Abstract
The crystal structure of the complete signaling complex formed between bone morphogenetic protein 2 (BMP-2) and the extracellular domains (ECDs) of its type I receptor [bone morphogenetic protein receptor type Ia (BMPR-Ia)-ECD] and its type II receptor [activin receptor type II (ActRII)-ECD] shows two fundamental structural constraints for receptor assembly. First, the homodimeric BMP-2 ligand assembles two pairs of each receptor symmetrically, where each of the receptor ECDs does not make physical contact. Therefore, conformational communication between receptor ECDs, if any, should be propagated through the central ligand. Second, the type I and II receptor interfaces of the complex, when compared with those of binary complexes such as BMP-2/BMPR Ia–ECD, BMP-7/ActRII-ECD, and activin/ActRIIb-ECD, respectively, show there are common sets of positions repeatedly used by both ligands and receptors. Therefore, specificity-determining amino acid differences at the receptor interfaces should also account for the disparity in affinity of individual receptors for different ligand subunits. We find that a specific mutation to BMP-2 increases its affinity to ActRII-ECD by 5-fold. These results together establish that the specific signaling output is largely determined by two variables, the ligand–receptor pair identity and the mode of cooperative assembly of relevant receptors governed by the ligand flexibility in a membrane-restricted manner.
Keywords: activin receptor, receptor assembly
The TGF-β superfamily encompasses extracellular ligands involved in a diverse range of biological functions. Family members include TGF-β, bone morphogenetic proteins (BMPs), and growth and differentiation factors, along with activins and inhibins. Found in almost all cell types, these proteins are involved in numerous cell processes, including bone and joint development, cell proliferation and differentiation, and dorsal/ventral patterning (1). Because of their ubiquitous nature, TGF-β proteins are associated with a variety of diseases ranging from skeletal abnormalities and differentiation to metabolic (2) disorders and play critical roles in neoplastic development and stem cell differentiation (3).
TGF-β ligands are synthesized as inactive precursor molecules composed of an N-terminal prodomain and a C-terminal mature domain. To become active, the mature domain must be cleaved by a serine endoprotease, such as furin in the case of BMP-4 and TGF-β1 (4, 5). The overall architecture of the mature domain is conserved as a covalently disulfide-linked dimer throughout the superfamily. Each ligand subunit contains three intradisulfide bonds, forming the characteristic “cystine knot” motif (6). Some members of the TGF-β superfamily are known to form both homo- and heterodimers in vivo, with activin and inhibin being the best-studied prototype examples (7, 8).
To initiate its intracellular signaling cascade, the ligand recruits two sets of receptors, named type I and type II. The receptors are structurally conserved, each comprising an extracellular domain (ECD), a single transmembrane domain, and a large intracellular kinase domain. The ligand first binds two copies of its high-affinity receptor. The high-affinity receptor is generally the type II receptor, but for BMP-2, it is the type I receptor [BMP type Ia (BMPR-Ia), also known as ALK3]. After the binding of the high-affinity receptor, the lower-affinity receptor is then able to bind. How this sequential binding is achieved, and how it affects the overall signaling output, remain important structural questions. Each ligand subunit contributes to recruiting one of each type of receptor to form the complex. Once the complex is complete, a six-polypeptide chain complex (two subunits of the ligand and two pairs of each receptor type) is formed, and the constitutively active type II receptor is able to phosphorylate the type I receptor. In turn, the type I receptors phosphorylate SMAD proteins, initiating a signaling cascade to target DNA in the nucleus (9–12).
To date, nearly 40 TGF-β family members have been isolated in the human genome, with which five type II and seven type I receptors interact (13). To compensate for the disparity in the numbers between receptors and ligands, the receptors have the ability to bind multiple ligands. Activin receptor type II (ActRII), for example, is able to bind BMP-2 as a lower-affinity receptor but binds BMP-7 and activin with high affinity (14). Affinity is a measure of the energetic stability of the protein assembly, whereas the specificity reflects the relative affinity. Several studies identified critical residues on ActRII (15), activin (16), and BMP-2 (17) that disrupt the assembly of the ligand–receptor complex. Conversely, a single-residue change can increase the binding of GDF-5 to BMPR-Ia by >10-fold (18), or three residues can alter the specificity among TGF-β ligands to TGF-β receptor II (19). However, the structural basis for the overlapping specificities of many receptors at the atomic level could not be fully addressed because of the absence of the ternary six-chain complex structure.
Recently, several binary structures of the ligand in complex with its high-affinity receptor ECD have been determined: BMP-2/BMPR-Ia-ECD (20), TGF-β3/TGF-βRII-ECD (21), BMP-7/ActRII-ECD (22), and activin/ActRIIb-ECD (23, 24). Based on these structures, we have proposed the “wing-spread” hypothesis, in which the receptor ECDs do not contact one another, and the increased affinity of the lower-affinity receptor for the ligand bound to high-affinity receptors is largely governed by the 2D constraints of the receptors embedded in the membrane (23). Combined with the inherent ligand flexibility, this wing-spread hypothesis explains the sequential binding of both receptor types without molecular contacts between receptor ECDs. However, without knowledge of the ternary structure, we cannot rule out the possibility that conformational changes in the ligand can increase its affinity for the lower-affinity receptors.
Here we present the previously uncharacterized structure of the ternary complex representing the signaling competent complex of BMP-2 bound to the entire ECDs of both its type I receptor, BMPR-Ia, and its type II receptor, ActRII, refined to a resolution of 2.2 Å. Using the structural information of this BMP-2/BMPR-Ia-ECD/ActRII-ECD complex, we characterize residues that modulate receptor–ligand affinity and illustrate how binding specificities are encoded at each receptor-binding interface independently, and how the mode of receptor recruitment in the membrane can contribute to defining the binding affinity of both types receptors.
Results and Discussion
Structure of the Ternary Complex of BMP2/BMPR-Ia-ECD/ActRII-ECD.
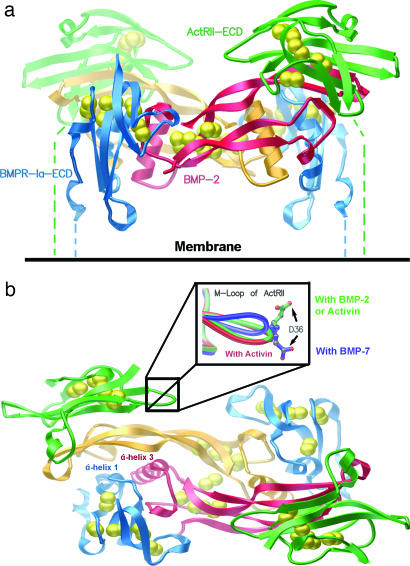
The structure of BMP-2/BMPR-Ia-ECD/ActRII-ECD represents the signaling-competent TGF-β ternary complex (Fig. 1). The asymmetric unit contains one pair of the complex halves: one BMP-2 monomer bound to one BMPR-Ia-ECD and one ActRII-ECD. Consequently, the two molecules are related by a twofold noncrystallographic symmetry axis. For BMP-2, residues 12–114 and 11–114 (of 1–114) were seen in the electron density maps for monomers A and D, respectively. For the two BMPR-Ia-ECD molecules, residues 34–118 and 33–120 (of 1–129) were refined as molecules B and E, whereas residues 8–99 and 7–99 (of 1–102) were resolved for ActRII-ECD molecules C and F. The ternary structure includes electron density for the L3 loop (residues 66–70) of BMPR-Ia, where there was no electron density in the binary structure of BMP-2/BMPR-Ia-ECD (20).
Fig. 1.
Ternary complex of BMP-2/BMPR-Ia-ECD/ActRII-ECD shows noncontacting assembly of receptors. (a) Ternary complex of BMP-2/BMPR-Ia-ECD/ActRII-ECD as seen in the membrane. BMP-2 dimer subunits are shown in red and orange, displaying the butterfly conformation. The two BMPR-Ia-ECDs are shown in blue, and the ActRII-ECDs are shown in green. Dashed lines represent C-terminal residues connecting to the membrane-spanning segments of the receptors. Cystines are shown as yellow spheres. (b) A top view of the complex. The color scheme is the same as in a. Labeled are α-helix 3 of BMP-2 and α-helix 1 of BMPR-Ia. (Inset) The M loop of the ActRII from BMP-2/ActRII-ECD/BMPR-Ia-ECD (green), BMP-7/ActRII-ECD (blue), and activin/ActRIIb-ECD (red) structures.
The overall architecture of the ternary complex is similar to the composite model of the ternary complex (22) generated by combining the crystal structures of BMP-7/ActRII-ECD and BMP-2/BMPR-Ia-ECD. Within each subunit of the BMP-2 dimer, the four β-strands form two antiparallel β-sheets (Fingers 1 and 2) extending outward from the dimer core like wings, in a curved fashion, giving the dimer a butterfly-like shape (Fig. 1a). Bound to the outside concave face of Fingers 1 and 2 is one ActRII-ECD molecule adopting the prototypical three-finger toxin fold (25) composed of six β-strands forming three fingers. As with the BMP-2, the three curved fingers in the ActRII create a complementary binding surface to the BMP-2 fingers (Fig. 1b). Unlike the ActRII interface, however, one BMPR-Ia interface is generated by both BMP-2 monomers. The majority of the interface is created by the α-helix 1 of BMPR-Ia (Fig. 1b, blue), packed against the prehelix loop and α-helix 3 of BMP-2 (Fig. 1b, red). The remainder of the interface is composed of mostly hydrophobic contacts between the concave face of the other subunit of BMP-2 (Fig. 1b, orange) with residues near the C-terminal end of α-helix 1 on BMPR-Ia (Fig. 1b, blue). In the current BMP-2/BMPR-Ia-ECD/ActRII-ECD structure, the C termini between ActRII-ECD and BMPR-Ia-ECDs are ≈35 Å apart. However, because of missing residues in the structure, e.g., ≈30 N-terminal residues of BMPR-Ia-ECD and ≈10 C-terminal residues connected to the transmembrane segment of the receptor, we cannot completely eliminate the possibility of additional interactions in the ternary complex of the full-length receptor’s ECDs.
BMP-2/ActRII Interface.
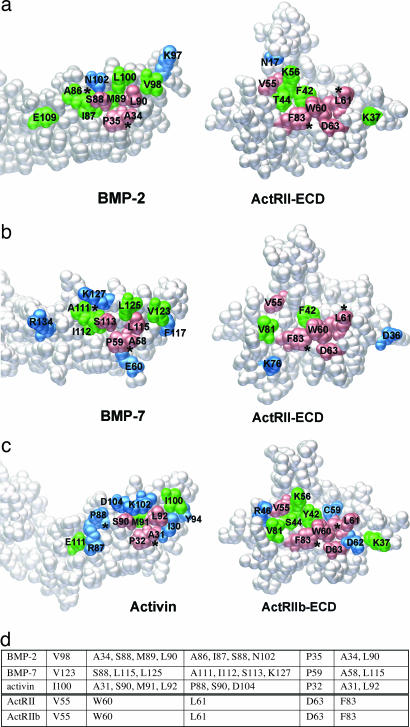
The BMP-2/ActRII-ECD interface represents the unprecedented structural view of a TGF-β ligand bound to its lower-affinity receptor (Fig. 2). This binding orientation of the concave face of one ActRII binding to the convex “knuckle” region of one BMP-2 monomer is consistent with what has previously been seen in both BMP-7/ActRII-ECD (22) and activin/ActRIIb-ECD (23, 24). The 670-Å2 surface buried upon binding is comparable to the other type II interfaces, 660 Å2 for BMP-7/ActRII-ECD and 774 Å2 for activin/ActRIIb-ECD. The interface is mostly hydrophobic in nature and is composed of 12 residues from BMP-2 and 10 residues from ActRII based on the criteria of ≤4 Å for any interatomic distance between two chains (Fig. 2a). Previous studies have shown three residues, Phe-42, Trp-60, and Phe-83, on ActRII to be important for binding ligands (15). These residues form a hydrophobic core at the BMP-2/ActRII-ECD interface, contacting Ala-34, Pro-35, Ser-88, Met-89, and Leu-90 from BMP-2. These five amino acids forming the hydrophobic core are identical in all three ligands, except that Val in BMP-7 (equivalent to Met-89) lies outside the interface (Fig. 2b). Given that this interface forms either the high- or lower-affinity interface with affinities varied by ≈50-fold, we conclude that specific nonconserved residues outside the hydrophobic core must contribute to defining the ligand specificity (Fig. 2 a–c, blue).
Fig. 2.
Ligand-binding interfaces of type II receptors show a nearly common set of interface positions. Peeled-away interfaces of BMP-2/ActRII-ECD (a), BMP-7/ActRII-ECD (b), and activin/ActRIIb-ECD (c) pairs shown as space-filling model. Orientation is as in Fig. 1a. Colored are those in contact between the two chains, for identical (pink), highly conserved (green), and nonconserved (blue) residues. Asterisks denote reference contact points between the two molecules. (d) List of the five identical residues in ActRII and ActRIIb and their contacting amino acids on three ligands.
To analyze any conformational changes occurring within the ternary structure, the ActRII-ECD of the ternary complex were compared with those of BMP-7/ActRII-ECD (Fig. 2b) and activin/ActRII-ECD (Fig. 2c). Cα rms differences are small, yielding an average of 0.678 and 0.584 Å, respectively. However, the M loop (residues 33–39), conserved only among activin-binding receptors (22), shows significant movement between the complexes with BMP-7 and -2. For the M loop of ActRII, the average Cα rms deviation was 2.878 Å, with a maximum deviation of 5.65 Å for Asp-34 (Fig. 1b Inset). The reason for this shift in the M loop may be the opposite charge occupying the structurally equivalent position on the ligand. In the BMP-2/ActRII interface, a polar contact is formed by the side chain of Glu-109 in BMP-2 and the side chain of Lys-37 in ActRII. In contrast, at the BMP-7/ActRII-ECD interface, the contact is formed by the side chain of Arg-134 of BMP-7 (equivalent to Glu-109 in BMP-2) and the side chain of Asp-36 of ActRII. To make this charge pair interaction, the backbone of the M loop undergoes a conformational change, becoming further exposed toward the solvent, which in turn shifts the side chain of Asp-36 into the BMP-7/ActRII interface (Fig. 1b Inset, green to blue). In activin, Glu-111 (equivalent to Glu-109 in BMP-2) interacts with Lys-37 of ActRIIb, resulting in the M loop being in the same position as in the BMP-2/ActRII-ECD interface (Fig. 1b Inset, red). Although the conformation of the M loop may play a role in modulating the receptor–ligand affinity, there must be additional interactions besides the M loop conformation to define ActRII as the high- or lower-affinity receptor interfaces for activin or BMP-2, respectively.
BMP-2/BMPR-Ia Interface.
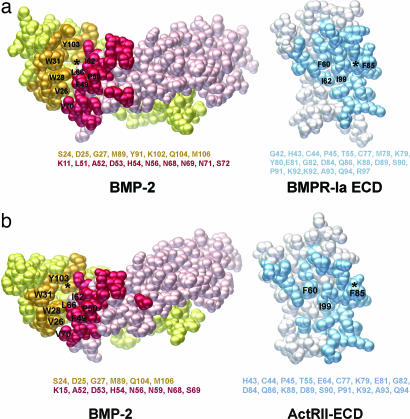
The BMP2/BMPR-Ia-ECD interface of the ternary complex consists of one BMPR-Ia molecule contacting both BMP-2 monomers (Fig. 3). The buried surface area is 1,217 Å2, formed by 25 residues from BMPR-Ia (Fig. 3a, blue) and 12 residues from one BMP-2 subunit (Fig. 3a, orange) and 16 residues from the other BMP-2 subunit (Fig. 3a, red). This interface corresponds closely with that of the binary complex of BMP-2/BMPR-Ia-ECD (Fig. 3b). When the BMP-2/BMPR-Ia-ECD complex is aligned with the ternary complex, the average Cα rms deviation is 0.784 Å, with up to a 1.6-Å deviation seen in the prehelix loop region (residues 52–56). In the alignment, this deviation is largely offset by a corresponding shift in the BMPR-Ia molecule to maintain proper receptor–ligand contacts and orientation. Compared with free and BMPR-Ia-ECD-bound BMP-2, the ternary complex illustrates that BMP-2 is a rigid ligand and does not undergo significant conformational change upon receptor binding. This is in contrast to a flexible ligand, such as activin, which has been crystallized bound to ActRIIb-ECD in several conformations, none of which display the intact type I receptor interface (23, 24).
Fig. 3.
Ligand-binding interfaces of type I receptors are formed at the junction between two ligand monomers. (a) Peeled-away interfaces of BMP-2/BMPR-Ia-ECD of the ternary complex. Residues from two BMP-2 subunits (red and orange) and those from BMPR-Ia-ECD (blue) are shown in the standard orientation (Fig. 1a). (b) Peeled-away interfaces of BMP-2/BMPR-Ia-ECD of the binary complex (20). Only those amino acids that were discussed for two pockets are labeled. All other residues are listed below.
The majority of the interface residues are hydrophobic in nature and form two main pockets. The first pocket is formed by residues Phe-49 and Pro-50 in the prehelix loop of BMP-2 (Fig. 3a, red) interacting with Ile-62, Phe-60, and Ile-99 of BMPR-Ia (Fig. 3a, blue). The second pocket is formed by α-helix 3 of one BMP-2 (Ile-62, Leu-66, and Val-70) subunit (Fig. 3a, red), α-helix 1 of BMPR-Ia (Fig. 3a, blue), and the convex face of the second BMP2 (Val-26, Trp-28, Trp-31, and Tyr-103) subunit (Fig. 3a, orange). The residues form a large cavity 18.6 Å long and 8.5 Å wide in which Phe-85 of BMPR-Ia fits (Fig. 3a, blue). This structural arrangement was previously described as a “knob-into-hole” binding motif as a key feature of the all type I binding sites (20). Like the type II receptor interface, the ternary structure demonstrates no allosteric conformational changes occurring at the type I binding site as a result of type II receptor ECD binding.
Enhanced Affinity of BMP2 for ActRII.
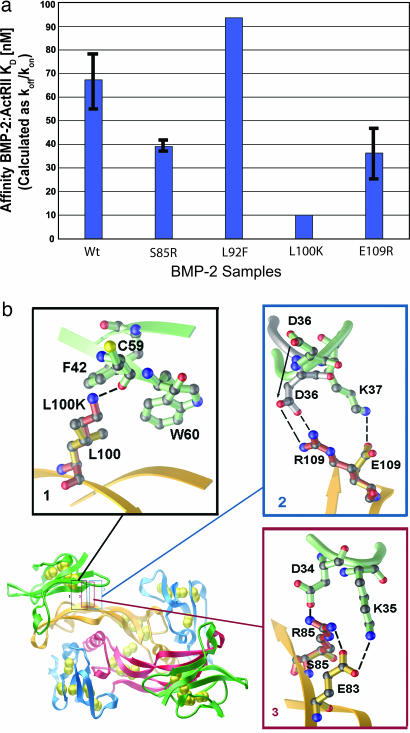
One unique characteristic of TGF-β receptors is their ability to bind multiple TGF-β ligands with varying affinity. Previously, we have reported that ActRII-ECD (or ActRIIb-ECD) binds both BMP7 and activin with high affinity, ≈1 nM (22, 23). To dissect how BMP-2 and -7 share ActRII as their respective lower- and high-affinity receptors (Fig. 2 a and b), we attempted to select several ligand-specific amino acids of BMP-2 to change the affinity to be like BMP-7 or activin. First, the ligand side of BMP-7/ActRII-ECD was aligned to the BMP-2 of the ternary complex. Using the alignments, two mutations, L92F and E109R (all BMP-2 numbering), were individually made to mimic the BMP-7/ActRII interface. Additionally, two mutations, S85R and L100K, were made to mimic the activin/ActRIIb interface after alignment with activin/ActRIIb-ECD. Leu-92 lies at one end of the hydrophobic core necessary for type II receptor binding, whereas Ser-85 and Glu-109 fall on the periphery of the interface (Fig. 2). Lys-102 in activin (Leu-100 in BMP-2) has been shown to be critical for its binding to ActRII (16).
Using surface plasmon resonance (Biacore) with ActRII-ECD immobilized on the chip surface, we find BMP-2wt affinity ranged from 50 to 80 nM (Fig. 4a), similar to the previously reported affinity of 38–50 nM (17, 22). BMP-2L100K showed the tightest binding, with a 5-fold increase in affinity compared with the wild type, whereas mutants BMP2S85R and BMP2E109R showed a smaller 1.5- to 2-fold increase (Fig. 4a). BMP2L92F did not exhibit any significant change in the ActRII affinity. When BMPR-Ia-ECD was immobilized to the Biacore chip surface, all samples displayed the same affinity as wild type, indicating that the mutations affected only the type II receptor interface locally. It is currently unknown whether the increased binding of the BMP-2 mutants results in increased signaling output.
Fig. 4.
Receptor-binding specificity can be altered by local changes at its interface. (a) Biacore affinity data of BMP-2 mutants to ActRII-ECD. Affinities are shown in nanomolar concentrations calculated as koff/kon. (b) Closeups of the three BMP-2 mutants showing increased affinity for ActRII-ECD. Residues are shown in ball-and-stick representation for BMP-2 (orange) and ActRII-ECD (green). Shown in red is the modeled conformation of the mutant sidechains of BMP2S85R (1), BMP2L100K (2), and BMP2E109R (3). In box 2, the M loop based on BMP-7/ActRII-ECD structure is shown in gray. H bonds and charge pairs are shown as dashes (nitrogen, blue; oxygen, red; and sulfur, yellow).
Detailed stereochemistry of the interface shows that the changes in affinities of these mutants (L100K, E109R, and S85R) can be explained by local energetic gain. Leu-100 of BMP-2, located at the other end (opposite from Leu-92) of the hydrophobic core, is particularly interesting, because Lys-102 of activin (equivalent to Leu-100 in BMP2) forms a H bond with the backbone carbonyl of Cys-59 in ActRIIb (23). When Lys is modeled to replace Leu-100 of BMP-2, the same contact can be formed, using a geometrically favored rotamer, to Cys-59 of ActRII leaving a H bond distance of 2.8 Å (Fig. 4b, box 1). The Lys maintains most of the hydrophobic characteristics of the Leu (the same number of side-chain carbons) but gains an additional H bond. This predicted gain of binding energy can account for the increase in affinity.
The pronounced conformational difference of the M loop of the ActRII noted between the interfaces of BMP-7/ActRII-ECD and BMP-2/ActRII-ECD places the side-chain Lys-37 of ActRII charge-paired to Glu-109 in BMP-2 (Fig. 4b, box 2). Switching the charge from negative to positive, the BMP2E109R with the aforementioned conformational change of the M loop (Fig. 1b) now allows the Arg to form two H bonds with Asp-36 of ActRII, whereas the Glu-109 in BMP2wt could form only one H bond with Lys-37 of ActRII (Fig. 4b, box 2). Finally, Arg-87 of activin (Ser-85 in BMP-2) makes a charge pair to Asp-62 on Finger 2 of ActRIIb near the M loop along with several water-mediated contacts. Arg modeled in the BMP2S85R makes a strong H-bond network with Asp-34 and Lys-35 of ActRII as well as Glu-83 of BMP-2 (Fig. 4b, box 3). Although these are not the same contacts as in the activin/ActRIIb-ECD interface, the additional contacts would be expected to have the same effect by gaining favorable charge interaction. Compared with the wild type, the increased affinities of the mutants, BMP2L100K, BMP2E109R, and BMP2S85R, are all due to the decreased dissociation rates.
It is worth noting that our measurements of binding affinity most likely underestimate the actual effects of affinity changes, because the measurement was made between ligands and isolated receptor ECDs. Receptors are membrane-bound, and their orientations are two-dimensionally restricted. Because two pairs of type I and type II receptors will be locked on the ligand dimer for signaling (26), the overall binding affinity of the second high-affinity receptor as well as two lower-affinity receptors will be affected not only by the ligand affinity itself but also by the ligand flexibility between two monomers. Because the type I interface is formed at the junction between the monomers of a ligand, ligand flexibility is also an important variable in creating the type I binding interface (22, 23). To determine how the two factors, affinity and flexibility, contribute to the ultimate signaling output, it will be important to measure how the affinity-enhancing mutations (L100K, E109R, and S85R of BMP-2 or their reverse mutations introduced to a flexible ligand like activin) affect the signaling output.
Given that the structure of the ternary complex reveals no significant allosteric changes to account for the affinity changes at the receptor interfaces, independent receptor binding to each subunit of a ligand can be particularly important to understand the signaling mechanism by heterodimeric ligands such as BMP-2/BMP-7, because each subunit encodes different receptor specificity. For instance, the BMP-2/BMP-7 heterodimer indeed exhibits greater activity than the respective homodimers (27). More recently, the heterodimer of Dpp/Scw, BMP homologs in Drosophila, was discovered to be responsible in determining dorsal tissue types (28), whereas the heterodimer of BMP-7/GDF-7 is the acting component that influences the direction of commissural axons (29). Our experimental results now establish the ternary structure and thus the wing-spread hypothesis as the firm ground to address experimentally how the specific binding affinities encoded locally at individual receptor interfaces can coordinate the wide range of TGF-β responses under the common structural framework.
Materials and Methods
Protein Expression, Purification, and Crystallization.
The ECD of BMPR-Ia, residues 1–129, was expressed in Escherichia coli as a thioredoxin fusion protein and purified based on previously published strategies (30). The ECD of ActRII, residues 1–102, was expressed in Pichia pastoris as described (31). The mature wild-type BMP-2 was expressed in E. coli, purified, and refolded from inclusion bodies using a modified protocol (32).
All proteins were purified to homogeneity by reverse-phase chromatography. The samples were lyophilized and resuspended in 10 mM Na acetate, pH 4. The BMP-2 was then diluted 1:1 with 4× high salt/Tris/3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) buffer (4× 200 mM Tris, pH 7.9/2.8 M NaCl/7.2% CHAPS (HSTC)]. The binary complex of BMP-2 and BMPR-Ia-ECD was first formed by using a 1:2 molar ratio in 1× HSTC. This complex was purified over a Superdex 75 (Amersham Pharmacia) column and then concentrated by using a Vivaspin 6 concentrator with a 5-kDa cutoff (Sartorius). The ternary complex was generated by adding 2.2 molar equivalents of ActRII-ECD to the purified binary complex. The final protein concentration was adjusted to 10 mg/ml. BMP-2 mutants were generated by QuikChange (Invitrogen), expressed, and purified the same as the wild type.
Data Collection, Phasing, and Refinement.
The complex was crystallized by the hanging-drop vapor diffusion method in 4 M Na Formate/0.1 M Hepes, pH 7.5/3% dioxane. After 1 week at 4°C, hexagonal crystals grew to an average size of 100 × 100 × 40 μm in the space group P6522 with a = b = 104.0 Å and c = 362.5 Å. The BMP-2/BMPR-Ia-ECD/ActRII-ECD crystals were soaked in mother liquor with 15% xylitol (used as cyroprotectant) for <5 min before being flash-frozen in liquid nitrogen. Diffraction data were collected from two separate crystals, one collected at the Advanced Light Source on beamline 5-1, and the other collected at the Stanford Synchrotron Radiation Laboratory on beamline 9-2. Both data sets were independently scaled and integrated by using hkl 2000 (33). Molecular replacement was performed by using phaser (34) to solve the initial phases for the data from crystal one. The best solution was found by searching with two independent halves (one BMP-2 monomer with one BMPR-Ia-ECD) of the BMP-2/BMPR-Ia-ECD dimer (20). The model was refined by using refmac5 (34) interspersed with rounds of manual building in o (35). The model was refined to 2.5 Å by using data from crystal one and then further extended to 2.2 Å by using data from crystal two (Table 1). The final structure yielded an R factor of 22.4%, with a free R factor of 25.9%. The overall geometry of the structure was good as determined by procheck (36), with 84.8% of the residues in the most favorable regions and none in the disallowed regions, as evaluated by Ramachandran plot.
Table 1.
Data collection and refinement statistics
| Crystal 1 | Crystal 2 | |
|---|---|---|
| Beamline | ALS 5-1 | SSRL 9-2 |
| Number of reflections | 332,362 | 454,504 |
| Unique reflections | 39,363 | 62,054 |
| Resolution, Å | 2.50 | 2.15 |
| Final resolution shell, Å | 2.59–2.50 | 2.23–2.15 |
| Average I/σI | 24.8 (3.2) | 18.8 (4.7) |
| Rsym, % | 6.8 (40.4) | 8.5 (40.2) |
| Completeness, % | 94.7 (96.2) | 96.4 (89.4) |
| Rcryst, % | 19.9 | 22.4 |
| Rfree, % | 24.1 | 25.9 |
| rms deviation | ||
| Bonds, Å | 0.013 | 0.018 |
| Angles, ° | 1.44 | 1.66 |
Surface Plasmon Resonance (Biacore) Affinity Studies.
The affinity of the BMP-2 mutants to ActRII-ECD was monitored by using a Biacore 3000 machine, and the data were analyzed by using biaevalution software Ver. 4.1 (Biacore). Using primary amine coupling, ActRII-ECD was immobilized on a CM5 chip in flow cell 3. Flow cell 1 was left blank as a negative control, whereas BMPR-Ia-ECD was immobilized to flow cell 4 as a positive control. They were immobilized for 5 min at a flow rate of 10 μl per min and at a concentration of 50 μg/ml in 10 mM Na acetate, pH 4.0. The experiments were performed at 40 μl per min in 20 mM Tris·HCl, pH 7.9/200 mM NaCl/0.36% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate/0.005% P20. A minimum of four concentrations, plus a zero concentration, were run per sample for kinetic analysis, and the data were fit by using a 1:1 Langmuir binding with no bulk refractive shift.
Acknowledgments
We thank Kent Baker for assisting in the x-ray data analysis, Jason Greenwald and Ezra Wiater for numerous discussions, the staff at Advanced Light Source and Stanford Synchrotron Radiation Laboratory for x-ray data collection, Elizabeth Komives for Biacore data collection, and the Chapman Foundation Fellowship (G.P.A.) and National Institutes of Health (W.W.V. and S.C.) for support.
Abbreviations
- BMP
bone morphogenetic protein
- ActRII
activin receptor type II
- BMPR-Ia
BMP receptor type Ia
- ECD
extracellular domain.
Footnotes
Conflict of interest statement: W.W.V. is a cofounder, consultant, equity holder, and member of the Board of Directors of Acceleron Pharma, to which ActRII has been licensed by the Salk Institute. S.C. is a member of the scientific advisory board for Acceleron Pharma.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2GOO).
References
- 1.Massague J., Blain S. W., Lo R. S. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 2.Chen C., Grzegorzewski K. J, Barash S., Zhao Q., Schneider H., Wang Q., Singh M., Pukac L., Bell A. C., Duan R., et al. Nat. Biotechnol. 2003;21:294–301. doi: 10.1038/nbt795. [DOI] [PubMed] [Google Scholar]
- 3.Morrison S. J., Shah N. M., Anderson D. J. Cell. 1997;88:287–298. doi: 10.1016/s0092-8674(00)81867-x. [DOI] [PubMed] [Google Scholar]
- 4.Cui Y., Jean F., Thomas G., Christian J. L. EMBO J. 1998;17:4735–4743. doi: 10.1093/emboj/17.16.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubois C. M., Laprise M. H., Blanchette F., Gentry L. E., Leduc R. J. Biol. Chem. 1995;270:10618–10624. doi: 10.1074/jbc.270.18.10618. [DOI] [PubMed] [Google Scholar]
- 6.McDonald N. Q., Hendrickson W. A. Cell. 1993;73:421–424. doi: 10.1016/0092-8674(93)90127-c. [DOI] [PubMed] [Google Scholar]
- 7.Vitt U. A., Hsu S. Y., Hsueh A. J. Mol. Endocrinol. 2001;15:681–694. doi: 10.1210/mend.15.5.0639. [DOI] [PubMed] [Google Scholar]
- 8.Mathews L. S. Endocr. Rev. 1994;15:310–325. doi: 10.1210/edrv-15-3-310. [DOI] [PubMed] [Google Scholar]
- 9.Attisano L., Wrana J. L. Cytokine Growth Factor Rev. 1996;7:327–339. doi: 10.1016/s1359-6101(96)00042-1. [DOI] [PubMed] [Google Scholar]
- 10.Massague J. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 11.Shi Y., Massague J. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 12.Wrana J. L., Attisano L., Wieser R., Ventura F., Massague J. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 13.Manning G., Whyte D. B., Martinex R., Hunter T., Sudarsanam S. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 14.Feng X. H., Derynck R. Annu. Rev. Cell Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 15.Gray P. C., Greenwald J., Blount A. L., Kunitake K. S., Donaldson C. J., Choe S., Vale W. J. Biol. Chem. 2000;275:3206–3212. doi: 10.1074/jbc.275.5.3206. [DOI] [PubMed] [Google Scholar]
- 16.Wuytens G., Verschueren K., de Winter J. P., Gaijendran N., Beek L., Devos K., Bosman F., de Waele R., Andries M., van den Eijnden-van Raaij A. J., et al. J. Biol. Chem. 1999;274:9821–9827. doi: 10.1074/jbc.274.14.9821. [DOI] [PubMed] [Google Scholar]
- 17.Kirsch T., Nickel J., Sebald W. EMBO J. 2000;19:3314–3324. doi: 10.1093/emboj/19.13.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nickel J., Kotzsch A., Sebald W., Mueller T. D. J. Mol. Biol. 2005;349:933–947. doi: 10.1016/j.jmb.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 19.De Crescenzo G., Hinck C. S., Shu Z., Zuniga J., Yang J., Tang T., Baardsness J., Mendoza V., Sun L., Lopez-Casillas F., et al. J. Mol. Biol. 2006;355:47–62. doi: 10.1016/j.jmb.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 20.Kirsch T., Sebald W., Dreyer M. K. Nat. Struct. Biol. 2000;7:492–496. doi: 10.1038/75903. [DOI] [PubMed] [Google Scholar]
- 21.Hart P. J., Deep S., Taylor A. B., Shu Z., Hinck C. S, Hinck A. P. Nat. Struct. Biol. 2002;9:203–208. doi: 10.1038/nsb766. [DOI] [PubMed] [Google Scholar]
- 22.Greenwald J., Groppe J., Gray P., Wiater E., Kwiatowski W., Vale W., Choe S. Mol. Cell. 2003;11:605–617. doi: 10.1016/s1097-2765(03)00094-7. [DOI] [PubMed] [Google Scholar]
- 23.Greenwald J., Vega M. E., Allendorph G. P., Fischer W. H., Vale W., Choe S. Mol. Cell. 2004;15:485–489. doi: 10.1016/j.molcel.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 24.Thompson T. B., Woodruff T. K., Jardestzky T. S. EMBO J. 2003;22:1555–1566. doi: 10.1093/emboj/cdg156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenwald J., Fischer W. H., Vale W. W., Choe S. Nat. Struct. Biol. 1999;6:18–22. doi: 10.1038/4887. [DOI] [PubMed] [Google Scholar]
- 26.Knaus P., Sebald W. Biol. Chem. 2001;382:1189–1195. doi: 10.1515/BC.2001.149. [DOI] [PubMed] [Google Scholar]
- 27.Israel D. L., Nove J., Kerns K. M., Kaufman R. J., Rosen V., Cox K. A., Wozney J. M. Growth Factors. 1996;13:291–300. doi: 10.3109/08977199609003229. [DOI] [PubMed] [Google Scholar]
- 28.Shimmi O., Umulis D., Othmer H., O’Connor M. B. Cell. 2005;120:873–886. doi: 10.1016/j.cell.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butler S. J., Dodd J. Neuron. 2003;38:389–401. doi: 10.1016/s0896-6273(03)00254-x. [DOI] [PubMed] [Google Scholar]
- 30.Kirsch T., Nickel J., Sebald W. FEBS Lett. 2000;468:215–219. doi: 10.1016/s0014-5793(00)01214-x. [DOI] [PubMed] [Google Scholar]
- 31.Greenwald J., Le V., Corrigan A., Fischer W., Komives E., Vale W., Choe S. Biochemistry. 1998;37:16711–16718. doi: 10.1021/bi981939o. [DOI] [PubMed] [Google Scholar]
- 32.Groppe J., Rumpel K., Economides A. N., Stahl N., Sebald W., Affolter M. J. Biol. Chem. 1998;273:29052–29065. doi: 10.1074/jbc.273.44.29052. [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Collaborative Computational Project, Number 4. Acta Crystallogr. D. 1994;50:760–776. [Google Scholar]
- 35.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Acta Crystallogr. A. 1991;47:100–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 36.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M.. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]