Abstract
We report a general combinatorial approach to identify optimal substrates of a given protease by using quantitative kinetic screening of cellular libraries of peptide substrates (CLiPS). A whole-cell protease activity assay was developed by displaying fluorescent reporter substrates on the surface of Escherichia coli as N-terminal fusions. This approach enabled generation of substrate libraries of arbitrary amino acid composition and length that are self-renewing. Substrate hydrolysis by a target protease was measured quantitatively via changes in whole-cell fluorescence by using FACS. FACS enabled efficient screening to identify optimal substrates for a given protease and characterize their cleavage kinetics. The utility of CLiPS was demonstrated by determining the substrate specificity of two unrelated proteases, caspase-3 and enteropeptidase (or enterokinase). CLiPS unambiguously identified the caspase-3 consensus cleavage sequence DXVDG. Enteropeptidase was unexpectedly promiscuous, but exhibited a preference for substrates with the motif D/ERM, which were cleaved substantially faster than the canonical DDDDK recognition sequence, widely used for protein purification. CLiPS provides a straightforward and versatile approach to determine protease specificity and discover optimal substrates on the basis of cleavage kinetics.
Keywords: caspase-3, enteropeptidase, FACS, peptidase
Proteolytic enzymes play central roles in most biological processes. At the same time, proteases are implicated in the pathogenesis of diverse diseases ranging from viral and bacterial infections (1) to cancer and neurodegenerative diseases (2, 3). The biological functions of proteases, or peptidases, are largely determined by the spectrum of biologically occurring substrates. Frequently, substrate specificity has been inferred by comparison and alignment of known, physiological substrates (4). Given this information, individually synthesized fluorogenic peptide substrates have been used extensively to measure protease activity (5). More recently, the substrate specificity of a small group of proteases has been characterized in greater detail by using combinatorial peptide libraries (6, 7). Library-based approaches to characterize specificity have contributed substantially to our understanding of protease function (6, 7). Nevertheless, quantitative characterization of protease specificity and substrate cleavage kinetics remains a bottleneck in basic and applied biological research (6, 8).
Among combinatorial methods, the “substrate phage” approach has been used to define the substrate specificities of ≈30 proteases and has been recently reviewed (6). In this approach, a library of peptide substrates is displayed on the surface of filamentous bacteriophage (9) as fusions between a surface-anchoring affinity domain and the display scaffold protein, typically the phage minor coat protein GPIII. The substrate library then is allowed to bind to a solid surface via the affinity domain. Phage liberated from the surface by proteolytic cleavage are recovered and amplified by infecting Escherichia coli, and the entire process is repeated multiple times. Typically, substrate phage clones are then isolated, and their DNA is sequenced to identify potential substrate motifs. Because substrate cleavage kinetics cannot be readily measured in the phage format, kinetic characterization to quantify the substrate specificity generally involves preparation of soluble, fluorogenic substrates by using recombinant (10) or synthetic methods (11).
To enable direct kinetic characterization, combinatorial fluorogenic substrate libraries (7) can be synthesized by using solid-phase synthesis methods or light-directed parallel synthesis (12). Array-based methods enable quantitative measurements of cleavage kinetics for many substrates in parallel (13). However, these methods are typically restricted to consideration of a comparatively small number of substrates, because complete investigation of just three random substrate positions requires construction of high-density arrays of 8,000 members (13). Solution-phase fluorogenic substrate libraries also are useful for profiling specificity on the basis of catalytic turnover. For example, pooled substrate libraries can be synthesized with four randomized positions and a fluorogenic leaving group (Fl) in place of the P1′ residue (e.g., XXXX-Fl). The rate and extent of conversion of this pooled mixture of ≈160,000 unique substrates can be quantified by using fluorimetry, although cleavage kinetics for individual sequences and cooperative phenomena between different residues cannot be identified directly. Alternatively, two of four nonprime positions can be fixed, yielding as many as 400 potential sublibraries (7). Recently, the pooled, fluorogenic substrate library method has been extended to identify preferred amino acids at the P1′ site (14). Fluorogenic substrate libraries have proven useful to profile the specificity of several proteases and to aid the design of inhibitors for the treatment of viral infections and type II diabetes (15, 16).
Previously reported experimental methods have contributed substantially to our understanding of proteolytic function. Yet, given the explosion of the number of uncharacterized proteolytic enzymes, new approaches are needed urgently to determine protease specificity rapidly, accurately, and quantitatively. Additionally, the capability to identify kinetically optimal substrates for a given protease or biological tissue would be especially valuable for the development of proteolytically activated switches for diagnostic imaging and therapeutic applications (17). We sought to develop a general methodology to quantitatively determine substrate specificity that combines simple library construction and manipulation in bacteria with quantitative analysis and screening on the basis of catalytic turnover. Here we report the development of cellular libraries of peptide substrates (CLiPS) and the application of this approach to quantitatively determine the substrate specificity of proteases. Our results demonstrate that CLiPS will be broadly useful for characterizing proteases and in the development of optimal substrates for technological applications.
Results
Development of a Whole-Cell Assay for Peptidase Activity.
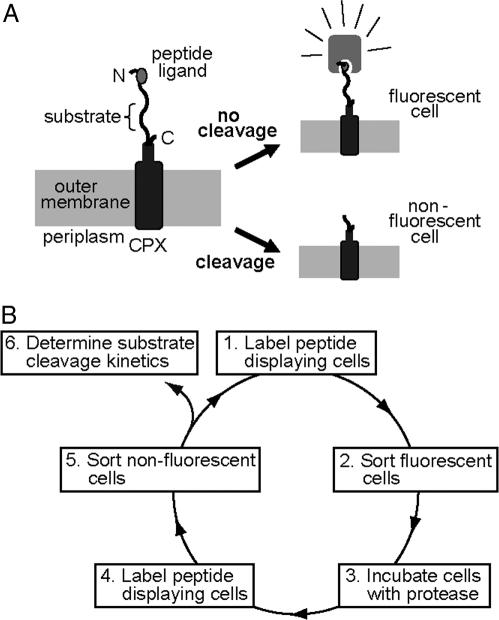
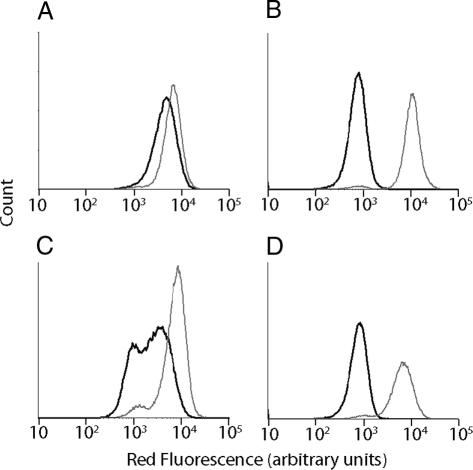
Given the utility of FACS as a quantitative library screening tool, we sought to assay for peptidase activity by displaying reporter substrates on the surface of Esherichia coli (Fig. 1A). Reporter substrates were designed consisting of a peptide ligand that binds the fluorescent probe streptavidin-conjugated phycoerythrin (SA-PE) and a peptide substrate oriented such that cleavage removes the SA-PE-binding ligand from the cell surface. In this way, protease activity toward a given substrate would be detectable by monitoring whole-cell fluorescence by using FACS. Reporter substrates were displayed on E. coli by using circularly permutated outer membrane protein X (CPX), which presents both N- and C termini on the cell surface, enabling presentation of passenger peptides as nonconstrained, terminal fusions (18). As a control, a substrate-reporter display vector was constructed incorporating a known enteropeptidase cleavage site (DDDDK) flanked by flexible peptide linker sequences, as “spacers,” allowing protease access to the substrate, and a SA-PE-binding peptide ligand. Cells displaying the substrate were fluorescently labeled with SA-PE, resulting in a >20-fold increase in mean fluorescence intensity over background autofluorescence, as measured by flow cytometry (Fig. 2). Incubation of this cell population with enteropeptidase before labeling with SA-PE resulted in a ≈20-fold decrease in mean fluorescence intensity (Fig. 2B), whereas a negative control cell population displaying the sequence GGQSGQ exhibited minimal change in fluorescence (Fig. 2A). These results demonstrated that enzymatic cleavage of reporter substrates could be detected as a decrease in fluorescence intensity of cells by using FACS and hydrolysis is not due to cleavage outside of the designated substrate region.
Fig. 1.
CLiPS. (A) Display of reporter substrates, consisting of a substrate peptide and fluorescent-probe peptide ligand, on the surface of E. coli as fusions to the N terminus of circularly permuted outer membrane protein OmpX (CPX). Substrate cleavage results in a reduction of cellular fluorescence as detected by flow cytometry. (B) Substrate libraries are screened by depleting the library pool of clones that do not display a peptide and then enriching clones with hydrolyzed substrates.
Fig. 2.
Measurement of substrate conversion by FACS. Flow cytometry analysis of bacterial cell populations displaying either linker (GGSGGS) (A) or canonical substrate (DDDDK) (B) before (gray line) and after (black line) treatment with enteropeptidase. During library screening for enteropeptidase substrates, cell populations collected from screen 1A (C) and screen 3B (D) were analyzed by flow cytometry before (gray line) and after (black line) enteropeptidase treatment. The loss of fluorescence due to treatment, shown by the shift in the black line, demonstrates enrichment of enteropeptidase substrates after screen 3B (D).
To extend this single-cell substrate cleavage assay to identify optimal substrates for a given protease, a substrate library was constructed in E. coli by combinatorial randomization of six sequential amino acid positions within the substrate region (Fig. 1A). This CLiPS had a theoretical diversity of 6.4 × 107 unique amino acid sequences. The constructed library contained 1.5 × 108 independent transformants. Thus, this library is expected to include all possible 5-mer and 4-mer substrate sequences with >95% and 99% confidence limits, respectively, assuming a random distribution (19). Using the whole-cell activity assay, a screening methodology was devised to isolate library members displaying substrates cleaved by a given protease and, thereby, identify optimal substrates.
Determination of Enteropeptidase and Caspase-3 Specificity by Using CLiPS.
To demonstrate the general utility of CLiPS, the 6-mer substrate library was screened to identify optimal substrates for two unrelated proteases: caspase-3 and enteropeptidase. These proteases recognize the canonical substrates DEVD↓ (20) and DDDDK↓ (4), respectively. Caspase-3 was chosen to validate CLiPS, because specificity has been investigated extensively by using both substrate phage and fluorogenic substrates (21, 22). In contrast, enteropeptidase specificity is less well characterized and has been investigated primarily by using individually synthesized, fluorogenic substrate variants (23). For each protease, optimal substrates were identified by performing a two-step screen for hydrolysis (Fig. 1B). First, library members that display the affinity epitope were purified by sorting (Fig. 1B), thereby removing library members that do not display substrates (i.e., members with stop codons and frameshift mutations). The resulting library population was amplified by growth, treated with protease, labeled with SA-PE, and cells with reduced fluorescence resulting from substrate hydrolysis were collected (Fig. 1B). After three cycles of screening for enteropeptidase substrates, >95% of the enriched library displayed reporter substrates and exhibited cleavage similar to the canonical substrate (Fig. 2 C and D). Therefore, a final sort was performed to identify enteropeptidase substrates that hydrolyzed more rapidly than the canonical substrate.
In applications that involve complex protease-containing mixtures, such as cellular lysates or tissue extracts, we anticipated that specificity could be identified by removing substrates that are cleaved by an appropriate background mixture. For this reason, we investigated whether substrates of a target protease can be determined in the presence of cell lysates. Nonspecifically cleaved substrates were depleted from the library first by incubation with E. coli lysate protein that does not contain the target protease, caspase-3 (Fig. 1B). Subsequently, cells displaying specifically cleaved substrates were isolated after incubating the library with lysate from E. coli-expressing caspase-3 (Fig. 1B). This process ensured that cleavage during screening was due to caspase-3 activity and not endogenous E. coli proteolytic activity. Two cycles of screening resulted in the enrichment of library members exhibiting caspase-3 dependent cleavage. Incubation of the enriched library with caspase-3 containing lysates, but not caspase-3-free lysates, resulted in a reduction of the mean fluorescence intensity of the population, as measured by flow cytometry (data not shown). Single clones from the enriched library were isolated from the remaining population by plating. Thus, CLiPS was capable of identifying caspase-3-specific substrates in the presence of a complex mixture.
Characterization of Substrate Cleavage Kinetics.
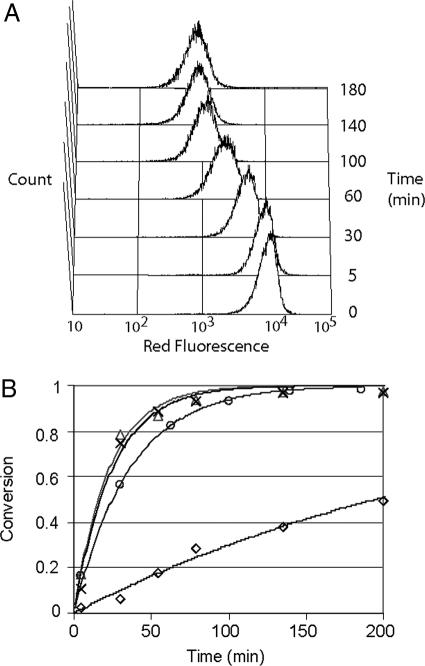
The use of multicopy substrate display on whole cells enabled simple and direct quantitative characterization of cleavage kinetics. Consequently, flow cytometry was used to rank individual isolated clones on the basis of substrate conversion, and those clones exhibiting >50% conversion were identified by DNA sequencing (Tables 1 and 2). Substrates efficiently cleaved by caspase-3 revealed a strong substrate consensus of DxVDG (Table 1), in agreement with the known specificity of caspase-3. The substrates identified for enteropeptidase shared a consensus sequence of D/ERM, indicating a substrate preference at the P1′ position (Table 2). Interestingly, enteropeptidase substrates identified by CLiPS were cleaved more rapidly than the canonical sequence, DDDDK (Table 2). Four isolated clones with high conversion were investigated further to quantify cleavage kinetics. Clones exhibiting multiple arginine residues were excluded to avoid substrates that may have multiple cleavage sites. Individual substrate displaying clones (e.g., EP4.1; EP, enteropeptidase) exhibited uniform substrate turnover (Fig. 3A), as determined by flow cytometry. In this way, the extent of conversion for each clone could be determined at several different time points and fit to a Michaelis–Menton model (Fig. 3B). The observed second-order rate constant (kcat/KM) for the most rapidly cleaved substrate (EP4.3 SGDRMW) was 13-fold greater than that for the canonical substrate DDDDK (Table 3).
Table 1.
Caspase-3 substrates identified by using CLiPS with a 6-mer library
| Substrate | P5 | P4 | P3 | P2 | P1 | P1′ | P2′ | P3′ | P4′ | Conversion |
|---|---|---|---|---|---|---|---|---|---|---|
| Canonical | q | D | E | V | D | G | g | q | s | 0.95 ± 0.03 |
| CS 2.7 | s | D | G | V | D | G | W | g | g | 0.95 |
| CS 2.14 (4)* | s | D | V | V | D | G | W | g | g | 0.94 ± 0.03 |
| CS 2.23 | s | D | G | V | D | G | V | g | g | 0.93 |
| CS 2.11 | g | g | s | L | D | T | W | T | A | 0.81 |
| CS 2.20 (2) | L | D | T | V | D | R | g | g | q | 0.79 ± 0.01 |
| CS 2.59 | s | D | S | T | D | S | G | g | g | 0.79 |
| CS 2.1 | g | s | Q | V | D | G | V | G | g | 0.75 |
| CS 2.26 | g | s | E | V | D | G | R | H | g | 0.75 |
| CS 2.56 | s | T | E | V | D | G | P | g | g | 0.75 |
| CS 2.47 | g | s | E | V | D | G | G | W | g | 0.74 |
| CS 2.10 | T | D | G | T | D | G | g | g | q | 0.72 |
| CS 2.62 | Q | D | G | V | D | T | g | g | q | 0.70 |
| CS 2.2 | g | s | E | V | D | G | S | R | g | 0.67 |
| CS 2.4 | g | s | Y | V | D | G | V | V | g | 0.64 |
| CS 2.33 | S | D | F | V | D | R | V | g | g | 0.59 |
| CS 2.36 (2) | g | s | M | V | D | G | A | M | g | 0.56 ± 0.05 |
| Consensus | X | D | X | V | D | G |
*Number of clones with identical sequence.
Table 2.
Enteropeptidase substrates identified by using CLiPS with a 6-mer library
| Substrate | P5 | P4 | P3 | P2 | P1 | P1′ | P2′ | P3′ | P4′ | Conversion |
|---|---|---|---|---|---|---|---|---|---|---|
| Canonical | D | D | D | D | K | g | g | q | s | 0.15 ± 0.08 |
| EP 4.3 | s | S | G | D | R | M | W | g | g | 0.97 ± 0.01 |
| EP 4.6 | s | S | G | E | R | M | M | G | g | 0.93 ± 0.03 |
| EP 4.7 | g | s | D | D | R | R | A | G | g | 0.91 ± 0.03 |
| EP 4.8 | V | R | D | Y | R | M | g | g | q | 0.87 ± 0.04 |
| EP 3.6 | g | s | s | D | R | A | R | V | W | 0.86 ± 0.05 |
| EP 4.1 | s | V | D | Y | R | F | L | g | s | 0.84 ± 0.02 |
| EP 4.2 | M | H | G | E | R | M | g | g | s | 0.84 ± 0.02 |
| EP 2.5 | M | S | G | E | R | M | g | g | s | 0.84 ± 0.03 |
| EP 4.10 | g | s | S | E | R | A | A | A | G | 0.78 ± 0.02 |
| EP 4.9 | s | V | L | D | R | W | M | g | g | 0.72 ± 0.05 |
| EP 4.4 | s | E | Y | D | R | Q | L | g | s | 0.71 ± 0.01 |
| EP 2.2 | A | A | V | E | R | W | g | g | s | 0.69 ± 0.14 |
| Consensus | X | X | D/E | R | M | X |
Fig. 3.
Enteropeptidase substrate cleavage kinetics. (A) Time-dependent substrate conversion for clone EP 4.1 (VDYRFL) measured by FACS. (B) Average conversion for cell surface displayed enteropeptidase substrates identified by using CLiPS: VDYRFL (○), SGDRMW (▵), and SGERMM (×) with canonical DDDDK (◇). Data were fit to Michaelis–Menton model, which is shown as a line for each substrate.
Table 3.
Comparison of kinetic constants determined by using CLiPS, fluorescent protein FRET substrates, or synthetic peptides
| Substrate | Sequence |
kcat/KM, μM−1min−1 |
||
|---|---|---|---|---|
| CLiPS | CyPet-YPet | Synthetic | ||
| Linker | GGSGGS | — | 0.19 ± 0.03 | — |
| Canonical | DDDDK | 1.3 ± 0.2 | 57 ± 6 | 7.1 ± 0.7 |
| EP4.1 | VDYRFL | 9.7 ± 0.2 | 520 ± 80 | — |
| EP4.2 | MHGERM | 12.4 ± 1.4 | 480 ± 30 | — |
| EP4.3 | SGDRMW | 18.0 ± 9.3 | 1,020 ± 120 | 36 ± 15 |
| EP4.6 | SGERMM | 14.7 ± 3.3 | 710 ± 40 | — |
To determine how cleavage kinetics (kcat/KM) measured by using surface displayed reporter substrates relate to those measured in solution, two independent approaches were applied to measure kcat/KM for soluble substrates. Because enteropeptidase is often used to remove peptide affinity tags, substrates were assayed in the context of a fusion protein. Specifically, fluorogenic substrates were constructed by using fluorescent proteins that exhibit FRET (CyPet and YPet) (24) and were used to determine protease cleavage kinetics as described in ref. 25. CyPet-YPet substrates for enteropeptidase having recognition sequences of DDDDKG, GGSGGS, or four sequences identified by CLiPS (EP4.1, EP4.2, EP4.3, or EP4.6) were constructed, expressed in E. coli, and purified. Substrate conversion by enteropeptidase was measured in real-time by fluorimetry and fit to Michaelis–Menton kinetics (Table 3). In relative agreement with whole-cell assays, the CLiPS substrate, SGDRMW, cleaved at a rate 17-fold faster than DDDDK. Absolute values of kcat/KM for cell-surface-tethered and soluble substrates differed systematically, but importantly, the relative ranking of the cleavage rates of individual substrates was identical in either context. To further confirm the improved hydrolysis rate for the SGDRMW substrate, relative to DDDDK, fluorogenic peptide substrates were synthesized and cleavage was measured by using fluorimetry (Table 3). The kcat/KM of the CLiPS-identified substrate SGDRMW was >5-fold higher than that of DDDDK. Collectively, these results demonstrate that whole-cell fluorescence assays provide a reliable means to quantitatively measure and rank cleavage kinetics of individual substrate sequences and that CLiPS enables identification of substrates with improved cleavage kinetics.
Discussion
CLiPS provides a general and quantitative approach to determine protease specificity and discover optimal substrates. CLiPS differs in several important respects from previously reported combinatorial methods to determine protease specificity, including substrate phage and fluorogenic substrate libraries. In contrast with phagemid or phage substrate libraries, 103 to 104 copies of the substrate are displayed on the surface of a single cell, enabling quantitative, real-time measurement of substrate conversion for a given clone. Importantly, use of single-cell fluorescence as an indicator of substrate conversion enables quantitative library screening. Likewise, whole-cell fluorescence measurements enable calculation of substrate cleavage kinetics for isolated clones, eliminating the need to prepare soluble substrates by using synthetic or recombinant methods. Finally, bacterial libraries can be manipulated with relative ease and amplified indefinitely by growth without introducing measurable library bias (26). Application of CLiPS to caspase-3 and enteropeptidase enabled rapid library screening to identify a spectrum of rapidly cleaved substrates and direct determination of individual substrate kinetic constants in the cell display format. Given the simplicity of library manipulation and screening, CLiPS provides a scalable solution to rapidly characterize proteases.
Although the primary physiological function of enteropeptidase is to activate trypsin, knowledge of enteropeptidase's recognition sequence, DDDDK, and its tolerance for various amino acids at P1′ (27) have contributed to widespread use of this enzyme in protein purification applications. Typically, only the catalytic subunit is used because it exhibits an activity similar to the full-length protein (28). Enteropeptidase specificity has been investigated previously by comparing natural substrate sequences and by measurement of the hydrolysis rates of synthetic fluorogenic peptides (4, 23, 29, 30). However, substrate specificity has not been characterized in detail by screening combinatorial peptide libraries, despite the obvious importance of identifying unwanted secondary cleavage sites in fusion proteins (23). Application of CLiPS to enteropeptidase revealed a remarkably broad substrate specificity, with a strong preference for arginine at P1 and one or more Asp or Glu residues at P2 or P3. Although libraries constructed by using NNS codons are expected to have a 3:1 ratio of Arg to Lys residues consistent with codon usage, the observed ratio (12:0) indicates a preference for arginine over lysine at the P1 position. Interestingly, the P1′ position in those substrates with the highest levels of conversion was predominantly occupied by methionine, indicating that the positions carboxyl terminal to the scissile bond influence activity. CLiPS resulted in identification of a rapidly cleaved enteropeptidase substrate (SGDRMW), exhibiting a 17-fold faster cleavage than the DDDDK substrate, in the context of a fusion protein. The contextual dependence of substrate cleavage rates could reflect differences in substrate conformation and accessibility that are known to influence proteolysis (31, 32). Previously, others have observed differences in the rates of cleavage of surface-tethered peptide substrates and free substrates in solution prompting the use of relative kcat/KM values for comparisons (14). Nevertheless, the rapidly cleaved substrate SGDRMW identified here may prove useful in protein purification applications, because less enzyme or shorter reactions times could be used to harvest desired proteins, thereby minimizing unwanted hydrolysis that occurs at secondary sites (33).
Caspase-3, an “executioner” caspase of the apoptotic cascade, hydrolyzes a large number of unique substrates that carry out the apoptotic program (34). Given the importance of this enzyme in biology, substrate specificity has been investigated by using several different approaches, including a 4-mer substrate phage library (21) and by comparison of the activities toward a panel of fluorogenic substrates differing by single residues (22). Collectively, these studies have clearly identified a consensus cleavage sequence of DxV/LDG. Secondary analysis of substrate phage clones by using synthetic fluorogenic peptides revealed that substrates with the highest conversion also possessed a DxVD consensus (21). Consistent with these results, positional scanning with synthetic peptides indicated a preference for aspartic acid and glycine at the P4 and P1′ positions, respectively (22). In the present study, two rounds of screening with unpurified caspase-3-containing samples yielded an unambiguous consensus (DxVDG) consistent with previous reports. These results demonstrate that CLiPS does not require purified enzyme preparations, because two-step screening favors removal of substrates cleaved by endogenous E. coli proteases to identify target-specific sequences. Such an approach may prove useful for identifying tissue or disease-specific protease substrates (35) that indicate the presence of protease markers with high sensitivity or that enhance the specificity of therapeutics or imaging agents (17).
In this study, all of the identified clones exhibited protease specific loss of fluorescence. It is possible specificity determination for proteases with highly nonspecific or very weak activity could be challenging because cleavage potentially could occur outside of the substrate region or insufficient substrate conversion could complicate sorting. In such cases, linker sequence modifications or changes in the substrate surface density and reaction conditions could be beneficial. The ability to monitor cleavage kinetics on the cell surface during the course of a reaction enables rational adjustment of display level or reaction conditions to suit different proteases. We have shown that CLiPS can be used to characterize protease activity by displaying substrates on the external surface of E. coli. This method also should be extendable to other cell types, including yeast and mammalian cells, by using appropriate cell-surface display technologies (36). Eukaryotic hosts might prove useful, because posttranslational modifications also have been shown to influence substrate specificity (37). The utilization of FACS enables screening of 107 substrate sequences in <1 h. Many commercially available cell sorters enable isolation of positive clones into microwell plates, enabling downstream clone characterization to be performed in a high-throughput manner. Although the gap between the number of discovered and characterized proteases is growing, CLiPS provides an efficient and quantitative means to bridge this gap.
Materials and Methods
Reagents and Strains.
SA-PE (Invitrogen), the catalytic subunit light chain of enteropeptidase (EP) (New England Biolabs), oligonucleotides (Operon Biotechnologies, Huntsville, AL), synthetic peptides (New England Peptide, Gardner, MA), and Ni-NTA resin (Qiagen, Valencia, CA) were used without modifications. Plasmid pET-23bc3 containing procaspase-3 was obtained from American Type Culture Collection (ATCC no. 99625). E. coli strain MC1061 was used for all experiments (38). All bacterial growth was performed at 37°C with vigorous shaking in LB broth, supplemented with 34 μg/ml chloramphenicol, unless another antibiotic is specified.
Plasmid Vector and Library Construction.
Construction of a control plasmid (pBSX) expressing a GGQSGQ linker and the previously identified streptavidin-binding peptide CX72-S8 was performed as described in ref. 18. Plasmids encoding surface displayed peptide substrates for enteropeptidase (DDDDK) and caspase-3 (DEVD) were constructed as follows. Primers 1 and 2 were used with primer 3 (EP substrate pBSEPX) and primer 4 (caspase-3 substrate pBSCSX) (Table 4, which is published as supporting information on the PNAS web site) to amplify the DNA fragments encoding an in-frame fusion of the streptavidin-binding peptide (WCHPMWEVMCLR), the substrate sequence flanked by flexible linkers, and a circularly permuted outer membrane protein X (CPX) (18). Products were digested with SfiI and ligated to similarly digested pBAD33. A substrate library of the form X6, where X is any amino acid, was constructed by using PCR with a synthetic oligonucleotide incorporating NNS codons (primer 6) along with primers 1 and 5 (Table 4). The product was digested with SfiI and ligated into a similarly digested pBAD33 plasmid. Transformation of the plasmid library into electrocompetent MC1061 yielded 1.5 × 108 colony forming units.
Fluorescent proteins exhibiting FRET (24) (CyPet and YPet) were used to construct fluorogenic protease substrates. To reduce background hydrolysis lysines at the C terminus of CyPET and the N terminus of YPet were removed with primers 7 and 8, respectively. Substrates EP4.1, EP4.2, EP4.3, and EP4.6 identified for enteropeptidase by CLiPS were amplified with YPet by using reverse primer 9 with forward primers 10, 11, 12, and 13, respectively (Table 4). The canonical EP substrate was amplified as a fusion to YPet by using primers 9 and 14 (Table 4). As a negative control, a GGSGGS linker was substituted for the 6-aa substrate sequence by using primers 9 and 15 (Table 4). These products were digested with KpnI and SphI and ligated to a similarly digested plasmid containing CyPET to yield plasmids pBC21Y, pBC41Y, pBC42Y, pBC43Y, pBCEPY, and pBCGSY.
Substrate Library Screening.
For screening and clone analysis, overnight cultures were subcultured by dilution into fresh medium (1:50) and grown for 2 h at 37°C. The subculture was induced with 0.04% arabinose and incubated with shaking at room temperature. Cell aliquots were washed with PBS (pH 8.0), and optical density at 600 nm (OD600) was measured to estimate cell concentration. Cells (108) were pelleted by centrifugation, the supernatant was removed, and the cells were resuspended in reaction buffer (10 μl). After addition of the enzyme, the reaction mixture was incubated at room temperature on a rotary shaker (60 rpm). Cells were removed and diluted 100-fold in PBS to stop the reaction, pelleted by centrifugation, and resuspended in PBS containing SA-PE (50 nM). After incubation at room temperature (1 h), cells were washed with PBS and analyzed or sorted by using a FACSAria cell sorter (Becton Dickinson).
For enteropeptidase cleavage assays, cultures were induced for 2 h. The reaction buffer for enteropeptidase was 50 mM Tris-Cl (pH 8.0) supplemented with 20 mM NaCl/2 mM CaCl2. Three complete cycles of sorting for cleaved substrates were performed by alternating between sorting cells that display substrates in the absence of any added enzyme and sorting cells with hydrolyzed substrates after enzyme treatment. To remove clones from the library pool that did not properly display the substrate and binding peptide (e.g., stop codons or frame-shift mutations), cells were sorted (1A, 2A, and 3A) after 1 h incubation in the reaction buffer without enzyme. Sorts for enteropeptidase hydrolysis (1B, 2B, 3B, and 4) were performed after reactions with 2.9 nM of light-chain enteropeptidase for 22, 18, 4, and 1 h, respectively. A population with the canonical substrate, MC1061/pBSEPX, was used to set the sorting gates during each of the first three rounds, and the fourth round sort gate was set to isolate substrates that hydrolyzed faster than the canonical substrate. The enriched library pool was plated, and individual clones were assayed for substrate conversion by using a 1-h reaction with and without 2.9 nM EP.
For caspase-3 assays, cultures were induced for 3 h, reactions were carried out in PBS (pH 8.0), and all sorting was performed after 5-h reactions. The human pro-caspase-3 gene was amplified by PCR from pET23bc3 digested with EcoRI and KpnI and ligated into similarly digested pBAD30-yielding pB30CS. Overnight cultures of MC1061/pBAD30 and MC1061/pB30CS were subcultured 1:100, grown in LB with 50 μg/ml carbenicillin for 2 h and induced for 5 h at 37°C. From these cultures, soluble protein was isolated by using B-PER-II Bacterial Protein Extraction Reagent (Pierce). Protein extracts were dialyzed in PBS to remove detergent. In an effort to preferentially detect caspase-3 activity over potential endogenous protease activity, 1 μl of soluble protein from MC1061/pBAD30 culture was added to the induced library in sorting cycles 1A and 2A. An induction time of 3 h was found to increase resolution between fluorescent and nonfluorescent cells and was used for experiments with caspase-3. Reaction buffers used were based on known compatible reaction buffers, and reaction times were based on cleavage observed for positive control bacteria displaying known substrates. Background hydrolysis of the regions flanking the substrate site (i.e., a clone displaying the GGQSGQ linker and streptavidin-binding peptide) was measured under each reaction condition to ensure that hydrolysis occurred in the designated substrate region. Sorting for caspase-3 hydrolysis (1B and 2B) was performed after adding 1 μl of soluble protein prepared from MC1061/pB30CS. The enriched library pool was plated, and individual clones were assayed for specific conversion by using 5-h reactions with soluble protein preparations with and without caspase-3.
Recombinant and Synthetic Protein Substrate Reaction Conditions.
Overnight cultures of MC1061 transformed with the FRET substrate-encoding expression vectors described above were subcultured 1:50 and grown for 2 h. The cultures were induced by the addition of arabinose to 0.04% wt/vol and incubated at room temperature for 16 h with shaking. Soluble protein was prepared by using BPER-II as above, and fusion proteins with C-terminal 6x-His tags were purified by using Ni-NTA resin (Qiagen). Reactions were performed in 100 μl of enteropeptidase reaction buffer with ≈0.5 μM fusion protein, as determined by using the extinction coefficient of YPet at 514 nm (24) and Beer's law. Enteropeptidase was added to each reaction to a final concentration of 0.29 nM, and fluorescence emission at 475 nm (cyan) and 527 nm (yellow) was monitored upon excitation at 433 nm by using a Safire fluorimeter (Tecan, Durham, NC).
Synthetic substrates, using Edans and Dabcyl as donor and quencher, were obtained from New England Peptide for the canonical substrate, Dabcyl-DDDDKGG-(E-Edans)-amide, and EP4.3, Dabcyl-SGDRMW(E-Edans)-amide. Reactions were performed in enteropeptidase reaction buffer with between 1 and 4 μM of each peptide as determined by using the extinction coefficient of Dabcyl at 468 nm of 32,000 M−1cm−1. Enteropeptidase was added to each reaction to a final concentration of 0.29 nM, and fluorescence emission at 495 nm was monitored upon excitation at 340 nm by using a Safire fluorimeter.
Kinetic Data Analysis.
The extent of conversion of cell surface displayed peptide substrates was measured directly by using flow cytometry to measure changes in mean fluorescence of clonal cell populations upon protease treatment. Specifically, for each sample, conversion was determined by flow cytometry analyses with the relationship
where (FL−) is the fluorescence after incubating without enzyme, (FL+) is fluorescence after incubation with enzyme, and (FL0) is fluorescence of unlabeled cells. For the recombinant FRET protein reactions, conversion was calculated by dividing the ratiometric FRET signal (yellow fluorescence/cyan fluorescence) by the FRET ratio that results from complete cleavage (25). For synthetic peptide reactions, conversion was calculated by dividing the fluorescence increase by the fluorescence change due to complete hydrolysis. The reported enteropeptidase KM for the physiological substrate tripsinogen is 17 μM (28) but 600 μM for short DDDDK peptides (39). Given that KM for short peptides is much larger than the substrate concentrations that were used (<5 μM), the Michaelis–Menton model simplifies to
allowing substrate conversion to be expressed as
where [S] is the substrate concentration, [E] is enzyme concentration, and t is time. To determine the second-order rate constant (kcat/KM), the time-dependent conversion for each substrate was fit to Eq. 3. Reported values represent the average kcat/KM and SD of three experiments.
Supplementary Material
Acknowledgments
We thank Paul Bessette and Claudia Gottstein for critically reading the manuscript. This project was supported, in part, by University of California Toxic Substances Research and Training Program Grant SB-030104 and National Science Foundation CAREER Award BES-0449399.
Abbreviations
- CLiPS
cellular libraries of peptide substrates
- EP
enteropeptidase
- SA-PE
streptavidin-conjugated phycoerythrin.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office. G.G. is a guest editor invited by the Editorial Board.
References
- 1.Duesbery N. S., Webb C. P., Leppla S. H., Gordon V. M., Klimpel K. R., Copeland T. D., Ahn N. G., Oskarsson M. K., Fukasawa K., Paull K. D., Vande Woude G. F. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- 2.Coussens L. M., Fingleton B., Matrisian L. M. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 3.Haass C., De Strooper B. Science. 1999;286:916–919. doi: 10.1126/science.286.5441.916. [DOI] [PubMed] [Google Scholar]
- 4.Bricteux-Gregoire S., Schyns R., Florkin M. Comp. Biochem. Physiol. B. 1972;42:23–39. doi: 10.1016/0305-0491(72)90055-7. [DOI] [PubMed] [Google Scholar]
- 5.Yaron A., Carmel A., Katchalski-Katzir E. Anal. Biochem. 1979;95:228–235. doi: 10.1016/0003-2697(79)90210-0. [DOI] [PubMed] [Google Scholar]
- 6.Deperthes D. Biol. Chem. 2002;383:1107–1112. doi: 10.1515/BC.2002.119. [DOI] [PubMed] [Google Scholar]
- 7.Harris J. L., Backes B. J., Leonetti F., Mahrus S., Ellman J. A., Craik C. S. Proc. Natl. Acad. Sci. USA. 2000;97:7754–7759. doi: 10.1073/pnas.140132697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marnett A. B., Craik C. S. Trends Biotechnol. 2005;23:59–64. doi: 10.1016/j.tibtech.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 9.Matthews D. J., Wells J. A. Science. 1993;260:1113–1117. doi: 10.1126/science.8493554. [DOI] [PubMed] [Google Scholar]
- 10.Cloutier S. M., Chagas J. R., Mach J. P., Gygi C. M., Leisinger H. J., Deperthes D. Eur. J. Biochem. 2002;269:2747–2754. doi: 10.1046/j.1432-1033.2002.02960.x. [DOI] [PubMed] [Google Scholar]
- 11.McCarter J. D., Stephens D., Shoemaker K., Rosenberg S., Kirsch J. F., Georgiou G. J. Bacteriol. 2004;186:5919–5925. doi: 10.1128/JB.186.17.5919-5925.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fodor S. P., Read J. L., Pirrung M. C., Stryer L., Lu A. T., Solas D. Science. 1991;251:767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- 13.Salisbury C. M., Maly D. J., Ellman J. A. J. Am. Chem. Soc. 2002;124:14868–14870. doi: 10.1021/ja027477q. [DOI] [PubMed] [Google Scholar]
- 14.Barrios A. M., Craik C. S. Bioorg. Med. Chem. Lett. 2002;12:3619–3623. doi: 10.1016/s0960-894x(02)00786-2. [DOI] [PubMed] [Google Scholar]
- 15.Leiting B., Pryor K. D., Wu J. K., Marsilio F., Patel R. A., Craik C. S., Ellman J. A., Cummings R. T., Thornberry N. A. Biochem. J. 2003;371:525–532. doi: 10.1042/BJ20021643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marnett A. B., Nomura A. M., Shimba N., Ortiz de Montellano P. R., Craik C. S. Proc. Natl. Acad. Sci. USA. 2004;101:6870–6875. doi: 10.1073/pnas.0401613101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang T., Olson E. S., Nguyen Q. T., Roy M., Jennings P. A., Tsien R. Y. Proc. Natl. Acad. Sci. USA. 2004;101:17867–17872. doi: 10.1073/pnas.0408191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rice J. J., Schohn A., Bessette P. H., Boulware K. T., Daugherty P. S. Protein Sci. 2006;15:825–836. doi: 10.1110/ps.051897806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bosley A. D., Ostermeier M. Biomol. Eng. 2005;22:57–61. doi: 10.1016/j.bioeng.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Barrett A. J., Rawlings N. D., Woessner J. F. Handbook of Proteolytic Enzymes. San Diego: Academic; 2004. [Google Scholar]
- 21.Lien S., Pastor R., Sutherlin D., Lowman H. B. Protein J. 2004;23:413–425. doi: 10.1023/b:jopc.0000039555.92058.51. [DOI] [PubMed] [Google Scholar]
- 22.Stennicke H. R., Renatus M., Meldal M., Salvesen G. S. Biochem. J. 2000;350:563–568. [PMC free article] [PubMed] [Google Scholar]
- 23.Likhareva V. V., Mikhailova A. G., Vaskovsky B. V., Garamin S. K., Rumsh L. D. Lett. Pept. Sci. 2002;9:71–76. [Google Scholar]
- 24.Nguyen A. W., Daugherty P. S. Nat. Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 25.Felber L. M., Cloutier S. M., Kundig C., Kishi T., Brossard V., Jichlinski P., Leisinger H. J., Deperthes D. Biotechniques. 2004;36:878–885. doi: 10.2144/04365PT04. [DOI] [PubMed] [Google Scholar]
- 26.Bessette P. H., Rice J. J., Daugherty P. S. Protein Eng. Des. Sel. 2004;17:731–739. doi: 10.1093/protein/gzh084. [DOI] [PubMed] [Google Scholar]
- 27.Hosfield T., Lu Q. Anal. Biochem. 1999;269:10–16. doi: 10.1006/abio.1998.3084. [DOI] [PubMed] [Google Scholar]
- 28.Light A., Fonseca P. J. Biol. Chem. 1984;259:13195–13198. [PubMed] [Google Scholar]
- 29.Light A., Savithri H. S., Liepnieks J. J. Anal. Biochem. 1980;106:199–206. doi: 10.1016/0003-2697(80)90138-4. [DOI] [PubMed] [Google Scholar]
- 30.Matsushima M., Ichinose M., Yahagi N., Tsukada-Kato S., Miki K., Omata M., Kim Y. T., Ito H., Takahashi T., Sakurai Y., et al. J. Biochem. (Tokyo) 1999;125:947–951. doi: 10.1093/oxfordjournals.jbchem.a022373. [DOI] [PubMed] [Google Scholar]
- 31.Hazebrouck S., Machtelinckx-Delmas V., Kupiec J. J., Sonigo P. Biochem. J. 2001;358:505–510. doi: 10.1042/0264-6021:3580505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coombs G. S., Bergstrom R. C., Madison E. L., Corey D. R. J. Biol. Chem. 1998;273:4323–4328. doi: 10.1074/jbc.273.8.4323. [DOI] [PubMed] [Google Scholar]
- 33.Sharma A., Khoury-Christianson A. M., White S. P., Dhanjal N. K., Huang W., Paulhiac C., Friedman E. J., Manjula B. N., Kumar R. Proc. Natl. Acad. Sci. USA. 1994;91:9337–9341. doi: 10.1073/pnas.91.20.9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholson D. W. Cell Death Differ. 1999;6:1028–1042. doi: 10.1038/sj.cdd.4400598. [DOI] [PubMed] [Google Scholar]
- 35.Cloutier S. M., Kundig C., Gygi C. M., Jichlinski P., Leisinger H. J., Deperthes D. Tumor Biol. 2004;25:24–30. doi: 10.1159/000077720. [DOI] [PubMed] [Google Scholar]
- 36.Boder E. T., Wittrup K. D. Nat. Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 37.Vlach J., Hennecke S., Amati B. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Casadaban M. J., Cohen S. N. J. Mol. Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 39.Lu D., Futterer K., Korolev S., Zheng X., Tan K., Waksman G., Sadler J. E. J. Mol. Biol. 1999;292:361–373. doi: 10.1006/jmbi.1999.3089. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.