Abstract
Ras-mediated vulval development in C. elegans is inhibited by the functionally redundant sets of class A, B, and C synthetic Multivulva (synMuv) genes. Three of the class B synMuv genes encode an Rb/DP/E2F complex that, by analogy with its mammalian and Drosophila counterparts, has been proposed to silence genes required for vulval specification through chromatin modification and remodeling. Two class A synMuv genes, lin-15A and lin-56, encode novel nuclear proteins that appear to function as a complex. We show that a third class A synMuv gene, lin-8, is the defining member of a novel C. elegans gene family. The LIN-8 protein is nuclear and can interact physically with the product of the class B synMuv gene lin-35, the C. elegans homolog of mammalian Rb. LIN-8 likely acts with the synMuv A proteins LIN-15A and LIN-56 in the nucleus, possibly in a protein complex with the synMuv B protein LIN-35 Rb. Other LIN-8 family members may function in similar complexes in different cells or at different stages. The nuclear localization of LIN-15A, LIN-56, and LIN-8, as well as our observation of a direct physical interaction between class A and class B synMuv proteins, supports the hypothesis that the class A synMuv genes control vulval induction through the transcriptional regulation of gene expression.
THE retinoblastoma (Rb) gene was the first tumor-suppressor gene to be cloned, and the Rb pathway has been found to be a frequent target of inactivation in many human cancers (Nevins 2001). The nematode Caenorhabditis elegans possesses a single homolog of Rb, lin-35, which functions in the inhibition of both cellular proliferation and differentiation (Lu and Horvitz 1998; Boxem and van den Heuvel 2001). The class A synthetic Multivulva (synMuv) genes function redundantly with the lin-35 Rb gene to inhibit Ras-mediated vulval induction. The analysis of the class A synMuv genes may further our understanding of activities that interact with the Rb pathway in the regulation of cell-fate determination and in the prevention of oncogenic transformation.
The vulva of the C. elegans hermaphrodite is the conduit through which embryos are expelled and is also the point of entry for sperm after mating with a male. The vulva is formed by the descendants of three of six equipotent cells, P(3–8).p. These six cells are all able to express any one of three fates: the 1° vulval fate, the 2° vulval fate, and the 3° nonvulval fate. Vulval development is induced by activation of a receptor tyrosine kinase (RTK)/Ras pathway (Kornfeld 1997; Sternberg and Han 1998). During wild-type larval development, signaling from the anchor cell of the somatic gonad activates an RTK/Ras pathway, causing P6.p to adopt the 1° vulval fate and directly or indirectly causing P5.p and P7.p to adopt the 2° vulval fate. P(5–7).p then divide to produce 22 cells that migrate and fuse to form the toroidal vulva. P3.p, P4.p, and P8.p normally express the 3° nonvulval fate, dividing once and fusing with the hypodermis. Loss of RTK/Ras pathway signaling results in the expression of the nonvulval 3° fate by P(5–7).p and thus in a Vulvaless (Vul) phenotype. Vul animals lack a functional vulva and are consumed by their internally hatched progeny. Ectopic activation of RTK/Ras pathway signaling results in the expression of 1° or 2° vulval fates by P3.p, P4.p, and P8.p, causing a Multivulva (Muv) phenotype. The extra vulval tissue produced in Muv animals forms ectopic ventral protrusions.
Class A, B, and C synMuv genes act to antagonize RTK/Ras function in potential vulval cells (Ferguson and Horvitz 1989; Ceol and Horvitz 2004). As a result of a functional redundancy among these three classes of synMuv genes, only hermaphrodites mutant in two sets of genes exhibit the synMuv phenotype. Genetic screens and targeted studies have identified at least four class A synMuv genes: lin-8, lin-15A, lin-38, and lin-56; at least 17 class B synMuv genes: lin-9, lin-13, lin-15B, lin-35, lin-36, lin-37, lin-52, lin-53, lin-54, lin-61, dpl-1, efl-1, hda-1, hpl-2, let-418, mep-1, and tam-1; and four class C synMuv genes: trr-1, mys-1, epc-1, and ssl-1 (Horvitz and Sulston 1980; Ferguson and Horvitz 1985, 1989; Lu and Horvitz 1998; Hsieh et al. 1999; Solari and Ahringer 2000; von Zelewsky et al. 2000; Ceol and Horvitz 2001, 2004; Couteau et al. 2002; Unhavaithaya et al. 2002; Thomas et al. 2003; X. Lu, M. M. Harrison, P. W. Sternberg and H. R. Horvitz, unpublished results). A subset of the class B synMuv genes encode proteins that mediate histone modification, chromatin remodeling, and transcriptional repression. In particular, efl-1, dpl-1, lin-35, lin-53, hda-1, let-418, and hpl-2 encode C. elegans homologs of E2F, DP, Rb, the Rb-associated protein RbAp48, a class I histone deacetylase (HDAC), the Mi-2 chromatin-remodeling ATPase, and heterochromatin protein 1 (HP1), respectively (Lu and Horvitz 1998; Solari and Ahringer 2000; von Zelewsky et al. 2000; Ceol and Horvitz 2001; Couteau et al. 2002). The mammalian homologs of LIN-53 RbAp48, HDA-1 HDAC, and LET-418 Mi-2 are components of the histone deacetylase and chromatin-remodeling NuRD complex, while HP1 has been shown to function as a histone H3 methyl-lysine-9-binding protein (Knoepfler and Eisenman 1999; Richards and Elgin 2002). Because of their molecular identities, the synMuv B genes are thought to antagonize RTK/Ras function in the vulva by silencing transcription of vulval specification genes through chromatin modification and remodeling. LIN-35 Rb is likely to play a pivotal role in this process, as evidence suggests that mammalian pRb mediates the association of the sequence-specific heterodimeric transcription factor DP/E2F with the NuRD complex components HDAC1 and RbAp48 as well as with a histone H3 K9 methyltransferase (Nicolas et al. 2000; Nielsen et al. 2001; Zhang and Dean 2001). The resultant likely recruitment of Mi-2 and HP1 may induce a facultative heterochromatic state around the targeted genes, preventing transcription. The class C synMuv genes encode a putative C. elegans Tip60/NuA4-like histone acetyltransferase complex; it has not yet been determined if this putative complex acts in transcriptional activation or repression (Ceol and Horvitz 2004).
The class A synMuv genes may inhibit vulval development through the regulation of transcription. Of the four known class A synMuv genes, two—lin-56 and lin-15A—have been cloned and encode novel nuclear proteins that likely associate in a functional complex in vivo (Clark et al. 1994; Huang et al. 1994; E. M. Davison, A. M. Saffer, L. S. Huang, J. DeModena, P. W. Sternberg and H. R. Horvitz, unpublished results). Furthermore, LIN-56 and LIN-15A share a novel C2CH motif related to the THAP domain, shown in the human protein THAP1 to possess zinc-dependent sequence-specific DNA-binding activity in vitro (Clouaire et al. 2005; E. M. Davison, A. M. Saffer, L. S. Huang, J. DeModena, P. W. Sternberg and H. R. Horvitz, unpublished results). Here we report our characterization of a third class A synMuv gene, lin-8.
MATERIALS AND METHODS
Strains and general techniques:
C. elegans strains were cultivated on NGM agar seeded with Escherichia coli strain OP50 as described by Brenner (1974) and were grown at 20° unless otherwise indicated. Bristol strain N2 was used as the wild-type strain. The mutant alleles used in this study are listed below, and a description of each can be found in Riddle et al. (1997) unless noted otherwise:
LG I: lin-35(n745).
LG II: lin-8(n111, n2376, n2378, n2403, n2731, n2738, n2739, n2741) (Thomas et al. 2003), dpy-10(e128).
LG III: lin-52(n771) (Ferguson and Horvitz 1989; Thomas et al. 2003).
LG X: lin-15B(n744, n2245).
pPK5363 is a Tc1-transposon insertion polymorphism on LG II found in the NL7000 but not in the N2 strain (Korswagen et al. 1996). nIs128 contains a GFP transgene integrated into LG II (H. T. Schwartz and H. R. Horvitz, unpublished results). In addition, the following deficiencies were used: ccDf1, ccDf2, and ccDf11 (Chen et al. 1992).
Deletion and polymorphism mapping:
To test complementation of lin-8 with the deficiencies ccDf1, ccDf2, and ccDf11, lin-8(n111) dpy-10(e128); lin-15B(n744) hermaphrodites were mated with ccDf/+ males. The Muv phenotype of the non-Dpy male progeny, half of which should possess lin-8 in trans to the relevant deficiency, was scored. In the ccDf1 and ccDf2 crosses, 0/88 and 0/84 male offspring, respectively, exhibited ventral protrusions, whereas 20/63 male offspring of the ccDf11 cross exhibited ventral protrusions.
The left endpoint of ccDf1 was defined relative to the physical map by determining if cosmid sequences from the interval between sup-9 and lin-31 could be amplified by polymerase chain reaction (PCR) from ccDf1 homozygous inviable embryos. A drop of chitinase solution (20 mg/ml chitinase, 50 mm NaCl, 70 mm KCl, 2.5 mm MgCl2, 2.5 mm CaCl2) was placed over single inviable embryos, which were promptly transferred to 10 μl of lysis buffer (50 mm KCl, 10 mm Tris, pH 8.2, 2.5 mm MgCl2, 0.45% NP-40, 0.45% Tween-20, 0.01% gelatin, 60 μg/ml proteinase K) and subsequently frozen at −80°. Embryos were thawed and lysed by incubation at 60° for 1 hr. Proteinase K was inactivated by incubation at 95° for 15 min. The lysate from each inviable embryo was used for three PCR reactions: the test amplification; amplification of a sequence from cosmid F34D6, which served as a positive control for successful lysis; and amplification of lin-31, which served as a negative control to confirm identification of each egg as a ccDf1 homozygote. Sequences from cosmids B0454, ZC239, F39E9, and W10G11 but not from cosmids M151 or F19B10 were successfully amplified from ccDf1 homozygous inviable embryos, placing the left endpoint of ccDf1 between W10G11 and M151. The primers used were as follows: F34D6.7, 5′-CACCTGAAGATTCAAGTTTAG-3′; F34D6.11, 5′-GTGTGAGCTCAGCAGCTTC-3′; B0454 Fwd, 5′-GGTTCTCGTTAGCTGAGTGG-3′; B0454 Rev, 5′-GTACGGAGCCAAGATCATACG-3′; ZC239 Fwd, 5′-GCAGAGACGTTGGATCCTAGC; ZC239 Rev, 5′-CTTCAGGAGTCGGTGAACTCG-3′; F39E9 Fwd, 5′-CAGTCTCAGGCTAGACTTGG-3′; F39E9 Rev, 5′-GCTGAGCAGATCTCGAATGG-3′; W10G11 Fwd1, 5′-GCTTCCACATTCAGTGAAGG-3′; W10G11 Rev1, 5′-CAAGCCAGAAGAGCAAGTCG-3′; W10G11 N1, 5′-CGAGATGTAAGCTCAGTATGG-3′; M151 Fwd, 5′-CATCGGTCTCCCATAGTTACC-3′; M151 Rev, 5′-GCTCTGGCTGCTCGAGTTCC-3′; F19B10 Fwd, 5′-CTGAAGCATTGGCTCAGAGG-3′; F19B10 Rev, 5′-CGTCATTGATGGACCATGTGC-3′; lin-31 Fwd1, 5′-GCTATTCAGGACTCTGACG-3′; lin-31 Rev1, 5′-CCTTCCCAGGACGATCG-3′.
The pPK5363 polymorphism is an insertion of the Tc1 transposon into cosmid C17F4 present in the NL7000 but not in the N2 strain (Korswagen et al. 1996). To map lin-8 against pPK5363, lin-8(n111) dpy-10(e128)/NL7000; lin-15B(n744) males were crossed with lin-8(n111) dpy-10(e128); lin-15B(n744) hermaphrodites, and resulting Muv non-Dpy and Dpy non-Muv hermaphrodite progeny were picked and used to establish homozygous recombinant lines. PCR employing Tc1-specific (5′-GCTGATCGACTCGATGCCACGTCG-3′) and C17F4-specific (5′-CCATCAACGAGTACGATACG-3′) primers was used to determine if polymorphism pPK5363 was present. Of the Muv non-Dpy chromosomes, 4/13 carried pPK5363, and of the Dpy non-Muv chromosomes, 2/14 carried pPK5363. These results placed lin-8 to the left of pPK5363, which is itself to the left of dpy-10.
Transgenic animals:
Germline transformation by micro-injection was performed as described by Mello et al. (1991). The coinjection marker pRF4 was injected at a concentration of 80 ng/μl. Transgenic animals were identified using the Roller phenotype generated by expression of the rol-6(su1006) dominant allele from pRF4. Experimental constructs were injected at a concentration of 20 or 50 ng/μl.
Antibody preparation, immunoblotting, and immunocytochemistry:
Anti-LIN-8 antibodies were generated using purified maltose-binding protein (MBP)-LIN-8(aa 1–386) fusion protein. The crude antisera were subsequently affinity purified against the GST-LIN-8 (aa 1–386) fusion protein as described by Koelle and Horvitz (1996) and then preadsorbed against an acetone precipitate of proteins prepared from lin-8(n2731) mixed-stage worms, essentially as described by Harlow and Lane (1988). Affinity-purified and preadsorbed anti-LIN-8 antibodies HM2247 were used at a dilution of 1:1000 for Western blots. Samples for Western analysis were prepared by Dounce homogenization of mixed-stage worms in hypotonic lysis buffer (10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 1 mm DTT, 250 mm sucrose) containing 1× protease inhibitor cocktail (800 μg/ml benzamidine HCl, 500 μg/ml phenanthroline, 500 μg/ml aprotinin, 500 μg/ml leupeptin, 500 μg/ml pepstatin A, 50 mm phenylmethylsulfonyl fluoride; BD Biosciences, Franklin Lakes, NJ) as well as phosphatase inhibitors (0.2 mm sodium orthovanadate, 50 mm sodium fluoride, 1 μm microcystin-LR).
Affinity-purified and preadsorbed anti-LIN-8 antibodies HM2247 were used at a dilution of 1:100 for immunocytochemistry. Anti-α-tubulin mouse monoclonal antibodies DM1A (Sigma, St. Louis) and MH27 (Francis and Waterston 1991), which recognizes the apical borders of C. elegans epithelial cells, were used as positive controls for immunocytochemistry at 1:100 and 1:1000 dilutions, respectively. Embryos were fixed in 0.8% paraformaldehyde for 20 min as described by Guenther and Garriga (1996). Larvae and adults were fixed in 2% paraformaldehyde for 15 min, essentially as described by Finney and Ruvkun (1990). Images were obtained using a Zeiss LSM510 laser confocal microscope and software and processed using Adobe Photoshop software.
Two-hybrid and in vitro binding experiments:
The yeast two-hybrid screen of a C. elegans cDNA library was performed as described by Walhout and Vidal (2001). Full-length lin-35 Rb was used as bait. A total of 1.4 × 106 colonies of the C. elegans AD-wrmcDNA library (Walhout et al. 2000b) were screened. Interaction of LIN-35 Rb and LIN-8 could not be tested in the reverse orientation, as LIN-8 was found to self-activate when fused to the Gal4 DNA-binding domain.
The full-length and partial MBP-LIN-8 constructs were made by subcloning appropriate portions of the lin-8 cDNA into vector pMAL-c2 (NEB, Beverly, MA). The GST-LIN-8 (aa 175–285) construct was made by subcloning the appropriate portion of the lin-8 cDNA into vector pGEX-2T (Amersham Biosciences, Piscataway, NJ). MBP and GST fusion constructs were expressed in E. coli BL21(DE3) cells (Studier et al. 1990) and purified using amylose resin (NEB) or glutathione Sepharose 4B (Amersham Biosciences), respectively, as recommended by the manufacturers. The constructs encoding LIN-35 Rb (aa 1–555) and LIN-35 Rb (aa 270–961) have been described previously (Lu and Horvitz 1998) and were used as templates for in vitro synthesis of 35S-labeled protein (TNT-coupled reticulocyte lysate system, Promega, Madison, WI). In vitro binding experiments were otherwise performed as described by Reddien and Horvitz (2000), and formation of protein complexes was analyzed by SDS-PAGE and autoradiography.
RESULTS
LIN-8 defines a family of novel C. elegans proteins:
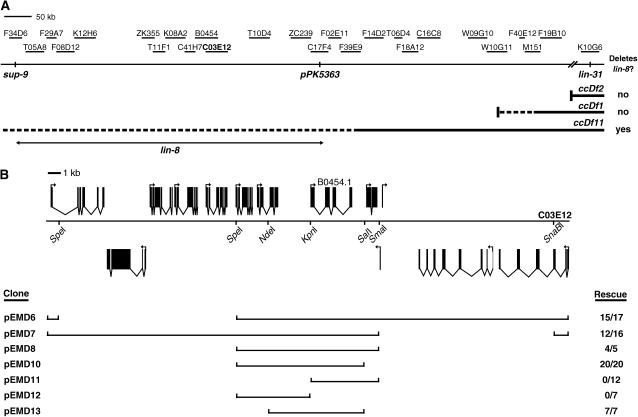
The class A synMuv gene lin-8 was originally identified through the chance recovery of a lin-8(lf); lin-9(lf) double mutant in a screen by S. Brenner (personal communication) for animals abnormal in morphology or behavior; the Muv phenotype of this strain was later shown to be synthetic, as it required the presence of two unlinked mutations, lin-8(n111) and lin-9(n112) (Horvitz and Sulston 1980). An additional eight alleles of lin-8 have since been identified in two independent screens for synMuv A genes (Thomas et al. 2003). lin-8 was previously mapped to the 7-MU interval between sup-9 and lin-31 on chromosome II (Ferguson and Horvitz 1985). We used the deficiencies ccDf1, ccDf2, and ccDf11, each of which deletes lin-31 (Chen et al. 1992), to more precisely locate lin-8 on the physical map. We performed complementation tests and found that of these three deficiencies, only ccDf11 deletes the lin-8 locus. As the sup-9 locus resides in cosmid F34D6 (Perez de la Cruz et al. 2003) and the left endpoints of ccDf1, ccDf2, and ccDf11 had been defined approximately by experiments using PCR (data concerning the left endpoints of ccDf2 and ccDf11 were generously provided to us by C. A. Spike and R. K. Herman), we placed lin-8 between the cosmids F34D6 and M151 (Figure 1A). Further mapping using the polymorphism pPK5363 placed lin-8 between cosmids F34D6 and C17F4. We injected cosmids from this interval into lin-8(n111); lin-15B(n744) animals and obtained rescue of their synMuv phenotype with cosmid C03E12 as well as with a 7.5-kb subclone of C03E12 (Figure 1B). This minimal rescuing fragment contains a single intact predicted gene, B0454.1 (C. elegans Sequencing Consortium 1998). We determined the sequences of the nine alleles of lin-8 and found all to contain mutations within the B0454.1 open reading frame (ORF) (Table 1). Furthermore, RNA-mediated interference (RNAi) of B0454.1 in lin-15B(n744) animals resulted in a synMuv phenotype, and expression of the B0454.1 ORF under the control of the two C. elegans heat-shock promoters (Stringham et al. 1992) shortly after L1 lethargus efficiently rescued the synMuv phenotype of lin-8(n2731); lin-15B(n744) animals (data not shown). We conclude that lin-8 and B0454.1 are equivalent.
Figure 1.
Cloning of lin-8. (A) Physical map of the genomic region containing the lin-8 locus. Deficiency and polymorphism mapping placed lin-8 between cosmids F34D6 and C17F4. lin-8 rescuing cosmid C03E12 is shown in boldface type. Solid lines indicate regions known to be deleted by the deficiencies; dashed lines indicate regions that may be deleted by the deficiencies. (B) Transformation rescue of lin-8. (Top) The predicted open reading frames within rescuing cosmid C03E12 and (bottom) the subclones derived from this cosmid. (C) Sequence of the LIN-8 protein. The region sufficient for interaction with LIN-35 Rb is underlined.
TABLE 1.
Sequences of lin-8 mutations
| Allele | Wild-type codon | Mutant codon | Substitution |
|---|---|---|---|
| n111 | CTG | CCG | L20P |
| n2741 | GTG | ATG | V68M |
| n2376 | GAG | AAG | E148K |
| n2378 | CGC | TGC | R154C |
| n2403 | GAG | AAG | E164K |
| n2724 | GAG | AAG | E164K |
| n2738 | TGG | TAG | W79amber |
| n2731 | CAA | TAA | Q113ochre |
| n2739 | AGA | TGA | R304opal |
Amino acid substitutions are indicated as wild-type residue, residue number, and mutant residue. Mutated bases are underlined.
lin-8 encodes a novel, acidic protein of 386 amino acids (Figure 1C). Sequencing of six lin-8 cDNA clones (courtesy of Yuji Kohara) verified the exon/intron junctions predicted by GENEFINDER (Edgley et al. 1997). Extensive database searches [using PSI-BLAST (Altschul et al. 1997), PROSITE (Falquet et al. 2002), Pfam (Bateman et al. 2002), and SMART (Schultz et al. 2000)] with the LIN-8 protein sequence have revealed no significant canonical motifs and no apparent sequence homologs in other species. LIN-8 is, however, a member of a family of 17 C. elegans proteins (Figure 2). This family was independently detected by the Pfam protein families database and is referred to there as DUF278 (Bateman et al. 2002). While the scores are very weak, LIN-8 identifies the most distant family member in BLAST searches and vice versa. The biological roles of the remaining 16 family members are not known. Several of the family members, including LIN-8, possess an N-terminal proline-rich region (Figure 2) containing at least one PxxP motif, the core sequence to which SH3 domains can bind (Kay et al. 2000).
Figure 2.
LIN-8 defines a family of C. elegans proteins. Alignment of LIN-8 with the other 16 members of the LIN-8 family; all of these proteins are from C. elegans. Solid background indicates identity with LIN-8 in at least three additional family members. Arrowheads indicate positions of missense mutations found in the indicated lin-8 alleles. The proline-rich N-terminal motif and the region of LIN-8 sufficient for interaction with LIN-35 Rb are indicated by solid lines.
Characterization of lin-8 alleles:
To identify null alleles of lin-8 as well as residues important for LIN-8 function, we characterized all nine independently isolated lin-8 alleles (Tables 1 and 2). Three lin-8 alleles—n2731, n2738, n2739—contain nonsense mutations (Table 1). The remaining six lin-8 alleles—n111, n2376, n2378, n2403, n2724, n2741—contain missense mutations (Table 1). Two of the missense alleles, n2403 and n2724 (Thomas et al. 2003), contain the identical nucleotide change; only n2403 was subsequently used for quantitative studies. Four of the five amino-acid residues altered in the missense alleles are conserved in several LIN-8 family members (Figure 2).
TABLE 2.
lin-8 allele strengths
| Penetrance of Muv phenotype (%)
|
||||
|---|---|---|---|---|
|
lin-15B(n2245)
|
lin-52(n771)
|
|||
| Genotypeab | 15° (n) | 20° (n) | 15° (n) | 20° (n) |
| lin-8(n111) | 29 (94) | 100 (109) | 49 (98) | 99 (143) |
| lin-8(n2741) | 1 (98) | 100 (101) | 15 (96) | 99 (146) |
| lin-8(n2376) | 98 (99) | 100 (103) | 80 (96) | 100 (155) |
| lin-8(n2378) | 99 (98) | 100 (101) | 88 (97) | 100 (142) |
| lin-8(n2403)c | 92 (101) | 100 (104) | 98 (100) | 100 (107) |
| lin-8(n2738) | 68 (96) | 100 (103) | 96 (97) | 100 (139) |
| lin-8(n2731) | 77 (74) | 100 (104) | 99 (97) | 100 (146) |
| lin-8(n2739) | 76 (100) | 100 (102) | 63 (95) | 97 (119) |
| lin-8(n2376) dpy-10(e128)/ccDf11 | 96 (135) | ND | ND | ND |
| lin-8(n2738) dpy-10(e128)/ccDf11 | 95 (127) | ND | ND | ND |
| lin-8(n2731) dpy-10(e128)/ccDf11 | 94 (183) | ND | ND | ND |
| lin-56(n2728)d | 100 (127) | 100 (127) | ND | ND |
| lin-56(n3355)d | 100 (124) | 100 (100) | ND | ND |
Penetrance of Muv phenotype (%), percentage of animals with at least one pseudovulva on their ventral sides. n, number of animals scored. ND, not determined.
Animals homozygous for either a lin-8(lf) or a lin-56(lf) allele were raised at either 15° or 20° for at least three generations before scoring.
To generate lin-8(lf)/ccDf11; lin-15B(n2245) animals, dpy-10(e128); lin-15B(n2245) hermaphrodites were mated with ccDf11/nIs128 males. The resulting non-Dpy non-GFP ccDf11/dpy-10(e128); lin-15B(n2245) male offspring were then crossed with lin-8(lf) dpy-10(e128); lin-15B(n2245) hermaphrodites, and the Muv phenotype of any non-Dpy cross progeny of this mating was scored. All crosses were carried out at 15°.
Since n2403 and n2724 contain the same amino acid change, only n2403 was analyzed.
The n2728 allele contains a deletion of the entire lin-56 locus. The n3355 allele contains an early nonsense mutation within the lin-56 coding sequence (E. M. Davison, A. M. Saffer, L. S. Huang, J. DeModena, P. W. Sternberg and H. R. Horvitz, unpublished results).
The nine lin-8 alleles are not easily distinguishable in combination with the strong canonical synMuv B allele lin-15B(n744) (data not shown). The synMuv phenotype is inherently temperature sensitive: both its penetrance and its expressivity are usually greater in mutants raised at 20° than at 15° (Ferguson and Horvitz 1989). We therefore quantitated the penetrance of the synMuv phenotype associated with each lin-8 allele in combination with two weak synMuv B alleles, lin-15B(n2245) (Thomas et al. 2003) and lin-52(n771) (Ferguson and Horvitz 1989), at both 15° and 20° (Table 2). The missense alleles can be placed in three categories on the basis of phenotypic strength: weak (n2741), intermediate (n111), and strong (n2376, n2378, n2403). The three strong missense alleles all mutate charged amino acids in a cluster of residues conserved in many members of the LIN-8 family. Of the nonsense mutations, n2731 and n2738 appeared to be substantially stronger than n2739 when tested in combination with lin-52(n771). Both n2731 and n2738 are predicted to truncate more than two-thirds of the wild-type LIN-8 protein, whereas n2739 is predicted to leave more than two-thirds of the wild-type LIN-8 protein intact.
On the basis of their molecular lesions, we considered n2731 and n2738 to be candidate null alleles of the lin-8 locus. Neither nonsense allele, however, inactivated the synMuv A pathway to the same extent as did loss of the class A synMuv gene lin-56 in the lin-15B(n2245) mutant background (Table 2). We also observed that the strong missense mutations n2376, n2378, and n2403 appeared more penetrant for the synMuv phenotype than either n2731 or n2738 in combination with lin-15B(n2245) (Table 2). To more stringently determine if n2731 and n2738 were null alleles of the lin-8 locus, we compared the penetrances of the synMuv phenotype of hermaphrodites homozygous for each allele with hermaphrodites heterozygous for each allele and for ccDf11, a deficiency that deletes the lin-8 locus (see Figure 1A). The penetrances of the synMuv phenotype for both lin-8(n2731); lin-15B(n2245) and lin-8(n2738); lin-15B(n2245) homozygotes were weaker than those of lin-8(n2731)/ccDf11; lin-15B(n2245) and lin-8(n2738)/ccDf11; lin-15B(n2245) heterozygotes, respectively (Table 2). One possible interpretation of these observations is that neither n2731 nor n2738 completely eliminates lin-8 function. However, the ccDf11 deficiency also eliminates several other LIN-8 family members: B0454.9, C41H7.3, C41H7.4, C41H7.5, C41H7.6, F14D2.2, F54D10.3, K08A2.1, and K08A2.4. We suspect that n2731 and/or n2738 are null alleles of lin-8 and that the penetrances of their synMuv phenotypes are enhanced by a decrease in the dosage of one or more of the lin-8 family genes deleted by ccDf11. This latter hypothesis is supported by the observation that no LIN-8 protein is detected in n2738 protein extracts (see below).
Unlike mutants carrying the nonsense mutations n2731 and n2738, mutants carrying the missense mutation n2376 had an equally penetrant phenotype when homozygous as when heterozygous over the ccDf11 deficiency (Table 2), suggesting that n2376 may be a null allele of lin-8. However, n2376 results in the production of stable LIN-8 protein at least in extracts from mixed-stage animals (see below). Furthermore, lin-8(n2376) retains wild-type lin-8 function in another assay (H. T. Schwartz and H. R. Horvitz, unpublished results). Thus, it is likely that the n2376 allele is not a null allele of lin-8 but is instead specifically defective for lin-8 synMuv A function.
LIN-8 is a nuclear protein expressed in many cells:
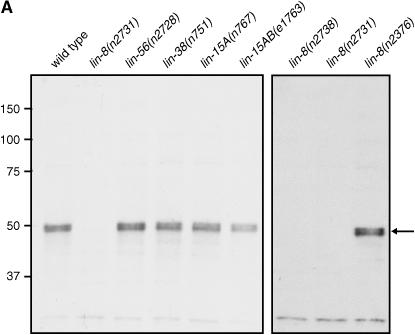
To determine the expression pattern and localization of the LIN-8 protein, we generated a rabbit polyclonal antibody against a fusion of full-length LIN-8 with maltose-binding protein (MBP-LIN-8). The affinity-purified and preadsorbed antibody recognized an apparent doublet of ∼50 kD in wild-type but not in lin-8(n2731) or lin-8(n2738) protein extracts analyzed by Western blots (Figure 3A); the predicted size of the LIN-8 protein is 44 kD. As the two LIN-8 proteins are approximately equal in their levels and the six lin-8 cDNA clones (courtesy of Yuji Kohara) that we analyzed are identical in sequence, we suspect that the LIN-8 protein may be post-translationally modified. The n2376 missense mutation does not destabilize the full-length LIN-8 protein (Figure 3A).
Figure 3.
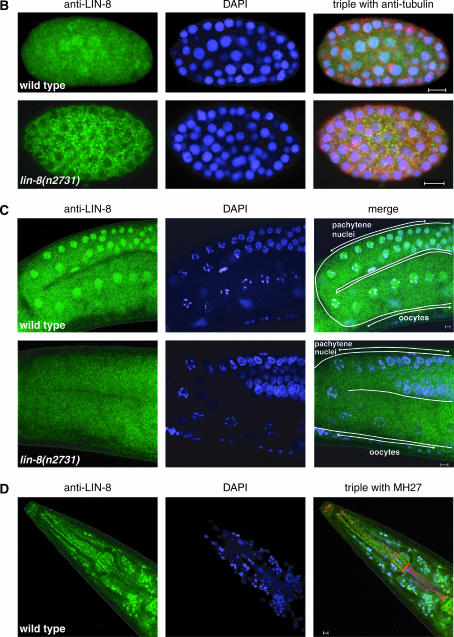
LIN-8 protein is expressed broadly and localized in nuclei. (A) Western analysis of protein extracts from wild-type, lin-8(lf), lin-56(n2728), lin-38(n751), lin-15A(n767), and lin-15AB(e1763) mixed-stage worms probed with affinity-purified and preadsorbed anti-LIN-8 antibody. The position of the apparent LIN-8 protein doublet is indicated by the arrow. The molecular weights of marker proteins are indicated at left in kilodaltons. (B–D) Whole-mount staining of wild-type and lin-8(lf) animals with affinity-purified and preadsorbed anti-LIN-8 antibodies (green), as well as DAPI (blue) to visualize DNA. Staining with antitubulin antibody (red) is shown as a fixation control in embryos. Staining with the MH27 antibody (red), which recognizes the apical borders of C. elegans epithelial cells, is shown as a fixation control in adults. Bars, 5 μm. (B) LIN-8 staining is observed in multiple nuclei in the wild-type but not the lin-8(n2731) embryo. (C) LIN-8 staining is present in the wild-type but not in the lin-8(n2731) gonad in pachytene nuclei and in oocytes. (D) LIN-8 staining is observed in multiple nuclei in the wild-type adult head.
Since lin-8 functions with lin-15A, lin-38, and lin-56 to inhibit vulval development (Ferguson and Horvitz 1989; Thomas et al. 2003), we analyzed the impact of loss-of-function mutations in these class A synMuv genes on the LIN-8 protein. By Western blot analysis, neither the levels nor the electrophoretic mobility of LIN-8 appears to be altered in lin-56(lf), lin-15A(lf), or lin-38(lf) mutants (Figure 3A). This result contrasts with that for the class A synMuv proteins LIN-15A and LIN-56, which are dependent on each other, but not on lin-8 or lin-38, for wild-type levels (E. M. Davison, A. M. Saffer, L. S. Huang, J. DeModena, P. W. Sternberg and H. R. Horvitz, unpublished results).
We used the same anti-LIN-8 antibody for whole-mount staining of worms. LIN-8 appeared to be predominantly localized to nuclei (Figure 3, B–D). We observed LIN-8 expression in many cells in embryos, larvae, and adults, and LIN-8 staining was particularly prominent in the germline as well as in neuronal nuclei of the head (Figure 3, B–D). Although diffuse within the syncytium of the distal gonad arms, LIN-8 was specifically associated with germ cell nuclei during the pachytene stage and was also localized to oocyte nuclei (Figure 3C). No anti-LIN-8 staining was observed in any of these somatic or germ cell nuclei in lin-8(n2731) animals at any stage (Figure 3, B and C; data not shown). Background staining in the larval midbody of lin-8(n2731) animals was too high to examine LIN-8 expression in vulval cells.
LIN-8 interacts with LIN-35 Rb in vitro:
We performed a yeast two-hybrid analysis of a C. elegans cDNA library using full-length lin-35 Rb as bait and identified LIN-8 as a potential LIN-35 Rb interactor (data not shown). The Gal4-based screen made use of three reporter genes: GAL1∷HIS3, GAL1∷lacZ, and SPAL10∷URA3 (Fields and Song 1989; Walhout and Vidal 2001). Of 1.4 × 106 transformants, we identified 11 clones that grew on selective medium in the presence of 3-aminotriazole. Further analysis revealed that 6 of these 11 clones also expressed β-galactosidase and were able to grow in the absence of uracil. All 6 clones that tested positive for expression of all three reporter genes were found to contain the B0454.1 open reading frame encoding LIN-8. Neither LIN-35 nor LIN-8 interacted in the yeast two-hybrid system with any of 29 other vulval proteins tested (Walhout et al. 2000a; data not shown). Western blot analyses (data not shown) indicate that LIN-35 is expressed at wild-type levels in lin-8(n2731) worms and that LIN-8 is expressed at wild-type levels in lin-35(n745) worms, which lack LIN-35 protein (Lu and Horvitz 1998).
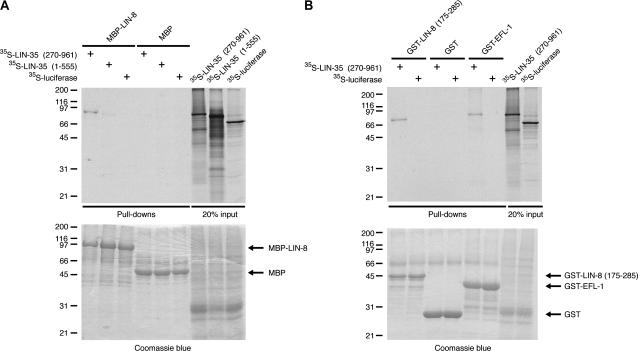
To test the hypothesis that LIN-8 and LIN-35 Rb can interact and, if so, to identify the region of LIN-35 Rb required for the interaction, we sought to determine if LIN-8 could associate in vitro with two different fragments of LIN-35 Rb. The A/B pocket domain of mammalian pRb, p107, and p130 mediates association with many interacting proteins (Morris and Dyson 2001), and in C. elegans is contained within a portion of LIN-35 Rb sufficient for interaction in in vitro pull-down experiments with LIN-53 RbAp48, HDA-1 HDAC, DPL-1 DP, and EFL-1 E2F (Lu and Horvitz 1998; Ceol and Horvitz 2001). An [35S]methionine-labeled N-terminal LIN-35 Rb fragment (aa 1–555), which lacks an intact pocket domain, failed to show any significant association with a full-length MBP-LIN-8 fusion protein in in vitro pull-down assays (Figure 4A). By contrast, an [35S]methionine-labeled C-terminal LIN-35 Rb fragment (aa 270–961), which contains the pocket domain, interacted with MBP-LIN-8 (Figure 4A). LIN-8 and LIN-35 Rb are thus capable of interacting in both yeast two-hybrid and in vitro pull-down assays. Furthermore, these observations suggest that, as with its other C. elegans interactors (Lu and Horvitz 1998; Ceol and Horvitz 2001), LIN-35 Rb associates with LIN-8 through its C terminus, possibly via the pocket domain.
Figure 4.
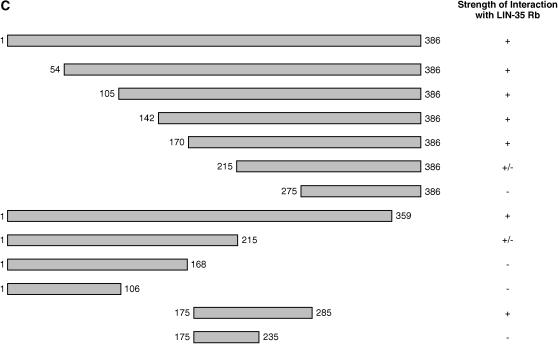
LIN-8 interacts with LIN-35 Rb in vitro. (A) LIN-35 Rb (aa 270–961) but neither LIN-35 Rb (aa 1–155) nor luciferase interacts with full-length MBP-LIN-8 fusion protein. None of the constructs interacts with MBP alone. Twenty percent of the 35S-labeled proteins used in the binding reactions are shown. Coomassie blue staining indicates that approximately equal amounts of full-length MBP-LIN-8 and MBP were used in the binding reactions. The molecular weights of marker proteins are indicated at the left in kilodaltons. (B) LIN-35 Rb (aa 270–961) but not luciferase interacts with GST-LIN-8 (aa 175–285) and GST-EFL-1. Neither construct binds to GST alone. Twenty percent of the 35S-labeled protein used in the binding reactions are shown. Coomassie blue staining indicates that approximately equal amounts of MBP, GST, and fusion proteins were used in the binding reactions. The molecular weights of marker proteins are indicated at the left in kilodaltons. (C) Summary of the LIN-8 fragments used for in vitro pull-down experiments and of their ability to interact with LIN-35 Rb (aa 270–961). +, wild-type interaction; +/−, interaction detected but weaker than that of wild type; −, no interaction.
We next identified the domain of LIN-8 required for interaction with the C-terminal LIN-35 Rb fragment using an in vitro pull-down assay. Progressive deletions of N- and C-terminal LIN-8 residues revealed that amino acids 170–359 of LIN-8 were necessary for interaction with LIN-35 Rb (data not shown; Figure 4C). Furthermore, amino acids 175–285 of LIN-8 were sufficient for interaction with the C-terminal LIN-35 Rb fragment (Figure 4, B and C). Several viral proteins interact with the pocket domain of pRb via an LXCXE motif (Harbour and Dean 2000); this sequence is not contained within amino acids 175–285 of LIN-8. Amino acids 175–285 of LIN-8 share a small region of similarity with other LIN-8 family members (Figure 2). None of the lin-8 missense mutations resides within the interaction domain, and thus no existing mutation compromises the predicted association between LIN-35 Rb and LIN-8 by directly affecting the interaction domain.
DISCUSSION
The class A synMuv genes function redundantly with the C. elegans homolog of the mammalian tumor suppressor pRb to inhibit Ras-mediated vulval development. We have shown that the class A synMuv gene lin-8 encodes a novel nuclear protein that not only functions redundantly but also physically interacts with C. elegans Rb. Given these observations, we propose that the class A synMuv genes act in transcriptional regulation. Further characterization of the mechanism by which the proteins of the class A synMuv pathway act may well reveal molecular processes that interact with the mammalian Rb tumor-suppressor pathway in both the regulation of cell fate and the prevention of tumorigenesis.
Class A synMuv genes may regulate transcription:
The two previously cloned synMuv A genes—lin-15A and lin-56—encode novel nuclear proteins that share a novel C2CH motif also found in the synMuv B proteins LIN-15B and LIN-36, as well as in HIM-17, a protein required for meiotic recombination and histone H3 lysine-9 methylation in the germline (Reddy and Villeneuve 2004; E. M. Davison, A. M. Saffer, L. S. Huang, J. DeModena, P. W. Sternberg and H. R. Horvitz, unpublished results). This C2CH motif is likely related to the THAP domain (Roussigne et al. 2003; Clouaire et al. 2005) and has been proposed to mediate interaction with chromatin or chromatin-associated proteins (Reddy and Villeneuve 2004; E. M. Davison, A. M. Saffer, L. S. Huang, J. DeModena, P. W. Sternberg and H. R. Horvitz, unpublished results). The THAP domain of the human protein THAP1 has been shown to possess zinc-dependent sequence-specific DNA-binding activity in vitro (Clouaire et al. 2005). It has therefore been proposed that the synMuv A proteins inhibit vulval development through the regulation of transcription (E. M. Davison, A. M. Saffer, L. S. Huang, J. DeModena, P. W. Sternberg and H. R. Horvitz, unpublished results).
The nuclear localization of LIN-8 and the physical association between LIN-8 and LIN-35 Rb is consistent with the hypothesis that LIN-8 is present at the sites of transcriptional repressor complexes. How might the proteins encoded by these three synMuv A genes modulate transcriptional activity? Three general mechanisms seem possible. First, like some synMuv B proteins (Lu and Horvitz 1998; Solari and Ahringer 2000; von Zelewsky et al. 2000; Ceol and Horvitz 2001; Couteau et al. 2002), the synMuv A proteins may impact chromatin structure. Second, the synMuv A proteins may have more direct functions in regulating the initiation, elongation, or termination of transcription. Third, the synMuv A proteins may mediate the localization of target genes to nuclear subdomains where their transcription could be coordinately and efficiently regulated. The localization of genes to the vicinity of centromeric heterochromatin, for example, may contribute to transcriptional repression in Drosophila and mammals (Gasser 2001).
Physical interaction between synMuv A and B proteins:
Indirect physical interaction between class A and class B proteins was proposed by Walhout et al. (2000a), who found that the synMuv A protein LIN-15A shared interactors with the synMuv B proteins LIN-36 and LIN-37 in the yeast two-hybrid system. As RNAi analysis has thus far not revealed a role for any of these shared interactors in the class A or class B synMuv pathways or in antagonism of these pathways (C. J. Ceol and H. R. Horvitz, unpublished results), the significance of this observation remains unclear.
The direct interaction between LIN-8 and LIN-35 Rb in vitro described in this article suggests that LIN-8 and LIN-35 Rb may associate in vivo. The biological role of such an interaction is unclear. If the putative interaction between LIN-8 and LIN-35 Rb were to facilitate lin-8 and/or lin-35 Rb function in vulval development, then one might expect to observe both synMuv A and synMuv B activity associated with one or both of these genes. However, a reduction of lin-8 function does not result in a synMuv phenotype in combination with a loss of the function of the synMuv A genes lin-15A, lin-38, or lin-56 (Ferguson and Horvitz 1989; Thomas et al. 2003), suggesting that lin-8 does not possess class B synMuv activity. Similarly, a loss of lin-35 Rb function does not result in a synMuv phenotype in combination with a loss of the function of the synMuv B genes lin-36, lin-37, or lin-15B (Ferguson and Horvitz 1989), suggesting that lin-35 Rb does not possess class A synMuv activity.
Proteins that physically interact often work together directly in the same biological process. By contrast, synthetic genetic interactions between null alleles of two genes usually indicate that the genes affect a biological process through separate mechanisms. Although lin-8 and lin-35 Rb function in the parallel synMuv A and synMuv B pathways, respectively, the proteins that they encode physically interact in vitro. If the interaction between LIN-8 and LIN-35 Rb is biologically important, then three models could explain why neither lin-8 nor lin-35 Rb appears to possess both synMuv A and synMuv B activity. First, the functional consequence of the LIN-8/LIN-35 Rb interaction may be redundant with another process in vulval development. For example, LIN-8 may be independently localized to the promoters of vulval specification genes by both LIN-35 Rb and another protein. Second, LIN-8 and LIN-35 Rb may function together in the vulva, but in some process not required for vulval development. Third, lin-8 and lin-35 Rb may act together but not in the vulva. The widespread expression of lin-8 and lin-35 Rb (Lu and Horvitz 1998) indicates that they could function together in other tissues. Mutation of lin-35 Rb has indeed been shown to result not only in the synMuv B phenotype but also in defects in cell cycle progression (Boxem and van den Heuvel 2001; Fay et al. 2002; Garbe et al. 2004), in defects in pharyngeal morphogenesis (Fay et al. 2003), and in severely reduced expression of a muscle-cell-specific gfp reporter gene from repetitive transgene arrays (the Tam phenotype) (Hsieh et al. 1999). Although current evidence suggests that lin-8 does not function with lin-35 Rb in the regulation of either cell cycle progression or transgene expression (Hsieh et al. 1999; Boxem and van den Heuvel 2002; Garbe et al. 2004; E. C. Andersen and H. R. Horvitz, unpublished observations), the possibility remains that lin-8 and lin-35 Rb act together in the developing pharynx or in processes not yet analyzed.
Partial redundancy in the LIN-8 family:
The nonsense alleles n2731 and n2738 appeared to be null alleles on the basis of their molecular lesions and lack of LIN-8 protein, yet, by comparison to loss of the class A synMuv gene lin-56 and by comparison to a deficiency that removes the lin-8 locus, did not appear to have lost all synMuv A pathway activity. By contrast, three of the lin-8 missense alleles impaired synMuv A activity almost to the extent seen upon loss of lin-56. One of these three lin-8 missense alleles, n2376, acted like a null by deficiency analysis but did not destabilize full-length LIN-8 protein (at least in extracts from mixed-stage animals). One hypothesis to account for these observations is that LIN-8 normally functions as part of a protein complex and that other family members can partially replace LIN-8 activity within the complex in its absence. Specifically, in mutants that lack LIN-8 protein, closely related family members may partially substitute for LIN-8. ccDf11, the deletion used for deficiency analysis, removes both lin-8 and several lin-8 family members. The predicted partial replacement of LIN-8 by other LIN-8 family members may therefore be reduced in lin-8(null)/ccDf11 heterozygotes as compared to lin-8(null) homozygotes. The strong missense alleles may encode stable LIN-8 proteins that inactivate other family members either by direct interaction or by competition with a partner. A similar phenomenon has been observed in Saccharomyces cerevisiae for the MAP kinases Fus3 and Kss1, which in wild-type yeast regulate the mating pheromone response and filamentation pathways, respectively (Madhani et al. 1997). In the complete absence of Fus3, Kss1 provides substitute MAPK activity for the mating pheromone response pathway and thus only a slight reduction in mating efficiency is observed. By contrast, when Fus3 is rendered catalytically inactive by a missense mutation, Kss1 cannot substitute in the mating pheromone response pathway and a much stronger mating defect results. A comparable model has been proposed for the histone deacetylase family in Drosophila, as missense but not null mutations of the histone deacetylase HDAC1 dominantly suppress silencing caused by position-effect variegation (Mottus et al. 2000).
The biological roles of the remaining 16 lin-8 family members are not known. Their similarity to lin-8 suggests that these genes are also likely to have roles in transcriptional regulation, perhaps with other components or in other cells. The high degree of similarity shared by the lin-8 family members also suggests that redundancy may have prevented their identification in genetic screens: more than one family member may have to be inactivated for a mutant phenotype to be apparent.
Interactions within the class A synMuv pathway:
LIN-56 and LIN-15A are dependent on each other for wild-type protein levels and likely form a functional complex in vivo (E. M. Davison, A. M. Saffer, L. S. Huang, J. DeModena, P. W. Sternberg and H. R. Horvitz, unpublished results). Mutation of lin-8 or lin-38 does not perturb the expression or localization of either LIN-56 or LIN-15A, indicating that neither lin-8 nor lin-38 is normally required for formation or stability of the putative LIN-56/LIN-15A complex. In this article, we demonstrate that lin-56, lin-15A, and lin-38 do not appear to be required for expression of LIN-8. These results form a basis upon which our understanding of both the roles of individual components of the class A synMuv pathway and the interactions among these components can be further expanded.
Implications for human cancer:
Mammalian tumorigenesis requires deregulation of cell proliferation, cell differentiation, and apoptosis and is thus an inherently synthetic process requiring multiple mutations in the proto-oncogene and tumor-suppressor pathways controlling these biological activities (Hanahan and Weinberg 2000). The Rb tumor-suppressor pathway likely plays a critical role in preventing oncogenic transformation, as its inactivation is observed in many human cancers (Nevins 2001). That the class A synMuv genes function redundantly with the C. elegans Rb pathway suggests that mammalian counterparts of the synMuv A genes may well possess tumor-suppressor activity. We hope that characterization of the mechanism by which the class A synMuv genes function will lead to greater understanding of processes that act with the mammalian Rb pathway both in cell-fate determination and in protection from oncogenic transformation and that the redundancy of the class A, B, and C synMuv genes in regulating C. elegans vulval cell fates will serve as a model for the etiology of other synthetic processes, such as tumorigenesis, the manifestation of which requires multiple mutations.
Acknowledgments
We thank Craig Ceol and Hillel Schwartz for critical reading of the manuscript and members of the Vidal lab for their support. We are grateful to Caroline Spike and Bob Herman for data concerning the left endpoints of the ccDf1, ccDf2, and ccDf11 deficiencies. We thank Beth Castor for help with DNA sequence determination, Na An for strain management, Yuji Kohara for EST cDNA clones, and Alan Coulson and the C. elegans Sequencing Consortium for cosmid clones and sequences. The deficiency strains were provided by Theresa Stiernagle of the Caenorhabditis Genetics Center, which is supported by the National Institutes of Health (NIH) National Center for Research Resources. This work was supported by NIH grant GM24663. E.M.D. was supported by a Howard Hughes Medical Institute predoctoral fellowship. H.R.H. is an Investigator of the Howard Hughes Medical Institute.
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession no. DQ150101.
References
- Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller et al., 2002. The Pfam proteins families database. Nucleic Acids Res. 30: 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxem, M., and S. van den Heuvel, 2001. lin-35 Rb and cki-1 Cip/Kip cooperate in developmental regulation of G1 progression in C. elegans. Development 128: 4349–4359. [DOI] [PubMed] [Google Scholar]
- Boxem, M., and S. van den Heuvel, 2002. C. elegans class B synthetic Multivulva genes act in G1 regulation. Curr. Biol. 12: 906–911. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium, 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018. [DOI] [PubMed] [Google Scholar]
- Ceol, C. J., and H. R. Horvitz, 2001. dpl-1 DP and efl-1 E2F act with lin-35 Rb to antagonize Ras signaling in C. elegans vulval development. Mol. Cell 7: 461–473. [DOI] [PubMed] [Google Scholar]
- Ceol, C. J., and H. R. Horvitz, 2004. A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev. Cell 6: 563–576. [DOI] [PubMed] [Google Scholar]
- Chen, L., M. Krause, B. Draper, H. Weintraub and A. Fire, 1992. Body-wall muscle formation in Caenorhabditis elegans embryos that lack the MyoD homolog hlh-1. Science 256: 240–243. [DOI] [PubMed] [Google Scholar]
- Clark, S. G., X. Lu and H. R. Horvitz, 1994. The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics 137: 987–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouaire, T., M. Roussigne, V. Ecochard, C. Mathe, F. Amalric et al., 2005. The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc. Natl. Acad. Sci. USA 102: 6907–6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau, F., F. Guerry, F. Müller and F. Palladino, 2002. A heterochromatin protein 1 homolog in Caenorhabditis elegans acts in germline and vulval development. EMBO Rep. 3: 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgley, M. L., C. A. Turner and D. L. Riddle, 1997. On-line C. elegans resources, pp. 1059–1062 in C. elegans II, edited by D. L. Riddle, T. Blumenthal, B. J. Meyer and J. R. Priess. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Falquet, L., M. Pagni, P. Bucher, N. Hulo, C. J. Sigrist et al., 2002. The PROSITE database: its status in 2002. Nucleic Acids Res. 30: 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay, D. S., S. Keenan and M. Han, 2002. fzr-1 and lin-35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes Dev. 16: 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay, D. S., E. Large, M. Han and M. Darland, 2003. lin-35/Rb and ubc-18, an E2 ubiquitin-conjugating enzyme, function redundantly to control pharyngeal morphogenesis in C. elegans. Development 130: 3319–3330. [DOI] [PubMed] [Google Scholar]
- Ferguson, E. L., and H. R. Horvitz, 1985. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics 110: 17–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, E. L., and H. R. Horvitz, 1989. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics 123: 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields, S., and O. Song, 1989. A novel genetic system to detect protein-protein interactions. Nature 340: 245–246. [DOI] [PubMed] [Google Scholar]
- Finney, M., and G. Ruvkun, 1990. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63: 895–905. [DOI] [PubMed] [Google Scholar]
- Francis, R., and R. H. Waterston, 1991. Muscle cell attachment in Caenorhabditis elegans. J. Cell Biol. 114: 465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe, D., J. B. Doto and M. V. Sundaram, 2004. Caenorhabditis elegans lin-35/Rb, efl-1/E2F and other synthetic multivulva genes negatively regulate the anaphase-promoting complex gene mat-3/APC8. Genetics 167: 663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser, S. M., 2001. Positions of potential: nuclear organization and gene expression. Cell 104: 639–642. [DOI] [PubMed] [Google Scholar]
- Guenther, C., and G. Garriga, 1996. Asymmetric distribution of the C. elegans HAM-1 protein in neuroblasts enables daughter cells to adopt distinct fates. Development 122: 3509–3518. [DOI] [PubMed] [Google Scholar]
- Hanahan, D., and R. A. Weinberg, 2000. The hallmarks of cancer. Cell 100: 57–70. [DOI] [PubMed] [Google Scholar]
- Harbour, J. W., and D. C. Dean, 2000. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 14: 2393–2409. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and D. P. Lane, 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Horvitz, H. R., and J. E. Sulston, 1980. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics 96: 435–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, J., J. Liu, S. A. Kostas, C. Chang, P. W. Sternberg et al., 1999. The RING finger/B-Box factor TAM-1 and a retinoblastoma-like protein LIN-35 modulate context-dependent gene silencing in Caenorhabditis elegans. Genes Dev. 13: 2958–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L. S., P. Tzou and P. W. Sternberg, 1994. The lin-15 locus encodes two negative regulators of Caenorhabditis elegans vulval development. Mol. Biol. Cell 5: 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay, B. K., M. P. Williamson and M. Sudol, 2000. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14: 231–241. [PubMed] [Google Scholar]
- Knoepfler, P. S., and R. N. Eisenman, 1999. Sin meets NuRD and other tails of repression. Cell 99: 447–450. [DOI] [PubMed] [Google Scholar]
- Koelle, M. R., and H. R. Horvitz, 1996. EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell 84: 115–125. [DOI] [PubMed] [Google Scholar]
- Kornfeld, K., 1997. Vulval development in Caenorhabditis elegans. Trends Genet. 13: 55–61. [DOI] [PubMed] [Google Scholar]
- Korswagen, H. C., R. M. Durbin, M. T. Smits and R. H. A. Plasterk, 1996. Transposon Tc1-derived, sequence-tagged sites in Caenorhabditis elegans as markers for gene mapping. Proc. Natl. Acad. Sci. USA 93: 14680–14685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, X., and H. R. Horvitz, 1998. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell 95: 981–991. [DOI] [PubMed] [Google Scholar]
- Madhani, H. D., C. A. Styles and G. R. Fink, 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91: 673–684. [DOI] [PubMed] [Google Scholar]
- Mello, C. C., J. M. Kramer, D. Stinchcomb and V. Ambros, 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, E. J., and N. J. Dyson, 2001. Retinoblastoma protein partners. Adv. Cancer Res. 82: 1–54. [DOI] [PubMed] [Google Scholar]
- Mottus, R., R. E. Sobel and T. A. Grigliatti, 2000. Mutation analysis of a histone deacetylase in Drosophila melanogaster: missense mutations suppress gene silencing associated with position effect variegation. Genetics 154: 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins, J. R., 2001. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 10: 699–703. [DOI] [PubMed] [Google Scholar]
- Nicolas, E., V. Morales, L. Magnaghi-Jaulin, A. Harel-Bellan, H. Richard-Foy et al., 2000. RbAp48 belongs to the histone deacetylase complex that associates with the retinoblastoma protein. J. Biol. Chem. 275: 9797–9804. [DOI] [PubMed] [Google Scholar]
- Nielsen, S. J., R. Schneider, U.-M. Bauer, A. J. Bannister, A. Morrison et al., 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412: 561–565. [DOI] [PubMed] [Google Scholar]
- Perez de la Cruz, I., J. Z. Levin, C. Cummins, P. Anderson and H. R. Horvitz, 2003. sup-9, sup-10 and unc-93 may encode components of a two-pore K+ channel that coordinates muscle contraction in Caenorhabditis elegans. J. Neurosci. 23: 9133–9145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien, P. W., and H. R. Horvitz, 2000. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat. Cell Biol. 2: 131–136. [DOI] [PubMed] [Google Scholar]
- Reddy, K. C., and A. M. Villeneuve, 2004. C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell 118: 439–452. [DOI] [PubMed] [Google Scholar]
- Richards, E. J., and S. C. R. Elgin, 2002. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108: 489–500. [DOI] [PubMed] [Google Scholar]
- Riddle, D. L., T. Blumenthal, B. J. Meyer and J. R. Priess (Editors), 1997. C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Roussigne, M., S. Kossida, A.-C. Lavigne, T. Clouaire, V. Ecochard et al., 2003. The THAP domain: a novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem. Sci. 28: 66–69. [DOI] [PubMed] [Google Scholar]
- Schultz, J., R. R. Copley, T. Doerks, C. P. Ponting and P. Bork, 2000. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28: 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solari, F., and J. Ahringer, 2000. NURD-complex genes antagonise Ras-induced vulval development in Caenorhabditis elegans. Curr. Biol. 10: 223–226. [DOI] [PubMed] [Google Scholar]
- Sternberg, P. W., and M. Han, 1998. Genetics of RAS signaling in C. elegans. Trends Genet. 14: 466–472. [DOI] [PubMed] [Google Scholar]
- Stringham, E. G., D. K. Dixon, D. Jones and E. P. M. Candido, 1992. Temporal and spatial expression patterns of the small heat shock (hsp16) genes in transgenic Caenorhabditis elegans. Mol. Biol. Cell 3: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier, F. W., A. H. Rosenberg, J. J. Dunn and J. W. Dubendorff, 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185: 60–89. [DOI] [PubMed] [Google Scholar]
- Thomas, J. H., C. J. Ceol, H. T. Schwartz and H. R. Horvitz, 2003. New genes that interact with lin-35 Rb to negatively regulate the let-60 ras pathway in Caenorhabditis elegans. Genetics 164: 135–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya, Y., T. H. Shin, N. Miliaras, J. Lee, T. Oyama et al., 2002. MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell 111: 991–1002. [DOI] [PubMed] [Google Scholar]
- von Zelewsky, T., F. Palladino, K. Brunschwig, H. Tobler, A. Hajnal et al., 2000. The C. elegans Mi-2 chromatin-remodelling proteins function in vulval cell fate determination. Development 127: 5277–5284. [DOI] [PubMed] [Google Scholar]
- Walhout, A. J. M., and M. Vidal, 2001. High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24: 297–306. [DOI] [PubMed] [Google Scholar]
- Walhout, A. J. M., R. Sordella, X. Lu, J. L. Hartley, G. F. Temple et al., 2000. a Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287: 116–122. [DOI] [PubMed] [Google Scholar]
- Walhout, A. J. M., G. F. Temple, M. A. Brasch, J. L. Hartley, M. A. Lorson et al., 2000. b GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328: 575–592. [DOI] [PubMed] [Google Scholar]
- Zhang, H. S., and D. C. Dean, 2001. Rb-mediated chromatin structure regulation and transcriptional repression. Oncogene 20: 3134–3138. [DOI] [PubMed] [Google Scholar]