Abstract
The homing endonuclease I-CreI recognizes a site in the gene encoding the 23S rRNA of Chlamydomonas reinhardtii. A very similar sequence is present in the 28S rRNA genes that are located on the X and Y chromosomes of Drosophila melanogaster. In this work we show that I-CreI expression in Drosophila is capable of causing induced DNA damage and eliciting cell cycle arrest. Expression also caused recombination between the X and Y chromosomes in the heterochromatic regions where the rDNA is located, presumably as a result of a high frequency of double-strand breaks in these regions. Approximately 20% of the offspring of males expressing I-CreI showed exceptional inheritance of X- and Y-linked markers, consistent with chromosome exchange at rDNA loci. Cytogenetic analysis confirmed the structures of many of these products. Exchange between the X and Y chromosomes can be induced in males and females to produce derivative-altered Y chromosomes, attached-XY, and attached-X chromosomes. This method has advantages over the traditional use of X rays for generating X-Y interchanges because it is very frequent and it generates predictable products.
I-CreI is a site-specific homing endonuclease from Chlamydomonas reinhardtii (Thompson et al. 1992). It recognizes a conserved consensus sequence and generates a double-strand break. The 22-bp recognition site for I-CreI occurs in the 23S rRNA gene of the Chlamydomonas chloroplast (Durrenberger and Rochaix 1993). I-CreI can function in human cells (Monnat et al. 1999) or in Drosophila melanogaster, where it is used routinely during in vivo gene-targeting techniques (Rong et al. 2002) when the I-CreI recognition sequences are introduced into a transgene. As we now show, I-CreI also recognizes endogenous sequences in the Drosophila genome. A sequence similar to the conserved Chlamydomonas recognition sequence can be found, with a 17/22-bp match, in the gene for the Drosophila 28S rRNA. None of the differences between the Chlamydomonas and Drosophila sequences prevent cutting by I-CreI in vitro (Argast et al. 1998).
When we first expressed I-CreI in Drosophila, there were strong indications that endogenous genomic sequences were cut. A 1-hr, 37° heat shock that normally does not affect viability (Golic and Lindquist 1989; Rong and Golic 2000) caused almost complete lethality for flies carrying the heat-inducible 70I-CreI transgene (Rong et al. 2002). Two genetic observations suggested that this lethality was caused by DNA damage to the sex chromosomes. First, females that were heterozygous white+/white displayed frequent eye-color mosaicism after I-CreI expression. Mitotic exchange between the two X's could produce such mosaicism for the white+ gene (Stern 1936), and since double-strand breaks could cause such recombination (Friesen 1936), we predicted that an I-CreI recognition site was on the X chromosome. Second, X/Y males were observed to be more sensitive to I-CreI expression than were X/X females. If exchange occurred between the sex chromosomes, then exchanges between the X and Y would be more likely to produce sex chromosome aneuploidy than exchanges between two X's. For these reasons, it seemed likely that exchange was occurring between the two sex chromosomes at a genetic element common to both, such as the rDNA loci.
The ribosomal DNA genes [rDNA; genetically defined as the bobbed (bb) loci] are genetically redundant, being found in clusters on the X and on the Y chromosomes (Lindsley and Zimm 1992). The X-linked rDNA locus is within the pericentric heterochromatin of the left (long) arm, while the rDNA locus of the Y chromosome is within the short arm. Within the rDNA, individual transcription units are found in both orientations (Robbins and Swanson 1988). Each cistron contains the genes for the 2S, 5.8S, 18S, and 28S rRNAs, with ∼100–200 cistrons found in each cluster. A copy of either the X-linked or the Y-linked array is typically sufficient for full function in Drosophila.
To confirm that I-CreI lethality and mosaicism were the result of I-CreI-mediated double-strand breaks in the rDNA, we investigated whether chromosome rearrangements between sex chromosomes could be recovered after expression of I-CreI. The results of those experiments are reported here. When we induced I-CreI expression in flies carrying marked sex chromosomes, we found that a large fraction of their offspring had rearranged sex chromosomes. Following I-CreI expression, we recovered products of X-Y exchange, Y-Y exchange, and X-C(1;Y) exchange. These chromosomes had the expected genetic properties and cytological structures. These results show that I-CreI expression is an efficient method for producing a variety of experimentally useful rearranged sex chromosomes in Drosophila.
Generation of DNA damage by I-CreI was further confirmed by a strong reduction in the mitotic index in larval neuroblasts after its expression. This result suggests that I-CreI expression can also be a useful method for inducing controlled DNA damage for investigations of cell cycle checkpoints and DNA repair.
MATERIALS AND METHODS
Drosophila stocks and strains:
The I-CreI-expressing lines were of genotype v, P{v+, 70I-CreI} and w1118; P{v+, 70I-CreI} Sb/TM6, Ubx. The X chromosomes were either y1 or y1 w1118. The Y chromosomes were BSYy+, y+Y, and Y,P{RSw, w+}10A (referred to as Yw+ in this work). We used y w/BSY and C(1)DX, y f/Y during the course of the crosses and for balancing the exchanged chromosomes in stocks. Since we did not use the forked marker, we simply refer to it as C(1)DX, y in the text and tables. The RSw-10A chromosome was generated in our lab (Golic et al. 1998), and all others were obtained from the Bloomington Drosophila Stock Center and are described by Lindsley and Zimm (1992).
Heat-shock regimen:
Flies of the appropriate genotype (see results) were allowed to lay eggs for 3 days, after which the adults were transferred to new food. The 0- to 3-day-old embryos and larvae were heat-shocked at 36° by immersing the vial in a circulating water bath for 1 hr. The cotton plugs were pushed down in the vials to prevent the larvae from escaping the heat shock by crawling up the walls of the vial. Progeny were scored on days 14 and 18 after the vial was set.
Cytological analysis:
Larvae of the appropriate sex (male for X-Y exchanges and female for X-X exchanges) were dissected in 0.7% sodium chloride. Brains were hypotonically shocked for 10 min in 0.5% sodium citrate and then were transferred to fresh methanol:acetic acid:water (in 11:11:2 proportions). After 2 min, the brains were moved to dichlorodimethylsilane-treated coverslips containing a drop of fresh 45% acetic acid. The brains were squashed and the slides frozen on dry ice. We removed the coverslips with a razor blade and immediately immersed the slides in 95% ethanol at −20°. After 15 min, we allowed the slides to air dry at room temperature. To stain the neuroblast chromosomes, the slides were rehydrated in 1× PBS with 1 mg/ml 4′,6-diamidino-2-phenylindole. The neuroblasts were mounted, and we used a Zeiss Axiophot microscope and an Axiocam digital camera to visualize and photograph the mitotic neuroblast chromosomes. Images were taken with Axiocam software and postprocessed for brightness and contrast using Adobe Photoshop 7 on a G4 iMac.
Statistical analyses:
Statistical analyses were performed using Excel: Mac v.X (Microsoft) or Instat 3.0a (GraphPad Software). Table 1 was analyzed using chi square contingency tables. We compared X, I-CreI/Y without heat shock to the same genotype with heat shock: χ2 = 48.4, d.f. = 1, P < 0.001; the null hypothesis states that the frequency of mitotic (anaphase plus metaphase) figures are the same. Table 2 was analyzed using chi square contingency tables. We compared the number of progeny expected from meiotic nondisjunction between females outcrossed to y w/y w (21 + 14 of 5099, or 0.69%) and those outcrossed to C(1)DX, y/Y (8 + 4 of 4064, or 0.3%) after heat shock: χ2 = 6.78, d.f. = 1, P = 0.009; null hypothesis states that the frequencies of nondisjunctional progeny are identical.
TABLE 1.
Mitotic index without and with I-CreI expression
| Neuroblasts
|
|||||
|---|---|---|---|---|---|
| Genotype | Heat shock | Mitotic | Nonmitotic | Total | % |
| X/Y | − | 60 | 1282 | 1342 | 4.5 |
| X/Y | + | 36 | 788 | 824 | 4.4 |
| X,I-CreI/Y | − | 67 | 1118 | 1185 | 5.7 |
| X,I-CreI/Y | + | 7 | 1143 | 1150 | 0.6 |
Mitotic index (metaphase plus anaphase) of cells without (−) and with (+) heat-shock-induced expression of I-CreI (X, I-CreI/Y). X/Y lacks the 70I-CreI transgene and serves as the control.
TABLE 2.
Normal, exceptional, and nondisjunctional progeny without and with I-CreI expression
| Female progeny
|
Male progeny
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mother's genotype | Heat shock | y B+ | y B | y+ B+ | y+ B | y+ B | y+ B+ | y B | y B+ | Total | % exceptions |
| y w | − | 1500 | 0 | 0 | 1 | 1371 | 0 | 0 | 0 | 2872 | 0.03 |
| y w | + | 2316 | 97 | 224 | 21 | 1868 | 441 | 118 | 14 | 5099 | 17.9 |
| C(1)DX, y | − | 24 | 1 | 5 | 1401 | 0 | 1 | 0 | 1830 | 3262 | 1.0 |
| C(1)DX, y | + | 8 | 73 | 304 | 1181 | 4 | 177 | 424 | 1893 | 4064 | 24.4 |
Counts and frequencies of exceptional progeny without (−) and with (+) heat-shock-induced expression of I-CreI in y w/BSYy+ males. Males were outcrossed to either y w/y w or C(1)DX, y/Y females.
RESULTS
Mitotic arrest or delay in response to I-CreI expression:
As described in the Introduction, we had several reasons to believe that I-CreI could generate double-strand breaks in the rDNA. This type of DNA damage typically produces cell cycle arrest until the damage can be repaired. As confirmation that DNA double-strand breaks were produced by I-CreI expression, we assayed the mitotic index of larval neuroblast cells shortly after heat-shock-induced expression of 70I-CreI. Cell cycle arrest should be apparent as a reduction in the fraction of cells undergoing mitosis. Third instar larvae of genotype X, 70I-CreI/Y were heat-shocked at 36°, returned to room temperature for 1 hr, and then larval brains were dissected, fixed, and stained to look for mitotic figures. As a control, we dissected brains from the same genotype prior to heat shock and from larvae without the 70I-CreI transgene, both before and after heat shock. The results are shown in Table 1 and clearly indicate that I-CreI expression causes a dramatic drop in mitotic index (P < 0.001; see Statistical analyses in materials and methods). The most likely explanation for this phenotype is the rapid induction of I-CreI, followed by efficient DNA cleavage and the induction of DNA damage checkpoints and cell cycle arrest prior to mitosis.
Chromosomes used to examine exchange:
The X and Y chromosomes used in this study are shown in Figure 1. Briefly, the X chromosome was normal in structure, but carried either recessive yellow (y) and white (w) mutations or y alone. For the purpose of discussion, we refer to the region between the centromere of the X and the rDNA locus as Xh (X-heterochromatin), and the region distal to the rDNA locus as Xe (X-euchromatin). Most genetic information on the X, except the centromere and the rDNA locus itself, lies within Xe.
Figure 1.
Structures of the chromosomes used in this study. The X chromosome has a normal structure and is mutant for yellow (y) and white (w). Yw+ is a normally structured Y chromosome that has a P{white+} transposed near the telomere of the short arm (“w+” triangle). y+Y is a Y chromosome of normal structure but has a portion of the X chromosome (including yellow+, y+) translocated to the telomere of the long arm. BSYy+ has X chromosome material, including BarStone (BS) and y+, translocated to the long and short arms, respectively. Centromeres are indicated by constrictions, euchromatin by thin lines, and heterochromatin by rectangles. The blocks of rDNA are indicated by the shaded portions of the heterochromatin. Xe, Xh, Yl, and Ys are chromosomal segments discussed in the text. The male fertility loci—kl-1, kl-2, kl-3, kl-5, ks-1, and ks-2—are also shown.
The Y chromosomes were basically normal in structure with the following exceptions. The Yw+ chromosome carried a w+-containing P-element transposed to a site near the short-arm telomere (Golic et al. 1998). The y+Y chromosome had the tip of the X chromosome (including the y+ locus) appended to the long arm of the Y. The BSYy+ chromosome contained X chromosome-derived material on both the long and the short arm (Williamson 1976; Lindsley and Zimm 1992). The X-ray irradiation that generated this chromosome introduced not only the BS and y+ markers, but also two additional blocks of sex chromosome heterochromatin, presumably containing rDNA arrays, on the long arm just proximal to BS and on the short arm just proximal to y+ (Gatti and Pimpinelli 1983). The short-arm fertility factors (ks-1 and ks-2) are located distal to the normal site of rDNA on this and on normal Y chromosomes. We refer to the region of the Y that lies distal to the normal rDNA as Ys (Y-short) and to the remainder of the normal Y chromosome as Yl (Y-long), since it greatly simplifies our discussion. Yl includes all the long-arm fertility factors, the centromere, and a small portion of the short arm. This nomenclature differs from the accepted YS and YL terminology (in uppercase letters) where the boundary of YS and YL is defined by the centromere, and not by the rDNA locus. All three of these Y chromosomes are wild type for rDNA function and fertility genes (kl-1, kl-2, kl-3, kl-5, ks-1, ks-2) and behave in all respects as normal Y chromosomes.
X-Y exchange:
We first tested whether I-CreI expression in males could cause exchange between the rDNA regions of X and Y chromosomes. For this experiment we made use of the BSYy+ chromosome. Normally, when a y/BSYy+ male is mated to y/y females, the daughters will be yellow Bar+ and the sons will be yellow+ BarS. In the control experiment (heat-shocked males with no 70I-CreI transgene), this was observed. There was only one exceptional offspring, a y+ BS female, among nearly 3000 regular offspring (0.03%; Table 2). This most likely arose by meiotic nondisjunction of the sex chromosomes in either a male or a female parent. Such exceptional offspring typically arise at a rate of ∼0.05–0.1% (Bridges 1916a,b).
Crosses to free-X females:
To test the ability of I-CreI to induce breakage and exchange in rDNA, we crossed y w/BSYy+; Sb 70I-CreI/+ males, in which I-CreI expression had been induced by heat shock, to y w/y w females. The production of progeny with exceptional phenotypes by these males was increased ∼500-fold to 17.9% of all progeny (Table 2). Most of the exceptional progeny inherited only one or the other of the dominant markers from the BSYy+ chromosome, and these included daughters that inherited one of these two markers, indicating that some form of chromosomal exchange was responsible for producing most of the exceptional offspring.
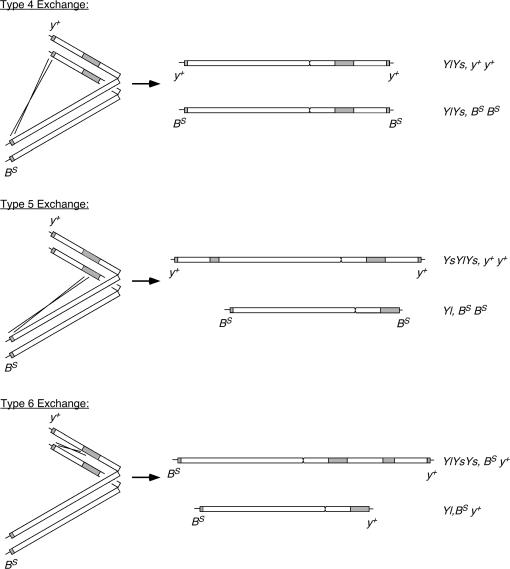
A large number of different exchange events are possible with four sites of rDNA in the parental males, and at least six different types of exceptional progeny were produced solely on the basis of the inheritance of the BS and y+ markers. We considered nine types of exchange that could generate the exceptional progeny: three interchromosomal and six intrachromosomal (types 1–3 and types 4–9, respectively; Figure 2). Excluded from our analyses are multiple exchanges and exchange events without phenotypic consequence, such as exchanges between homologous rDNA loci of sister chromatids. Also excluded are exchanges that would generate dicentric or acentric chromosomes, such as exchanges that may occur between rDNA subunits lying in opposite orientation. It is likely that such events do occur, but excluding their consideration at this point simplifies our discussion.
Figure 2.
Predicted exchange types. Nine types of simple exchange between regions of rDNA are predicted (excluding dyscentric exchanges, i.e., those that generate dicentric chromosomes; Lindsley 1955). Types 1–3 involve interchromosomal exchange between X and Y chromosomes. Types 4–6 involve intrachromosomal exchange between sister chromatids of the marked Y. Types 7–9 involve intrachromosomal exchange within one chromatid of the Y.
The three interchromosomal exchange types (types 1–3) can produce exceptional BarS or yellow+ daughters (XeYl, XeYlYs, or XeYsYl). The BarS or yellow+ sons (with Xh derivatives) can also be produced by these interchromosomal exchanges or by types 4 and 5 of intrachromosomal exchange. The yellow+ BarS daughters and yellow Bar+ sons could result from nondisjunction, which would require an increase of severalfold over the control level. Alternatively, the yellow Bar+ sons might also be produced by intrachromosomal exchanges, such as those designated as types 7 and 8, that could produce ring Y chromosomes [R(Y)].
Further tests confirmed that many, if not all, of the speculated exchanges did occur. Phenotypic observations and genetic behavior revealed at least two types of yellow+ Bar+ males, some of which must have been produced by intrachromosomal exchange. The first type appeared to carry only a portion of the Y chromosome. The duplication of the achaete-scute complex (AS-C) included with y+ on the tip of Ys produces a Hairy wing phenotype (Lindsley and Zimm 1992). Nine independent yellow+ male progeny that exhibited this Hairy wing phenotype were selected at random, and when mated, all were sterile. This was expected for the y+ derivatives produced by type 1 or type 3 exchange, because such chromosomes lack, at least, the long-arm fertility factors, and males carrying only this Ys fragment will be sterile. These sterile males also typically exhibited a variegated yellow/yellow+ phenotype, probably a reflection of position-effect variegation imposed on the y+ gene because of its proximity to Y heterochromatin. This variegation was not seen with BSYy+, but is likely visible in these sterile sons because they lack Yl and, as a consequence, position effects are enhanced because of the reduced heterochromatic content of the genome (Gowen and Gay 1934).
The second type of yellow+ Bar+ son exhibited an extreme Hairy wing phenotype and no y+ variegation. Three of three randomly selected males were fertile. They probably represent the products of type 4 or type 5 exchanges, with at least one complete complement of fertility factors and two copies of the y+ duplication, including the AS-C.
At least two, possibly three, types of yellow BarS sons were recovered. We randomly sampled the progeny of 10 I-CreI-expressing males. Of their pooled yellow BarS sons, 20 showed a variegated BarS/Bar+ phenotype, 6 showed an extremely strong BarS phenotype, and 27 displayed the normal BarS phenotype. We crossed 36 of these yellow BarS sons, and all were sterile. It is likely that the BarS-variegated sterile males carried the smaller product of a type 2 exchange, with BarS variegating because a position effect on BS was revealed owing to a reduction of heterochromatin in the genome of these males. The extreme BarS males were sterile and probably carried the Yl, BSBS product of a type 5 exchange. Although a chromosome with an extreme BarS phenotype could result from type 4 exchange, we expect such events to produce fertile males. It may be that our sampling was insufficient to recover these events: among the yellow BarS sons, the extreme BarS phenotypes were not frequent and we tested only three such males.
All expected phenotypic classes of exceptional females were also recovered. Only one type of yellow+ Bar+ daughter was expected: that bearing an attached-XY produced by a type 2 exchange. Most of the yellow+ Bar+ daughters had a mild Hairy wing phenotype and did not show variegation of y+. A few (7/224 = 3%) did exhibit y+ variegation, although it is not clear that the chromosome that they carry is any different from that of the nonvariegators. In some genetic backgrounds y+Y chromosomes are known to exhibit y+ variegation (Hadorn et al. 1970), and it is possible that these are simply the visible cases of infrequent variegation. It is also possible that a second round of exchange between the two rDNA blocks on the right arm of the attached-XY produced by type 2 exchange could delete a portion of the Y and alter the context of the appended y+ gene or the heterochromatic content of the genome, causing variegation of y+.
The yellow BarS daughters were of generally uniform appearance, with a BarS phenotype somewhat less severe than that of X/BSYy+ males. These were presumably produced by type 1 or type 3 exchange.
Finally, it is worth commenting further on the high frequency of yellow+ BarS females and yellow Bar+ males. These are the expected phenotypes for sex chromosome nondisjunction occurring in either parent. Only two of the yellow Bar+ males were mated: one was sterile and one was fertile. Thus, at least some of these exceptional males are fertile and carry a complete Y chromosome, likely males carrying a ring-Y produced by type 7 exchange. It is also possible that the frequency of X ↔ Y nondisjunction is elevated in these males. Disjunction of the X and Y depends on chromosome pairing mediated by elements in the rDNA loci (McKee and Karpen 1990). If the rDNA complement of the X or the Y were damaged by I-CreI-mediated double-strand breaks, it is conceivable that nondisjunction would result because of a decreased pairing fidelity. Thus, we think it likely that males that have expressed I-CreI have increased X ↔ Y nondisjunction. This issue could be resolved by further testing of the yellow+ BarS females, but this has not been done.
Several of the X-Y exchange events diagrammed would not produce exceptional progeny and would not have been recognized in this experiment. Type 6 and type 9 exchanges would produce yellow+ BarS sons, as would one of the two predicted products from exchanges of types 7 and 8. The Yl products from types 6 and 9 exchange might be recovered with appropriate crosses to initially provide the Ys fertility factors in trans. Recognition and recovery of the other exchange products that differ from the original BSYy+ only by the internal arrangement of the arms would be difficult, but would be possible with extensive cytological analysis.
Crosses to attached-X females:
We carried out a mating of similarly heat-shocked y w/BSYy+; Sb 70I-CreI/+ males to C(1)DX, y/Y females. With this cross, partial Y chromosomes should be recoverable in females, and attached-XY chromosomes recoverable in males with an extra Y to provide any missing fertility factors. In the control cross, using males without 70I-CreI, 24 yellow Bar+ females were produced. These were probably mostly XXX metafemales, as judged by their phenotype of crumpled wings. The five yellow+ Bar+ daughters were almost certainly a result of a single germline mitotic exchange, as they were all offspring of a single male. The two remaining exceptions, a yellow BarS female and a yellow+ Bar+ male were also offspring of a single male, apparently representing reciprocal products of spontaneous X-Y exchange.
In the crosses with 70I-CreI expression, we measured 24.4% exceptional progeny. Only a small fraction of these (12/4064 = 0.3%; Table 2) could have resulted from nondisjunction, again showing that breakage-induced exchange is the dominant source of exceptions. As in the previous experiment, all predicted exceptional phenotypes were recovered.
The yellow BarS daughters were of at least two types: 10 showed consistent BS variegation, while 3 showed a very strong BarS phenotype, stronger than the typical BarS phenotype. These likely represent the Y-derivatives produced by type 2 exchange for the B variegators or the Y-derivatives produced by types 4 and 5 exchange with two copies of BS for the very strong BarS phenotypes. The remaining 60 appeared Bar or weakly variegated for BS and were not analyzed further. The yellow+ Bar+ daughters were sometimes variegated for y+, usually showing a very mild Hairy wing phenotype. Since the Hairy wing phenotype produced by the AS-C-containing y+ duplication is reduced in females carrying attached-X chromosomes (Lindsley and Zimm 1992), these probably represent flies carrying the Y-derivatives with one or two copies of y+. We did not quantify the frequency of variegating yellow+ or Hairy wing phenotypes in these flies because of the unreliability of these mild phenotypes.
Strikingly, a few percent (∼6%) of both the regular and the exceptional daughters had a bobbed phenotype, as judged by the simultaneous occurrence of both etched tergites and small bristles, likely betraying a deficiency of rDNA cistrons. The C(1)DX chromosome carries no rDNA itself, and these females rely entirely on the Y chromosome inherited from their fathers for this function (Ashburner et al. 2004). This result indicates that I-CreI expression can result in deletions within the rDNA through either intrachromosomal deletion or unequal sister-chromatid exchange. The bobbed females cannot have resulted from a few preexisting rDNA-deficient Y chromosomes segregating in the stock, because the progeny of a single male were mostly bobbed+, with only occasional bobbed flies.
In these crosses all exceptional types of sons were also observed. There was little or no variation in y+ or BS expression in these flies.
We note that the two phenotypes expected as a result of sex chromosome nondisjunction are present at a much lower rate in these crosses than in the crosses to free-X females (0.3% vs. 0.69%, respectively, P = 0.009; see Statistical analyses in materials and methods), in spite of a somewhat higher overall recovery of exceptional progeny. This is most likely a consequence of the expected death of three-quarters of the progeny produced by X ↔ Y nondisjunction in males when mated to C(1)DX/Y females, whereas none of the nondisjunction sperm produce lethal zygotic genotypes when crossed to y w/y w females. In part, this result may also be attributed to the fact that sex chromosome nondisjunction in C(1)DX/Y females does not generate progeny of exceptional phenotype, although, on the basis of the control cross to free-X females, female nondisjunction accounted for only a small fraction of the yellow+ BarS daughters and yellow Bar+ sons in those matings.
Cytology of rearranged chromosomes:
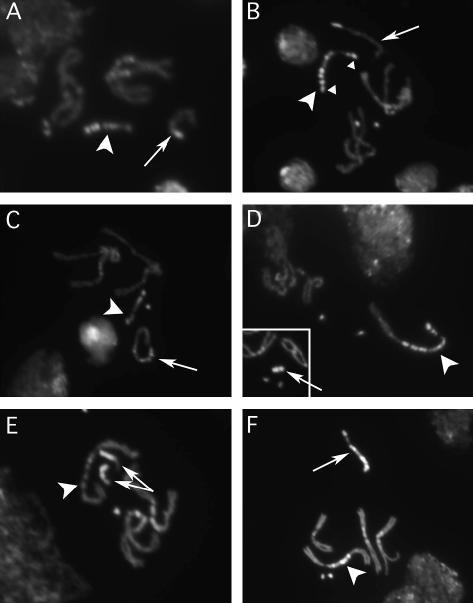
To confirm the existence of the expected chromosome rearrangements, we examined the mitotic metaphase cytology of several such rearrangements. We did not attempt to generate stable stocks of all derivatives generated in this study, but chose to retain a subset of derivatives with phenotypes expected for some exchange products. After selecting chromosomes that behaved genetically as expected for products of type 1, 2, or 3 exchanges (Figure 2), we prepared mitotic figures to observe the structure of both X- and Y-derivatives. In some cases (e.g., Figure 3, C and E) we performed crosses to generate males that carried reciprocal exchange chromosomes, enabling the analysis of both in the same preparation. Normal X (arrow) and Y (arrowhead) chromosomes are shown in Figure 3A, and the BSYy+ chromosome is shown in Figure 3B with small triangles indicating the material appended to the tip of each arm during its construction.
Figure 3.
Cytology of recombinant chromosomes. Mitotic chromosomes from larval neuroblasts. (A) Normal-structure X (arrow) and Y (arrowhead) chromosomes. (B) Karyotype containing a normal X chromosome (arrow) and the BSYy+ (arrowhead) chromosome. Triangles indicate chromatin added to the BSYy+ chromosome during its construction. (C) An attached-XY chromosome product of a type 1 exchange. The arrow marks the YsXh chromosome and the arrowhead marks the XeYl chromosome. (D) An attached-XY chromosome product of a type 2 exchange. An arrowhead marks the XeYlYs chromosome; the Xh chromosome [prepared from a C(1)DX/Xh, BS female] is indicated in the inset by an arrow. (E) A C(1;Y) chromosome product of a type 3 exchange. Two Xh chromosomes are marked with arrows (one is likely from a neighboring nucleus), and the C(1;Y) chromosome is marked with an arrowhead. (F) A C(1)RM chromosome generated as diagrammed in Figure 4. The C(1)RM chromosome is indicated by an arrowhead, and the normal Y is indicated by an arrow. In A–F, the autosomal karyotype is normal.
A type 1 exchange was expected to generate a short Y chromosome derivative, consisting of Ys marked with y+ attached to Xh. We selected 11 chromosome stocks that could represent either type 1 or type 3 exchanges. Of those, 4 were confirmed to be products of type 1 exchanges cytologically, and the remainder were products of type 3 exchanges. A chromosome with a structure indicative of a type 1 exchange is indicated by an arrowhead in Figure 3C. The complementary exchange product, Xe attached to Yl and marked by BS, is indicated by an arrow. Since males with the karyotype shown in Figure 3C possess an entire Y chromosome, albeit split into two different chromosomes, we expected them to be fertile and all were.
Type 2 exchange was expected to generate two chromosome derivatives: one very short one consisting of just Xh and the BS duplication from the tip of Yl and the other consisting of the entire Y chromosome plus Xe. We cytologically analyzed chromosomes from five chromosome stocks that behaved like products of type 2 exchanges, and all had this structure. The short chromosome, taken from a BarS female, is indicated with an arrow in the inset in Figure 3D (the small dots are chromosomes 4), and the longer XeYlYs chromosome, taken from a fertile yellow+ male, is indicated by an arrowhead.
The type 3 exchanges were expected to generate chromosomes similar to those of type 2: a small chromosome consisting of Xh and the end of Ys and another consisting of the Y chromosome and Xe. In the type 3 exchange, the Xh chromosome is marked with the yellow+ duplication and the XeYsYl chromosome with BS. Since the yellow+ end of the BSYy+ is longer than the BS fragment, we expected that the Xe y+ chromosome would be slightly longer than the BS chromosome produced by type 1 exchange (Figure 3E, arrows). Of 11 chromosomes that behaved genetically as type 1 or 3 exchanges, 7 were demonstrated to be products of type 3 exchanges cytologically. The expected structure of these chromosomes (Figure 2) was further confirmed by the fertility of yellow males, indicating that they contain all the normal Y sequence on their large XeYsYl chromosomes.
Y-Y exchanges:
It should be possible to detect I-CreI-induced exchange between two Y chromosomes by using males with two Y chromosomes that differ by markers on either side of a region of rDNA and screening for recombinants. To investigate the possibility of Y-Y exchange, we heat-shocked y w/y+Y/Yw+; 70I-CreI/+ males and crossed them to y w/y w females. We were interested in exchange events that generated y+Yw+ chromosomes (for other work) and so collected yellow+ white+ sons that represented potential Y-Y exchange events. Some of these progeny were expected to carry recombinant Y chromosomes with both markers (y w/y+Yw+), while others were expected to be males that received two paternal Y chromosomes and a maternal X chromosome (y w/y+Y/Yw+). Progeny with the desired recombinant chromosomes could be distinguished by three criteria. First, since the white+ gene on the Yw+ variegates (Golic et al. 1998; Maggert and Golic 2000), a y w/Yw+ male has very little white+ expression. In contrast, a y w/y+Y/Yw+ male shows greater w+ expression as a result of suppression of position-effect variegation by his extra Y chromosome. Second, a y w/y+Y/Yw+ male mated to y w females produces sons of three types: yellow+ white, yellow white+, and yellow+ white+, but a y w/y+Yw+ male produces only yellow+ white+ sons. Third, the y w/y+Y/Yw+ male also frequently transmits the marked Y chromosomes to daughters, giving female progeny with yellow+ or white+ phenotypes. In contrast, a y w/y+Yw+ male produces only yellow white female progeny. Thus, the desired y w/y+Yw+ male has relatively low expression of white+, exhibits linkage between y+ and w+, and shows sex-limited inheritance of these genes. We crossed 36 yellow+ white+ males with low levels of white+ expression to y w/y w females. Of these, two gave only yellow white daughters and yellow+ white+ sons. We concluded that these carried the desired recombinant y+Yw+ chromosomes. This result demonstrates that I-CreI is also useful for mediating exchange between two Y chromosomes.
I-CreI-mediated exchange in females:
To test whether I-CreI-mediated exchange can occur in females, we chose to generate a new attached-X chromosome by exchange between a normal-sequence X chromosome (marked with y) and an attached-XY or C(1;Y) (Figure 3D). On the basis of its genetic and cytological properties, this C(1;Y) has the structure expected for the chromosome produced by a type 2 exchange and is marked y w·y+ as diagrammed in Figure 4.
Figure 4.
Construction of an attached-X chromosome. Exchange between a normal-sequence X chromosome and C(1;Y) to generate C(1)RM is diagrammed.
To generate the attached-X chromosome, y w+/C(1;Y), y w·y+; I-CreI/+ females were heat-shocked and mated to y w/BSY males. Progeny with a new attached-X chromosome were expected to appear as exceptional BarS daughters, with the reciprocal product producing exceptional Bar+ sons. We scored the offspring of 53 individual females. Among 1570 daughters, we recovered 10 BarS daughters. Cytological analysis of one such chromosome revealed the structure of XeYlXe (Figure 3F, Figure 4). Another attached-X with the structure XeYlYsXe was also possible, but we did not specifically screen for this chromosome. We also recovered 6 Bar+ sons that were sterile and not investigated further.
These reverse-metacentric attached-X chromosomes [C(1)RM] were initially w/w+. Since they were generated, they all have produced only matroclinous daughters and patroclinous sons, confirming that they are attached-X chromosomes. However, a fraction of the female progeny were white, indicating that recombination occurred between the two Xe arms to generate w/w homozygotes, a normal behavior for C(1)RM chromosomes with heterozygous arms (Anderson 1925).
I-CreI-induced exchange is limited to the sex chromosomes:
Although I-CreI consensus sites were not found in the Drosophila genome at any site other than at the rDNA loci, we investigated genetically whether chromosome exchange could occur elsewhere in the genome in response to DNA damage induced by expression of I-CreI. Since meiotic recombination does not normally occur in males in Drosophila (Ashburner et al. 2004), we monitored whether I-CreI-mediated exchange would occur on autosomes 2 and 3 in males.
The test for recombination on chromosome 2 was carried out by crossing y w; 70I-CreI/TM3, Sb virgins to al1 sp2 males and heat-shocking the progeny at 36° for 1 hr. aristaless (al) and speck (sp) are located at the tips of opposite arms of chromosome 2, so recombination anywhere along chromosome 2 was expected to produce al + and + sp recombinant chromosomes. Daughters were heterozygous for yellow and white on the X chromosome, allowing us to confirm that 70I-CreI was induced by the presence of yellow patches on the abdomen and white sectors in the eyes of females. The al sp/+ +; I-CreI/+ heterozygous sons were outcrossed to females that were heterozygous for Sternopleural/SM1, Cy al sp. We scored the SM1-bearing progeny and observed 1063 nonrecombinant (al sp or + +) progeny. We recovered no recombinant progeny, indicating that I-CreI was unable to induce exchange on chromosome 2 between aristaless and speck.
We independently tested chromosome 3 exchange in a similar fashion, using a chromosome that carried recessive mutations for roughoid (ru), hairy (h), Kinked (Ki), rotund (rn), pink (p), rough (ro), and claret (ca). Progeny of ru h Ki rn p ro ca/70I-CreI heterozygous males crossed to ru1 h1 Ki1 rnroe-1 pp ro1 ca1 females fell into two classes exclusively: 1606 wild-type flies and 959 that exhibited all recessive phenotypes. We conclude that neither chromosome 2 nor chromosome 3 possesses sequences recognized by the I-CreI enzyme.
DISCUSSION
Compound chromosomes, like translocations, are rearrangements involving genetic elements joined to separate centromeres. Compound chromosomes are distinct in that they involve whole arms of homologous chromosomes. The generation of compounds of the X and Y relies critically upon exchanges that occur in heterochromatin. The reasons for this are straightforward. The only genes that are clearly located in X heterochromatin are the multicopy rDNA, which are also found on the Y. Thus, rearrangement breakpoints in X heterochromatin are readily tolerated and move essentially the entire chromosome as one piece. Rearrangements involving the Y are, by necessity, a result of breaks in heterochromatin because the Y is entirely heterochromatic. Furthermore, because the Y is entirely dispensable except for male fertility, and because Y aneuploidy is well tolerated, segments of the Y linked to the X have often been used to provide regions in which rearrangement breakpoints may occur to generate more complex compound chromosomes (Novitski 1954).
Exchanges to generate rearrangements between the X and Y have been achieved classically in one of two ways: spontaneously or by induction with ionizing radiation. Spontaneous exchanges between the X and the Y tend to occur in regions of shared homology, specifically between X heterochromatin and the base of the short arm of the Y, where the rDNA is located on each chromosome (Neuhaus 1937; Lindsley 1955; Maddern 1981; Williams et al. 1989). Induced exchanges also occur in these regions (Lucchesi 1965) but can occur in other regions with appreciable frequency (Ashburner et al. 2004). The frequencies of X-Y exchange in males typically range from ∼0.01% occurring spontaneously up to ∼2% occurring after irradiation.
The work presented here shows that I-CreI may also be used to generate rearrangements between the X and Y chromosomes, and with nearly 10-fold greater efficiency than has been previously achieved. Rearrangements occurring between rDNA regions of the X and BSYy+ arose in ∼20% of the progeny of an I-CreI-expressing male. Special schemes were not required to avoid aneuploidy of X chromosome segments because exchange occurred in heterochromatin. Many of the predicted X and Y exchange events were recovered and verified genetically and/or cytologically. I-CreI is known to generate double-strand breaks in the 23S rRNA gene of Chlamydomonas, and it is almost certainly the case that the sex chromosome rearrangements that we produced result from double-strand breaks in the closely related 28S gene of Drosophila. We confirmed that I-CreI causes DNA damage since its expression results in a large reduction in the mitotic index, an expected response to double-strand breaks.
New compound-XY and compound-X chromosomes were produced in these experiments. For instance, we designed and generated a novel C(1)RM chromosome that shows the properties of typical reverse-metacentric compound-X chromosomes (Novitski 1954). Attached-X chromosomes that exist and are in use typically have been kept in stock for decades. Some experiments may require the generation of attached-X chromosomes with heterozygous markers or with specific markers. Attached-X chromosomes are especially useful in meiotic studies, because they allow half-tetrad analysis of meiotic exchanges. However, to be useful in studies of exchange, such chromosomes should be heterozygous for markers along their length so that recombination can be assayed. It is difficult to maintain heterozygosity on attached-X chromosomes, and this has limited their use for this purpose. This work shows that, with I-CreI, it is possible to build new heterozygous attached-X chromosomes with relative ease as needed. The efficient generation of exchanges with I-CreI makes it quite feasible to routinely produce compound sex chromosomes.
Several types of exchange products were predicted and confirmed cytologically. It is likely that chromosome derivatives other than those described in Figure 2 were also produced. Dicentric chromosomes may be generated by unequal sister-chromatid exchange between rDNA repeats in opposite directions or by nonhomologous end-joining. Such dicentric chromosomes break in mitosis and, in the male germline, are subject to de novo telomere addition (Ahmad and Golic 1998). Thus, some of the exceptional progeny that we recovered may carry the products of dicentric breakage. The high frequency of exchange products suggests that multiple exchange events probably also occurred and were transmitted, but we did not specifically screen for such chromosomes.
Although our X-Y exchange experiments used a Y chromosome with three regions of rDNA, I-CreI exchanges should also occur between normal X and Y chromosomes. Exchange events could be recovered if, for instance, the Y chromosome was appropriately marked, perhaps with P-element insertions. In this fashion we produced a doubly marked Y by using I-CreI to mediate exchange between two singly marked Y chromosomes. Although our sample size was limited, the efficiency of this exchange appears to be on the order of 5%. Y chromosomes normally do not undergo meiotic recombination, and I-CreI represents the most efficient way of generating recombinant Y chromosomes.
The primary limitation on the variety of X-Y rearrangements that could be recovered in our experiments was the location of rDNA regions where I-CreI makes breaks. It may be possible to expand the range of chromosome rearrangements that can be recovered through the use of P-element insertions carrying the canonical recognition site for I-CreI (Rong et al. 2002) or carrying Drosophila rDNA (Karpen et al. 1988; McKee and Karpen 1990).
As an alternative to either spontaneous recombination or radiation-induced exchange, the maternal-effect mutation Rex (Ribosomal exchange) may be used to induce X-Y recombination at rates up to ∼3% (Rasooly and Robbins 1991). Rex also causes chromosomal exchange between regions containing rDNA and can produce expansion and contraction of the rDNA repeats (Robbins 1981). The rDNA arrays of Drosophila include repeated retrotransposon elements—the R1 and R2 elements; the origin, function, and regulation of the R2 element in the rDNA genes has been reviewed recently (Eickbush and Eickbush 2003). It is possible that Rex represents a mutation that derepresses these elements, allowing for an increased incidence of DNA damage in these loci, as has been proposed (Hawley and Marcus 1989; Robbins and Pimpinelli 1994). We note that I-CreI has effects that are very similar to those of Rex, and it is likely that these effects arise from the same cause—the generation of double-strand breaks within the rDNA.
A large number of homing endonucleases are known (Burt and Koufopanou 2004). Among these, there are likely to be others that recognize sequences normally found in the Drosophila genome, and they might be similarly used to generate rearrangements between endogenous sites. Other techniques that might also produce chromosome rearrangements at specific loci are being developed. Double-strand breaks can be generated at many specific sites in the genome with engineered site-specific nucleases. Carroll and co-workers (Bibikova et al. 2002) have designed novel zinc-finger nucleases that can target specific sequences, and have used these to generate double-strand breaks within the yellow gene. Another approach has been to alter the specificity of existing nucleases that recognize complex sites. Stoddard and co-workers (Chevalier et al. 2002) have altered the DNA recognition of a chimeric homing endonuclease (I-CreI and I-DmoI), creating a synthetic enzyme, E-DreI, that has altered DNA specificity.
The use of nucleases designed to recognize existing sites has the potential to generate specific chromosome rearrangements between normal genomic sequences. Another approach has been to use transposons, or sequences carried by transposons, to generate rearrangements between the sites of transposon insertion. In Drosophila, chromosome rearrangements between defined sites in euchromatin have also been generated during the mobilization of transposons (Lim 1979, 1988; Berg et al. 1980; Engels and Preston 1984; Gray et al. 1996) by FLP-FRT-mediated site-specific recombination (Golic and Golic 1996; Ryder et al. 2004; Thibault et al. 2004), by expression of I-SceI to make double-strand breaks simultaneously at two different sites (Egli et al. 2004; Y. S. Rong, personal communication), and by Cre-lox site-specific recombination (Egli et al. 2004). However, these methods all rely on the insertion of foreign sequences and, as yet, do not fulfill a need for the efficient generation of rearrangements in heterochromatin. It may be possible to extend the use of site-specific recombinases or I-SceI to produce rearrangements between heterochromatic sites by generating collections of elements that carry those target sites at heterochromatic locations (Konev et al. 2003). Until that time, I-CreI expression serves for the efficient production of compound sex chromosomes.
Beyond the demonstrated utility of this technique in generating chromosome translocations, I-CreI may also be used as a tool for studying the responses to DNA damage (Royou et al. 2005). Because the induction of I-CreI by heat shock causes a large number of breaks at a defined location, it causes rapid and efficient cell cycle arrest. I-CreI expression provides an alternative to the standard use of X rays for the induction of damage, and in some respects is preferable, because the nature of the cellular damage is more precisely defined than when ionizing radiation is used.
Acknowledgments
We thank Mary Golic and William F. Ryan III for technical assistance. This work was supported by National Institutes of Health grant GM-065604. Keith Maggert was supported by National Research Service Award Postdoctoral Fellowship GM-65777.
References
- Ahmad, K., and K. G. Golic, 1998. The transmission of fragmented chromosomes in Drosophila melanogaster. Genetics 148: 775–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, E. G., 1925. Crossing over in a case of attached X chromosomes in Drosophila melanogaster. Genetics 10: 403–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argast, G. M., K. M. Stephens, M. J. Emond and R. J. Monnat, Jr., 1998. I-PpoI and I-CreI homing site sequence degeneracy determined by random mutagenesis and sequential in vitro enrichment. J. Mol. Biol. 280: 345–353. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., K. G. Golic and R. S. Hawley, 2004. Drosophila: A Laboratory Handbook, Ed. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Berg, R., W. R. Engels and R. A. Kreber, 1980. Site-specific X-chromosome rearrangements from hybrid dysgenesis in Drosophila melanogaster. Science 210: 427–429. [DOI] [PubMed] [Google Scholar]
- Bibikova, M., M. M. Golic, K. G. Golic and D. Carroll, 2002. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161: 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges, C. B., 1916. a Non-disjunction as proof of the chromosome theory of heredity. Genetics 1: 1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges, C. B., 1916. b Non-disjunction as proof of the chromosome theory of heredity. Genetics 1: 107–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt, A., and V. Koufopanou, 2004. Homing endonuclease genes: the rise and fall and rise again of a selfish element. Curr. Opin. Genet. Dev. 14: 609–615. [DOI] [PubMed] [Google Scholar]
- Chevalier, B. S., T. Kortemme, M. S. Chadsey, D. Baker, R. J. Monnat et al., 2002. Design, activity, and structure of a highly specific artificial endonuclease. Mol. Cell 10: 895–905. [DOI] [PubMed] [Google Scholar]
- Durrenberger, F., and J. D. Rochaix, 1993. Characterization of the cleavage site and the recognition sequence of the I-CreI DNA endonuclease encoded by the chloroplast ribosomal intron of Chlamydomonas reinhardtii. Mol. Gen. Genet. 236: 409–414. [DOI] [PubMed] [Google Scholar]
- Egli, D., E. Hafen and W. Schaffner, 2004. An efficient method to generate chromosomal rearrangements by targeted DNA double-strand breaks in Drosophila melanogaster. Genome Res. 14: 1382–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush, D. G., and T. H. Eickbush, 2003. Transcription of endogenous and exogenous R2 elements in the rRNA gene locus of Drosophila melanogaster. Mol. Cell. Biol. 23: 3825–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels, W. R., and C. R. Preston, 1984. Formation of chromosome rearrangements by P factors in Drosophila. Genetics 107: 657–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen, H., 1936. Auslosung von crossing-over bei Drosophila mannchen durch rontgenisierung der imago. Genetica 18: 187–192. [Google Scholar]
- Gatti, M., and S. Pimpinelli, 1983. Cytological and genetic analysis of the Y chromosome of Drosophila melanogaster. Chromosoma 88: 349–373. [Google Scholar]
- Golic, K. G., and M. M. Golic, 1996. Engineering the Drosophila genome: chromosome rearrangements by design. Genetics 144: 1693–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic, K. G., and S. Lindquist, 1989. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell 59: 499–509. [DOI] [PubMed] [Google Scholar]
- Golic, K. G., M. M. Golic and S. Pimpinelli, 1998. Imprinted control of gene activity in Drosophila. Curr. Biol. 8: 1273–1276. [DOI] [PubMed] [Google Scholar]
- Gowen, J. W., and E. H. Gay, 1934. Chromosome constitution and behavior in ever-sporting and mottling in Drosophila melanogaster. Genetics 19: 189–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, Y. H., M. M. Tanaka and J. A. Sved, 1996. P-element-induced recombination in Drosophila melanogaster: hybrid element insertion. Genetics 144: 1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadorn, E., R. Gsell and J. Schultz, 1970. Stability of a position-effect variegation in normal and transdetermined larval blastemas from Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 65: 633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley, R. S., and C. H. Marcus, 1989. Recombinational controls of rDNA redundancy in Drosophila. Annu. Rev. Genet. 23: 87–120. [DOI] [PubMed] [Google Scholar]
- Karpen, G. H., J. E. Schaefer and C. D. Laird, 1988. A Drosophila rRNA gene located in euchromatin is active in transcription and nucleolus formation. Genes Dev. 2: 1745–1763. [DOI] [PubMed] [Google Scholar]
- Konev, A. Y., C. M. Yan, D. Acevedo, C. Kennedy, E. Ward et al., 2003. Genetics of P-element transposition into Drosophila melanogaster centric heterochromatin. Genetics 165: 2039–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, J. K., 1979. Site-specific instability in Drosophila melanogaster: the origin of the mutation and cytogenetic evidence for site-specificity. Genetics 93: 681–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, J. K., 1988. Intrachromosomal rearrangements mediated by hobo transposons in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 85: 9153–9157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley, D. L., 1955. Spermatogonial exchange between the X and Y chromosomes of Drosophila melanogaster. Genetics 40: 24–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley, D. L., and G. G. Zimm, 1992. The Genome of Drosophila melanogaster. Academic Press, San Diego.
- Lucchesi, J. C., 1965. The nature of induced changes between the attached-X and Y chromosomes of Drosophila melanogaster. Genetics 51: 209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddern, R. H., 1981. Exchange between the ribosomal RNA genes of the X and Y chromosomes in Drosophila melanogaster males. Genet. Res. 38: 1–7. [DOI] [PubMed] [Google Scholar]
- Maggert, K. A., and K. G. Golic, 2000. The Y chromosome of Drosophila melanogaster exhibits chromosome-wide imprinting. Genetics 162: 1245–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee, B. D., and G. H. Karpen, 1990. Drosophila ribosomal RNA genes function as an X-Y pairing site during male meiosis. Cell 61: 61–72. [DOI] [PubMed] [Google Scholar]
- Monnat, R. J., A. F. M. Hackmann and M. A. Cantrell, 1999. Generation of highly site-specific DNA double-strand breaks in human cells by the homing endonucleases I-PpoI and I-CreI. Biochem. Biophys. Res. Commun. 255: 88–93. [DOI] [PubMed] [Google Scholar]
- Neuhaus, M. E., 1937. Additional data on crossing-over between X and Y chromosomes in Drosophila melanogaster. Genetics 22: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitski, E., 1954. The compound X chromosomes in Drosophila. Genetics 39: 127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasooly, R. S., and L. G. Robbins, 1991. Rex and a suppressor of Rex are repeated neomorphic loci in the Drosophila melanogaster ribosomal DNA. Genetics 129: 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins, L. G., 1981. Genetically induced mitotic exchange in the heterochromatin of Drosophila melanogaster. Genetics 99: 443–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins, L. G., and S. Pimpinelli, 1994. Chromosome damage and early developmental arrest caused by the Rex element of Drosophila melanogaster. Genetics 138: 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins, L. G., and E. E. Swanson, 1988. Rex-induced recombination implies bipolar organization of the ribosomal RNA genes of Drosophila melanogaster. Genetics 120: 1053–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, Y. S., and K. G. Golic, 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018. [DOI] [PubMed] [Google Scholar]
- Rong, Y. S., S. W. A. Titen, H. B. Xie, M. M. Golic, M. Bastiani et al., 2002. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 16: 1568–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royou, A., H. Macias and W. Sullivan, 2005. The Drosophila Grp/Chk1 DNA damage checkpoint controls entry into anaphase. Curr. Biol. 15: 334–339. [DOI] [PubMed] [Google Scholar]
- Ryder, E., F. Blows, M. Ashburner, R. Bautista-Llacer, D. Coulson et al., 2004. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167: 797–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, C., 1936. Somatic crossing over and segregation in Drosophila melanogaster. Genetics 21: 625–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault, S. T., M. A. Singer, W. Y. Miyazaki, B. Milash, N. A. Dompe et al., 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283–287. [DOI] [PubMed] [Google Scholar]
- Thompson, A. J., X. Yuan, W. Kudlicki and D. L. Herrin, 1992. Cleavage and recognition pattern of a double-strand-specific endonuclease (I-CreI) encoded by the chloroplast 23S rRNA intron of Chlamydomonas reinhardtii. Gene 119: 247–251. [DOI] [PubMed] [Google Scholar]
- Williams, S. M., J. A. Kennison, L. G. Robbins and C. Strobeck, 1989. Reciprocal recombination and the evolution of the ribosomal gene family of Drosophila melanogaster. Genetics 122: 617–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, J. H., 1976. The genetics of the Y chromosome, pp. 667–699 in Genetics and Biology of Drosophila, Vol. 3b, edited by M. Ashburner and E. Novitski. Academic Press, San Diego.