21 = p2q1 − d, and M22 = p2q2 + d. Given in parentheses below each expected value is the value for the frequencies in the experimental population used in the QTL analysis, where p1 = 0.5 and q1 = 0.5 (where p2 = 1 − p1 and q2 = 1 − q1) and d is zero.
The genetic variance of a trait (in this case trait X) is given by 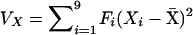 and the genetic covariance between traits X and Y [cov(X, Y)] is given by
and the genetic covariance between traits X and Y [cov(X, Y)] is given by 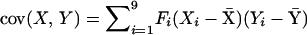 , where
, where 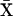 and
and 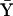 are the mean genotypic values of the two traits and have the values
are the mean genotypic values of the two traits and have the values 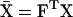 and
and 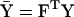 .
.
For simplicity, we focus on the genotype frequencies of our experimental population (an F2 intercross), where all alleles are at equal frequency (0.5) and there is no linkage disequilibrium between pairs of loci that are not physically linked on a chromosome. (Because of the high level of linkage disequilibrium within chromosomes in the F2 population we are not able to analyze interactions between loci on the same chromosome.) For this population, the expected genetic variances of the two traits and the covariance between them contributed by two loci were derived using the genotypic values in Equation 2 and the frequencies from Table 1. The genetic variance of trait X is given by the equation
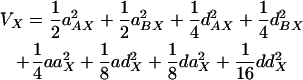 |
(3) |
(an analogous equation can be written for trait Y). The covariance between traits is given by
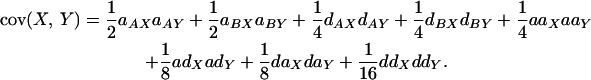 |
(4) |
The structures of the variances and covariance are analogous in that both can be partitioned into two classes that reflect the types of genetic effects contained in the terms: single-locus and epistatic effects. Consider the covariance equation (Equation 4); the first class of effects (Equation 4, line 1) results from cases where either locus on its own affects the expression of both traits. This class contains four terms corresponding to the additive and dominance effects of each locus (i.e., where a single locus affects the expression of both traits). The first two terms, 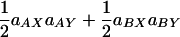 , correspond to the additive genetic covariance while the next two terms,
, correspond to the additive genetic covariance while the next two terms, 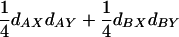 , correspond to the dominance genetic covariance between the traits. We refer to this class collectively as single-locus effects. These are the covariance terms that result from the familiar single-locus pleiotropic effects. Note that a locus must show the same type of effect on both traits (i.e., must have either an additive effect or a dominance effect on both traits). If the sign of the effect of a locus on the two traits is the same, its contribution to the covariance will be positive and if the sign of effects is opposite, its contribution will be negative. We refer to the former as positive pleiotropy and to the latter as negative pleiotropy. Thus, one can see that the sign of pleiotropy is not related to the sign of the QTL effects, only to the sign of the product of their effects. The second class (Equation 4, line 2) results from cases where the pair of loci has an epistatic effect on both traits. There are four terms in this class, corresponding to the four types of epistasis. Note that, as with single-locus effects, both traits must show the same pattern of epistasis for epistasis to contribute to the genetic covariance between the traits. Together, these four terms correspond to the epistatic genetic covariance between traits.
, correspond to the dominance genetic covariance between the traits. We refer to this class collectively as single-locus effects. These are the covariance terms that result from the familiar single-locus pleiotropic effects. Note that a locus must show the same type of effect on both traits (i.e., must have either an additive effect or a dominance effect on both traits). If the sign of the effect of a locus on the two traits is the same, its contribution to the covariance will be positive and if the sign of effects is opposite, its contribution will be negative. We refer to the former as positive pleiotropy and to the latter as negative pleiotropy. Thus, one can see that the sign of pleiotropy is not related to the sign of the QTL effects, only to the sign of the product of their effects. The second class (Equation 4, line 2) results from cases where the pair of loci has an epistatic effect on both traits. There are four terms in this class, corresponding to the four types of epistasis. Note that, as with single-locus effects, both traits must show the same pattern of epistasis for epistasis to contribute to the genetic covariance between the traits. Together, these four terms correspond to the epistatic genetic covariance between traits.
From Equation 4 we can see that single-locus additive effects have the greatest impact on the covariance when examined on a per term basis. Dominance and epistatic effects have a smaller impact, with dd epistasis having a smaller effect than the forms of epistasis that contain an additive component in the interaction.
The contribution of pleiotropic loci involved in multiple epistatic interactions to the covariance between traits was also derived using this same genetic model. These kinds of effects occur when a single locus affects a pair of traits, but its effect on each trait depends on its interaction with a different locus. For example, if locus A interacts with locus B to affect trait X while locus A interacts with locus C to affect trait Y we would consider locus A to be pleiotropic since it affects both X and Y, but its pleiotropic effect is modified by two other loci. These types of effects could contribute considerable complexity to the genetic architecture of pleiotropy, but they do not contribute to the covariance between traits in the population studied here. As a result, we do not present the derivation of these covariances or discuss these effects here.
MATERIALS AND METHODS
QTL analysis—the population and traits:
The house mice used in this study were the F2 progeny of F1 hybrids obtained from an original intercross of the Large (LG/J) and Small (SM/J) inbred strains. All mice were bred under conditions previously described (Cheverud et al. 1996), and eventually 535 individuals were sacrificed at 70 days of age, and their spleens were removed for DNA extraction. A total of 76 polymorphic microsatellites on the 19 autosomes were scored in these mice (Routman and Cheverud 1995), with their positions determined through the use of the MAPMAKER 3.0b program (Lander et al. 1987; Lincoln et al. 1992) as previously given (Cheverud et al. 1996; Leamy et al. 1997). Altogether, these 76 loci defined a total of 55 intervals between loci with an average interval length of 27.5 cM.
Leamy et al. (1999) originally measured a total of nine skull traits in these mice from both the early developing neurocranial vault and the face area that completes its growth much later in postnatal life (Moss 1973; Moore 1981; Bogin 1988). We made use of eight of these same traits, including the cranial vault width (CVW), height (CVH), and length (CVL) that represent the early developing group. We used the facial length (FCL), height (FCH), and width (FCW) and zygomatic arch width (ZYW) and length (ZYL) to represent the late-developing group. Leamy et al. (1999) also measured basicranial length (BCL), but this trait was not used because it cannot clearly be classified into either the early or the late-developing group. Precise descriptions of these traits, as well as a calculation of the amount of measurement error involved, are given in Leamy et al. (1999).
Single-QTL analyses:
After appropriate adjustment of all skull traits for potential effects of sex, dam, block, and litter size (see Leamy et al. 1999), we ran separate QTL analyses for the three early developing skull traits and for the five late-developing skull traits. We used the interval mapping approach described by Haley and Knott (1992), except that multivariate canonical correlation rather than regression techniques were used to allow us to simultaneously analyze all traits in each group and thus identify QTL pleiotropically affecting more than one of these traits. This canonical correlation approach to QTL mapping has been previously described (for example, Workman et al. 2002) and is outlined below.
Index values were assigned to genotypes following the (Cockerham 1954; see also Goodnight 2000; Kao and Zeng 2002) model of genetic effects, which corresponds to the values shown in Equation 2 (with the LG/J allele as the “+” allele and the SM/J as the “−” allele). Genotypic index values were imputed every 2 cM between flanking microsatellite markers, using the previously calculated recombination percentages on each chromosome (see Cheverud et al. 1996) and the equations in Haley and Knott (1992). Canonical correlation of the index values with the skull traits was then performed at each position 2 cM apart on all chromosomes.
For each of the 19 autosomes, the canonical correlation analyses generated F-values with their associated probabilities that were converted to a linear scale by logarithmic transformation [LPR = log 10(1/Prob.)] to make the results comparable to LOD scores obtained via maximum-likelihood analysis (Lander and Botstein 1989). QTL were considered significant when the largest LPR value on a given chromosome exceeded the 5% (and 1%) threshold values calculated from 1000 permutations for each chromosome. An experimentwise threshold value that ensured no greater than 5% type I error in QTL testing across the entire genome also was obtained from the 50th (5%) highest LPR scores that were observed on any chromosome during each of 1000 iterations (Churchill and Doerge 1994). QTL were assigned to the position of the largest LPR value on a chromosome. If a QTL was found on any chromosome, the presence of two QTL was also tested for (see Leamy et al. 1999 for details).
Sex-specific QTL effects also were tested for by first assigning a code for the sex (males = 1, females = 2) of each individual. A whole-genome scan for the skull traits (in each of the separate groups) was then run as before, but for the interaction of sex with the additive and dominance genotypic index values (but also partialing the main effects due to genotypic values and sex). Chromosomewise and experimentwise threshold LOD values again were estimated by permutation tests, and chromosomes with significant LOD values were assumed to contain a QTL whose effects differed in the two sexes. No significant sex effects on the expression of these QTL were found, however, so sexes were combined in all subsequent analyses.
Once the positions of all QTL on each chromosome were established, we ran multivariate regressions of the traits on the additive and dominance genotypic index values for the QTL at that site to test for significance for each trait and to estimate additive (a) and dominance (d) genotypic values. For each chromosome with two QTL, tests of significance were done with canonical correlation where effects of the other QTL were partialed out, and then regressions were run that included the genotypic index values at the site of the QTL not being analyzed. For each trait found to be significant in the F-test from the regression, the a- and d-values were tested for significance using individual t-tests.
If each of two or more traits reached significance at the 5% level for a given QTL, it was assumed that a QTL was pleiotropically affecting these traits. Because of the developmental relationship among the skull traits being analyzed, we assume that cases where QTL affect multiple traits are the result of pleiotropic effects rather than close linkage of separate genes. However, it is possible that common QTL positions for multiple traits could result from linkage between independent genes affecting trait expression. For this suite of traits it appears that the most parsimonious assumption is for pleiotropy rather than linkage due to their shared developmental pathways. The general implications of the analyses presented here apply equally well in either case since closely linked genes will act as a single QTL in this population and thus understanding their contribution to patterns of covariation should depend on the net influences of the linkage group and not necessarily on individual gene effects.
Epistasis analysis:
We tested for significant interactions among each pair of QTL for the early and late-developing trait groups, using the canonical correlation of the epistatic index values from the Cockerham model (shown in Equation 2) with the skull traits, partialing out the single-locus additive and dominance effects of both of the loci. Locations that showed a significant interaction between QTL were then analyzed using multivariate regressions of the traits on the additive, dominance, and epistatic genotypic index values for the pair of QTL to test for significance for each trait and to estimate the four epistatic genotypic values (Equation 1).
We used the conventional 5% significance level without adjustment for multiple comparisons since the locations for epistasis testing were chosen because of their significant main effects but without any prior knowledge of potential epistatic effects (Goodman 1998; Thompson 1998). As was the case for single-locus pleiotropy, if two or more traits reached significance for a given pair of QTL, it was assumed that these QTL were pleiotropically affecting these traits. For those pairs of QTL reaching significance, testing for the individual significance of each of the four genotypic epistasis terms (aa, ad, da, and dd) was done via individual t-tests.
Estimation of covariances:
To specifically assess the impact of QTL on trait integration in the early and late-developing skull groups, we computed seven [phenotypic, additive genetic, dominance genetic, one-locus genetic (additive + dominance), two-locus genetic (epistatic), total genetic (one- + two-locus genetic), and environmental] sets of covariances for each pair of traits. Phenotypic covariances were calculated for the traits not adjusted in any way, and environmental covariances were calculated from the residuals of a regression of each trait on all total genetic effects that reached significance in the QTL analyses.
To calculate the additive, dominance, single-locus, epistatic, and total genetic covariances, we first obtained residuals from regressions of each of the individual traits on its significant genetic effects. Variances/covariances calculated from these residuals then were subtracted from the corresponding phenotypic variances/covariances to yield for each the various genetic variances and covariances of the traits. Correlations for each pair of traits also were calculated for each of the seven genetic and environmental categories by division of the appropriate covariances by the square root of the product of the variances.
Expected variances and covariances from the two-locus model:
The additive, dominance, and epistatic QTL effects estimated by multiple regression were used to calculate the expected variances and covariances of the skull traits in the early and late-developing groups using Equations 3 and 4. These equations give the variance and covariance contributed by each locus or two-locus combination, but many loci and interacting pairs may contribute to each variance or covariance. Because the individual variances and covariances are additive, the overall expected variances and covariances are calculated as the sum of the components contributed by each locus or two-locus combination.
Correspondence between predicted and estimated variances and covariances:
The utility of the model was evaluated by examining the relationship between the expected variances and covariances (genetic, additive, dominance, and epistatic) from the genetic model and the variances and covariances estimated from the multivariate regression procedure using a model II regression (reduced major axis, RMA) and correlations. Together, these two statistical approaches allow us to describe the shape of the bivariate distribution. The regression indicates the form of the relationship between the observed and expected values while correlation indicates the strength of the relationship. In a model II regression, if the genetic model predicts the correct relative values of the variances or covariances, the slope (β) will be 1 and the constant (intercept) (c) will be 0. Deviations from a slope of 1 indicate that the model predicts relative values (i.e., relative to the mean) that are either smaller (when β > 1) or larger (β < 1) than those estimated using the regression residuals approach. Because variance and covariance values can be difficult to interpret, we present the regression constants as proportional to the estimated values (i.e., these were calculated by dividing the constants by the overall means of the estimated values). For example, a value of 0.1 would indicate that the constant is 10% of the average estimated value. A nonzero constant indicates that the covariance values are, on average, larger or smaller than predicted. The relationship between the estimated and predicted genetic correlations was also analyzed using RMA regressions and correlations.
RESULTS
Single-QTL effects:
QTL for the early (designated E) and for the late-developing traits (designated L) are listed in Table 2 and are symbolized by numbers and extensions that indicate chromosome number and whether they are the first or second QTL on that chromosome.
TABLE 2.
QTL for the early and late-developing skull traits
| QTL | Site | Early developing traits | QTL | Site | Late-developing traits |
|---|---|---|---|---|---|
| E1 | 110 | CVW (d), CVH (a, d), CVL (a) | L1.1 | 52 | FCL (a), FCW (a, d), ZYL (d) |
| E2 | 12 | CVL (a) | L1.2 | 92 | FCH (a), ZYW (a) |
| E3 | 44 | CVW (a), CVH (d), CVL (a, d) | L2 | 0 | FCH (a, d), FCW (a), ZYW (a), ZYL (a, d) |
| E4 | 62 | CVH (d), CVL (a, d) | L3 | 44 | FCH (a), FCW (a), ZYW (a), ZYL (a, d) |
| E5 | 104 | CVW (a) | L4.1 | 36 | FCL (a, d), ZYW (a, d) |
| E6.1 | 30 | CVW (a), CVH (d), CVL (d) | L4.2 | 68 | FCH (a), ZYL (d) |
| E6.2 | 90 | CVL (a) | L6 | 80 | FCL (a), FCH (a), ZYW (a), ZYL (a) |
| E7.1 | 24 | CVW (a), CVH (a, d), CVL (a, d) | L7 | 58 | FCH (a, d), ZYW (a), ZYL (a) |
| E7.2 | 76 | CVH (a) | L8 | 18 | FCL (a), FCW (a), ZYW (a), ZYL (a) |
| E9 | 46 | CVW (d), CVH (d), CVL (d) | L9.1 | 2 | ZYW (d) |
| E10.1 | 22 | CVW (d), CVH (d) | L9.2 | 40 | FCH (a) |
| E10.2 | 44 | CVH (d), CVL (d) | L10 | 64 | FCL (a), FCH (a), FCW (a), ZYL (a) |
| E11.1 | 34 | CVW (a) | L11 | 90 | FCL (a), ZYL (a) |
| E11.2 | 88 | CVH (a), CVL (a) | L12.1 | 8 | ZYL (a) |
| E12.1 | 30 | CVH (d) | L12.2 | 58 | FCL (a), FCW (a) |
| E12.2 | 54 | CVH (a) | L13 | 70 | FCH (a), ZYW (a, d), ZYL (a, d) |
| E13 | 2 | CVW (a) | L14 | 32 | FCL (a), FCH (a), FCW (a), ZYW (a), ZYL (a) |
| E14 | 50 | CVW (a), CVH (a), CVL (a) | L15 | 50 | FCL (a), FCH (a, d), ZYL (a) |
| E15 | 46 | CVW (a) | L17 | 8 | |
| E17 | 6 | CVH (a) | L19 | 4 | FCL (a), FCW (d), ZYL (d) |
| E19 | 26 |
QTL are shown with their locations (Site) in centimorgans from the first marker on each chromosome. Each QTL is designated E or L for the early or late-developing traits and with a number and extension indicating its chromosome number and whether it is the first or second QTL on that chromosome. Significant (P < 0.05) additive (a) and dominance (d) effects are indicated in parentheses.
For the early developing traits, 21 QTL were identified. Pleiotropy is present for 10 of these QTL, with 6 QTL affecting all three traits and 4 QTL affecting two of the three traits. E19 reached multivariate significance in the canonical correlation runs, but not univariate significance for any of the three traits. This likely indicates a combination of effects that is in contrast to the pattern of phenotypic correlation among these traits. These 21 QTL contribute 33, 29, and 31%, respectively, of the variance of CVW, CVH, and CVL. The majority QTL have positive additive genotypic (a) values for the three traits. This suggests that the LG/J allele at these QTL results in larger skull traits than the SM/J allele. Significant d-values are fairly prevalent (18 of 36 total possibilities) and most are positive.
For the late-developing traits, 20 QTL were identified. Again 1 QTL (L17) reached multivariate but not univariate significance for any of the five traits. Pleiotropy is present for 16 of the 20 QTL. Thus pleiotropy for the QTL affecting the late-developing skull traits appears more prominent, although this is primarily due to the fact that five rather than three traits are being used.
For late-developing traits, individual QTL contributed 19, 29, 32, 30, and 35%, respectively, of the variance for FCL, FCH, FCW, ZYW, and ZYL. Most QTL show positive additive effects but 14 QTL showed significant dominance. On the basis of the relative magnitude of dominance effects in the early vs. late QTL, it appears that dominance is less important for the QTL for the late-developing traits compared with those for the early developing traits.
Epistatic effects:
Tests for pairwise combinations of the 21 QTL affecting the early developing skull traits showed that a total of 29 combinations reached significance at the 5% level (Table 3). This is much higher than the 10 or 11 expected by chance alone (for P = 0.05). All of these 29 pairs of QTL affected only a single trait. This result is significantly different (χ2 = 18.2, d.f. = 1, P < 0.001) from the ratio of single-locus pleiotropy described above, suggesting that epistatic pleiotropy is much less common than single-locus QTL pleiotropy within this trait group. All but 2 (E12.1 and E13) of the 21 QTL are involved in significant epistatic interactions for one or more of the early developing skull traits. One QTL (E2) is involved in six significant interactions, and several other QTL appear in a large number of interactions (e.g., E7.1 is in five interactions). The importance of E2 is apparent, and even though this QTL by itself significantly affected only CVL (Table 2), its interactions with other QTL primarily affect CVH. These 29 epistatic combinations contribute 11, 14, and 14%, respectively, to the variance of CVW, CVH, and CVL. A model including both the single-locus and significant epistatic effects of the 21 loci accounts for 39, 36, and 40%, respectively, of the variance of CVW, CVH, and CVL.
TABLE 3.
Epistasis for the QTL affecting the early developing skull traits
| QTL-QTL | Traits affected | QTL-QTL | Traits affected |
|---|---|---|---|
| E1-E7.2 | CVW (ad) | E6.2-E11.1 | CVL (dd) |
| E1-E10.1 | CVH (da) | E7.1-E11.2 | CVW (da) |
| E1-E10.2 | CVH (aa) | E7.1-E14 | CVL (aa) |
| E2-E4 | CVH (ad) | E7.1-E15 | CVL (aa) |
| E2-E6.1 | CVH (aa) | E7.2-E15 | CVL (da) |
| E2-E7.1 | CVH (ad, da) | E7.2-E17 | CVW (ad) |
| E2-E10.1 | CVH (dd) | E9-E11.2 | CVH (dd) |
| E2-E10.2 | CVL (ad) | E9-E19 | CVW (aa) |
| E2-E19 | CVH (da) | E10.1-E12.2 | CVW (ad) |
| E3-E7.1 | CVH (da) | E10.1-E14 | CVW (da) |
| E3-E15 | CVH (da) | E10.2-E14 | CVH (da) |
| E5-E9 | CVL (aa) | E11.1-E12.2 | CVW (dd) |
| E5-E15 | CVL (ad) | E12.2-E14 | CVW (aa) |
| E6.1-E10.2 | CVH (dd) | E15-E19 | CVL (ad, da) |
| E6.1-E17 | CVL (ad) |
Epistatic components (aa, ad, da, and dd) significant at the 5% level also are given in Table 3 for each early developing trait reaching overall significance for epistasis. All four epistatic combinations are represented, their frequencies of 3 (aa), 6 (ad), 5 (da), and 2 (dd) not being significantly different from a 1:2:1 (ad and da are combined because they differ only in the arbitrary order of the loci and, therefore, represent the same type of genetic effects) ratio (χ2 = 2.38, d.f. = 2, P = 0.30).
For the late-developing skull traits, a total of 38 pairs of QTL reached statistical significance for overall epistasis (Table 4). Fourteen of these pairs are pleiotropic and significantly affect more than one trait. This ratio of 24:14 is significantly different (χ2 = 11.4, d.f. = 1, P < 0.001) from the comparable 3:16 ratio for single-locus pleiotropy. Thus epistatic pleiotropy for the late-developing skull traits also is not as common as the pleiotropy generated from individual QTL. All 20 QTL are involved in significant epistatic interactions for one or more of the late-developing skull traits. These 38 epistatic combinations contribute 10, 11, 23, 26, and 14%, respectively, to the variance of FCL, FCH, FCW, ZYW, and ZYL. A model including both the single-locus and significant epistatic effects of the 20 loci accounts for 38, 34, 49, 44, and 45%, respectively, of the variance of FCL, FCH, FCW, ZYW, and ZYL.
TABLE 4.
Epistasis for the QTL affecting the late-developing skull traits
| QTL-QTL | Characters affected | QTL-QTL | Characters affected |
|---|---|---|---|
| L1.1-L2 | ZYW (aa, da), ZYL (da) | L8-L17 | ZYL (dd) |
| L1.1-L4.1 | FCH (ad) | L9.1-L12.2 | FCL (aa) |
| L1.1-L7 | ZYW (dd) | L9.1-L13 | FCW (aa, dd) |
| L1.1-L10 | ZYL (aa, da) | L9.1-L15 | ZYW (dd) |
| L1.1-L13 | FCH (ad, da), FCW (ad, dd) | L9.2-L10 | FCW (dd) |
| L1.2-L10 | ZYL (aa) | L9.2-L11 | FCW (ad, da), ZYW (ad, da) |
| L1.2-L13 | FCW (ad, dd), ZYW (ad, dd) | L9.2-L12.1 | ZYL (aa, dd) |
| L2-L9.2 | ZYL (da, dd) | L9.2-L12.2 | FCL (dd) |
| L3-L14 | ZYW (ad) | L9.2-L13 | FCW (dd), ZYW (da, dd) |
| L3-L17 | FCL (aa, ad) | L9.2-L19 | ZYW (da), ZYL (aa) |
| L4.1-L9.1 | FCW (aa, da, dd), ZYW (aa) | L10-L11 | FCL (ad), ZYL (dd) |
| L4.1-L12.1 | ZYW (dd) | L10-L12.1 | FCH (dd), FCW (ad, dd) |
| L4.2-L7 | ZYW (dd) | L10-L15 | FCW (da, dd), ZYW (da) |
| L4.2-L9.1 | FCW (ad) | L11-L14 | ZYW (da), ZYL (da) |
| L4.2-L14 | ZYL (dd) | L11-L19 | FCL (aa), ZYL (aa) |
| L6-L12.1 | FCL (da) | L12.1-L17 | ZYW (aa) |
| L7-L10 | FCH (dd), ZYL (da) | L12.2-L13 | FCH (ad, dd) |
| L8-L10 | FCH (dd) | L12.2-L17 | FCL (da), FCH (da), ZYW (da, dd), ZYL (dd) |
| L8-L14 | FCW (aa, dd) | L13-L14 | ZYW (ad) |
Epistatic components (aa, ad, da, and dd) significant at the 5% level for the late-developing traits (Table 4) show a pattern similar to that for the early developing traits. The totals over all five traits for the aa, ad, da, and dd epistatic types, respectively, are 14, 14, 19, and 27. These numbers do not deviate significantly (χ2 = 5.43, d.f. = 2, P = 0.066) from a 1:2:1 ratio, despite the overabundance of dd and an underabundance of aa.
Trait covariances:
Table 5 shows the phenotypic, genetic, and environmental covariances of each pair of traits in the early and late-developing skull groups derived from the analysis of single-locus effects and two-locus epistasis of the 21 QTL found to significantly affect the early developing and the 20 QTL affecting late-developing skull traits. Table 5 also includes covariances and the correlations (in parentheses) calculated from these covariances, using the corresponding estimates of the genetic and environmental variances. Early and late-developing traits show very similar patterns of trait correlations, with the average values of all seven of the correlations shown in Table 5 being nearly identical for the two sets of traits. For both sets of traits, all of the genetic effects contribute to trait integration and produce very similar average correlation values. In contrast, the environmental correlations are much smaller than the genetic correlations and tend to reduce the average level of integration of the traits, making the phenotypic correlations smaller than any of the genetic correlations.
TABLE 5.
Covariances and correlations of traits in the early and late-developing groups
| Covariances and correlations
|
|||||||
|---|---|---|---|---|---|---|---|
| Phenotypic | Genetic | Additive | Dominance | Single locus | Epistatic | Environmental | |
| Early developing | |||||||
| CVW-CVH | 0.0142 (0.50) | 0.0074 (0.69) | 0.0049 (0.71) | 0.0027 (0.85) | 0.0065 (0.74) | 0.0025 (0.67) | 0.0068 (0.38) |
| CVW-CVL | 0.0192 (0.40) | 0.0108 (0.57) | 0.0064 (0.51) | 0.0041 (0.89) | 0.0090 (0.59) | 0.0044 (0.78) | 0.0084 (0.29) |
| CVH-CVL | 0.0246 (0.52) | 0.0131 (0.73) | 0.0081 (0.78) | 0.0050 (0.88) | 0.0117 (0.81) | 0.0051 (0.79) | 0.0115 (0.39) |
| Means | 0.0193 (0.47) | 0.0104 (0.66) | 0.0065 (0.67) | 0.0039 (0.87) | 0.0091 (0.72) | 0.0040 (0.73) | 0.0089 (0.35) |
| Late developing | |||||||
| FCL-FCH | 0.0100 (0.34) | 0.0055 (0.61) | 0.0039 (0.62) | 0.0010 (0.76) | 0.0045 (0.65) | 0.0016 (0.50) | 0.0045 (0.22) |
| FCL-FCW | 0.0117 (0.33) | 0.0091 (0.70) | 0.0057 (0.70) | 0.0009 (0.96) | 0.0061 (0.70) | 0.0031 (0.58) | 0.0026 (0.12) |
| FCL-ZYW | 0.0396 (0.46) | 0.0196 (0.66) | 0.0146 (0.77) | 0.0014 (0.51) | 0.0155 (0.73) | 0.0088 (0.65) | 0.0200 (0.37) |
| FCL-ZYL | 0.0436 (0.49) | 0.0230 (0.73) | 0.0147 (0.70) | 0.0031 (0.87) | 0.0163 (0.70) | 0.0100 (0.95) | 0.0207 (0.37) |
| FCH-FCW | 0.0063 (0.55) | 0.0035 (0.76) | 0.0023 (0.76) | 0.0005 (0.96) | 0.0025 (0.74) | 0.0019 (1.06) | 0.0027 (0.41) |
| FCH-ZYW | 0.0141 (0.52) | 0.0069 (0.65) | 0.0042 (0.60) | 0.0015 (0.83) | 0.0050 (0.62) | 0.0039 (0.85) | 0.0072 (0.43) |
| FCH-ZYL | 0.0111 (0.39) | 0.0053 (0.48) | 0.0034 (0.42) | 0.0017 (0.83) | 0.0042 (0.46) | 0.0021 (0.59) | 0.0058 (0.34) |
| FCW-ZYW | 0.0182 (0.55) | 0.0116 (0.77) | 0.0077 (0.83) | 0.0022 (1.05) | 0.0084 (0.83) | 0.0063 (0.79) | 0.0066 (0.37) |
| FCW-ZYL | 0.0141 (0.41) | 0.0095 (0.59) | 0.0049 (0.47) | 0.0019 (1.19) | 0.0057 (0.50) | 0.0057 (0.91) | 0.0046 (0.25) |
| ZYW-ZYL | 0.0558 (0.68) | 0.0338 (0.92) | 0.0209 (0.89) | 0.0063 (0.91) | 0.0230 (0.85) | 0.0194 (1.21) | 0.0221 (0.48) |
| Means | 0.0224 (0.47) | 0.0128 (0.69) | 0.0210 (0.68) | 0.0020 (0.89) | 0.0091 (0.68) | 0.0063 (0.81) | 0.0097 (0.34) |
Shown are covariances of each pair of traits in the early and late-developing skull groups calculated from phenotypic, genetic (single-locus effects + epistatic), additive genetic, dominance genetic, single-locus (additive and dominance effects), epistatic, and environmental covariances. Correlations in parentheses were calculated by standardizing covariances with the appropriate variances of the traits.
Two-locus model predictions and fit with covariance estimates:
Expected single-locus (additive, dominance, and total), epistatic, and total genetic covariances and correlations are presented in Table 6. These values were calculated using the coefficients from the multiple regression in the QTL analysis and Equation 4. Table 6 also includes a count of the number of positive and negative components contributing to each covariance. These correspond to the numbers of loci or epistatic combinations that show either positive or negative pleiotropy on that pair of traits. The total counts of positive and negative terms are listed in the rows of means and do not represent mean counts, but totals for the covariances included in the means. These counts demonstrate that the vast majority of pleiotropic effects (84 of 88) contribute positively to covariances between traits (with all of the epistatic pleiotropic effects contributing positive components to the covariances). This directionality of effects may be interpreted as consistent with the pattern of genetic integration, where the sign of pleiotropic effects tends to be similar within trait groups (see below).
TABLE 6.
Expected covariances and correlations within early and late-developing trait groups
| Total genetic | Single-locus | Additive | Dominance | Epistatic | |
|---|---|---|---|---|---|
| Early developing | |||||
| CVW-CVH | 0.0647 (0.14) [5:0] | 0.0065 (0.37) [5:0] | 0.0026 (0.26) [2:0] | 0.0039 (0.56) [3:0] | 0.0 (0.0) [0:0] |
| CVW-CVL | 0.0067 (0.09) [4:0] | 0.0067 (0.24) [4:0] | 0.0058 (0.31) [3:0] | 0.0009 (0.09) [1:0] | 0.0 (0.0) [0:0] |
| CVH-CVL | 0.0188 (0.23) [9:0] | 0.0188 (0.63) [9:0] | 0.0080 (0.50) [4:0] | 0.0108 (0.80) [6:0] | 0.0 (0.0) [0:0] |
| Means | 0.0106 (0.15) [18:0] | 0.0106 (0.41) [18:0] | 0.0055 (0.36) [9:0] | 0.0052 (0.48) [10:0] | 0.0 (0.0) [0:0] |
| Late developing | |||||
| FCL-FCH | 0.0025 (0.09) [4:1] | 0.0015 (0.16) [3:1] | 0.0015 (0.18) [3:1] | 0.0 (0.0) [0:0] | 0.0010 (0.11) [1:0] |
| FCL-FCW | 0.0056 (0.14) [5:0] | 0.0056 (0.49) [5:0] | 0.0056 (0.53) [5:0] | 0.0 (0.0) [0:0] | 0.0 (0.0) [0:0] |
| FCL-ZYW | 0.0162 (0.17) [5:0] | 0.0132 (0.45) [4:0] | 0.0114 (0.43) [4:0] | 0.0018 (0.64) [1:0] | 0.0031 (0.08) [1:0] |
| FCL-ZYL | 0.0191 (0.19) [7:0] | 0.0175 (0.54) [6:0] | 0.0175 (0.62) [6:0] | 0.0 (0.0) [0:0] | 0.0016 (0.05) [1:0] |
| FCH-FCW | 0.0024 (0.17) [6:0] | 0.0018 (0.39) [4:0] | 0.0018 (0.43) [4:0] | 0.0 (0.0) [0:0] | 0.0007 (0.12) [2:0] |
| FCH-ZYW | 0.0066 (0.18) [7:1] | 0.0057 (0.49) [6:1] | 0.0057 (0.55) [6:1] | 0.0 (0.0) [0:0] | 0.0009 (0.07) [1:0] |
| FCH-ZYL | 0.0050 (0.13) [6:2] | 0.0050 (0.38) [6:2] | 0.0047 (0.43) [6:2] | 0.0002 (0.12) [1:0] | 0.0 (0.0) [0:0] |
| FCW-ZYW | 0.0167 (0.32) [9:0] | 0.0076 (0.52) [4:0] | 0.0076 (0.58) [4:0] | 0.0 (0.0) [0:0] | 0.0091 (0.40) [5:0] |
| FCW-ZYL | 0.0084 (0.16) [7:0] | 0.0084 (0.52) [7:0] | 0.0075 (0.54) [5:0] | 0.0009 (0.48) [2:0] | 0.0 (0.0) [0:0] |
| ZYW-ZYL | 0.0347 (0.26) [10:0] | 0.0261 (0.64) [7:0] | 0.0240 (0.69) [7:0] | 0.0021 (0.35) [1:0] | 0.0087 (0.18) [3:0] |
| Means | 0.0117 (0.18) [66:4] | 0.0092 (0.46) [52:4] | 0.0087 (0.50) [50:4] | 0.0005 (0.16) [5:0] | 0.0025 (0.10) [14:0] |
Shown are expected covariances of each pair of traits in the early and late-developing skull groups calculated using the covariance model (Equation 4) and the regression coefficients from the QTL analysis. Correlations in parentheses were calculated by standardizing covariances with the appropriate expected variances (calculated using Equation 3) of the traits. Shown in brackets are the numbers of positive and negative components [positive:negative] that sum to give the overall covariance. For example, in the case of the single-locus covariances these numbers correspond to the numbers of loci with positive and negative pleiotropic effects contributing to the covariance.
The total genetic covariances predicted from the model are strongly correlated with those estimated from the multivariate regression procedure (r = 0.94) and the RMA regression indicates that the values predicted from the model were very similar to the estimated values (β = 0.92, c = 0.13). Single-locus covariances predicted from the model were also strongly correlated with those estimated from the multivariate regression procedure (r = 0.93) and the RMA regression indicated generally good agreement between the predicted and measured values (β = 0.82, c = 0.14). However, the predicted additive genetic covariances showed a much closer match than did the dominance covariances (r = 0.95 for the additive vs. r = 0.56 for the dominance). This pattern was also reflected in the RMA regression, which indicated that, overall, the predicted additive covariances were closer to the estimated covariances than were the dominance covariances (β = 0.86, c = 0.12 for the additive vs. β = 0.58, c = 0.63 for the dominance). This pattern is primarily the result of the fact that the model predicted a large number of zero-dominance covariances (6 of 13), but none of the estimated dominance covariances were zero. Predicted and estimated epistatic covariances were also strongly correlated (r = 0.70), although less so than the single-locus values. The RMA regression slope indicates some lack of agreement between the two sets of values (β = 1.51, c = 0.50), which is due to the fact that the model predicts a number of zero covariances (6 of 13), while none were zero in the covariance analysis.
To simplify the presentation of the model results we do not present the predicted variances for the traits calculated from the model. The estimated single-locus, additive, dominance, epistatic, and total genetic variances are all highly correlated with the values predicted from the model (r = 0.96, 0.99, 0.83, 0.99, and 0.98, respectively). However, the RMA regression slopes indicate that the model predicted larger variances than were observed (β = 0.61, 0.68, 0.50, 0.36, and 0.28 for the single-locus, additive, dominance, epistatic, and total genetic variances, respectively). All of the regression constants were near zero except for the dominance regression constant, which had a value of c = 0.21.
The covariances and variances can be combined to examine the relationship between the predicted and estimated correlations. Correlations are often of interest because they are easy to interpret and may be seen as a critical measure of the strength of integration of traits. However, because they are composite terms, they are prone to inflation of errors. This is clearly seen in the pattern of correlations estimated here. The predicted and estimated single-locus, additive, and epistatic correlations show only moderate to low correlations (r = 0.44, 0.36, and 0.27, respectively). The predicted and estimated dominance correlations show almost no relationship, with a slightly negative correlation between the two (r = −0.21, which is not significantly different from zero). Again, this is primarily due to the fact that a large number of the dominance correlations were expected to be zero, but none were estimated as zero. The predicted and expected total genetic correlations showed a much stronger correlation (r = 0.71), suggesting that, when all of the genetic information is combined, the overall patterns of estimated values are strongly correlated with those predicted.
DISCUSSION
There has been a great deal of interest in understanding the contribution of various forms of physiological genetic effects (e.g., additive, dominance, and epistatic) to patterns of genetic variation within populations. With the emergence of the QTL paradigm there has been a rapid advancement in our understanding of the contribution of physiological genetic effects to the genetic architecture of trait variation. However, the genetic architecture of covariation between traits remains largely unknown (but see Cheverud 2004). As is the case with genetic variances, genetic covariances can be the result of single-locus additive or dominance effects and may also be produced by epistatic interactions between loci (Cheverud et al. 2004). While this follows logically from quantitative genetic theory (Mode and Robinson 1959), few theoretical or empirical analyses have examined the contribution of epistatic interactions to the genetic architecture of covariances between traits. Cheverud et al. (2004) discuss the closely related concept of differential epistasis, which occurs when epistatic interactions between pairs of loci are different for pairs of traits. However, in their analysis of differential epistasis, they did not explicitly examine epistatic pleiotropy or differential epistasis per se, but rather inferred the possible presence of differential epistasis from cases where the relationship between two traits varies with the genotype at a particular locus (see also Cheverud 2004). The two-locus population genetic model presented herein provides a framework for explicitly examining the contribution of epistatic interactions to covariation between traits. This model also allowed us to interpret the QTL analysis results in terms of their influence on patterns of genetic covariances and corresponding patterns of integration.
Pleiotropy and covariation:
It is often assumed that pleiotropic alleles necessarily contribute to covariation and that, in the presence of pleiotropy, the only way to have no covariation between traits is to have a balance of positive and negative pleiotropy. However, the results of the model clearly demonstrate that it is possible to have unbalanced pleiotropy (e.g., all positive pleiotropy) and no covariation. For single-locus effects, this lack of covariation can be achieved only when a locus shows an additive effect on one trait and a dominance effect on a second trait. This scenario does not contribute to covariation between traits in this population of intercross mice (where there is linkage equilibrium across chromosomes and equal allele frequencies) because the effects of the genotypes on the two traits are orthogonal. However, one would clearly consider this locus pleiotropic since it affects the expression of two different traits. The contributions of epistatic pleiotropic effects to covariances between traits show this same basic phenomenon: if the effects are orthogonal, then they do not contribute to a covariance between traits since the effects of a locus on the two traits are independent of each other. Since all genetic effects are orthogonal under the Cockerham (1954) model, a pair of traits must show the same pattern of epistasis for epistasis to contribute to their covariance.
In our QTL analysis we see the importance of the distinction between pleiotropic effects of loci and patterns of pleiotropy that lead to trait covariances. Although we found a high degree of pleiotropy (10 of 20 loci show significant effects on multiple traits in the early group and 16 of 19 in the late group; see Table 2), there are many examples of pleiotropic QTL that do not contribute to covariances between traits. For example, L4.2 is pleiotropic, affecting the expression of FCH and ZYL, but because it has an additive effect on FCH and dominance effect on ZYL, the single-locus effect of L4.2 does not contribute to a covariance between these traits.
It is important to keep in mind that we have analyzed the genetics of a particular population with a defined set of allele frequencies (i.e., an F2 population with equal frequencies of pairs of alleles at each locus) and that the contribution of various physiological genetic effects (e.g., physiological dominance or epistasis) to variances and covariances is expected to change with allele frequencies (see Cheverud and Routman 1995). As a result, in populations where the two alleles at each locus are not at equal frequency, many of the genetic effects that are orthogonal in our study population are not statistically independent. Thus, when allele frequencies at a pair of interacting loci are not 0.5, these nonorthogonal components can contribute to covariation between traits, allowing the covariances to change as allele frequencies change.
While the contributions of epistatic effects to covariances have a straightforward statistical explanation in terms of whether genetic effects on a pair of traits are orthogonal, it may be more intuitive to think about the epistatic effects as making the sign of pleiotropic effects of a locus variable or even unpredictable (Cheverud et al. 2004). That is, one can consider the net pleiotropic effects of a locus as a weighted average of the pleiotropic effects that the locus has on the various genetic backgrounds at the other loci it interacts with. For example, locus A may have pleiotropic effects on traits X and Y but the magnitude or sign of its pleiotropic effect on each trait may be modified by its interaction with locus B. This can make the overall pleiotropic effect of locus A variable across genetic backgrounds at locus B. At the extreme, this may even result in a net pleiotropy of zero when the effect of locus A has no net pleiotropic effect when averaged across the genetic background of locus B. This dependence of the pleiotropic effects of a locus on the genetic background of other loci can provide a means by which the pleiotropic effects of a locus can evolve and contribute to changes in the pattern of covariation between traits (Cheverud et al. 2004). This issue will be discussed in a more general model elsewhere.
Implications for integration:
Cheverud (1984) has shown that the genetic covariance due to pleiotropy is the weighted average of positive and negative pleiotropy at individual loci with pleiotropic effects. This basic result holds for epistatic pleiotropy, where the net covariance is the weighted average of the single-locus and epistatic effects on the covariance. Since it is ultimately single loci that are pleiotropic, we can consider the epistatic interactions as altering the pleiotropic effects of loci (Cheverud 2004). As a result, the weighted average of positive and negative pleiotropic effects must first be made within a locus across genetic backgrounds to determine the net sign of the covariance contributed by pleiotropic effects of a locus. The weighted average within a locus can then be used in the weighted average across loci to determine the net overall covariance. This implies that the occurrence of epistatic pleiotropy can allow for the sign of pleiotropic effects of individual loci to change with allele frequencies, which alter the weighted average pleiotropic effect of a locus (Cheverud 2004). Therefore, the presence of epistatic pleiotropy provides a means by which patterns of covariance structure and integration may evolve.
The results of the QTL analysis demonstrate that, for cases where a pair of loci shows epistatic effects, the epistasis does not alter the overall sign of single-locus pleiotropic effects of the loci since nearly all epistatic pleiotropic effects are of the same sign as these single-locus effects (positive). As a result, while epistasis can introduce variation into the sign or magnitude of the pleiotropic effects of these loci when viewed across genetic backgrounds, its net effect is to contribute positive components to covariances produced by loci in this population. Although there are a number of cases where loci that show a single-locus effect on one trait show an epistatic effect on a second trait that is of opposite sign, these do not contribute to covariances.
Across loci, it is clear that single-locus effects contribute to patterns of integration since all 18 single-locus effects contribute positive covariance components to early developing traits and 52 of 56 components are positive for the late-developing traits (Table 6). The model suggests that these single-locus components are largely responsible for the correlations between traits, but this general trend is not supported by the estimated covariances (Table 5). The estimated covariances show that although total genetic correlations were significantly larger than phenotypic correlations [which is a typical finding (Leamy 1977; Cheverud 1988)], genetic correlations due to single-locus effects were not significantly different from those due to epistatic effects.
The model shows that epistatic effects contribute relatively small covariance components (Table 6), and, along with their correspondingly large effect on trait variances, they are expected to make small contributions to trait correlations overall. Thus, as assessed by correlations, the epistatic effects of QTL found for the skull traits appeared to have only minor effects on their integration. If one uses the overall genetic correlations as measures of genetic integration, one might conclude that epistatic pleiotropy tends to reduce integration since genetic correlations would be larger in the absence of epistatic effects. However, since they show a strong directionality of effect when present and also often contribute considerable covariation one might conclude that the epistatic effects appear to have evolved to maintain patterns of genetic integration.
The observed pattern of pleiotropy in the skull is likely to have evolved as a correlated response to selection on adult body weight since the SM/J and LG/J lines were formed by directional selection on 60-day body weight rather than by covariation or integration in the skull per se. Our QTL analysis shows that the overwhelming majority of the pleiotropy detected is positive (see Table 6), suggesting that the selection regime may have favored positive pleiotropy and perhaps trait integration. However, because the overwhelming majority of single-locus QTL effects are positive (20 of 23 additive effects and 15 of 18 dominance effects are positive for early developing traits while 43 of 48 additive effects and 12 of 14 dominance effects are positive for late-developing traits), there is little opportunity for single-locus pleiotropy to be negative. As a result, we cannot determine whether selection favored positive pleiotropy because of trait integration or whether the predominance of positive pleiotropy is simply a secondary outcome of a selection regime that produced a pattern of positive genetic effects as a correlated response to directional selection on trait means (see Orr 1998).
In contrast to single-locus effects, the signs of epistatic effects are not strongly biased toward being positive or negative (14 of 31 epistatic effects are positive for early developing traits while 31 of 74 epistatic effects are positive for late-developing traits), but we still find that all epistatic pleiotropy is positive (Table 6). Therefore, the pattern of epistatic pleiotropy may show a signature of selection for positive pleiotropy and perhaps genetic integration. This suggests that epistatic pleiotropy could play a role in the evolution of integration, but it is unclear how epistatic effects could evolve to contribute to the evolution of integration. This requires further theoretical work.
Acknowledgments
We thank Charles Goodnight, Chris Klingenberg, Paula Kover, and Joshua Mutic for insightful discussions during the development of this work. We also thank Karl Broman and two anonymous reviewers for helpful comments on this article. This work was supported in part by funds provided by the University of North Carolina at Charlotte to L.J.L., by National Science Foundation (NSF) grants DEB-0236956 and DEB-0315901 and grants from the Natural Environment Research Council and the Biotechnology and Biological Sciences Research Council (United Kingdom) to J.B.W., and by NSF grant DEB-9726433 and National Institutes of Health grant DK52514 to J.M.C.
References
- Bogin, B., 1988. Patterns of Human Growth. Cambridge University Press, Cambridge, UK.
- Cheverud, J. M., 1982. Phenotypic, genetic, and environmental morphological integration in the cranium. Evolution 36: 499–516. [DOI] [PubMed] [Google Scholar]
- Cheverud, J. M., 1984. Quantitative genetics and developmental constraints on evolution by selection. J. Theor. Biol. 110: 155–171. [DOI] [PubMed] [Google Scholar]
- Cheverud, J. M., 1988. A comparison of genetic and phenotypic correlations. Evolution 42: 958–968. [DOI] [PubMed] [Google Scholar]
- Cheverud, J. M., 2000. Detecting epistasis among quantitative trait loci, pp. 58–81 in Epistasis and the Evolutionary Process, edited by J. B. Wolf, E. D. I. Brodie and M. J. Wade. Oxford University Press, New York.
- Cheverud, J. M., 2001. The genetic architecture of pleiotropic relations and differential epistasis, pp. 411–433 in The Character Concept in Evolutionary Biology, edited by G. P. Wagner. Academic Press, San Diego.
- Cheverud, J. M., 2004. Modular pleiotropic effects of quantitative trait loci on morphological traits, pp. 132–153 in Modularity in Development and Evolution, edited by G. Schlosser and G. P. Wagner. University of Chicago Press, Chicago.
- Cheverud, J. M., and E. J. Routman, 1995. Epistasis and its contribution to genetic variance components. Genetics 139: 1455–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud, J. M., E. J. Routman, F. A. M. Duarte, B. Van Swinderen, K. Cothran et al., 1996. Quantitative trait loci for murine growth. Genetics 142: 1305–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud, J. M., E. J. Routman and E. J. Irschick, 1997. Pleiotropic effects of individual gene loci on mandibular morphology. Evolution 51: 2006–2016. [DOI] [PubMed] [Google Scholar]
- Cheverud, J. M., T. H. Ehrich, T. T. Vaughn, S. F. Koreishi, R. B. Linsey et al., 2004. Pleiotropic effects on mandibular morphology II: differential epistasis and genetic variation in morphological integration. J. Exp. Zool. B Mol. Dev. Evol. 302B: 424–435 [DOI] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerham, C. C., 1954. An extension of the concepts of partitioning hereditary variance for analysis of covariances among relatives when epistasis is present. Genetics 39: 859–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, S. N., 1998. Multiple comparisons, explained. Am. J. Epidemiol. 147: 807–812. [DOI] [PubMed] [Google Scholar]
- Goodnight, C. J., 2000. Quantitative trait loci and gene interaction: the quantitative genetics of metapopulations. Heredity 84: 587–598. [DOI] [PubMed] [Google Scholar]
- Gromko, M. H., A. Briot, S. C. Jensen and H. H. Fukui, 1991. Selection on copulation duration in Drosophila melanogaster: predictability of direct response versus unpredictability of correlated response. Evolution 45: 69–81. [DOI] [PubMed] [Google Scholar]
- Haley, C. S., and S. A. Knott, 1992. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69: 315–324. [DOI] [PubMed] [Google Scholar]
- Kao, C.-H., and Z-B. Zeng, 2002. Modeling epistasis of quantitative trait loci using Cockerham's model. Genetics 160: 1243–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., and D. Botstein, 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., P. Green, J. Abrahamson, A. Barlow, M. Daley et al., 1987. MAPMAKER: an interactive active computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Leamy, L., 1977. Genetic and environmental correlations of morphometric traits in randombred house mice. Evolution 31: 357–369. [DOI] [PubMed] [Google Scholar]
- Leamy, L. J., E. J. Routman and J. M. Cheverud, 1997. A search for quantitative trait loci affecting asymmetry of mandibular characters in mice. Evolution 51: 957–969. [DOI] [PubMed] [Google Scholar]
- Leamy, L. J., E. J. Routman and J. M. Cheverud, 1999. Quantitative trait loci for early and late developing skull characters in mice: a test of the genetic independence model of morphological integration. Am. Nat. 153: 201–214. [DOI] [PubMed] [Google Scholar]
- Lincoln, S., M. Daly and E. Lander, 1992. Constructing genetic maps with MAPMAKER/EXP 3.0. Whitehead Institute Technical Report, Ed. 3. Whitehead Institute, Cambridge, MA.
- Mackay, T. F. C., 2001. The genetic architecture of quantitative traits. Annu. Rev. Genet. 35: 303–339. [DOI] [PubMed] [Google Scholar]
- Mode, C. J., and H. F. Robinson, 1959. Pleiotropism and the genetic variance and covariance. Biometrics 15: 518–537. [Google Scholar]
- Moore, W., 1981. The Mammalian Skull. Cambridge University Press, Cambridge, UK.
- Moss, M., 1973. A functional cranial analysis of primate craniofacial growth, pp. 191–208 in Craniofacial Biology of the Primates, Vol. 3, edited by M. R. Zingeser. Symposium IVth International Congress on Primatology. Karger, Basel, Switzerland.
- Orr, H. A., 1998. Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics 149: 2099–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, P. C., M. C. Whitlock and K. Fowler, 2001. Inbreeding changes the shape of the genetic covariance matrix in Drosophila melanogaster. Genetics 158: 1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff, D. A., 1997. Evolutionary Quantitative Genetics. Chapman & Hall, New York.
- Routman, E. J., and J. M. Cheverud, 1995. Polymorphism for PCR-analyzed microsatellites: data for two additional inbred mouse strains and the utility of agarose gel electrophoresis. Mamm. Genome 6: 401–404. [DOI] [PubMed] [Google Scholar]
- Sing, C. F., J. H. Stengard and S. L. R. Kardia, 2003. Genes, environment, and cardiovascular disease. Arterioscler. Thromb. Vasc. Biol. 23: 1190–1196. [DOI] [PubMed] [Google Scholar]
- Thompson, J. R., 1998. Invited commentary: re: multiple comparisons and related issues in the interpretation of epidemiologic data. Am. J. Epidemiol. 142: 801–806. [DOI] [PubMed] [Google Scholar]
- Whitlock, M. C., P. C. Phillips and K. Fowler, 2002. Persistence of changes in the genetic covariance matrix after a bottleneck. Evolution 56: 1968–1975. [DOI] [PubMed] [Google Scholar]
- Wolf, J. B., E. D. Brodie, III and M. J. Wade, 2000. Epistasis and the Evolutionary Process. Oxford University Press, New York.
- Workman, M. S., L. J. Leamy, E. J. Routman and J. M. Cheverud, 2002. Analysis of QTL effects on the size and shape of mandibular molars in mice. Genetics 160: 1573–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]


