Abstract
The tyrosine kinase inhibitors gefitinib (Iressa) and erlotinib (Tarceva) have shown anti-tumor activity in the treatment of non-small cell lung cancer (NSCLC). Dramatic and durable responses have occurred in NSCLC tumors with mutations in the tyrosine kinase domain of the epidermal growth factor receptor (EGFR). In contrast, these inhibitors have shown limited efficacy in glioblastoma, where a distinct EGFR mutation, the variant III (vIII) in-frame deletion of exons 2–7, is commonly found. In this study, we determined that EGFRvIII mutation was present in 5% (3/56) of analyzed human lung squamous cell carcinoma (SCC) but was not present in human lung adenocarcinoma (0/123). We analyzed the role of the EGFRvIII mutation in lung tumorigenesis and its response to tyrosine kinase inhibition. Tissue-specific expression of EGFRvIII in the murine lung led to the development of NSCLC. Most importantly, these lung tumors depend on EGFRvIII expression for maintenance. Treatment with an irreversible EGFR inhibitor, HKI-272, dramatically reduced the size of these EGFRvIII-driven murine tumors in 1 week. Similarly, Ba/F3 cells transformed with the EGFRvIII mutant were relatively resistant to gefitinib and erlotinib in vitro but proved sensitive to HKI-272. These findings suggest a therapeutic strategy for cancers harboring the EGFRvIII mutation.
Keywords: irreversible inhibitor, lung cancer, lung squamous cell carcinoma
Epidermal growth factor receptor (EGFR) is commonly overexpressed and mutated in many human malignancies and is often associated with aggressive phenotypes (1–3). Before the recent discovery of the somatic mutations in the EGFR kinase domain (4–8) in non-small cell lung cancers (NSCLC), deletions of the extracellular domain were considered the most frequent EGFR mutations in the different tumor types (9–13). These deletions have an activating effect on the receptor, giving cells expressing these truncated receptors a proliferative advantage. The most common truncated receptor is the variant III EGFR deletion mutant (EGFRvIII, delta 801EGFR, del2-7 EGFR), containing an in-frame deletion of exons 2–7 (801 bp) from the extracellular domain, initially characterized at the genomic level in glioblastoma.
Studies using immunohistochemical assays with EGFRvIII mutant-specific antibodies suggest that this mutation is present in multiple other tumor types, including NSCLC (10, 11, 14). However, because of EGFR's large and complex genomic structure (28 exons spanning ≈190 kb) and its large intron 1 (123 kb) where genomic deletions frequently occur, it has been difficult to assess and verify the existence of the EGFRvIII mutations at the genomic level. EGFRvIII mutations have been well demonstrated in glioblastoma, where they are present in >50% of glioblastomas with amplification of EGFR gene locus (12, 15, 16), but no genomic evidence for the existence of EGFRvIII mutations has been reported in NSCLC. Furthermore, the role of EGFRvIII mutation in the potential pathogenesis of NSCLC is unclear.
Here, we report that the EGFRvIII mutation is present in ≈5% of human lung squamous cell carcinoma (SCC) but not in adenocarcinoma and investigate the role of EGFRvIII mutant in lung tumorigenesis and tumor maintenance as well as its response to different EGFR small molecule inhibitors.
Results
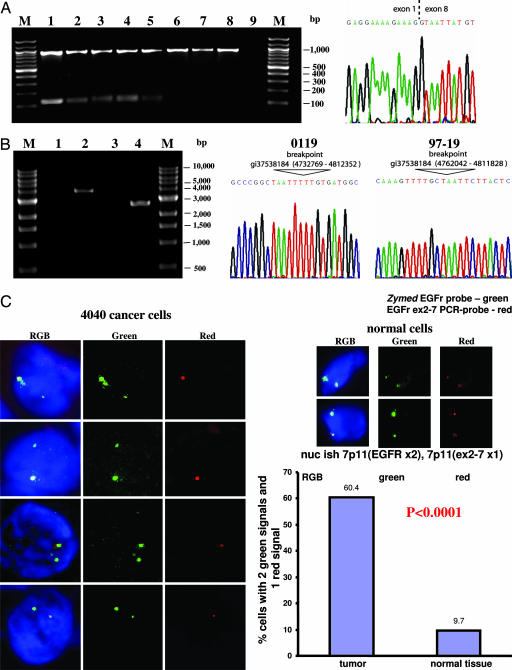
To determine the prevalence of EGFRvIII mutation in human NSCLC, 179 lung tumors frozen at time of the initial resection and verified to be adenocarcinomas (n = 123) or SCC (n = 56) by histology were harvested for RNA and subjected to RT-PCR analyses for the presence of unique EGFRvIII sequences (see Materials and Methods). None of the 123 adenocarcinoma RNA specimens showed evidence of the EGFRvIII 128-bp PCR transcript. Significantly, 5 of the 56 SCC RNA samples gave rise to a 128-bp PCR product specific for the EGFRvIII transcript (Fig. 1A Left). Sequencing confirmed that this product links exon 1 to exon 8 (Fig. 1A Right). As expected, this 128-bp band is not present in the RT-PCR analyses of the normal lung tissues from the corresponding five patients with the EGFRvIII-positive transcripts (Fig. 1A and data not shown), confirming that EGFRvIII is a somatic mutation in lung cancer patients.
Fig. 1.
Identification of EGFRvIII in human NSCLC at both RNA and genomic DNA levels. (A Left) Detection of WT EGFR (929 bp) and EGFRvIII (128 bp) in NSCLC by RT-PCR. Lanes are as follows: M, marker; 1, 97-19-tumor; 2, 4040-tumor; 3, 4050088A2-tumor; 4, 54943-tumor; 5, 0119-tumor; 6, 0119-normal tissue; 7, 088V-tumor; 8, 3811-tumor; 9, H2O. Lanes 7 and 8 serve as negative controls. (A Right) A representative sequencing data of the 128-bp band identifies the EGFRvIII transcript shown from 0119 tumor. (B) Long-range PCR products within the EGFR locus identified the EGFRvIII breakpoint in tumors 0119 (Left, lane 2) and 97-19 (Left, lane 4) but not in normal corresponding genomic DNA controls (Left, lane 1 for 0119 and lane 3 for 97-19, respectively). DNA sequence analyses of the PCR products reveal 79,583-bp and 49,785-bp interstitial deletions of EGFR locus in 0119 tumor (Center) and 97-19 tumor (Right), respectively. (C) FISH analysis (as described in Materials and Methods) confirmed the presence of EGFRvIII in 4040 frozen tissue sections. The EGFR probe spanning the entire locus was labeled with Cy3 (green), and the EGFR exon 2–7 probe was labeled with Cy5 (red). Statistical analysis showed that 60.4% of 4040 tumor cells analyzed have only one copy of exon 2–7 in comparison (Left) with 9.7% in normal tissue (Right). P < 0.0001 using Student's exact t tests.
To distinguish a genomic rearrangement from alternative splicing, long-range PCR amplifications were performed on the genomic DNA from all five SCC tumors positive for the EGFRvIII mutation by RT-PCR screening, using an antisense primer from exon 8 and a series of validated sense primers at ≈3-kb intervals within intron 1 of the EGFR genomic locus; only a rearranged locus could be amplified under these conditions (see Materials and Methods). PCR products suggestive of genomic DNA rearrangement within EGFR allele were identified in DNA from tumors 0119 and 97-19 but not in the corresponding negative controls (Fig. 1B). The sequences of these two PCR fragments confirmed the presence 79,583-bp and 49,785-bp interstitial deletions within the EGFR locus in the two samples, respectively, resulting in the EGFRvIII mutation (Fig. 1B Center and Right, respectively).
FISH analyses using a probe that spans the entire EGFR gene locus and another probe that is specific for a 10-kb region across exon 2 to exon 7 validated another one of five RT-PCR-positive samples (4040) to harbor the EGFRvIII mutation at the genomic level. In contrast to the two genomic DNA validated EGFRvIII containing tumors (0119 and 97-19) that harbor amplifications of the EGFR gene locus (Fig. 6 A and B, which is published as supporting information on the PNAS web site), cancer cells from the 4040 sample have two copies of the EGFR gene, with one of the two copies harboring the EGFRvIII mutation (Fig. 1C).
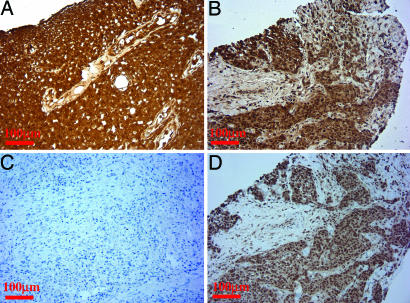
As an independent confirmation of EGFRvIII expression at the RNA levels, we performed quantitative real-time expression PCR analysis. Because there are no ideal methods to measure absolute expression levels of distinct genes using quantitative real-time RT-PCR (qRT-PCR), we measured the relative levels of EGFRvIII and total EGFR in human SCC samples, compared with that from an EGFRvIII-expressing, glioblastoma-positive control (see below). The relative expression of EGFRvIII compared with total EGFR in samples 97-19 [Δthreshold cycle (ΔCt) = −10.4 ± 0.9] and 0119 (ΔCt = −9.6 ± 0.3) and 4040 (ΔCt = −10.2 ± 0.6) was similar to that found in the glioblastoma sample (ΔCt = −10.8 ± 0.5). Furthermore, immunostaining using the EGFRvIII-specific antibody DH8.3 (17–19) demonstrated that EGFRvIII is expressed in all these three SCC tumor samples at a notable level on a per cell basis comparable with that of the glioblastoma sample confirmed to harbor EGFRvIII at both RNA and genomic DNA levels (Fig. 2 A and B and data not shown). Negative staining of DH8.3 was seen in an SCC tumor that is negative for EGFRvIII by RT-PCR screening (Fig. 2C and data not shown). The expression of EGFRvIII was accompanied by EGFR activation, as indicated by the positive immunostaining with phospho-EGFR (Y1068) antibody (Fig. 2D). Furthermore, these three EGFRvIII-positive SCC samples have WT K-ras by sequencing (data not shown). Thus, the combination of genomic DNA validation studies, FISH, quantitative real-time RT-PCR and immunostaining analyses established the prevalence of EGFRvIII mutation to be ≈3/56 (5%) in human SCC.
Fig. 2.
Immunostaining analyses of the lung SCC tumor samples positive for EGFRvIII. Immunostaining of EGFRvIII-specific antibody DH8.3 was preformed as described in Materials and Methods. (A) Glioblastoma sample positive for EGFRvIII from both RT-PCR and FISH analysis serves as positive control. (B) The three SCC tumors identified from the RNA and genomic DNA for the presence of EGFRvIII were positive for EGFRvIII staining. Representative immunostaining is shown for the 97-19 tumor. (C) The SCC tumor confirmed to be negative for EGFRvIII serves as negative control. (D) These SCC samples positive for EGFRvIII staining are also stained positive for phospho-EGFR (Y1068). Representative immunostaining photo is shown from a 97-19 tumor.
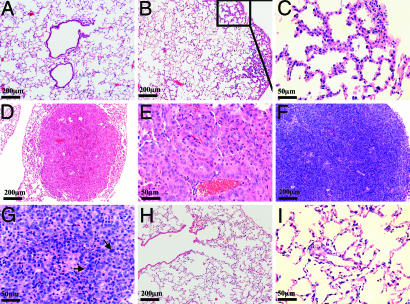
To examine the role of the EGFRvIII mutation in lung tumorigenesis and tumor maintenance in vivo, we generated a transgenic mouse with tetracycline-inducible expression of EGFRvIII. This transgenic mouse, Tet-op-EGFRvIII, was then crossed to the CCSP-rtTA mouse, which has been shown to target the expression of the reverse tetracycline transactivator protein (rtTA) to lung type II pneumocytes to generate bitransgenic mice that were subjected to doxycycline induction (20, 21). As expected, lung development was normal in monotransgenic Tet-op-EGFRvIII mice, which did not develop any detectable lung lesions after 4 months of doxycycline administration (Fig. 3A). Induction of EGFRvIII expression in the lungs of the bitransgenic mice Tet-op-EGFRvIII/CCSP-rtTA after doxycycline administration was confirmed at both RNA and protein levels (Fig. 7 A and B, which is published as supporting information on the PNAS web site). Interestingly, the endogenous mouse EGFR level is almost undetectable in these bitransgenic mice. One potential explanation is that the minimal baseline expression of EGFRvIII causes feedback inhibition of endogenous mouse EGFR expression. The expression of EGFRvIII leads to activation of AKT and extracellular signal-regulated kinase 1/2 (ERK1/2) in the signaling pathway (Fig. 7B). Within 6–8 weeks of doxycycline administration, Tet-op-EGFRvIII/CCSP-rtTA bitransgenic mice developed atypical adenomatous hyperplasia (AAH) (Fig. 3 B and C) (22, 23). Sustained doxycycline induction resulted in the growth of large lung adenomas after 12 weeks (Fig. 3 D and E) and adenocarcinomas after 16 weeks (Fig. 3F-G) in these bitransgenic mice. These lung tumor cells stained positive for phospho-EGFR, phospho-AKT, and phospho-MAPK as well as Ki67, confirming the activation of the EGFR pathway in vivo (Fig. 8, which is published as supporting information on the PNAS web site). Detailed analyses of the other organs in these mice did not reveal any other site with tumors (data not shown). Withdrawal of doxycycline for 7 days, after 12 weeks and 16 weeks of continuous doxycycline treatment, resulted in dramatic tumor regression as revealed by MRI (Fig. 9, which is published as supporting information on the PNAS web site) and histology (Fig. 3 H and I and data not shown). Strikingly, no tumor regrew even after a 6-week period of doxycycline withdrawal (Fig. 9), indicating that the lung tumor maintenance depended on the continued expression of EGFRvIII. Thus, EGFRvIII mutant is oncogenic in the lung compartment in vivo and is essential for lung tumor maintenance.
Fig. 3.
Expression of EGFRvIII in mouse lungs leads to the development of lung cancer in vivo. (A) Monotransgenic Tet-op-EGFRvIII mice had normal lungs with no lesions observable after 4 months of doxycycline administration. (B and C) Bitransgenic mice and Tet-op-EGFRvIII/CCSP-rtTA mice develop atypical adenomous hyperplasia (AAH) in the lungs after ≈6–8 weeks of doxycycline administration. (D and E) Large lung adenoma after 12 weeks of doxycycline treatment. (F and G) Adenocarcinomas after 16 weeks of doxycycline induction. The higher-magnification image (G) shows multiple giant and pleomorphic nuclei (arrows). (H and I) Tumor regression after doxycycline withdrawal. After 12 weeks of doxycycline administration, Tet-op-EGFRvIII/CCSP-rtTA mice with documented lung tumors by MRI were given fresh water without doxycycline for 1 week before lung harvest.
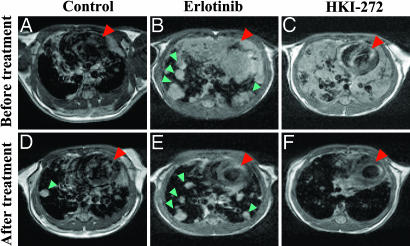
Treatment with small molecule EGFR tyrosine kinase inhibitors has demonstrated a clinical response in NSCLC patients bearing tumors with EGFR kinase domain mutations (4–8). However, the effectiveness of these inhibitors for the EGFRvIII mutation remains unknown. Thus, we sought to determine the activities of small molecule EGFR inhibitors on EGFRvIII-driven lung tumors in vivo using Tet-op-EGFRvIII/CCSP-rtTA, Ink4A/Arf−/− mice. The loss of Ink4A/Arf function in these mice, which recapitulates the frequently reported inactivation of the p16 pathway in human NSCLC (24, 25), allows more rapid development of lung tumors (26) (data not shown). Continuous doxycycline administration for >8 weeks, which caused lung lesions documented by means of MRI (Fig. 4 A–C). The mice were then treated with continued doxycycline and with daily oral administration of placebo vehicle, erlotinib (a reversible EGFR inhibitor), or HKI-272 (an irreversible EGFR/ERB2 inhibitor) at 50 mg/kg, a concentration that has been shown effective in xenograft models with lung cancer cells or breast cancer cells (27, 28). As expected, after 7 days of placebo treatment, the number and size of lung tumors increased (Fig. 4 A and D). Seven days of erlotinib treatment led to an average 45% (±17.6%) reduction in tumor volume in the three treated mice (Fig. 4 B and E), consistent with its activity against EGFRvIII at high concentrations (see below). The three HKI-272-treated mice experienced a more substantial tumor response, an average 88% (±10.4%) reduction in tumor volume, as measured in the restaging MRI (Fig. 4 C and F). Decrease of both total EGFRvIII and phospho-EGFR levels was observed after 1 week of erlotinib treatment, and these decreased levels were even more dramatic in the HKI-272 treatment group (Fig. 10, which is published as supporting information on the PNAS web site). Thus, HKI-272 has potent in vivo activity for tumors driven and maintained by EGFRvIII expression.
Fig. 4.
Treatment of the lung tumors in the Tet-op-EGFRvIII/CCSP-rtTA, Ink4A/Arf−/− mice with EGFR inhibitors. The Tet-op-EGFRvIII/CCSP-rtTA, Ink4A/Arf−/− mice given doxycycline water for >8 weeks were imaged with MRI (see Materials and Methods) before treatment with vehicle (A), erlotinib (B), or HKI-272 (C). The same mice were imaged again after 1 week of daily gavage with vehicle (D), erlotinib (E), and HKI-272 (F). All mice were given fresh water with doxycycline throughout the experiment. The red arrowheads point to the heart of each mouse. The green arrowheads point to the individual tumor nodules.
To quantitate the difference in sensitivity of EGFRvIII tumorigenesis to HKI-272 compared with erlotinib first noted in vivo, we used an in vitro system. Retroviruses containing EGFRvIII, the EGFR-L858R-activating kinase domain mutation, or EGFR-L858R-T790M with both activation and gefitinib/erlotinib treatment resistance mutation (29–31) were stably infected into Ba/F3 cells, a murine pro-B cell line that can be rendered growth factor-independent by introduction of the EGFR-L858R, EGFR-L858R-T790M mutants (29, 32), or other activated kinases (33). In contrast to WT EGFR (32), expression of one of three various EGFR mutants led to a comparable level of phosphorylated tyrosine activity (Fig. 11, which is published as supporting information on the PNAS web site) and transformed the Ba/F3 cells and rendered them IL-3-independent.
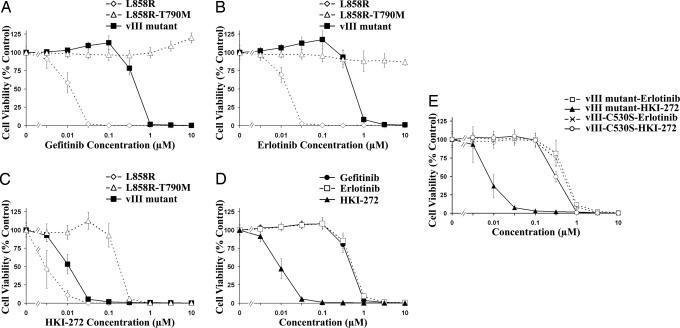
To test the drug sensitivity of these stable cell lines, we used at least two stable clones from each line with various expression levels of EGFR mutants for cell viability assays (MTS). Interestingly no correlation between the expression level and drug sensitivity was observed in this case (data not shown). Ba/F3 cells transformed with EGFRvIII were >40-fold more resistant to the reversible EGFR inhibitors gefitinib and erlotinib (Fig. 5 A and B; IC50 = 432 nM and 576 nM, respectively) compared with the EGFR-L858R mutants (IC50 of 10.8 nM and 12.5 nM, respectively) whereas EGFR-L858R-T790M-containing cells were resistant to gefitinib and erlotinib across all concentrations tested.
Fig. 5.
HKI-272 is effective in growth inhibition of Ba/F3 cells transformed with EGFRvIII, EGFR-L858R, and EGFR-L858R-T790M but fail to inhibit the Ba/F3 cells transformed with EGFRvIIIC530S. The sensitivity of various EGFR mutant-transformed Ba/F3 cells to EGFR inhibitors in the absence of IL-3 was determined by the MTS assay. The percentage of cell viability is shown relative to untreated controls. Results are indicated as mean ± SD. IC50 shown in the text was determined from these dose-response curves by using the xlfit 4 software package (IDBS). (A and B) EGFRvIII mutant (IC50 = 432 and 576 nM) is more resistant than EGFR-L858R mutant (IC50 = 10.8 and 12.5 nM) to gefitinib (A) or erlotinib (B), respectively. (C) HKI-272 inhibits growth of both EGFRvIII- and L858R-T790M-transformed Ba/F3 cells (IC50 = 9.4 nM and 179 nM, respectively). (D) EGFRvIII-transformed Ba/F3 cells are more sensitive to HKI-272 than to gefitinib or erlotinib treatment. (E) EGFRvIIIC530S-transformed Ba/F3 cells became resistant to HKI-272 treatment (IC50 = 327 nM).
In contrast, Ba/F3 cells expressing EGFRvIII mutants are highly sensitive to HKI-272, (Fig. 5 C and D; IC50 = 9.4 nM), as are Ba/F3 cells expressing EGFR-L858R (IC50 = 3.5 nM); as reported (30), we also found that EGFR-L858R-T790M-expressing cells are sensitive to HKI-272 (IC50 = 180 nM). The growth inhibition responses of different EGFR mutant Ba/F3 cell clones to erlotinib and HKI-272 are consistent with the reduction in levels of phospho-EGFR, phospho-AKT, and phospho-ERK1/2 after inhibitor treatment (Fig. 12, which is published as supporting information on the PNAS web site). These data confirm the greater sensitivity of EGFRvIII-driven tumors to HKI-272 treatment noted in vivo and suggest that HKI-272 could be of greater therapeutic efficacy than gefitinib or erlotinib in tumors harboring EGFRvIII mutations.
Irreversible inhibitors of EGFR such as CL-387,785 and HKI-272 are believed to block the kinase enzymatic function by covalently binding to Cys-797 residue in the EGFR (corresponding to Cys-530 in the EGFRvIII truncated protein) (28, 34). We generated the C530S mutation in the background of the EGFRvIII mutation and introduced EGFRvIII-C530S into Ba/F3 cells (Fig. 11). As expected, the Ba/F3 cells are transformed and became independent of IL-3 with similar phosphotyrosine levels in comparison with other EGFR mutants, implying that the kinase activity of the mutated EGFRvIII protein is preserved. However, these EGFRvIII-C530S-transformed cells are now resistant to HKI-272 (IC50 of ≈327 nM as compared with the IC50 = 9.4 nM for the EGFRvIII-transformed Ba/F3 cells) (Fig. 5E). Interestingly, these EGFRvIII-C530S-transformed cells have retained similar sensitivity to erlotinib as EGFRvIII-transformed cells (IC50 of 500 nM), suggesting that the C530S mutation does not interfere with the erlotinib's mechanism of action. Thus, these results further support the molecular mechanism of action for HKI-272 and suggest that resistance to HKI-272 can be developed in transformed cells harboring EGFRvIII.
Discussion
This study revealed the presence of EGFRvIII mutation specifically in human lung SCC through RT-PCR analysis and subsequently confirmed by both the genomic DNA FISH analysis and/or breakpoint validation as well as EGFRvIII-specific antibody immunostaining. From both real-time RT-PCR and immunostaining analyses, we consistently observed a comparable level of EGFRvIII expression in these human lung SCC samples to that from a glioblastoma sample positive for EGFRvIII identified from both RT-PCR and FISH screening. These data indicate that EGFRvIII could potentially play an essential role in the human NSCLC tumor initiation and maintenance. Although determining the true prevalence of the EGFRvIII mutation in lung cancer is technically difficult, we estimate its prevalence in SCC to be at least 5%. Our data establish that the EGFRvIII mutations are found predominantly in human SCC, whereas the EGFR kinase domain mutations are found mainly in human adenocarcinomas. Future identification of the specific genetic mechanisms that drive the development of specific EGFR mutations in each histological cancer subtype would provide additional insight into lung tumorigenesis.
The murine data confirm that overexpression of EGFRvIII is oncogenic in the lung tissues. Further, the abrogation of EGFRvIII expression by withholding doxycycline causes regression of the lung tumors, demonstrating that these tumors are dependent on the activated EGFR pathway. Although the EGFRvIII mutation is found predominantly in human pulmonary SCC in our study, its expression in mouse lung in type II pneumocytes results in the development of lung adenocarcinoma. One potential explanation for this difference in the histological phenotype might be that the correct cell type for the development of SCC is not appropriately targeted in the mouse. Another equally plausible explanation might be that additional genetic changes are needed for these EGFRvIII-activated, transformed cells to progress/differentiate into squamous cell carcinoma. Recent genomic studies suggest that the genomic aberrations between human adenocarcinoma and SCC may be very similar, with p63 overexpression as one of the major distinct differences (35).
Finally, we showed that HKI-272 dramatically inhibits the growth of EGFRvIII-transformed cells in vitro and tumor growth in vivo. Gefitinib and erlotinib also inhibit the growth of cells harboring the EGFRvIII mutations although at a much higher IC50 than HKI-272. This partial activity may provide an explanation for the recently reported responses seen with erlotinib in a small percentage of non-adenocarcinoma NSCLC (36). We also showed that mutations in the cysteine 530 residue in EGFRvIII can confer resistance to HKI-272 sensitivity in EGFRvIII-transformed cells, providing a potential mechanism for the development of in vivo resistance in the clinical setting. It will be beneficial to conduct a phase I trial of HKI-272 to determine its dose, schedule, and safety profile. Our results suggest that HKI-272 may be more effective for treating cancers that harbor the EGFRvIII mutation, such as glioblastoma. Diagnostic genomic screening of patients for the EGFRvIII mutation may serve to identify patients who are likely to respond to HKI-272 or similar therapy.
Materials and Methods
Validation of EGFRvIII Mutant Expression at RNA and DNA Levels.
Total RNA samples or cRNA (37) were retrotranscribed into first-strand cDNA by using the SuperScript Fist-Strand Synthesis System following the manufacturer's protocol (Invitrogen). The region spanning EGFR exon 1 and exon 8 was then amplified by PCR. RT-PCR products, 128 bp (EGFRvIII) and/or 929 bp (WT EGFR), were visualized by 2% agarose gel electrophoresis. All of the positive clones were confirmed in at least three independent experiments. The EGFRvIII bands were then purified by using a Qia-Quick purification kit (Qiagen, Valencia, CA) and cloned into PCR4-TOPO (Invitrogen), and DNA sequencing was performed on two clones for each band. To determine the relative ratio of EGFRvIII to total EGFR at RNA level in three EGFRvIII-positive lung SCC samples in comparison with the EGFRvIII-positive glioblastoma sample, we performed quantitative real-time PCR using a PRISM 7500 Sequence Detection System (Applied Biosystems) and a QuantiTect SYBR Green PCR kit (Qiagen). β-Actin serves as internal control. The relative ratio of EGFRvIII to total EGFR is expressed as ΔCt = (the average of Ct from total EGFR) − (the average of Ct from EGFRvIII). Primer sequences are included in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Genomic DNA (20 ng) from EGFRvIII mutation-positive samples were used as template for long-range PCR by using the Expand Long Template PCR System following the manufacturer's protocol (Roche Applied Science, Indianapolis), with a series of validated sense primers with a 3-kb interval in EGFR intron 1 and one antisense primer in exon 8 (38). The breakpoints from 0119 and 97-19 were amplified by using, respectively, A7 and F14 sense primers with antisense primer in exon 8. PCR products were visualized by 0.8% agarose gel electrophoresis, and location of the breakpoints was obtained by both direct PCR sequencing and cloning into PCR4-TOPO (Invitrogen), followed by sequencing (for primers, protocol for both RT-PCR and long-range PCR, and FISH, see Supporting Materials and Methods).
Establishment of Ba/F3 Stable Cell Lines with Expression of Various EGFR Mutants.
Ba/F3 cells were cultured in RPMI medium 1640 supplemented with 10% FBS, 10% WEHI-3B-Sup, 100 units/ml penicillin, 100 μg/ml streptomycin, and 592 mg/liter l-glutamine at 37°C and 5% CO2 (33). Retroviral production in human embryonic kidney (HEK) 293T cells and infection into Ba/F3 cells were performed by using standard conditions as described (33). Polyclonal cell lines stably expressing EGFR mutants were established by using 2 μg/ml puromycin containing medium and cultured without WEHI-3B-Sup to obtain transformed clones. Pooled stable cells were used for experiments. All experiments were done with at least two stable cell lines with different expression levels of various EGFR mutants; representative data are shown.
EGFR Inhibitor Treatment in Various Ba/F3 Stable Cell Lines.
Various Ba/F3 stable cell lines (0.5–1.5 × 104 cells per well) were treated with either gefitinib (WuXi PharmaTech, Shanghai, China) or erlotinib (WuXi PharmaTech) or HKI-272 (Wyeth) at the final concentrations (0, 0.0033, 0.01, 0.033, 0.1, 0.33, 1, 3.3, and 10 μM). The concentration of DMSO in the medium was adjusted to 0.1%. After 72–76 h of incubation, viable cell numbers were measured by using 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS; CellTiter 96 Aqueous Non-Radioactive Cell Proliferation Kit; Promega) according to the manufacturer's protocol. Each essay consisted of eight replicate wells of each combination of cells and EGFR inhibitor concentration, and each assay was repeated at least three times. The drug concentration resulting in 50% cell viability was scored as the 50% inhibitory concentration (IC50), and the IC50 was determined from dose-response curve by using xlfit 4 (IDBS, Surrey, U.K.).
Mouse Cohorts.
R. A. DePinho (Dana–Farber Cancer Institute) generously provided Ink4A/Arf−/− and Tet-op-EGFRvIII mice. J. Whitsett (University of Cincinnati, Cincinnati) generously provided the CCSP-rtTA mice. These strains were intercrossed to produce experimental cohorts Tet-op-EGFRvIII/CCSP-rtTA and Tet-op-EGFRvIII/CCSP-rtTA, Ink4A/Arf−/− mice. All mice were housed in the pathogen-free environment at the Dana–Farber Cancer Institute. Mice were genotyped by PCR (primers and conditions are available upon request). To induce EGFRvIII expression, adult mice (6-week-old) were provided with fresh drinking water containing 2 mg/ml doxycycline (Sigma) and 5% (wt/vol) sucrose, whereas controls received a 5% sucrose solution without doxycycline.
Lung Preparation and Histology Analysis.
Mice were killed at the time indicated, and the trachea was cannulated. The left lobes were tied with suture thread and dissected. The right lobes of mice lungs were then fixed with neutral buffered 10% formalin overnight, embedded in paraffin, and sectioned at 5 μm. Hematoxylin/eosin (H&E) stains were performed in the Department of pathology in Brigham and Women's Hospital.
Immunohistochemical analysis was performed on formalin-fixed paraffin sections (5 μm) after antigen retrieval (10 min of boiling in 10 mM sodium citrate buffer followed by 20 min of antigen unmasking at room temperature). The primary antibodies and dilutions used in these studies were as follows: phospho-AKTSer-473 (1:25; Cell Signaling Technology, Beverly, MA), phospho-MAPKThr-202/Tyr-204 (1:50; Cell Signaling Technology), phospho-EGFR Y1068 (1:50; Cell Signaling Technology), and Ki67 (1:800; Abcam, Inc., Cambridge, MA). Immunostaining using EGFRvIII-specific antibody DH8.3 (1:20; a gift from W. Cavenee and F. Furnari (Ludwig Institute for Cancer Research, San Diego) was performed as described (17, 18).
EGFR Inhibitors Treatment in Vivo.
After having been continuously given doxycycline water for >8 weeks, the Tet-op-EGFRvIII/CCSP-rtTA, Ink4A/Arf−/− mice were subjected to MRI to determine total tumor burden. After initial imaging, either HKI-272 (Wyeth) or erlotinib (Biaffin, Kassel, Germany) was administered orally at 50 mg/kg daily. After 1 week of treatment, mice were imaged with MRI to determine the reduction in tumor volume. Both erlotinib and HKI-272 were formulated in 0.5% methocellulose-0.4% polysorbate-80 (Tween 80) and administered daily by gavage. For MRI and tumor volume measurement, see Supporting Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Webster Cavenee and Frank Furnari for generously contributing the EGFRvIII-specific antibody DH8.3 for immunostaining and C13 for Western blot analysis, Dr. Ronald DePinho for providing reagents and advice, and Dr. Jeffrey Whitsett for providing the CCSP-rtTA transgenic mice. Drs. Norman Sharpless, Richard Maser, and Bruce Johnson provided insightful comments on the manuscript. K.-K.W. is supported by National Institutes of Health (NIH) Grant K08AG 2400401, the Sidney Kimmel Foundation for Cancer Research, the Joan Scarangello Foundation to Conquer Lung Cancer, and the Flight Attendant Medical Research Institute. M.M. is supported by American Cancer Society Research Grant RSG-03-240-01-MGO, Joan's Legacy, the Emerald Foundation, the Flight Attendant Medical Research Institute, and Novartis Pharmaceuticals. T.S. is supported by a Career Development Award as part of the Dana–Farber/Harvard Cancer Center Specialized Program of Research Excellence (SPORE) in Lung Cancer and NIH Grant P20 CA90578. G.I.S. is supported by NIH Grants R01 CA 90687 and P20 CA90578.
Abbreviations
- EGFR
EGF receptor
- EGFRvIII
variant III EGFR deletion mutant
- NSCLC
non-small cell lung cancer
- SCC
squamous cell carcinoma
- rtTA
reverse tetracycline transactivator protein
- Ct
threshold cycle.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hynes N. E., Lane H. A. Nat. Rev. Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 2.Arteaga C. L. Exp. Cell Res. 2003;284:122–130. doi: 10.1016/s0014-4827(02)00104-0. [DOI] [PubMed] [Google Scholar]
- 3.Mendelsohn J., Baselga J. Oncogene. 2000;19:6550–6565. doi: 10.1038/sj.onc.1204082. [DOI] [PubMed] [Google Scholar]
- 4.Paez J. G., Janne P. A., Lee J. C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F. J., Lindeman N., Boggon T. J., et al. Science. 2004;304:497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 5.Lynch T. J., Bell D. W., Sordella R., Gurubhagavatula S., Okimoto R. A., Brannigan B. W., Harris P. L., Haserlat S. M., Supko J. G., Haluska F. G., et al. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 6.Pao W., Miller V., Zakowski M., Doherty J., Politi K., Sarkaria I., Singh B., Heelan R., Rusch V., Fulton L., et al. Proc. Natl. Acad. Sci. USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tokumo M., Toyooka S., Kiura K., Shigematsu H., Tomii K., Aoe M., Ichimura K., Tsuda T., Yano M., Tsukuda K., et al. Clin. Cancer Res. 2005;11:1167–1173. [PubMed] [Google Scholar]
- 8.Shigematsu H., Lin L., Takahashi T., Nomura M., Suzuki M., Wistuba II, Fong K. M., Lee H., Toyooka S., Shimizu N., et al. J. Natl. Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 9.Frederick L., Wang X. Y., Eley G., James C. D. Cancer Res. 2000;60:1383–1387. [PubMed] [Google Scholar]
- 10.Okamoto I., Kenyon L. C., Emlet D. R., Mori T., Sasaki J., Hirosako S., Ichikawa Y., Kishi H., Godwin A. K., Yoshioka M., et al. Cancer Sci. 2003;94:50–56. doi: 10.1111/j.1349-7006.2003.tb01351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moscatello D. K., Holgado-Madruga M., Godwin A. K., Ramirez G., Gunn G., Zoltick P. W., Biegel J. A., Hayes R. L., Wong A. J. Cancer Res. 1995;55:5536–5539. [PubMed] [Google Scholar]
- 12.Wong A. J., Ruppert J. M., Bigner S. H., Grzeschik C. H., Humphrey P. A., Bigner D. S., Vogelstein B. Proc. Natl. Acad. Sci. USA. 1992;89:2965–2969. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pao W., Miller V. A. J. Clin. Oncol. 2005;23:2556–2568. doi: 10.1200/JCO.2005.07.799. [DOI] [PubMed] [Google Scholar]
- 14.Garcia de Palazzo I. E., Adams G. P., Sundareshan P., Wong A. J., Testa J. R., Bigner D. D., Weiner L. M. Cancer Res. 1993;53:3217–3220. [PubMed] [Google Scholar]
- 15.Ekstrand A. J., Sugawa N., James C. D., Collins V. P. Proc. Natl. Acad. Sci. USA. 1992;89:4309–4313. doi: 10.1073/pnas.89.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuan C. T., Wikstrand C. J., Bigner D. D. Endocr. Relat. Cancer. 2001;8:83–96. doi: 10.1677/erc.0.0080083. [DOI] [PubMed] [Google Scholar]
- 17.Jungbluth A. A., Stockert E., Huang H. J., Collins V. P., Coplan K., Iversen K., Kolb D., Johns T. J., Scott A. M., Gullick W. J., et al. Proc. Natl. Acad. Sci. USA. 2003;100:639–644. doi: 10.1073/pnas.232686499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishikawa R., Sugiyama T., Narita Y., Furnari F., Cavenee W. K., Matsutani M. Brain Tumor Pathol. 2004;21:53–56. doi: 10.1007/BF02484510. [DOI] [PubMed] [Google Scholar]
- 19.Shinojima N., Tada K., Shiraishi S., Kamiryo T., Kochi M., Nakamura H., Makino K., Saya H., Hirano H., Kuratsu J., et al. Cancer Res. 2003;63:6962–6970. [PubMed] [Google Scholar]
- 20.Perl A. K., Tichelaar J. W., Whitsett J. A. Transgenic Res. 2002;11:21–29. doi: 10.1023/a:1013986627504. [DOI] [PubMed] [Google Scholar]
- 21.Tichelaar J. W., Lu W., Whitsett J. A. J. Biol. Chem. 2000;275:11858–11864. doi: 10.1074/jbc.275.16.11858. [DOI] [PubMed] [Google Scholar]
- 22.Nikitin A. Y., Alcaraz A., Anver M. R., Bronson R. T., Cardiff R. D., Dixon D., Fraire A. E., Gabrielson E. W., Gunning W. T., Haines D. C., et al. Cancer Res. 2004;64:2307–2316. doi: 10.1158/0008-5472.can-03-3376. [DOI] [PubMed] [Google Scholar]
- 23.Jackson E. L., Willis N., Mercer K., Bronson R. T., Crowley D., Montoya R., Jacks T., Tuveson D. A. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minna J. D., Fong K., Zochbauer-Muller S., Gazdar A. F. Cancer J. 2002;8(Suppl. 1):S41–S46. [PubMed] [Google Scholar]
- 25.Kaye F. J. Oncogene. 2002;21:6908–6914. doi: 10.1038/sj.onc.1205834. [DOI] [PubMed] [Google Scholar]
- 26.Fisher G. H., Wellen S. L., Klimstra D., Lenczowski J. M., Tichelaar J. W., Lizak M. J., Whitsett J. A., Koretsky A., Varmus H. E. Genes Dev. 2001;15:3249–3262. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang S., Armstrong E. A., Benavente S., Chinnaiyan P., Harari P. M. Cancer Res. 2004;64:5355–5362. doi: 10.1158/0008-5472.CAN-04-0562. [DOI] [PubMed] [Google Scholar]
- 28.Rabindran S. K., Discafani C. M., Rosfjord E. C., Baxter M., Floyd M. B., Golas J., Hallett W. A., Johnson B. D., Nilakantan R., Overbeek E., et al. Cancer Res. 2004;64:3958–3965. doi: 10.1158/0008-5472.CAN-03-2868. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi S., Boggon T. J., Dayaram T., Janne P. A., Kocher O., Meyerson M., Johnson B. E., Eck M. J., Tenen D. G., Halmos B. N. Engl. J. Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 30.Kwak E. L., Sordella R., Bell D. W., Godin-Heymann N., Okimoto R. A., Brannigan B. W., Harris P. L., Driscoll D. R., Fidias P., Lynch T. J., et al. Proc. Natl. Acad. Sci. USA. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pao W., Miller V. A., Politi K. A., Riely G. J., Somwar R., Zakowski M. F., Kris M. G., Varmus H. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang J., Greulich H., Janne P. A., Sellers W. R., Meyerson M., Griffin J. D. Cancer Res. 2005;65:8968–8974. doi: 10.1158/0008-5472.CAN-05-1829. [DOI] [PubMed] [Google Scholar]
- 33.Luwor R. B., Zhu H. J., Walker F., Vitali A. A., Perera R. M., Burgess A. W., Scott A. M., Johns T. G. Oncogene. 2004;23:6095–6104. doi: 10.1038/sj.onc.1207870. [DOI] [PubMed] [Google Scholar]
- 34.Discafani C. M., Carroll M. L., Floyd M. B., Jr, Hollander I. J., Husain Z., Johnson B. D., Kitchen D., May M. K., Malo M. S., Minnick A. A., Jr, et al. Biochem. Pharmacol. 1999;57:917–925. doi: 10.1016/s0006-2952(98)00356-6. [DOI] [PubMed] [Google Scholar]
- 35.Tonon G., Wong K. K., Maulik G., Brennan C., Feng B., Zhang Y., Khatry D. B., Protopopov A., You M. J., Aguirre A. J., et al. Proc. Natl. Acad. Sci. USA. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsao M. S., Sakurada A., Cutz J. C., Zhu C. Q., Kamel-Reid S., Squire J., Lorimer I., Zhang T., Liu N., Daneshmand M., et al. N. Engl. J. Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 37.Bhattacharjee A., Richards W. G., Staunton J., Li C., Monti S., Vasa P., Ladd C., Beheshti J., Bueno R., Gillette M., et al. Proc. Natl. Acad. Sci. USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frederick L., Eley G., Wang X. Y., James C. D. Neuro-oncol. 2000;2:159–163. doi: 10.1093/neuonc/2.3.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.