Abstract
Epstein-Barr virus (EBV) is the causative agent of infectious mononucleosis and is associated with several forms of cancer, including lymphomas and nasopharyngeal carcinoma. The EBV immediate-early protein BZLF1 functions as a transcriptional activator of EBV early gene expression and is essential for the viral transition between latent and lytic replication. In addition to its role in the EBV life cycle, BZLF1 (Z) also has profound effects upon the host cellular environment, including disruption of cell cycle regulation, signal transduction pathways, and transcription. In an effort to understand the nature of Z interactions with the host cellular environment, we have developed a Drosophila model of early EBV infection, where we have expressed Z in the Drosophila eye. Using this system, we have identified a highly conserved interaction between the Epstein-Barr virus Z protein and shaven, a Drosophila homolog of the human Pax2/5/8 family of genes. Pax5 is a well-characterized human gene involved with B-cell development. The B-cell-specific Pax5 also promotes the transcription of EBV latent genes from the EBV Wp promoter. Our work clearly demonstrates that the Drosophila system is an appropriate and powerful tool for identifying the underlying genetic networks involved in human infectious disease.
EPSTEIN-BARR virus (EBV) is a ubiquitous herpesvirus, having infected most of the world's adult population. Primary infection with EBV may result in infectious mononucleosis and predisposes certain individuals to cancer (Zur Hausen et al. 1970; Rickinson and Kieff 2001). EBV infects mainly two cell types: epithelial cells, where it undergoes lytic replication, and B cells, where it initially replicates in a lytic manner before it enters a latent or dormant state (Rickinson and Kieff 2001). B cells normally may differentiate into plasma cells, which secrete Ig, or memory B cells (Calame et al. 2003). During EBV lytic replication in B cells, the B cells have been shown to enter a plasma cell differentiation pathway (Niedobitek et al. 1997), yet EBV persists in its latent form in memory B cells (Babcock et al. 1998), indicating that the virus takes advantage of B-cell differentiation pathways to efficiently replicate and maintain the virus.
EBV replication is dependent upon its host's cellular environment; hence the virus encodes proteins that interact with and alter the function of cellular proteins, thus altering the intracellular environment to suit the virus. One such EBV protein is BZLF1, an immediate-early protein expressed during lytic replication. BZLF1 (Z) is a transcriptional activator that binds to and activates the promoters of early EBV genes (Chevallier-Greco et al. 1986; Kenney et al. 1989; Cox et al. 1990; Holley-Guthrie et al. 1990; Giot et al. 1991; Quinlivan et al. 1993). The DNA-binding domain of Z bears homology to the AP1 site binding proteins c-Jun and c-Fos (Farrell et al. 1989). Therefore, Z is able to bind to AP1 and AP1-like sites, which are present in the promoters of the EBV early genes (Urier et al. 1989; Kieff and Rickinson 2001). A loss of Z DNA-binding ability, such as with the Z mutant Z311 (Giot et al. 1991), leads to a loss of Z transactivation function and prevents EBV lytic replication from occurring (Giot et al. 1991).
BZLF1 has also been shown to interact with and/or modify the function of several key cellular proteins, including p53, promyelocytic leukemia protein (PML), CREB-binding protein (CBP), NF-κB, and signal transduction proteins such as the p38 and c-Jun N-terminal kinases (Adamson et al. 2000; Adamson and Kenney 2001; reviewed in Sinclair 2003). BZLF1 physically associates with the p53 protein, and this interaction has been shown to prevent p53 from activating p53-responsive promoters (Zhang et al. 1994; Mauser et al. 2002b). In addition, overexpression of p53 prevents BZLF1 transactivation of EBV early promoters (Zhang et al. 1994). BZLF1 also physically associates with the p65 subunit of NF-κB, and this association was found to inhibit EBV lytic replication (Gutsch et al. 1994). Interestingly, BZLF1 increased the levels of nuclear NF-κB, yet did not increase the levels of NF-κB binding to DNA and in fact inhibited NF-κB transactivation of IκBα and ICAM-1 promoters (Morrison and Kenney 2004). CBP also physically associates with BZLF1, and this interaction increases BZLF1 transactivation of EBV early genes as well as increases EBV lytic replication (Adamson and Kenney 1999). Expression of BZLF1 alters the cellular localization of the PML, from nuclear dots to a diffuse nuclear pattern, as well as alters the localization and morphology of mitochondria in the cytoplasm (Adamson and Kenney 2001; LaJeunesse et al. 2005). Since PML is part of the cell's response to viral invasion, the relocalization of PML by BZLF1 may enhance EBV lytic replication. BZLF1 overexpression has also been shown to affect the cell cycle. Although these effects seem to be cell type dependent, BZLF1 generally promotes arrest in the G1 or G2/M phases of the cell cycle (as reviewed in Sinclair 2003). The cell cycle arrest may occur through a variety of mechanisms, such as a p21 increase via C/EBPα or upregulation of p53 and p27 proteins (as reviewed in Sinclair 2003).
Since EBV is a human virus, the main means of studying the virus thus far has been through cell culture systems. A wealth of information about EBV has been gained via cell culture, yet we became interested in studying EBV protein function in the context of an intact organism by expressing EBV proteins in tissues that exist in their natural habitat and are composed of a variety of cell types. Such a system may better reflect how EBV proteins function in their natural human host.
The Drosophila model system has become a useful and powerful tool to study a variety of human genetic diseases (Rebay et al. 1993, 2000; Bonini 2001; LaJeunesse et al. 2001; Bonini and Fortini 2003; Schreiter et al. 2004; Bier 2005). The Drosophila genome possesses a remarkable level of homology with the human genome, with as many as 70% of genes conserved between the two species (Edgar and Lehner 1996; Bernards and Hariharan 2001). Furthermore, genetic techniques such as dominant second-modifier screens enable a genetic dissection of a target gene's cellular function, signal transduction patterns associated with the target gene, and the identification of genetically interacting genes that may encode proteins that physically interact with the target gene. However, no study on genes involved in EBV replication has been conducted using the Drosophila model system.
When a virus invades a host cell, it ectopically expresses an array of new foreign genes that alter and commandeer the host's cellular machinery to propagate its life cycle. In this article, we demonstrate the first use of the Drosophila model system to investigate the host cell/virus relationship found in early Epstein-Barr viral infection. Transgenic GMR∷Z flies that express a mutant dose-sensitive eye phenotype were generated, making them amenable to dominant second-site modifier genetic screens. Phenotypic characterization of GMR∷Z flies showed that ectopic expression of Z affected the differentiation of cone cells and pigment cells within the developing Drosophila retina. Using these GMR∷Z flies, we performed a preliminary candidate gene screen and identified shaven (sv; also known as sparkling) as a strong enhancer of the GMR∷Z phenotype. Interestingly, shaven encodes the Drosophila homolog to the human Pax2/5/8 genes (Fu and Noll 1997; Fu et al. 1998). shaven and the human Pax2/5/8 genes share sequence homology, including 88% identity and 91% similarity between their paired domains.
Human Pax5 is a B-cell-specific transcription factor that plays a key role in B-cell development. Pax5 is necessary for cells to commit to a B-cell lineage and activates the expression of several target genes, including CD19 and CD79a. Pax5 inhibits plasma cell differentiation, however, and must itself be inhibited for plasma cell differentiation to occur (Calame et al. 2003). Moreover, Pax5 plays an important role in the establishment of EBV latency in EBV-infected B cells by activating the latent promoter Wp (Tierney et al. 2000a,b). In this article, we show that Z physically and functionally interacts with human Pax5, suggesting that Z may promote lytic replication in B cells by inhibiting Pax5 function.
MATERIALS AND METHODS
Cell lines:
HeLa cells (ATCC), which are cervical carcinoma cells, were maintained in DMEM with 10% fetal bovine serum as well as bacterial and fungal inhibitors (50 units/ml penicillin, 50 units/ml streptomycin, 0.25 μg/ml amphotericin B). Jijoye cells (Shannon Kenney), which are EBV-positive Burkitt's lymphoma cells, were maintained in RPMI 1640 with 10% fetal bovine serum as well as bacterial and fungal inhibitors.
Plasmids:
The control vector is SVpIE, the Z expression vector contains the Z gene in the SVpIE vector, the Pax5 expression vector contains the human Pax5 cDNA in the EVRF2 vector (James Hagman), and the Pax5 reporter contains the Pax5-binding site from the CD19 promoter (5′-AGA ATG GGG CCT GAG GCG TGA CCA CCG C-3′) in triplicate in the E1b-chloramphenicol transferase (CAT) vector. Transfections were carried out by the calcium phosphate method (Sambrook et al. 1989).
Fly culture:
Flies were maintained at 20° in plastic vials on a medium of cornmeal, yeast, molasses, and agar with methyl 4-hydroxybenzoate added as a mold inhibitor. w1118 was used as the wild-type line.
P-element-mediated transformation:
cDNAs for BZLF1 and Z311 were cloned into the pGMR vector. Z311 has a mutation at amino acid 185 of BZLF1 in the DNA-binding domain of an alanine to a lysine. Germline transformations were performed using the standard P-element protocol (Rebay et al. 1993). For GMR∷Z, several independent lines were isolated, including GMR∷Z.12, all of which had an identical phenotype. For GMR∷Z311, several independent lines were isolated, including GMR∷Z311.1/TM3 and GMR∷Z311.8/CyO, all with a phenotype very similar to GMR∷Z.
Weak GMR∷Z:
To create a dose-sensitive GMR∷Z fly, we mobilized the GMR∷Z P element from one established GMR∷Z line by crossing GMR∷Z.12 males to Δ2-3 females (Δ2-3 flies express the transposase necessary for P-element transposition). The resulting male progeny were crossed to w1118 females and their resulting progeny were screened for a more wild-type (more white+) eye color. One line, referred to as weak GMR∷Z, was isolated and has a milder phenotype.
Fly crosses:
weak GMR∷Z males were crossed to svspa-pol (M. Noll) females. weak GMR∷Z females were crossed to GMR∷Rbf (I. Hariharan; Du et al. 1996), GMR∷dap2A (I. Hariharan; de Nooij et al. 1996), GMR∷p35 (2-1) (G. Rubin; Hay et al. 1994), or CyclEAR95 (I. Hariharan; Du et al. 1996) males. The resulting double-heterozygous progeny from each cross were examined for a modification of GMR∷Z eye phenotype, as compared to weak GMR∷Z heterozygotes or candidate gene heterozygotes.
DNA purification:
Plasmid DNA was purified through QIAGEN columns as described by the manufacturer.
CAT assays:
Cell extracts were prepared 48 hr post-transfection and incubated at 37° with [14C]chloramphenicol in the presence of acetyl coenzyme A as previously described (Gorman et al. 1982). The percentage acetylation of chloramphenicol was quantitated by thin-layer chromatography followed by PhosphorImager screening.
EMSAs:
Electromobility shift assays (EMSAs) were performed as previously described (Garner and Revzin 1981). The synthetic double-strand oligonucleotide used in the binding reactions was 5′-end labeled with 32P using the Klenow reaction. The Pax5 site consists of the oligonucleotides 5′-AGA ATG GGG CCT GAG GCG TGA CCA CCG C-3′. For the protein preparation, Jijoye cells were infected with adenovirus that expressed either the LacZ gene or the Z gene at a multiplicity of infection of 50. The cells were harvested 48 hr postinfection and nuclear extracts were prepared. The binding reactions were conducted in a buffer consisting of 10 mm HEPES pH 7.9, 100 mm KCl, 1 mm EDTA, 1 mm DTT, 0.1% BSA, and 10% glycerol. Two micrograms of protein were added to each reaction and incubated at room temperature for 10 min before the addition of labeled probe (20,000 cpm). A total of 100 ng of unlabeled Pax5 site was used as a competitor. After addition of the probe, the reactions were incubated 30 min at room temperature and loaded onto a 4% polyacrylamide gel and run in 0.25× Tris-borate-EDTA buffer at room temperature.
Immunoprecipitation:
HeLa cells were transfected with expression vectors for Z and Pax5, washed twice with cold 1× PBS, and resuspended in buffer 8 (25 mm HEPES pH 7.5, 100 mm NaCl, 5 mm MgCl2, 100 mm EDTA, 200 μg/ml BSA, 0.01% Tween-20, protease inhibitors). After brief sonication and centrifugation, the lysate was diluted to 500 μl with buffer 8. Two micrograms of anti-Pax5 antibody (Santa Cruz) was added and the mixture incubated for 2 hr at 4°. Protein G beads (Zymed, South San Francisco, CA) were added and incubated for 1 hr at 4°. The beads were washed five times with buffer 8, resuspended in SDS-PAGE loading dye, and loaded onto an SDS-PAGE gel.
Western blot analysis:
Immunoblot analysis was performed as previously described (Adamson and Kenney 1999). The anti-Z antibody (Argene) and anti-Pax5 (Santa Cruz) were used at a dilution of 1:500. The secondary antibodies [goat anti-mouse-HRP or donkey anti-goat-HRP (Jackson ImmunoResearch, West Grove, PA)] were used at a dilution of 1:20,000. SuperSignal (Pierce, Rockford, IL) was used for detection.
Immunostaining of imaginal discs:
Eye-antenna imaginal discs were immunostained as described (Wolff 2000). The anti-Z antibody (Argene) was used at a 1:50 dilution, the anti-Cut antibody (Developmental Studies Hybridoma Bank) was used at a 1:50 dilution, the anti-Sv antibody (Markus Noll) was used at a 1:20 dilution, and the anti-Elav antibody (Developmental Studies Hybridoma Bank) used at a 1:500 dilution. Each primary antibody was incubated with several (∼10) discs overnight. The secondary antibodies [donkey anti-mouse-CY3 and donkey anti-rabbit-FITC (Jackson ImmunoResearch)] were used at a 1:2000 dilution and were incubated with the discs for 2 hr. Discs were mounted in antifade media (Molecular Probes, Eugene, OR).
Acridine-orange staining of imaginal discs:
Eye-antenna discs were dissected in S2 cell media and incubated in S2 cell media containing 0.0016 mm acridine orange (Sigma, St. Louis) for 5 min. The discs were rinsed with S2 cell media and mounted live for confocal microscopy.
BrdU labeling:
Eye-antenna discs were dissected in S2 cell media and incubated in 1 mg/ml BrdU solution in S2 cell media for 30 min. The discs were washed three times with 1× PBS, fixed in 4% paraformaldehyde/PBS for 30 min, washed three times with 1× PBS, and their DNA was denatured in 2N HCl/0.1% Triton X/PBS for 30 min. The discs were washed three times with PBS, blocked in 5% goat serum/1% BSA/0.1% Triton X/PBS for 30 min and incubated overnight at 4° in anti-BrdU antibody (1:1000 dilution; Phoenix Systems). The remainder of the protocol was carried out as for immunostaining of imaginal discs (Wolff 2000).
Immunocytochemistry:
HeLa cells were immunostained as previously described (Adamson and Kenney 2001), except that the incubation mix consisted of 1× TS (20 mm Tris base, 137 mm NaCl, pH 7.6) and 5% donkey serum, and the wash solution consisted of 1× TS only. The anti-Pax5 antibody (Santa Cruz) was used at a dilution of 1:100 and the anti-Z antibody (Argene) was used at a dilution of 1:50. Secondary antibodies [donkey anti-mouse-FITC and donkey anti-goat-CY3 (Jackson ImmunoResearch] were used at a 1:100 dilution. Hoechst stain was used to visualize nuclei.
RESULTS
BZLF1 expression produces a severe phenotype in Drosophila:
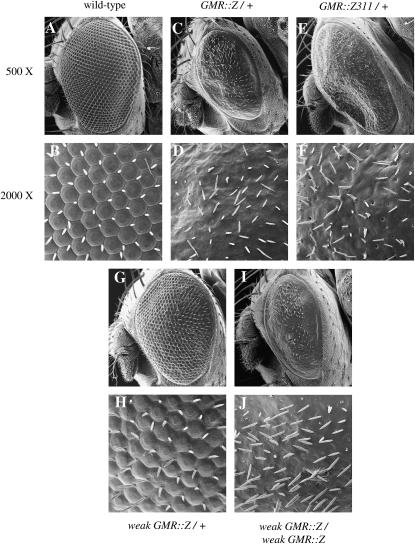
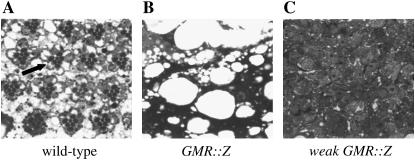
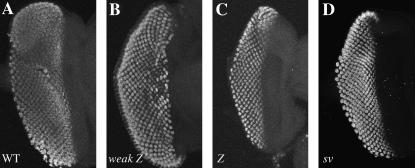
To generate BZLF1 transgenic flies, we cloned the coding region of Z into the Drosophila P-element eye-specific expression transformation vector pGMR (glass-mediated response) (Hay et al. 1994), which expressed Z specifically in all cells posterior to the morphogenetic furrow in the developing (larval) eye (Figure 3A). GMR∷Z/+ adult flies expressed a very striking eye phenotype in which all structures except the interommatidial bristles were absent (Figure 1, C and D). The bristles were disorganized and scattered throughout the thin cuticle that remained. There was also a loss of pigment cells, since the GMR∷Z eyes had no pigmentation and were white in color. In addition, the GMR∷Z eye was slightly smaller than that of the wild-type control. We observed no difference in the phenotype between homozygous and heterozygous GMR∷Z flies, demonstrating that GMR∷Z lines are not dose sensitive (with the exception of weak GMR-Z; see below). Sectioning of the adult eye revealed that no recognizable eye structures were detectable in GMR∷Z eyes (Figure 2B), implying an absence of all normal eye cell types in the adult eye. This result indicated that Z is biologically active in Drosophila cells and suggested that experiments performed on the fly will be at least partially relevant to human cells.
Figure 3.
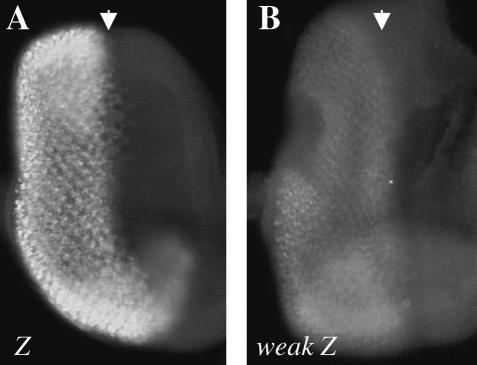
Weak GMR∷Z flies express less Z protein than GMR∷Z flies. Late third instar eye imaginal discs were immunostained for Z. (A) GMR∷Z/+ eye disc. (B) Weak GMR∷Z/+ eye disc. Disc immunostainings were performed concurrently and photographed with the same exposure times. Results were consistently reproducible. Arrows refer to the morphogenetic furrow.
Figure 1.
GMR∷Z and weak GMR∷Z adult eye phenotypes. Magnification is presented at both 500× and 2000×. (A and B) Wild-type eye. (C and D) GMR∷Z/+ eye. Note loss of ommatidia. (E and F) GMR∷ Z311/+ eye. Phenotype is similar to that of GMR∷Z. (G and H) Weak GMR∷Z/+ eye. Phenotype is intermediate between wild type and GMR∷Z. (I and J) Weak GMR∷Z/weak GMR∷Z homozygous eye. Phenotype is similar to that of GMR∷Z.
Figure 2.
Adult GMR∷Z eyes have no recognizable eye cells. Adult heads were stained with Giemsa and sectioned. (A) Wild-type eye. Arrow points to an ommatidium. (B) GMR∷Z/+ eye. (C) Weak GMR∷Z/+ eye.
Z is a transcription factor that binds to AP1 and AP1-like binding sites in DNA (Urier et al. 1989; Rickinson and Kieff 2001). To determine whether the GMR∷Z phenotype was due to rampant and promiscuous transcription via Z-binding sites (or via other similar sites within the Drosophila genome), we expressed the DNA-binding defective Z mutant Z311 in the Drosophila eye via the pGMR system. Z311 contains a mutation in the DNA-binding domain of Z that abolishes DNA binding (Giot et al. 1991). The loss of DNA-binding ability inhibits Z-mediated transactivation and consequently inhibits EBV lytic replication (Giot et al. 1991). GMR∷Z311/+ flies expressed a phenotype identical to GMR∷Z/+ flies (Figure 1, E and F), such that there was a loss of ommatidial structures and pigmentation, and only bristles and cuticle remained. This suggests that the Z-induced phenotypes are not due to unregulated transcriptional activation by Z, but rather to explicit protein-protein interactions between the Z protein and endogenous host proteins.
Since all of our GMR∷Z lines displayed an invariant and genetically unchangeable eye phenotype (not dose sensitive), one line with a moderate and dose-sensitive phenotype (referred to as weak GMR∷Z) was created by mobilizing the P element in one of our “strong” GMR∷Z lines. Weak GMR∷Z eye imaginal discs expressed less quantity of Z protein than GMR∷Z did (Figure 3B). While the GMR∷Z lines produced abundant Z protein, the weak GMR∷Z line produced a small amount of protein (compare Figure 3, A and B). Western analysis confirmed that weak GMR∷Z eye discs expressed greatly reduced levels of Z protein (data not shown).
The weak GMR∷Z phenotype is dose dependent; flies heterozygous for this insertion display a moderate eye phenotype (Figure 1, G and H), while flies homozygous for weak GMR∷Z appear similar to the original GMR∷Z flies (Figure 1, I and J). Sectioning of the weak GMR∷Z/+ adult eyes showed that, although some eye tissue was present, there were no recognizable ommatidial clusters (Figure 2C).
Z inhibits cell proliferation and increases cell death:
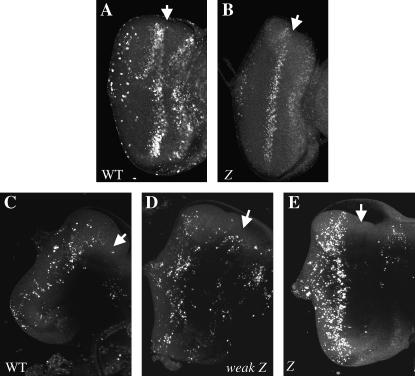
In human cells, Z has been shown to inhibit the cell cycle (Cayrol and Flemington 1996a,b; Mauser et al. 2002a; Sinclair 2003). To determine whether GMR∷Z inhibited the cell cycle, we examined cell proliferation via BrdU incorporation (Figure 4). GMR∷Z/GMR∷Z eye discs showed a significantly lower number of cells, when compared to wild-type eye discs, that had incorporated BrdU (Figure 4, A and B; Table 1). Therefore, fewer cells entered the cell cycle in GMR∷Z discs. This result is consistent with previously published data from human tissue culture cells showing that Z expression can arrest the cell cycle (Cayrol and Flemington 1996a,b; Mauser et al. 2002a). Moreover, this reduction in cell division may account for the slightly smaller size of the GMR∷Z discs.
Figure 4.
Homozygous GMR∷Z eye discs undergo less proliferation and more apoptosis than wild type. Late third instar eye imaginal discs were stained with BrdU (A and B) or acridine orange (C–E). (A) Wild-type eye disc. (B) GMR∷Z/GMR∷Z eye disc. (C) Wild-type eye disc. (D) Weak GMR∷Z/+ eye disc. (E) GMR∷Z/GMR∷Z eye disc. Arrows refer to the morphogenetic furrow of each disc.
TABLE 1.
Cell counts for acridine orange and BrdU labeling in wild-type and GMR∷Z eye imaginal discs
| Genotype/experiment | n | No. of cells labeled | % deviation from wild type |
|---|---|---|---|
| w1118 (wild type), BrdU | 10 | 159 ± 14 | |
| GMR∷Z/GMR∷Z, BrdU | 10 | 112 ± 12 | −30 |
| w1118 (wild type), acridine orange | 12 | 100 ± 16 | |
| GMR∷Z/GMR∷Z, acridine orange | 11 | 221 ± 42 | +221 |
We also examined whether the ectopic expression of Z affected cell viability by using acridine orange, a vital stain that is taken up by cells undergoing apoptosis (Figure 4) (Wolff 2000). We found that, while the level of cell death was comparable between wild-type and weak GMR∷Z/+ tissues (Figure 4, C and D), there was in increase in cell death in the GMR∷Z/GMR∷Z eyes (Figure 4E; Table 1), suggesting that, at least in part, the smaller size of the imaginal eye discs may also be due to a loss of cells via apoptosis. The levels of cell death and inhibition of the cell cycle are clearly dependent upon the level of Z protein present, since the low levels of Z in weak GMR∷Z flies did not induce these effects (Figure 4D; data not shown). However, the level of Z produced during lytic replication from an EBV genome is relatively high and is mimicked by the level of Z protein produced from transfected plasmids (our unpublished data). Thus we are confident that the level of Z protein produced in GMR∷Z flies is comparable to the level of Z protein produced by EBV. The above results, taken together, suggest that Z's arrest of the cell cycle and induction of apoptosis can account for the small imaginal eye discs observed in GMR∷Z flies. Moreover, the cell cycle arrest by Z supports our line of reasoning that Z's function within fly cells parallels what has been shown in human cells.
Z does not affect photoreceptor cells in the developing larval eye:
Since we observed a loss of specific eye cell types in GMR∷Z adult eyes, we investigated the fate of cell types in developing larval eye discs after the Z protein had been expressed yet before the pupal stage. The eye contains photoreceptor cells, cone cells, bristle cells, and pigment cells. To examine the effect of Z upon developing photoreceptor cells, we employed the anti-Elav antibody, which detects the photoreceptor-specific Elav protein (Robinow and White 1988). We immunostained third instar larval eye discs with the anti-Elav antibody and found that there was no loss of photoreceptor cell clusters at this early stage in GMR∷Z discs (Figure 5, A–C). To confirm this finding, we generated a fly line that contained the Z gene under the control of the upstream activating sequence (UAS) promoter element. We crossed these UAS∷Z flies to sev∷GAL4 flies, creating flies that produce GAL4 only in sevenless-containing (photoreceptor) cells. In these flies, the GAL4 binds to UAS and activates expression of Z only in the photoreceptor cells. No mutant phenotype was produced (data not shown). Therefore, even though Z is expressed in photoreceptor cells and appears to promote cell death in the imaginal discs (as in Figure 4), Z does not appear to negatively affect the photoreceptor cell precursors.
Figure 5.
GMR∷Z eye discs maintain their photoreceptors. Late third instar eye imaginal discs were immunostained with an anti-Elav antibody. Disc stainings were performed concurrently and photographed with the same exposure times. (A) Wild-type eye disc. (B) Weak GMR∷Z/+ eye disc. (C) GMR∷Z/GMR∷Z eye disc. (D) sv/sv eye disc.
Z prevents Cut expression by interfering with Shaven function:
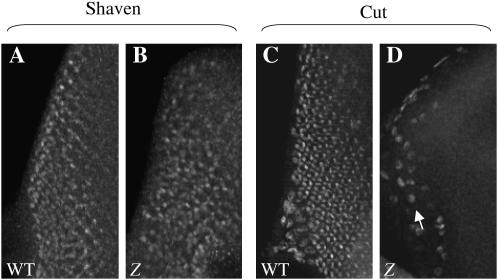
Since we found that Z does not affect developing photoreceptor cells in GMR∷Z discs, we turned our attention to the cone cells and bristle cells. As shown in Figure 1, bristles are present in GMR∷Z discs, albeit in an unorganized manner. Therefore Z does not appear to prevent bristle cell differentiation. To determine whether there was a disruption of cone cell development in GMR∷Z eye discs, we examined the expression of cut, a gene which is expressed in cone and bristle cells of the eye (Blochlinger et al. 1993). We immunostained third instar larval eye discs with the anti-Cut antibody and found that, although the presumptive bristle cells of GMR∷Z discs express Cut, the cone cells did not express Cut (Figure 6D). Thus the cut gene did not appear to be expressed in cone cells when Z was expressed.
Figure 6.
Cut protein levels, but not Sv protein levels, are reduced in GMR∷Z eye discs. (A and C) Wild-type eye discs. (B and D) GMR∷Z/GMR∷Z eye discs. Late third instar eye imaginal discs were immunostained for Sv (A and B) and Cut (C and D). Note the loss of Cut staining in the GMR∷Z disc (D), as compared to the wild-type disc (C). In D, the arrow refers to the peripodial cells, which are part of the membrane that surrounds the disc. Some of these cells show staining with the anti-Cut antibody, but the staining is not in the disc itself.
To elucidate the loss of cut expression in GMR∷Z eye discs, we examined an additional cone cell marker, shaven. sv is required for specification and differentiation of cone cells, primary pigment cells, and bristle cells in Drosophila and has been shown to transactivate Cut expression by activating the cut promoter (Fu and Noll 1997; Fu et al. 1998). We immunostained third instar larval discs with anti-Shaven antibody and found that, in contrast to the Cut results (where no Cut was present), the GMR∷Z cone cells expressed Shaven (Figure 6B). This result suggested to us that Z interfered with Shaven's transactivation function and therefore prevented cut gene expression, contributing to a loss of cone cell differentiation. The loss of the cone cells at this stage may lead to the subsequent disruption of the entire eye (Siddall et al. 2003).
shaven is a dose-sensitive modifier of GMR∷Z:
Using our phenotypic characterization of the GMR∷Z flies, we decided to perform a candidate gene screen to look for genetic interactors. The centerpiece of such genetic screens is the generation of a dose-dependent mutant phenotype, typically upon the ectopic expression of either the wild-type or the mutant target gene in a nonessential tissue such as the eye or wing (Rebay et al. 2000; LaJeunesse et al. 2001). A resulting mutant phenotype represents a defect in a mechanism associated with the target gene. For instance, expression of a dominant-negative form of the Drosophila neurofibromatosis homolog Merlin results in an overproliferation phenotype characteristic of loss of function for a tumor suppressor gene (LaJeunesse et al. 1998). Genes identified as enhancers of the sensitized phenotype can be thought of as genes that encode proteins that function as antagonists or negative regulators of the molecular mechanisms of which the target gene is a part, while suppressors of the phenotype can be thought of as genes that encode proteins that function as protagonists or positive regulators of the molecular mechanisms of which the target gene is a part. Such genetic screens have been used to identify genes associated with the molecular mechanisms underlying a number of inherited human genetic diseases, including neurofibromatosis II, neurodegenerative diseases, and obesity (Bonini 2001; LaJeunesse et al. 2001; Bonini and Fortini 2003; Schreiter et al. 2004; Bier 2005).
Using our dose-sensitive weak GMR∷Z line (since our strong GMR∷Z lines were not dose sensitive), we performed a candidate gene screen of genes known to affect proliferation, apoptosis, and differentiation in the developing eye. We crossed weak GMR∷Z to fly lines that overexpressed the Drosophila E2F/Dp, the Drosophila Rb (Rbf), the Drosophila p21 (dacapo), or the antiapoptosis baculovirus p35 gene in the eye, as described in materials and methods. We also crossed weak GMR∷Z to a cyclin E null allele (CyclEAR95). All of these crosses yielded flies whose eyes maintained the same phenotype as that of weak GMR∷Z eyes (data not shown). Therefore, the overexpression of a cell cycle promoter (E2F), cell cycle inhibitors (Rb, p21, and the null cyclin E allele), and even an antiapoptotic gene (p35) were unable to overcome the Z-induced phenotype.
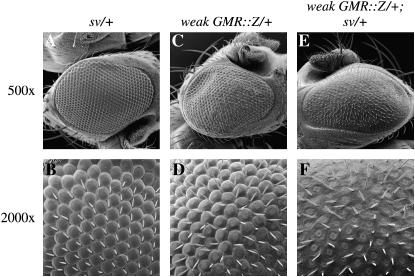
Since we found that Z may be preventing the differentiation of cone cells, we further examined the relationship between Z and sv and investigated whether there was a genetic interaction between weak GMR∷Z and sv. To do this, we crossed weak GMR∷Z to svspa-pol (a sv null allele that yields no functional Sv protein in the eye) (Fu and Noll 1997) and examined the heterozygous sv and GMR∷Z progeny (Figure 7). sv is recessive and flies heterozygous for sv mutations express a normal eye phenotype (Figure 7, A and B). However, flies heterozygous for svspa-pol and weak GMR∷Z displayed a strong GMR∷Z phenotype in which there was a loss of facets (Figure 7, E and F). Therefore, there is a genetic interaction between Z and sv, such that the presence of Z sensitizes the cells to a reduction in Sv levels. These results support our hypothesis that Z functions, at least in part, to disrupt the development of the Drosophila eye by interfering with sv function. This Z/Sv interaction likely contributes to the loss of differentiated cone cells.
Figure 7.
Genetic interaction between weak GMR∷Z and sv. (A and B) sv/+ eye. (C and D) weak GMR∷Z/+ eye. (E and F) weak GMR∷Z/+; sv/+ eye. The phenotype shown in F is much more severe than that of B and D.
Z inhibits human Pax5-mediated transactivation:
Since the goal of our study was to identify proteins that interact with Z in Drosophila and translate these findings to the normal hosts for EBV, humans, we next turned our attention to sv human homologs. The sv gene encodes a paired-box gene that is the homolog of the human Pax2/Pax5/Pax8 proteins (Fu and Noll 1997; Fu et al. 1998). Interestingly, Pax5 is a transcription factor that is expressed in B cells and central nervous system cells, while Pax2 is expressed in kidney and central nervous system cells and Pax8 is expressed in kidney, thyroid, and central nervous system cells (Chi and Epstein 2002). Pax5 is required for the development of B cells (Urbanek et al. 1994; Baker and Reddy 1995; Enver 1999; Mikkola et al. 2002) and has been shown to activate the EBV latent Wp promoter (Tierney et al. 2000a,b). We focused on this homolog in view of the fact that Pax5 and EBV share the same environment (B cells) in the human host's immune system. No connection had been previously established between Z and Pax5.
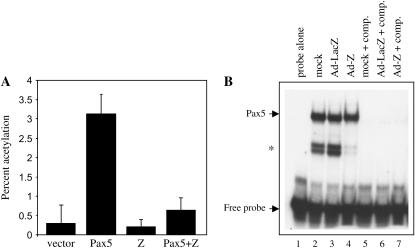
To determine whether Z altered Pax5 function in human cells, we examined Z's ability to disrupt human Pax5 transcriptional function. HeLa cells were transfected with Pax5 and Z expression plasmids and a reporter plasmid containing a Pax5-binding site upstream of a minimal promoter element and the CAT gene (Figure 8A). In repeated experiments, the addition of Z with Pax5 compromised the ability of Pax5 to drive CAT expression from the reporter construct, suggesting that the interaction that we observed between Z and Sv in Drosophila cone cells is also conserved between Z and Pax5 in human cells. Given these results, we sought to investigate the mechanism that Z may use to neutralize Pax5 function. To test whether Z altered the ability of Pax5 to bind DNA, we performed electromobility shift assays using several different Pax5-binding sites. We found that Z had no effect upon Pax5 binding to DNA, and an example of this is shown in Figure 8B. The levels of Pax5 binding in the presence of Z were identical to, if not more than, the levels of Pax5 binding in the presence of the control protein LacZ (compare in Figure 8B, lanes 3 and 4). These results suggest that Z may abrogate Pax5 transcriptional activity by a mechanism that does not alter DNA binding.
Figure 8.
Z decreases Pax5-mediated transactivation. (A) HeLa cells were transfected with expression plasmids as indicated, along with the Pax5-CAT reporter plasmid. CAT assays were performed in four separate experiments and are presented as the percentage of acetylation. (B) Electromobility shift assay. Jijoye cells were uninfected, infected with an adenovirus containing the control gene LacZ, or infected with an adenovirus containing the Z gene. Extracts of each were incubated with the CD19 gene Pax5-binding site. comp., cold CD19 site competitor added. (*) indicates partially degraded Pax5 protein that still binds to the CD19 site.
Z physically associates with and stabilizes Pax5 protein:
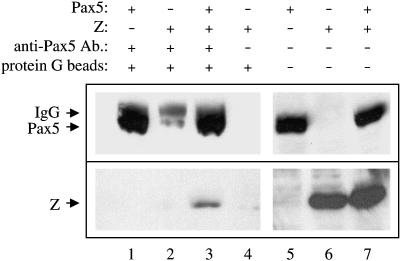
We next examined the ability of Z to physically associate with Pax5. We transfected HeLa cells (which do not contain endogenous Pax5 protein) with expression plasmids for Z and Pax5, co-immunoprecipitated the proteins with an anti-Pax5 antibody, and performed immunoblot analyses with anti-Z and anti-Pax5 antibodies (Figure 9). We found that the anti-Pax5 antibody immunoprecipitated the Pax5 protein (Figure 9, lanes 1 and 3) as well as co-immunoprecipitated the Z protein (Figure 9, lane 3). Z did not co-immunoprecipitate with the anti-Pax5 antibody in the absence of Pax5 protein (lane 2). Thus there is a physical interaction between Z and Pax5. In support of a Z/Pax5 physical interaction, we have previously found that Z is able to relocalize Pax5 protein within human cells. While Pax5 does not normally bind to chromosomes during mitosis (condensed chromosomes), we found that Pax5 is translocated to mitotic chromosomes when Z is present (Adamson 2005). We have shown that Z binds to chromosomes during mitosis and tethers several of its protein-binding partners to mitotic chromosomes (Adamson 2005).
Figure 9.
Z physically associates with Pax5. HeLa cells were transfected with expression plasmids for Z alone (lanes 2, 4, and 6), Pax5 alone (lanes 1 and 5) or Z plus Pax5 (lanes 3 and 7). Anti-Pax5 antibody was used to co-immunoprecipitate the Pax5/Z complex (lanes 1–3). An anti-Z antibody (bottom) and an anti-Pax5 antibody (top) were used for immunoblotting. The anti-Z antibody immunoblots revealed the co-immunoprecipitated Pax5/Z complex (lane 3), as well as the levels of Z protein present in the cell extracts used (lanes 6 and 7). The anti-Pax5 antibody immunoblots revealed the levels of Pax5 protein immunoprecipitated with the anti-Pax5 antibody (lanes 1–3), as well as the levels of Pax5 proteins present in the cell extracts used (lanes 5 and 7).
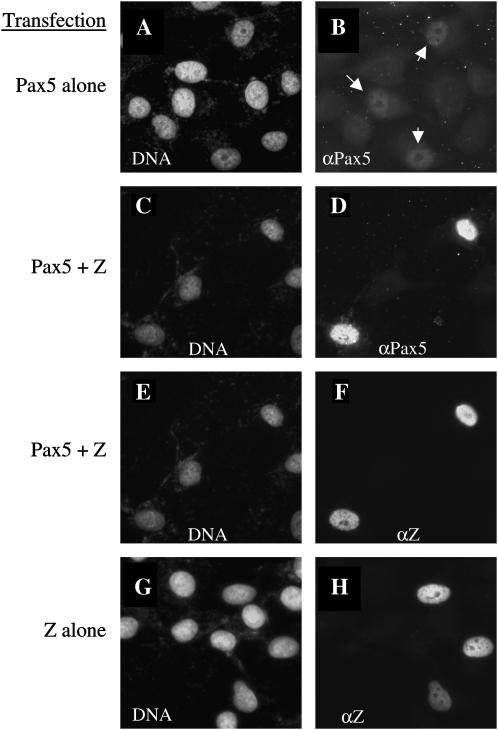
In addition, we discovered that expression of Z seemed to stabilize Pax5 protein levels. HeLa cells that coexpressed both Pax5 and Z proteins had much higher levels of Pax5 protein than cells that expressed the Pax5 protein alone (Figure 10, compare B and D). In a similar scenario, it was previously observed that Z stabilizes the p53 protein, while preventing p53 from transactivating its target promoters (Mauser et al. 2002b). These results, taken together, suggest that Z might interfere with Pax5 function by binding to Pax5 and preventing a critical interaction necessary for expression of Pax5 target genes.
Figure 10.
Z stabilizes Pax5 protein levels. HeLa cells were transfected with expression plasmids for Pax5 alone (A and B), Z plus Pax5 (C–F), or Z alone (G and H). Cells were immunostained with both anti-Z and anti-Pax5 antibodies, as well as with the DNA stain Hoechst. Arrows in B indicate cells that express Pax5, albeit at low levels. Note that when Pax5 is coexpressed with Z, Pax5 is present in high levels (D). All images were taken at the same time with identical exposure times.
DISCUSSION
In this article we demonstrate that the Drosophila model system can be a powerful discovery tool for new genes that are involved in viral pathogenesis. We have expressed the Epstein-Barr virus immediate-early protein Z in the Drosophila eye and found that Z induced a mutant eye phenotype. To establish the Drosophila system as a model for early EBV infection, it was necessary to determine whether the phenotype was biologically relevant and whether the phenotype was amenable to genetic manipulation. The expression of a mutant eye phenotype demonstrated that the EBV Z protein had biological activity within the context of a foreign cell type; however, whether this was relevant remained to be tested.
Z is a transcription factor that binds to and activates the promoters of early EBV genes (Chevallier-Greco et al. 1986; Kenney et al. 1989; Cox et al. 1990; Holley-Guthrie et al. 1990; Giot et al. 1991; Quinlivan et al. 1993) via AP1 and AP1-like binding sites (Urier et al. 1989; Rickinson and Kieff 2001). Given that Z functions as a transcriptional activator, one rationale for the GMR∷Z eye phenotype might have been simply rampant misexpression of endogenous Drosophila genes leading to aberrant development of the eye. To test this possibility, we examined the phenotypes expressed by a DNA-binding null mutant of Z referred to as Z311; Z311 cannot transactivate promoters. Ectopic expression of Z311 in the Drosophila eye produced the same phenotype as did wild-type Z and demonstrated that the phenotypes that we initially observed were not due to unregulated transcription but instead likely due to other protein-protein interactions.
In human cells, Z physically interacts with a number of different cellular factors and through these interactions affects the cell cycle and several different signal transduction pathways (Adamson and Kenney 1999, 2001; Adamson et al. 2000; Mauser et al. 2002b). In Drosophila cells, we examined the effect of Z on the cell cycle, specifically examining its effects upon cellular proliferation. In human cells, expression of Z has been shown to arrest the cell cycle (Mauser et al. 2002b), and likewise, the Drosophila eye imaginal disc cells that express Z have a reduced number of cells entering S phase. This result, along with the interesting phenotype, suggested to us that Z may be functioning in fly cells in a manner homologous to what has been demonstrated in human cells. Therefore, we believe that Drosophila is an appropriate system in which to hunt for proteins that physically and functionally interact with the Epstein-Barr virus Z protein.
The phenotypes expressed by all lines of GMR-Z (with the exception of weak GMR-Z) demonstrated no dose sensitivity. In these flies we observed a smoothened eye with disorganized bristles and a loss of pigment cells. However, we were able to identify a weakly expressing GMR∷Z line, which we refer to as weak GMR∷Z, that expressed reduced levels of Z protein. Heterozygous weak GMR∷Z eyes had a slightly smoothened eye phenotype with a reduction of some (but not all) pigment cells. However, homozygous weak GMR∷Z eyes expressed a phenotype that resembled the “strong” GMR∷Z eyes (smoothened with no pigment). This suggested to us that weak GMR∷Z would be a useful tool for identifying dominant second-site modifiers. With weak GMR∷Z we began a candidate-gene genetic screen to identify known genes or pathways that could modify the weak GMR∷Z phenotype. We looked for interactions between Z and E2F/Dp, Rbf, p21, cyclin E, p35, and shaven. The cell cycle promoters and inhibitors had no effect upon the weak GMR∷Z phenotype. The antiapoptotic baculovirus p35 protein also did not alter the weak GMR∷Z phenotype. The only candidate that did modify the weak GMR∷Z phenotype was the shaven mutant.
Characterization of the GMR∷Z eye phenotype revealed that expression of Z prevents the formation of lenses and pigmentation, presumably due to a loss of pigment and cone cells in the developing eye. This is strikingly similar to what has been described for loss of sv: expression of Z induces pigment cell and cone cell defects, as well as an increased number of apoptotic cells, all similar to sv loss of function (Siddall et al. 2003). Loss of Sv activity in eye imaginal disc cone cells leads to a disruption of signaling to the surrounding cells, which induces apoptosis of those surrounding cells (Siddall et al. 2003). Furthermore, in a manner similar to sv loss of function, we observed a reduction of the downstream activation of cut, suggesting that Z functions in the Drosophila eye, at least in part, by disrupting the normal transcriptional function of the Sv protein.
Given that the goal of this work was to establish the Drosophila model system as a tool for identifying genes involved in EBV virus/host cell interactions, after identifying the strong genetic interaction between Z and sv in the Drosophila eye, we wanted to determine whether this genetic interaction was biologically significant in human cells. Since a dose-dependent genetic interaction may reveal an underlying functional or physical interaction between the protein products of the two genes involved (Rebay et al. 2000; Lajeunesse et al. 2001), we wanted to determine whether Z disrupted the transcriptional activity of the human sv homolog, Pax5, and whether it did so through a physical interaction. Using co-immunoprecipitation, we showed that Z physically associated with Pax5. Moreover, we demonstrated that Z inhibited Pax5-mediated transcriptional activation in reporter assays, even though Z seemed to stabilize Pax5 protein levels. This is similar to what has been shown with the interaction between Z and p53, where, in Z-expressing cells, there is an increase in p53 protein levels (as well as an increase in p53 binding to p53 sites in DNA), yet there was an inhibition of p53 transactivation ability such that the p53 targets p21 and MDM2 were not induced (Mauser et al. 2002b). In regard to Pax5, Z may bind to and block the transactivation region of Pax5, so that even though Pax5 can still bind to DNA, it cannot recruit transcriptional machinery to a promoter.
The EBV Z protein has been shown to disrupt the function of several human genes involved in transcription, including CBP, p53, and NF-κB, through direct protein-protein interactions (Gutsch et al. 1994; Adamson and Kenney 1999; Mauser et al. 2002b). Our results suggest that Z binding to Pax5 may disrupt the ability of Pax5 to function as a transactivator, affecting both B-cell-specific genes and the EBV latent Wp promoter. It has been shown that EBV lytic replication occurs in B cells with a plasma cell morphology (Niedobitek et al. 1997), and in this state Pax5 must be inactive. The Blimp-1 protein has been shown to repress Pax5 expression during normal plasma cell differentiation (Calame et al. 2003). Z may also contribute to Pax5 inhibition during EBV lytic replication by binding to Pax5 and blocking its transactivation domain. In addition, Z may promote lytic replication in newly infected B cells, in lieu of latency establishment, by inhibition of Pax5 transactivation of the Wp promoter. Wp is the first latent promoter activated when EBV switches from lytic to latent replication, and Wp activates transcription of the latent EBNA2 and EBNA-LP genes (Tierney et al. 2000b). The inhibition of Pax5 by Z may prevent Wp activation and delay the onset of viral latency. The promotion of lytic replication would yield more viral particles and consequently more EBV-infected cells.
It is important to note that despite the fact that Epstein-Barr virus is a well-studied viral system, we have been able to identify a novel interaction between Z and Pax5 by using an innovative method. The effectiveness of this pilot project gives credence to the promise that larger-scale screens will be able to identify other genes involved in EBV's early commandeering of cellular machinery and will provide a richer and fuller picture of how a virus uses cellular machinery to propagate its own life cycle.
Acknowledgments
We thank Shannon Kenney for support of this work, Markus Noll for the anti-Shaven antibody, and James Hagman for the Pax5 expression vector. Both the anti-Elav antibody and the anti-Cut antibody were developed by G. M. Rubin and were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, Iowa. A.L.A. is supported by National Institutes of Health grant no. 1-R21-DE14602-01.
References
- Adamson, A. L., 2005. The Epstein-Barr virus BZLF1 protein associates with mitotic chromosomes. J. Virol. 79: 7899–7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson, A. L., and S. Kenney, 1999. The Epstein-Barr virus BZLF1 protein interacts physically and functionally with the histone acetylase CREB-binding protein. J. Virol. 73: 6551–6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson, A., and S. Kenney, 2001. The Epstein-Barr Virus (EBV) immediate-early protein, BZLF1, is SUMO-1-modified and disrupts PML bodies. J. Virol. 75: 2388–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson, A., D. Darr, E. Holley-Guthrie, R. A. Johnson, A. Mauser et al., 2000. Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 activate the ATF2 transcription factor by increasing the levels of phosphorylated p38 and c-Jun N-terminal kinases. J. Virol. 74: 1224–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock, G. J., L. L. Decker, M. Volk and D. A. Thorley-Lawson, 1998. EBV persistence in memory B cells in vivo. Immunity 9: 395–404. [DOI] [PubMed] [Google Scholar]
- Baker, S. J., and E. P. Reddy, 1995. B cell differentiation: role of E2A and Pax5/BSAP transcription factors. Oncogene 11: 413–426. [PubMed] [Google Scholar]
- Bernards, A., and I. K. Hariharan, 2001. Of flies and men—studying human disease in Drosophila. Curr. Opin. Genet. Dev. 11: 274–278. [DOI] [PubMed] [Google Scholar]
- Bier, E., 2005. Drosophila, the golden bug, emerges as a tool for human genetics. Nat. Rev. Genet. 6: 9–23. [DOI] [PubMed] [Google Scholar]
- Blochlinger, K., L. Y. Jan and Y. N. Jan, 1993. Postembryonic patterns of expression of cut, a locus regulating sensory organ identity in Drosophila. Development 117: 441–450. [DOI] [PubMed] [Google Scholar]
- Bonini, N. M., 2001. Drosophila as a genetic approach to human neurodegenerative disease. Parkinsonism Relat. Disord. 7: 171–175. [DOI] [PubMed] [Google Scholar]
- Bonini, N. M., and M. E. Fortini, 2003. Human neurodegenerative disease modeling using Drosophila. Annu. Rev. Neurosci. 26: 627–656. [DOI] [PubMed] [Google Scholar]
- Calame, K. L., K. I. Lin and C. Tunyaplin, 2003. Regulatory mechanisms that determine the development and function of plasma cells. Annu. Rev. Immunol. 21: 205–230. [DOI] [PubMed] [Google Scholar]
- Cayrol, C., and E. Flemington, 1996. a G0/G1 growth arrest mediated by a region encompassing the basic leucine zipper (bZIP) domain of the Epstein-Barr virus transactivator Zta. J. Biol. Chem. 271: 31799–31802. [DOI] [PubMed] [Google Scholar]
- Cayrol, C., and E. Flemington, 1996. b The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 15: 2748–2759. [PMC free article] [PubMed] [Google Scholar]
- Chevallier-Greco, A., E. Manet, P. Chavrier, C. Mosnier, J. Daillie et al., 1986. Both Epstein-Barr virus (EBV) encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an early EBV promoter. EMBO J. 5: 3243–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, N., and J. A. Epstein, 2002. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 18: 41–47. [DOI] [PubMed] [Google Scholar]
- Cox, M., J. Leahy and J. M. Hardwick, 1990. An enhancer within the divergent promoter of Epstein-Barr virus responds synergistically to the R and Z transactivators. J. Virol. 64: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nooij, J. C., M. A. Letendre and I. K. Hariharan, 1996. A cyclin-dependent kinase inhibitor, Dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell 87: 1237–1247. [DOI] [PubMed] [Google Scholar]
- Du, W., M. Vidal, J. E. Xie and N. Dyson, 1996. RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev. 10: 1206–1218. [DOI] [PubMed] [Google Scholar]
- Edgar, B., and C. Lehner, 1996. Developmental control of cell cycle regulators: a fly's perspective. Science 274: 1646–1652. [DOI] [PubMed] [Google Scholar]
- Enver, T., 1999. B-cell commitment: Pax5 is the deciding factor. Curr. Biol. 9: R933–R935. [DOI] [PubMed] [Google Scholar]
- Farrell, P., D. Rowe, C. Rooney and T. Kouzarides, 1989. Epstein-Barr virus BZLF1 trans-activator specifically binds to consensus Ap1 site and is related to c-fos. EMBO J. 8: 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, W., and M. Noll, 1997. The Pax2 homolog sparkling is required for development of cone and pigment cells in the Drosophila eye. Genes Dev. 11: 2066–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, W., H. Duan, E. Frei and M. Noll, 1998. shaven and sparkling are mutations in separate enhancers of the Drosophila Pax2 homolog. Development 125: 2943–2950. [DOI] [PubMed] [Google Scholar]
- Garner, M., and A. Revzin, 1981. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 9: 3047–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot, J.-F., I. Mikaelian, M. Buisson, E. Manet, I. Joab et al., 1991. Transcriptional synergy and interference between the EBV transcription factors EB1 and R require both the basic region and the activation domains of EB1. Nucleic Acids Res. 19: 1251–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman, C., L. Moffat and B. Howard, 1982. Recombinant genomes which express chloramphenicol acetyl transferase in mammalian cells. Mol. Cell. Biol. 2: 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsch, D. E., E. A. Holley-Guthrie, Q. Zhang, B. Stein, M. A. Blanar et al., 1994. The bZIP transactivator, BZLF1, of Epstein-Barr virus functionally and physically interacts with the p65 subunit of NFkB. Mol. Cell. Biol. 14: 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, B. A., T. Wolff and G. Rubin, 1994. Expression of baculovirus P35 prevents cell death in Drosophila. Development 120: 2121–2129. [DOI] [PubMed] [Google Scholar]
- Holley-Guthrie, E., E. Quinlivan, E. Mar and S. Kenney, 1990. The Epstein-Barr virus promoter for early antigen (EA-D) is regulated by the EBV transactivators, BRLF1 and BZLF1, in a cell specific manner. J. Virol. 64: 3753–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney, S., E. Holley-Guthrie, E.-C. Mar and M. Smith, 1989. The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J. Virol. 63: 3878–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff, E., and A. B. Rickinson, 2001. Epstein-Barr virus and its replication, pp. 2511–2573 in Fields Virology, Ed. 4, edited by D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin et al. Lippincott Williams & Wilkins, Philadelphia.
- LaJeunesse, D. R., B. M. McCartney and R. G. Fehon, 1998. Structural analysis of Drosophila merlin reveals functional domains important for growth control and subcellular localization. J. Cell Biol. 141: 1589–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaJeunesse, D. R., B. M. McCartney and R. G. Fehon, 2001. A systematic screen for dominant second-site modifiers of Merlin/NF2 phenotypes reveals an interaction with blistered/DSRF and scribbler. Genetics 158: 893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaJeunesse, D. R., K. Brooks and A. L. Adamson, 2005. Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 alter mitochondrial morphology during lytic replication. Biochem. Biophys. Res. Commun. 333: 438–442. [DOI] [PubMed] [Google Scholar]
- Mauser, A, E. Holley-Guthrie, D. Simpson, W. Kaufmann and S. Kenney, 2002. a The Epstein-Barr virus immediate-early protein BZLF1 induces both a G(2) and a mitotic block. J. Virol. 76: 10030–10037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauser, A., S. Saito, E. Appella, C. W. Anderson, W. T. Seaman et al., 2002. b The Epstein-Barr virus immediate-early protein BZLF1 regulates p53 function through multiple mechanisms. J. Virol. 76: 12503–12512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola, I., B. Heavey, M. Horcher and M. Busslinger, 2002. Reversion of B cell commitment upon loss of Pax5 expression. Science 297: 110–113. [DOI] [PubMed] [Google Scholar]
- Morrison, T. E., and S. C. Kenney, 2004. BZLF1, an Epstein-Barr virus immediate-early protein, induces p65 nuclear translocation while inhibiting p65 transcriptional function. Virology 328: 219–232. [DOI] [PubMed] [Google Scholar]
- Niedobitek, G., A. Agathanggelou, H. Herbst, L. Whitehead, D. H. Wright et al., 1997. Epstein-Barr virus (EBV) infection in infectious mononucleosis: virus latency, replication and phenotype of EBV-infected cells. J. Pathol. 182: 151–159. [DOI] [PubMed] [Google Scholar]
- Quinlivan, E., E. Holley-Guthrie, M. Norris, D. Gutsch, S. Bachenheimer et al., 1993. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter, BMRF1. Nucleic Acids Res. 21: 1999–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebay, I., R. G. Fehon and S. Artavanis-Tsakonas, 1993. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell 74: 319–329. [DOI] [PubMed] [Google Scholar]
- Rebay, I., F. Chen, F. Hsiao, P. A Kolodziej, B. H. Kuang et al., 2000. A genetic screen for novel components of the Ras/mitogen-activated protein kinase signaling pathway that interact with the yan gene of Drosophila identifies split ends, a new RNA recognition motif-containing protein. Genetics 154: 695–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickinson, A. B., and E. Kieff, 2001. Epstein-Barr virus, pp. 2575–2627 in Fields Virology, Ed. 4, edited by D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin et al. Lippincott Williams & Wilkin, Philadelphia.
- Robinow, S., and K. White, 1988. The locus elav of Drosophila melanogaster is expressed in neurons at all developmental stages. Dev. Biol. 126: 294–303. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual, Ed. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schreiter, K., A. Steuernagel, B. Rudolph, D. Rudolph, R. Wehr et al., 2004. DG175—an inner mitrochondrial transmembrane protein identified in a UCP-modifier screen in Drosophila. Obes. Res. 12: 171. [Google Scholar]
- Siddall, N. A., K. J. Behan, J. R. Crew, T. L. Cheung, J. A. Fair et al., 2003. Mutations in lozenge and D-Pax2 invoke ectopic patterned cell death in the developing Drosophila eye using distinct mechanisms. Dev. Genes Evol. 213: 107–119. [DOI] [PubMed] [Google Scholar]
- Sinclair, A. J., 2003. bZIP proteins of human gammaherpesviruses. J. Gen. Virol. 84: 1941–1949. [DOI] [PubMed] [Google Scholar]
- Tierney, R. J., H. E. Kirby, J. K. Nagra, J. Desmond, A. I. Bell et al., 2000. a Methylation of transcription factor binding sites in the Epstein-Barr virus latent cycle promoter Wp coincides with promoter down-regulation during virus-induced B-cell transformation. J. Virol. 74: 10468–10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney, R., H. Kirby, J. Nagra, A. Rickinson and A. Bell, 2000. b The Epstein-Barr virus promoter initiating B-cell transformation is activated by RFX proteins and the B-cell-specific activator protein BSAP/Pax5. J. Virol. 74: 10458–10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanek, P., Z. Q. Wang, I. Fetka, E. F. Wagner and M. Busslinger, 1994. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell 79: 901–912. [DOI] [PubMed] [Google Scholar]
- Urier, G., M. Buisson, P. Chambard and A. Sergeant, 1989. The Epstein-Barr virus early protein EB1 activates transcription from different responsive elements including AP-1 binding sites. EMBO J. 8: 1447–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, T., 2000. Histological techniques for the Drosophila eye, part I: larva and pupa, pp. 201–227 in Drosophila Protocols, edited by W. Sullivan, M. Ashburner and R. S. Hawley. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Zhang, Q., D. Gutsch and S. Kenney, 1994. Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol. Cell. Biol. 14: 1929–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zur Hausen, H., H. Schulte-Holthauzen, G. Klein, G. Henle, W. Henle et al., 1970. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature 228: 1956–1958. [DOI] [PubMed] [Google Scholar]