Abstract
Overexpression of Hairless (H) causes a remarkable degree of tissue loss and apoptosis during imaginal development. H functions as antagonist in the Notch-signaling pathway in Drosophila, and the link to growth and apoptosis is poorly understood. To further our insight into H-mediated apoptosis, we performed two large-scale screens for modifiers of a small rough eye phenotype caused by H overexpression. Both loss- and gain-of-function screens revealed known and new genetic interactors representing diverse cellular functions. Many of them did not cause eye phenotypes on their own, emphasizing a specific genetic interaction with H. As expected, we also identified components of different signaling pathways supposed to be involved in the regulation of cell growth and cell death. Accordingly, some of them also acted as modifiers of proapoptotic genes, suggesting a more general involvement in the regulation of apoptosis. Overall, these screens highlight the importance of H and the Notch pathway in mediating cell death in response to developmental and environmental cues and emphasize their role in maintaining developmental cellular homeostasis.
APOPTOSIS or programmed cell death is crucial to the correct development of all multicellular organisms. Moreover, dysfunction of apoptosis has been linked to pathologies such as cancer and neurodegeneration (Thompson 1995; Vaux and Korsmeyer 1999). The core mediators of apoptosis are members of the caspase family of cysteine proteases, triggering, when activated, the distinct cellular changes observed in dying cells (Hengartner 2000). Inhibitor of apoptosis proteins (IAPs) directly inhibit caspase activity and promote their ubiquitination and subsequent degradation (Palaga and Osborne 2002). A delicate balance between factors that activate and those that inhibit caspase activity determines cell death or survival. In Drosophila, induction of apoptosis relies on the function of at least one of the closely linked proapoptotic genes reaper (rpr), head involution defective (hid), and grim (Bangs and White 2000; Vernooy et al. 2000). Recently it was shown that the proapoptotic activity of the encoded proteins is caused by their ability to target IAPs for ubiquitin-mediated degradation as well as by a generalized inhibition of translation (reviewed in Palaga and Osborne 2002). Although rpr, hid, and grim may induce apoptosis through similar mechanisms, they are differentially expressed and therefore not redundant. For example, hid and rpr but not grim are expressed in nonneural tissues doomed to die during metamorphosis (Jiang et al. 1997). In contrast, rpr and grim eliminate supernumerary cells in the central nervous system, whereas hid acts on midline glia of the embryo (Grether et al. 1995; Chen et al. 1996; Robinow et al. 1997). These differences can be explained by a different regulation of these genes. It has been shown that rpr is a target of the tumor suppressor p53 and the ecdysone receptor signaling pathway (Brodsky et al. 2000; Jiang et al. 2000; Ollmann et al. 2000). Two independent studies revealed that MAPK signaling negatively regulates the function and expression of hid (Bergmann et al. 1998; Kurada and White 1998). These observations indicate that genes that regulate developmental decisions, such as members of the EGFR or ecdysone receptor pathways, also influence the apoptotic machinery (reviewed in Bangs and White 2000). Therefore it was not unexpected that the Notch-signaling pathway regulates apoptosis as well. The Notch pathway is highly conserved and promotes cell fate decisions through local cell-cell interactions (reviewed in Artavanis-Tsakonas et al. 1999; Schweisguth 2004). Originally it was associated with lateral inhibition processes, e.g., during embryonic central nervous system development. Later it was shown to regulate a vast array of patterning processes and cell fate decisions (reviewed in Artavanis-Tsakonas et al. 1999). More recently, the Notch-signaling pathway has been implicated in growth control in the developing eye and wing of Drosophila as an activation of Notch is linked to tissue overgrowth and the development of cancer (Go et al. 1998; Artavanis-Tsakonas et al. 1999; Giraldez and Cohen 2003; Chao et al. 2004).
Apart from an increase in cell proliferation, this could be partly a consequence of impaired apoptosis, since the Notch signal is required for the survival of cone cells in the Drosophila pupal eye (Go et al. 1998; Wech and Nagel 2005). Moreover, activation of Notch inhibits apoptosis mediated by presenilin, suggesting protection of neurons from apoptotic cell death by Notch signaling (Ye and Fortini 1999).
The core components of the N pathway include the transmembrane receptor Notch (N), the ligands Delta (Dl) and Serrate (Ser), and Suppressor of Hairless [Su(H)], which acts as a transcriptional switch on Notch target genes (reviewed in Schweisguth 2004). Hairless (H) acts as general antagonist by assembling a repressor complex together with Su(H) and corepressors on Notch target gene promoters (Morel et al. 2001; Barolo et al. 2002). In agreement with an antiapoptotic role of Notch signals, overexpression of H during imaginal development results in a pronounced reduction of tissue size and thus corresponds to an N loss-of-function phenotype (Go et al. 1998; our own observations). However, the underlying molecular mechanisms have remained largely elusive. Here we show that this effect can be at least partly explained by the induction of apoptosis. Correspondingly, the H-induced tissue loss can be rescued by antiapoptotic factors DIAP1 or baculoviral p35. To identify the molecular link between Notch signaling and apoptosis, we performed two large-scale genetic screens for modifiers of a small rough eye phenotype caused by H overexpression. At first, we screened 2290 enhancer-promotor (EP) lines from the Rørth collection plus a number of candidate upstream activation sequence (UAS) lines and identified 86 modifiers. Second, we searched through 214 overlapping deficiencies of the Drosophila genome and subsequently analyzed candidate loci. Altogether, our gain- and loss-of-function screens provided us with 112 different interactors that include genes previously implicated in proliferation and apoptosis, thus validating our strategy. In addition, we were able to assign for a number of known genes an as yet unknown role in apoptosis. Most interestingly, 15 of the identified interactors are not yet functionally described, raising the question of their molecular and genetic role in N signaling and the regulation of apoptosis.
MATERIALS AND METHODS
Drosophila stocks and maintenance:
Stocks were maintained on standard fly food at 18°. The collection of EP strains described by Rørth (1996) was obtained from Exelixis. The deficiency kit of overlapping deletions as well as other deficient chromosomes and mutant stocks were obtained from the Bloomington Stock Center. To simplify screening procedures, a recombinant chromosome (II) carrying GMR-Gal4 (Hay et al. 1997) and UAS-Hairless (Maier et al. 1999) was generated (GMR>H/CyO). At 25°, this stock shows an intermediate small rough eye phenotype, suited for the identification of enhancers and suppressors alike. GMR-Dmp53 (Ollmann et al. 2000), GMR-hid (Grether et al. 1995), GMR-rpr (White et al. 1996), and GMR-grim (Chen et al. 1996) flies ectopically express the respective gene under GMR control. Overexpression experiments in the wing were performed with biombmd65-Gal4 (Lecuit et al. 1996). UAS-lacZ was used as the control. For rescue of cell death, we used UAS-p35 (Hay et al. 1994) and UAS-DIAP1 (gift of A. Müller). UAS lines mentioned were obtained from the Bloomington Stock Center, apart from UAS-mastermind (gift of M. Muskavitch), UAS-widerborstDN (gift of S. Eaton), UAS-armadilloact (gift of S. Clevers), and UAS-JNKDN (gift of E. Kuranaga).
Gain-of-function screen:
GMR>H/CyO virgin female flies were mated to males from the EP collection. The progeny was scored under a dissecting microscope and selected on the basis of enhancement or suppression of the GMR>H/+ eye phenotype. Likewise, UAS constructs of candidate genes were co-overexpressed with GMR>H and the progeny inspected as well. Ten animals of the respective genotype were scored minimally and pursued further only if most of them exhibited the same modification. All modifiers were retested at least once. Crosses were carried out at 25°. For further validation of the results, overlapping deficiencies, and respective candidate mutant alleles were tested for reverse modification behavior.
Loss-of-function screen:
Males heterozygous for a deficiency or a given mutant allele were crossed to GMR>H/CyO virgin females and the F1 inspected for modification of the small eye phenotype in comparison with the siblings in a dissecting microscope. In the case of X chromosomal deficiences or mutations, reciprocal crosses were performed. Crosses were carried out at 25° as outlined above.
Phenotypic analyses:
For immuno-stainings, larval imaginal discs were fixed for 25 min in PBS + 4% paraformaldehyde at room temperature and washed several times in PBT (PBS + 0,1% Tween20). Primary antibody incubations were overnight at 4° after preincubation for 30 min in PBT + 4% normal goat serum at room temperature. We used rabbit anticleaved Caspase-3 (1:200; NEB Cell Signaling Technology) and rat anti-H (1:1000; Maier et al. 1999). After several washes in PBT, goat secondary antibodies coupled to fluorescein or Cy3 were added (1:200; Jackson ImmunoResearch, West Grove, PA). Imaginal discs were mounted in Vectashield (Vector Laboratories, Burlingame, CA). Pictures were taken on a Zeiss Axioskop linked Bio-Rad (Hercules, CA) MRC1024 confocal microscope. Wings and fly heads were analyzed by light microscopy and digitally photographed (Optronics, Pixera).
Molecular analysis:
EP insertions of candidate modifiers were positioned initially using the database resources of FlyBase (http://flybase.bio.indiana.edu/). In cases of unknown localization, inverse PCR and sequencing was performed according to the protocol given by E. J. Rehm (http://www.fruitfly.org/about/methods/inverse.pcr.html). For further confirmation, the respective EP line was crossed to GMR-Gal4 and/or ptc-Gal4 and in situ hybridizations with gene-specific probes were performed on eye- and/or wing-imaginal discs according to the protocol of de Celis et al. (1996). Probes were derived by PCR amplication of genomic DNA or cDNA (obtained from BioCAT, Heidelberg, Germany). For EP line 3139, we were unable to define the insertion via inverse PCR sequencing. Instead, the localization of the EP element was determined by in situ hybridization with a digoxygenin-labeled probe against white using a slightly modified protocol of Ashburner (1989).
RESULTS
Overexpression of the Notch antagonist H increases the rate of cell death in imaginal tissues:
H is a major antagonist of N signaling and increasing H activity is inversely correlated with N activity (reviewed in Schweisguth 2004). We and others observed that overexpression of H results in reduction of tissue size (Figure 1, D and J). This is accompanied by pronounced increase of apoptotic cells characterized by pyknotic nuclei, which are restricted to the area where ectopic H expression is induced in third instar larval discs (Figure 1, A–C and G–I). However, especially after overexpression of H in differentiating ommatidia, we observed a strong induction of caspase 3 activity in cells where H staining seemed extremely weak (Figure 1, A–C), which might be due to the fact that induction of caspase activity is correlated with reduced translation and degradation of the respective proteins. Comparable observations were already described after overexpression of activated Jun N-terminal kinase (JNK) (Adachi-Yamada and O'Connor 2002) and could be observed after overexpression of the known cell death inducer p53 (data not shown).
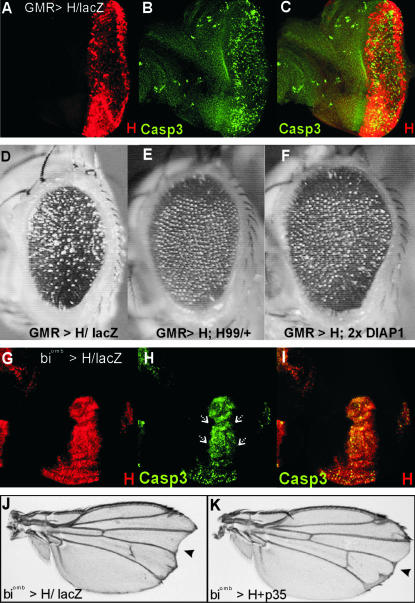
Figure 1.
H causes cell death during imaginal development. (A–C) Overexpression of H in differentiating cells of eye disc (red in A and C) induces cell death and activation of caspase 3 within the region where H is overexpressed (green in B and C). C is a merge of A and B. (D) The adults show a moderate small rough eye phenotype. (E) This can be suppressed in a Df(3L)H99 heterozygous background or with two additional copies of DIAP1 (F). (G–I) Likewise, overexpression of H within the central part of the wing blade (red) induces cell death executed by activation of caspase 3 (green; arrows in H). (J) In the adults, resultant wings are smaller and notched (arrowhead). (K) Loss of tissue and incisions can be rescued by simultaneous misexpression of baculovirus p35 (arrowhead). Genotypes are: GMR>H/UAS-lacZ (A–D); GMR>H, Df(3L)H99/+ (E); UAS-DIAP1, GMR>H, UAS-DIAP1 (F); biombmd65-Gal4, UAS-H/UAS-lacZ (G–J); biombmd65-Gal4/UAS-p35; UAS-H/+ (K).
Assuming a direct link between tissue loss and excess apoptosis, inhibition of cell death should suppress H overexpression phenotypes. Indeed, a co-overexpression of either Drosophila DIAP1 (Hay 2000) or the baculoviral caspase inhibitor protein p35 (Hay et al. 1994) with H largely rescued tissue loss in wing and eye (Figure 1, compare D with F and J with K). Moreover, halving the dose of the proapoptotic genes hid, rpr, and grim in a Df(3L)H99 heterozygous background also improved the phenotype (Figure 1E). These results strongly indicate that a major cause of tissue loss, which is observed as a result of the inhibition of N signaling, is increased apoptosis.
To gain insight into the molecular mechanisms of N involvement in apoptosis with the aim of isolating prospective mediators or regulators of this process, we designed genetic screens for modifiers of H-mediated tissue loss. As shown in Figure 1, both decrease of proapoptotic and increase of antiapoptotic factors is sufficient to modify H overexpression phenotypes, thus validating our strategy. To ease the screening procedure, we recombined the GMR-Gal4 driver construct with UAS-H (GMR>H). Animals heterozygous for GMR>H showed a moderate small rough eye phenotype, which can be easily scored in a dissecting microscope, allowing for the selection of enhancers as well as suppressors (Figure 2).
Figure 2.

Classification of enhancers and suppressors of GMR>H. (A) Wild-type eye showing regular size and arrangement of ommatidia. (B) Small rough eye phenotype caused by overexpression of H (GMR>H/lacZ). (C–F) Examples of suppressors are: (C) weak suppressor (S1) EP(2)816; (D) moderate suppressor (S2) EP(2)2518; (E) strong suppressor (S3) EP(X)1433; (F) suppressor with enlargement capacity (S4) EP(3)3704. Overgrowth of tissue is marked with arrows. (G–J) Examples for enhancers are: (G) weak enhancer (E1) EP(3)469; (H) moderate enhancer (E2) EP(3)3060; (I) strong enhancer (E3) EP(2)633; (J) extreme enhancer (E4) EP(3)3118.
Two major screens, a gain-of-function (GOF) screen and a loss-of-function (LOF) screen, were performed. We isolated suppressors that restored eye size and significantly ameliorated the ommatidial irregularity and also enhancers that further reduced the eye size and increased the rough appearance; in extreme cases such combinations even caused lethality. Figure 2 shows examples of the quality and strength of the effects and represents the different categories of modifiers that were recovered.
Gain-of-function screen:
In the gain-of-function screen, we co-overexpressed H together with 2290 individual lines from the EP collection (Rørth 1996) as well as 19 UAS constructs of candidate genes in the GMR pattern and screened the F1 progeny for modification of the eye phenotype. Subsequently, in a cross with GMR-GAL4, all modifiers were tested for their own overexpression phenotypes in the eye to distinguish possible additive effects from more specific interactions. In summary, 86 factors were recovered, namely 57 enhancers and 29 suppressors (Table 1 and supplementary Table S1 at http://www.genetics.org/supplemental/). This represents ∼4% of the total, a value close to that obtained in previous gain-of-function screens (Abdelilah-Seyfried et al. 2000; Kraut et al. 2001; Peña-Rangel et al. 2002; Tseng and Hariharan 2002; Bidet et al. 2003). Where available, deficient chromosomes, loss and gain of function, or dominant-negative alleles of candidate genes were obtained and tested for genetic interactions with GMR>H. In roughly half of the cases, gain and loss of function of the same locus showed opposite interactions.
TABLE 1.
Novel modifying EP or UAS lines of GMR>H
| Genea | Map | GOF linesb | GMRc | GMR>H/+d | Deficiency/allelee | Breakpointsf | GMR>H/+g | Cell deathh | Referencei |
|---|---|---|---|---|---|---|---|---|---|
| JNK pathway | |||||||||
| basket | 31B3 | UASDN | Little small | S1/2 | bsk1 | S1 | Not tested | ||
| misshapen | 62E6-7 | 549 | Wild type | E1 | msn06946 | S1 | Not tested | Kraut et al. (2001) | |
| GTPases, protein kinases and phosphatases | |||||||||
| raspberry | 9E1-2 | 1098 | Wild type | E1 | Df(1)HC133 | 9B9; 9F4 | S1 | Not tested | Bidet et al. (2003); Peña-Rangel et al. (2002) |
| CG2446 | 10D6-7 | 1503 | Little big | S1 | Df(1)m259-4 | 10C2; 10E2 | 0 | + | Abdelilah-Seyfried et al. (2000) |
| Casein kinase I | 11B7 | 1555 | Little big | S1 | Df(1)HF368 | 11A2; 11B9 | 0 | + | |
| CG5261 | 27F6 | 816 | Wild type | S1 | Df(2L)RF | 27E3-F; 28B3-4 | 0 | + | |
| connector enhancer of ksr | 54B9-11 | 576 | Little big | E1 | cnkE-2083 | E1 | Not tested | ||
| Drac2 | 66A1 | 3118 | Slits | E4 | Df(3L)pbl-X1 | 65F3; 66B10 | S1 | + | Bidet et al. (2003); Peña-Rangel et al. (2002); Tseng and Hariharan (2002) |
| Malic enzyme | 87C6-7 | 1250 | Little big | E1 | Df(3R)kar31 | 87C2; 87D1 | 0 | Not tested | Bidet et al. (2003); Peña-Rangel et al. (2002) |
| widerborst | 98A8 | 3113; 3559 | Little big | S2 | wdbdw | E1/2 | + | Abdelilah-Seyfried et al. (2000); Kraut et al. (2001) | |
| UASDN | E1/2 | ||||||||
| General transcriptional regulators/chromatin-remodeling factors | |||||||||
| nejire | 8F-9 | 1149; 1179; 1410 | Small | E2 | Df(1)C52; nejQ7 | 8E; 9C-D | S1 | + | Abdelilah-Seyfried et al. (2000); Peña-Rangel et al. (2002) |
| Dorsal switch | 14B15-16 | 355 | Wild type | S2 | Df(1)19 | 13F2-18; 14E | Lethal | + | Kraut et al. (2001); Peña-Rangel et al. (2002) |
| protein | Df(1)4b18 | 14B8; 14C1 | Lethal | ||||||
| longitudinals | 47A11-13 | 2537 | Wild type | E2 | Df(2R)E3363 | 47A; 47F | S1 | Not tested | Abdelilah-Seyfried et al. (2000) |
| lacking | |||||||||
| Rpd3 | 64B12 | 3672 | Wild type | E1 | Df(3L)GN24 | 63F6-7; 64C13-15 | S1 | Not tested | |
| Regena | 83B5-6 | 3713 | Wild type | E1 | Not tested | ||||
| Genes acting in protein transport or regulation of translation | |||||||||
| IGF-II mRNA | 9F5 | 1433 | Little big | S3 | Df(1)HC133 | 9B9; 9F4 | E1 | + | Kraut et al. (2001) |
| bind. protein | |||||||||
| blue cheese | 26A1 | 2299 | Little small | E2 | Mutants not available or not tested | + | Abdelilah-Seyfried et al. (2000); Kraut et al. (2001) | ||
| Sec 61α | 26D7-8 | 2567 | Wild type | E1 | Mutants not available or not tested | Not tested | |||
| eclair | 85E4 | 469 | Little small | E1 | Df(3R)GB104 | 85D12; 85E10 | S1 | Not tested | |
| CG11779 | 91F6-7 | 1123 | Big | S2 | Df(3R)Cha9 | 91C7-D1; 92A2 | 0 | + | |
| Novel genes with unknown function | |||||||||
| CG3600 | 2C1 | 1232 | Wild type | S1 | Df(1)sc8 | 1B; 3A3-C2 | Lethal | + | Tseng and Hariharan (2002) |
| CG12462 | 3,F4 | 1413 | Little small | E2 | Df(1)GA102 | 3D4-5; 3F7-8 | E1 | + | |
| CG11068 | 12D1-2 | 1595 | Little big | E3/4 | Df(1)HA92 | 12A6-7; 12D3 | S1 | Not tested | Tseng and Hariharan (2002) |
| CG32521 | 19F2-3 | 555 | Little big | S1 | Df(1)Q359 | 19E7; 19F6 | E1 | + | |
| CG17223 | 23C5 | 797 | Wild type | E2 | Df(2L)JS32 | 23C3-5; 23D1-2 | S1 | Not tested | |
| Df(2L)JS17 | 23C1-2; 23E1-2 | S1 | |||||||
| l(2)k10113 | 27F4-5 | 1221 | Little big | S1 | Df(2L)spd | 27D-E; 28C | 0 | + | Abdelilah-Seyfried et al. (2000); Peña-Rangel et al. (2002) |
| CG8788 | 45A11-12 | 2301 | Little small | E1 | Df(2R)w45 | 45A6-7; 45E2-3 | S1 | — | |
| l(2)05510 | 57A6 | 2356; 2587 | Pupal lethal | E4 | Df(2R)AA21 | 56F9-17; 57D11-12 | S1 | — | Abdelilah-Seyfried et al. (2000); Peña-Rangel et al. (2002); Tseng and Hariharan (2002) |
| CG17180 | 61C3 | 3104 | Little big | E2 | Df(3L)emc-E12 | 61A; 61D3 | 0 | Not tested | Bidet et al. (2003) |
| CG14959j | 63C2-3 | 3139 | Wild type | S1 | Df(3L)Awh2 | 63B10-11; 63E4-9 | 0 | — | Peña-Rangel et al. (2002); Tseng and Hariharan (2002) |
| CG7752 | 78C5-6 | 756 | Wild type | S2 | Df(3L)Pc-Mk | 78A2; 78C9 | E1 | + | |
| CG7552 | 88D1 | 666 | Little big | E1 | Df(3r)red-1 | 88B1; 88D3-4 | S1 | Not tested | Peña-Rangel et al. (2002); Tseng and Hariharan (2002) |
| CG5720 | 95F11-12 | 3716 | Little big | E1 | Df(3R)crb-F89 | 95D7-11; 95F15 | 0 | Not tested | Abdelilah-Seyfried et al. (2000); Bidet et al. (2003); Peña-Rangel et al. (2002) |
| CG15507 | 99B10 | 3084 | Wild type | E1 | Df(3R)01215 | 99A6; 99AC1 | S1 | Not tested | |
The identified modifiers were grouped according to their predicted molecular function and arranged by localization. Only those in which the EP element is inserted in sense orientation relative to the transcription unit are listed.
Genes are listed with their full names. Gene names are underlined if gain- and loss-of-function mutations showed the opposite genetic interaction with GMR>H.
EP lines are listed with their numbers. In some cases, a dominant-negative form (UASDN) of the respective gene was used.
Short description of the eye phenotype obtained after overexpression of EP or UASDN lines with GMR-Gal4.
Categories of phenotypes according to Figure 2; S, Suppressor; E, Enhancer. All crosses were maintained at 25°.
Name of deficiencies and loss-of-function alleles of candidate genes crossed with GMR>H. Deficiencies underlined in this column were also identified in the LOF screen.
Breakpoints of deficiencies according to FlyBase.
Phenotype of respective deficiency or mutant allele in trans over GMR>H categorized according to Figure 2; S, Suppressor; E, Enhancer; 0, no interaction. All crosses were maintained at 25°.
Modifiers were assayed for their influence on cell death processes. Suppressors of GMR>H were tested for their ability to rescue the eye phenotype caused by misexpression of p53 or proapoptotic genes. Enhancers of GMR>H as well as lines that caused lethality were subjected to a rescue experiment by simultaneously overexpressing DIAP1. Factors that showed modification are marked with (+) and with (−) for no interaction. For detailed results, see Table 2.
EP lines identified in other gain-of-function screens.
For EP(3)3139 and EP(3)3560 we were unable to identify the sequences responsible for the interaction. In situ hybridizations on chromosomes revealed that these lines have multiple insertions.
Table 1 and Table S1 (http://www.genetics.org/supplemental/) summarize the molecular, phenotypic, and genetic interaction data of the gain-of-function screen. Enhancers and suppressors were grouped according to their molecular functions. As expected from the rationale of our screen, we isolated several members the Notch-signaling pathway that behaved as enhancers as well as inhibitors of apoptosis that behaved as suppressors [thread (th), bantam (ban); Table S1]. Apart from a role in apoptosis, our analysis indicates an additional involvement of H in cell cycle progression or control (Table S1). This is in line with the idea that N signaling regulates cell division during development of the Drosophila eye (reviewed in Thomas 2005) and induces overproliferation after overexpression (Go et al. 1998). Three enhancers involved in ecdysone signaling were found (Table S1 and Figure 4, G–I), which might be expected since ecdysone triggers programmed cell death, for example, during metamorphosis (Jiang et al. 1997). Table S1 also lists miscellanous factors and the EP lines that are inserted in the antisense orientation. The following sections highlight some of our findings.
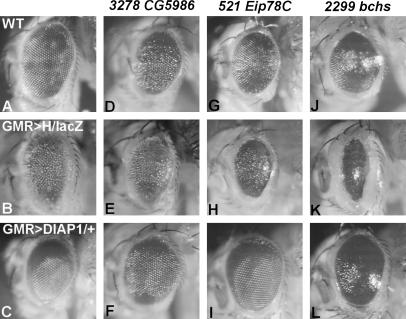
Figure 4.
Enhancers of GMR>H. Wild-type (A), GMR>H/lacZ (B), and DIAP1>GMR-Gal4 (C) are shown for comparison. Overexpression of EP(3)3278, EP(3)521, or EP(2)2299 with GMR-Gal4 caused somewhat smaller eyes (D, G, and J) and, in the case of EP(2)2299, also eyes with a glossy appearance (J). These phenotypes were rescued by simultaneous overexpression of DIAP1, arguing for an involvement in apoptosis (F–L). All three EP lines enhanced GMR>H, although to different degrees (E–K).
Wg-, Dpp-, and EGFR-signaling pathways:
Our screen identified components of the Wingless (Wg)- and Decapentaplegic (Dpp)-signaling pathways. Both pathways have well-characterized functions in eye development, cell growth, and proliferation (Giraldez and Cohen 2003; Voas and Rebay 2004). Activation of these pathways by overexpression either of the morphogens Wg and Dpp or of downstream components such as Pygopus, Armadillo, and the Dpp receptor Thick veins decreased eye size and induced the formation of necrotic black patches in GMR>H eyes (Table S1 at http://www.genetics.org/supplemental/; data not shown). At first sight this was surprising, as both pathways have been involved in protection from, rather than promotion of, apoptosis during Drosophila development (Pazdera et al. 1998). However, in the case of discontinuities in either morphogen gradient, so-called “morphogenetic apoptosis” is induced, which might explain the effects (Adachi-Yamada and O'Connor 2002; Ryoo et al. 2004). Accordingly, overexpression of Wg-pathway components caused much smaller eyes (Table S1). This is not observed for Dpp-pathway components and raises the possibility of more specific interactions with H.
Crosstalk between Notch and EGFR pathways has been amply documented in the past and was once more confirmed by our screen (Table S1 at http://www.genetics.org/supplemental/). EGFR signaling downregulates the proapoptotic factor Hid (Bergmann et al. 1998; Kurada and White 1998). In accordance, reducing the activity of EGFR signaling, for example, by overexpression of the negative regulator Anterior open or a dominant-negative form of EGFR enhanced the GMR>H phenotype as did a heterozygous mutation in the EGFR effector pointed (pntΔ88). Conversely, activation of EGFR signaling via overexpression of the activated EGFR, of Rhomboid, or of PointedP2 enlarged the eye size (Table S1).
The JNK-signaling pathway:
Genetic interactions between Notch- and JNK-signaling pathways have been described in the process of dorsal closure in the embryo (Zecchini et al. 1999). However, the JNK-signaling pathway also mediates morphogenetic apoptosis (Adachi-Yamada et al. 1999; Adachi-Yamada and O'Connor 2002; Ryoo et al. 2004). Indeed, our data suggest that H might induce apoptosis via the JNK pathway. For example, overexpression of the JNKKK Misshapen (Msn) specifically enhanced the GMR>H eye phenotype, whereas the overexpression of Msn alone did not affect eye morphology (see Table 1). Moreover, a dominant-negative form of the JNK Basket (Bsk) acted suppressive, as did a mutation in this gene (Table 1).
GTPases, protein kinases, and phosphatases:
The screen revealed, among others, Rac2 as a strong enhancer of GMR>H. Rac2 encodes a member of the Rho family of GTPases, known to contribute to the regulation of receptor-tyrosine-kinase- and JNK-signaling activity (Frost et al. 1996). Recently this activity was shown to be induced upon steroid- and radiation-induced cell death (Lee et al. 2003). Not suprisingly, overexpression of Rac2 results in slit eyes. However, the interaction with H seems not just additive, as a deficiency uncovering Rac2 acted suppressive (Table 1). Notably, we identified widerborst (wdb), which codes for the β′-regulatory subunit of protein phosphatase PP2A (Hannus et al. 2002), as a suppressor of H-mediated cell death (Table 1, Figure 3I). Remarkably, the rather weak wdbdw allele acted as a good enhancer (Table 1, Figure 3J), in agreement with the finding that PP2A-β′ is required for survival and protection from apoptosis in Drosophila S2 cells (Li et al. 2002) as well as in vivo (see Table 2).
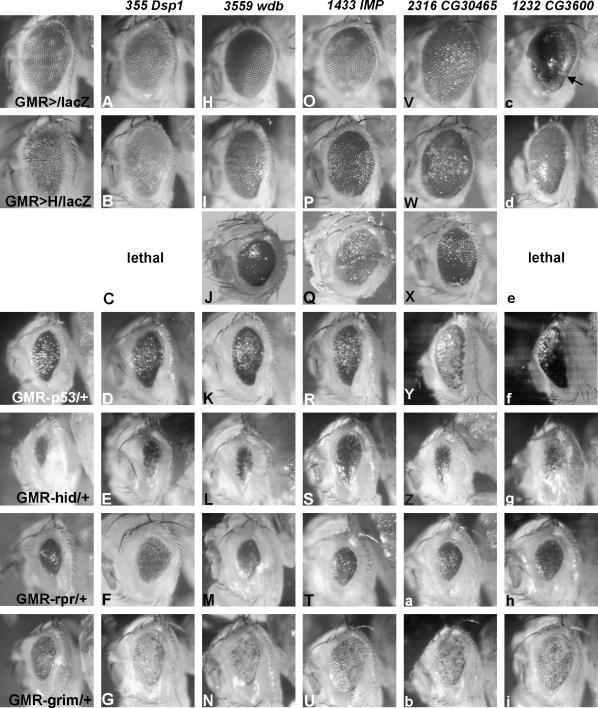
Figure 3.
Suppression of cell death by GMR>H suppressors. Five examples of GMR>H suppressors are shown, which generally rescue apoptosis. All eyes shown are from females. Control eyes for comparison are shown at the far left. Overexpression of the respective EP line in the GMR pattern is shown (A, H, O, V, c). (A–G) EP(X)355 drives expression of DspI and shows a weak rough eye phenotype after overexpression with GMR-Gal4 (A). It acts as moderate suppressor of GMR>H (B). Df(1)19 deleted DspI and caused lethality in combination with GMR>H (C). Simultaneous overexpression of DspI with proapoptotic factors GMR-p53 (D), GMR-hid (E), GMR-rpr (F), or GMR-grim (G) inhibited cell death in the eye. (H–N) Overexpression of Wdb with EP(3559) in the eye is phenotypically wild type (H) and suppressed GMR>H (I). Accordingly, the mutant wdbdw caused a moderate enhancement (J). Overexpression of wdb was able to suppress p53- and grim-mediated induction of cell death (K and N) but had little effect on GMR-hid and GMR-rpr (L and M). The strong suppressors EP(X)1433 (P), EP(2)2316 (W), and EP(X)1232 (d) also rescued the small rough eye phenotype of all tested cell death inducers (R–U, Y–b, f–i). Overexpression of EP(2)2316 and EP(X)1232 caused enlarged rough eyes (V, c), whereas GMR-Gal4 > EP(X)1433 showed a wild-type eye phenotype (O). In the case of GMR-Gal4> EP(X)1232, a black membrane, presumably a remnant of the pupal eye membrane, covered the adult eye (arrow in c). This is also visible in f.
TABLE 2.
H modifiers involved in cell death
| Gene | GOF/LOF | GMR-p53 | GMR-hid | GMR-rpr | GMR-grim |
|---|---|---|---|---|---|
| Factors with general influence | |||||
| anterior open | LOF | S | S | S | S |
| Arflike at 72A | LOF | E | E | E | E |
| CG3600 | GOF | S | S | S | S |
| CG30465 (antisense) | GOF | S | S | S | S |
| CG32521 | GOF | S | S | S | S |
| CG32737 | GOF | S | S | S | S |
| DER activated | GOF | S | S | S | S |
| Dorsal switch protein | GOF | S | S | S | S |
| LOF | E | E | E | E | |
| hemipterous | LOF | S | S | S | S |
| IGF-II mRNA bind protein | GOF | S | S | S | S |
| klumpfuss | LOF | S | S | S | S |
| pointed | GOF | S | S | S | S |
| puckered | LOF | E | E | E | E |
| reaper, hid, grim | LOF | S | S | S | S |
| Rho1 | LOF | E | E | E | E |
| thread | GOF | S | S | S | S |
| LOF | E | E | E | E | |
| Factors with influence on hid, rpr, and grim | |||||
| brahma | LOF | — | E | E | E |
| leonardo | LOF | S | — | S | S |
| nejire | LOF | — | S | S | S |
| sine oculis | LOF | — | E | E | E |
| Factors with specific influence on two or one proapoptotic gene | |||||
| p53 and grim | |||||
| CG2446 | GOF | S | — | — | S |
| CG5261 | GOF | S | — | — | S |
| Delta | LOF | E | — | — | E |
| Hairless | LOF | S | — | — | S |
| lilliputian | LOF | S | — | — | S |
| Notch | GOF | S | Lethal | Lethal | S |
| Suppressor of Hairless | GOF | S | — | — | S |
| widerborst | GOF | S | — | — | S |
| p53 and hid | |||||
| bantam | GOF | S | S | — | — |
| CG11779 | GOF | S | S | — | — |
| rhomboid | GOF | S | S | — | — |
| hid and reaper | |||||
| disc overgrown | GOF | — | S | S | — |
| nuclear fallout | GOF | — | S | S | — |
| reaper and grim | |||||
| Casein kinaseI | GOF | — | — | S | S |
| CG7752 | GOF | — | — | S | S |
| CG13791 (antisense) | LOF | — | — | S | S |
| Factors that influence one proapoptotic gene | |||||
| bazooka | LOF | — | — | S | — |
| Cyclin B | LOF | — | E | — | — |
| dishevelled | LOF | — | E | — | — |
| elbowB | GOF | S | — | — | — |
| l(2)k10113 | GOF | — | S | — | |
| tribbles | GOF | — | S | — | — |
| turtle | LOF | S | — | — | — |
| Enhancement and suppression | |||||
| defective proventriculus | LOF | S | E | E | — |
Suppressors identified in the gain-of-function screen (GOF) as well as all GMR>H modifiers from the loss-of-function screen (LOF) were crossed to flies overexpressing p53, hid, rpr, or grim in the GMR pattern. S, suppression; E, enhancement; −, no phenotypic change. Factors were subdivided into the following four categories: factors that generally influenced all proapoptotic genes and those that modified three, two, or just one factor. One factor, dev, enhanced p53 misexpression phenotypes, but rescued those of hid, rpr, and grim.
General transcriptional regulators and chromatin-remodeling factors:
Several factors involved in gene and chromatin regulation were identified (Table 1). As H functions via transcriptional repression of N target genes (Schweisguth 2004), these interactions might reveal novel components of the repression complex. For example, Dorsal switch protein I (DspI) was found as a moderate suppressor (Table 1 and Figure 3B). DspI functions as corepressor of Dorsal and furthermore as a rather general regulator in many tissues and at various stages of Drosophila development (Lehming et al. 1998; Mosrin-Huaman et al. 1998), raising the possibility of a specific interaction with H. Most interestingly, two deficiencies, both of which uncover the DspI locus, caused lethality in a GMR>H background (Table 1; Figure 3C), emphasizing the specificity of the H-DspI interaction. As further enhancer, we identified Rpd3, which encodes a product with histone deacetylase activity that binds to and activates the general corepressor protein Groucho (Chen et al. 1999). H mediates transcriptional repression by recruitment of Groucho (Barolo et al. 2002; our own observations), arguing for an involvement of Rpd3 in such a repressor complex.
Genes acting in protein transport or regulation of translation:
This group comprises three enhancers [Sec61α, Eclair, Blue cheese (Bchs)] and two suppressors [CG11779 and IGF-IImRNA-binding protein (IMP)]. Overexpression of either IMP or CG11779 suppressed general apoptosis as well (Tables 1 and 2; Figure 3, R–U) and the Bchs overexpression phenotype with glazed, smaller eyes was rescued by DIAP1 (Tables 1 and 2, Figure 4, J–L). It will be interesting to see what the role of these factors in apoptosis might be.
Novel factors with unknown function:
In the foregoing analysis we have concentrated on genes whose functions were already known, but one of the aims of this screen was to identify novel genes affecting H-mediated cell death. Altogether, we found a collection of 14 EP lines representing functionally uncharacterized loci, including nine enhancers and five suppressors. Of the former, six were confirmed by reciprocal interactions with respective overlapping deficiencies and three of the latter (Table 1), supporting the specificity of the genetic interaction with H. Most interestingly, most of the suppressors of the GMR>H small eye phenotype had a positive influence on cell death induced by p53, hid, rpr, or grim (Table 2 and Figure 3, V–i) and are currently under further genetic and molecular investigations.
Loss-of-function screen:
In a complementary set of experiments we searched for further modifiers by screening through an ordered collection of 214 chromosomal deficiencies that together uncover 75% of the genome and identified 41 deficiencies modifying the GMR>H phenotype (Table 3 and supplementary Table S2 at http://www.genetics.org/supplemental/). To confirm these interactions and refine the genomic regions, we tested >250 additional deficiency stocks and individual mutations of several candidate genes that map to the interacting deficiencies in an attempt to identify single loci. From these studies we identified mutations in 14 genes that act as dominant enhancers of GMR>H and mutations in 22 genes that act as suppressors (Table 3). Importantly, 13 deficiencies detected in our screen uncover modifiers found in the gain-of-function screen and showed the opposite influence on GMR>H (Table 2 and Table S2). Modifiers validated in this way were regulators of apoptosis [klumpfuss (klu), thread (th)] of cell proliferation [diminuitive (dm), dacapo (dap), Cyclin B (CycB)] and of cell growth or adhesion [fat (ft), inflated (if), multiple edematous wings (mew)]. We again recovered chromatin-remodeling factors [brahma (brm), smrter (smr), Rpd3] and the ubiquitin-conjugating enzyme crossbronx (Watts et al. 2003). Interestingly, mutations in Arf72A and Rho1 acted as enhancers of GMR>H—both encode small GTPases—whereas mutations in the kinase-encoding genes Rho-kinase (rok) and turtle acted as suppressors, arguing for an involvement of these factors in H-mediated cell death.
TABLE 3.
Deficiency screen for modifiers of GMR>H
| Name | Cytology | Modificationa | Candidate genesb | Modificationc | Cell deathd |
|---|---|---|---|---|---|
| Df(1)N-8 | 3C2-3; 3E3-4 | E2 | Notch(N5419) | Lethal | + |
| diminuitive (dm1) | Lethal | − | |||
| Df(1)C52 | 8E; 9C-D | S1 | nejire(nej3) | S1 | + |
| Df(1)v-N48 | 9F; 10C3-5 | E2 | dishevelled(dsh1) | E3 | + |
| Df(1)N105 | 10F7; 11D1 | S1 | smrter (smrG0361) | S1 | − |
| Df(1)N12 | 11D1-2; 11F1-2 | S1 | hemipterous (hepv39, v75) | S1 | + |
| multiple edematous wings (mewM6) | S1 | − | |||
| Df(1)sd72b | 13F1; 14B1 | S1 | scalloped(sdETX4) | S1 | − |
| Df(1)4b18 | 14B8; 14C1 | Lethal | Dorsal switch protein(Dsp1) | Lethal | + |
| Df(1)r-D1 | 14C2-4; 15B2-C1 | S2 | inflated (ifB2) | S1 | − |
| Rho-kinase (rok1) | S1 | − | |||
| Df(1)B25 | 15D3; 16A4-6 | E1 | Bar (B1) | E1 | − |
| bazooka (baz4) | S1 | + | |||
| Df(1)BK10 | 16A2; 16C7-10 | E1 | Bar (B1) | E1 | − |
| Df(1)HF396 | 18E1-2; 20E-F | S1 | amnesiac(amn1) | S1 | − |
| Df(2L)dp-79b | 22A2-3; 22D5-E1 | S1 | anterior open(aop1) | S1 | + |
| Df(2L)C144 | 22F3-4; 23C3-5 | S2 | lilliputian (lilliA17-2, 00632) | S2 | + |
| Df(2L)JS17 | 23C1-2; 23E1-2 | S1 | lilliputian (lilliA17-2, 00632) | S2 | + |
| Df(2L)sc19-8 | 24C2-8; 25C8-9 | S1 | echinoid (edk01102) | S1 | — |
| fat (ftG-rv) | S2 | — | |||
| turtle (tutlk14703) | S1 | + | |||
| Df(2L)sc19-4 | 25A5; 25E5 | E1 | thick veins (tkv1) | S1/2 | nt |
| Df(2R)cn9 | 42E; 44C | E1 | sine oculis (so1) | E1 | + |
| Df(2R)B5 | 46A; 46C | E1 | crossbronx (cbx05704) | S1 | — |
| dacapo (dap4) | S2 | — | |||
| Df(2R)X1 | 46C; 47A1 | S1 | crossbronx (cbx05704) | S1 | — |
| leonardo (leo07103) | S1 | + | |||
| Df(2R)Jp8 | 52F5-9; 52F10-53A1 | E2 | Rho1 (Rho1E.10) | E2 | + |
| Df(2R)X58-12 | 58D1-2; 59A | E2 | Cyclin B (CycB2) | E1 | + |
| defective proventriculus (dve01738) | S1 | + | |||
| Df(3L)HR232 | 63C1; 63D2 | S1 | sprouty (sty226) | S1/2 | — |
| Df(3L)GN24 | 63F6-7; 64C13-15 | S1 | Rpd3(Rpd304556) | S1 | − |
| Df(3L)vin2 | 67F2; 68D6 | S2 | klumpfuss (klu09036) | S1/2 | + |
| Df(3L)brm11 | 71F1-4; 72D1-10 | E2 | brahma (brm2) | E2/3 | + |
| thread(th4) | E2 | + | |||
| Arflike at 72A (Arf72AN6) | E2 | + | |||
| Df(3R)p712 | 84D4-6; 85B6 | E1/2 | puckered (pucE69) | E2 | + |
| Df(3R)e-N19 | 93B; 94A | E1 | Calcium/calmodulin dependent | ||
| protein kinase (CakiX-307) | E1 | − | |||
| Df(3R)TI-P | 97A; 98A1-2 | E1 | Serrate (SerRX102) | E1 | − |
Listed are those modifying deficiencies in which the responsible loci were identified. Underlined are deficiencies that affect genes also found in the gain-of-function screen and showing an opposite interaction with GMR>H. See Table S2 at http://www.genetics.org/supplemental/ for further modifying deficiencies where no candidates could be assigned.
Modifications were phenotypically categorized according to Figure 2: S, suppressors; E, enhancers.
Identified modifiers with full name (alleles used for the crosses are given in parentheses). Those genes also found in the gain-of-function screen are underlined. Candidate genes that did not interact with GMR>H are listed in Table S2.
Phenotypic classification is according to Figure 2: S, suppressors; E, enhancers.
Interaction with apoptosis-inducing factors were observed (+) or were not observed (−) (for details, see text and Table 2).
Several genes encoding signaling pathway components were identified. Apart from N, various components of the EGFR pathway were found, some of which had not yet come up in other screens [leonardo (leo), sprouty], highlighting a close link between N and EGFR signaling in the regulation of apoptosis (Miller and Cagan 1998; Wech and Nagel 2005). Isolation of several members of the JNK pathway in the loss-of-function screen supports our conclusion from the gain-of-function screen that H might mediate cell death via this pathway. For example, mutations in the JNK phosphatase puckered (puc), which encodes a negative regulator of JNK activity, enhanced GMR>H, whereas mutations in hemipterous (hep), the JNKK, were responsible for suppression by Df(1)N12.
In the cases of Df(1)RA2, Df(1)KA14, Df(1)HA85, Df(2L)ast2, Df(2L)sc19-4, Df(2R)w45-30n, Df(2R)017, Df(2R)AA21, Df(3L)M21, Df(3L)rdgC-co2, Df(3R)by10, Df(3R)by62, Df(3R)GB104, and Df(4)G, we were unable to recover the mutations responsible for the observed interaction with H. Three of them uncovered genes with yet-uninvestigated function (CG8788, l(2)05510, and mura) (see Table S2 at http://www.genetics.org/supplemental/).
Cell death involvement of interacting factors:
To inquire into a role of the isolated modifiers in cell death processes, we subjected them to further analyses, either rescue by DIAP1 or influence on proapoptotic factors.
Rescue of eye phenotypes by DIAP1:
Twenty-one factors reduced the eye size on their own or caused lethality upon overexpression with GMR-GAL4 (Table 1 and Table S1 http://www.genetics.org/supplemental/), suggesting a more general proapoptotic role. Most of them were subjected to a rescue experiment by a concurrent overexpression of DIAP1. In the majority of cases (12 of 18 tested), phenotypic rescue was observed. This result was expected for bchs (Figure 4, J–L), Drac2, Eip78C (Figure 4, G–I), fat facets (faf), Fmr1, Krüppel-homolog-1(Kr-h), and rpr, which have been connected before in one way or another with cell death. To our knowledge, however, such a connection was not yet demonstrated for other factors, including escargot (esg), nejire (nej), Cyp6μ1, and the novel factors CG12462 and CG5986 (Figure 4, D–F).
Analysis of influence on proapoptotic gene activity:
All interactors of the loss-of-function screens as well as the suppressors of the gain-of-function screen were tested for their ability to influence apoptosis caused by different cell death inducers, namely p53, hid, rpr, and grim. The rationale behind this screen was to further distinguish factors that influence cell death from factors that might influence other H-mediated processes, e.g., differentiation. Any of these cell death inducers causes a remarkable size reduction, irregularities of ommatidia, and in the case of grim, loss of pigmentation as well when overexpressed under GMR-control (Figures 3 and 5). The outcome of this experiment is summarized in Table 2 and a selection of interactions is shown in Figures 3–5.
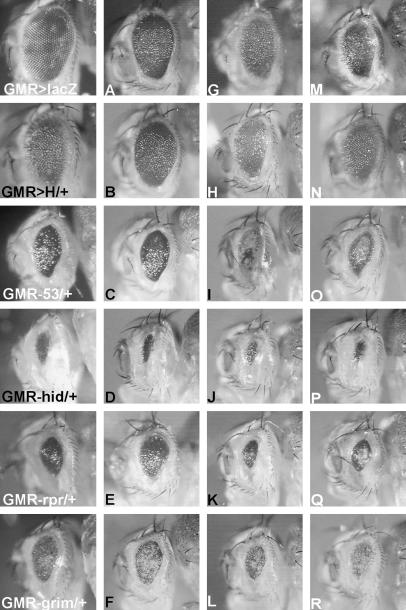
Figure 5.
Examples of modifiers identified in the deficiency screen. Far left panels show eyes for comparison. Df(2R)X1 was found as a suppressor of GMR>H (A) and uncovered leo07103 that suppresses not only GMR>H (B), but also proapoptotic activity of GMR-p53 (C), GMR-rpr (E), and GMR-grim (F), but not GMR-hid (D). Both Df(2R)Jp8 (G) and Df(3L)brm11 (M) enhanced GMR>H, and, among others, deleted Rho1 and Arflike at 72A, respectively. Mutant alleles had the same phenotypic effect (H and N). Moreover, both alleles enhanced apoptosis induced by p53, hid, rpr, and grim (I–L and O–R).
As expected, gain of the antiapoptotic factor th and loss of the proapoptotic factors rpr, hid, and grim had a general suppressive effect on all four cell death inducers and served as a positive control for our screen (Table 2). Interestingly, such a general influence was also observed for EGFR-pathway members aop, DERact, and pntP2, for JNK members puc and hep, as well as for Arf72A (Figure 5, O–R), Dsp1 (Figure 3, D–G), IMP (Figure 3, R–U), Rho1 (Figure 5, I–L), and the novel factors CG3600 (Figure 3, f–i), CG30465 (Figure 3, Y–b), CG32521, and CG32737, arguing for a rather direct role in the regulation of apoptosis.
However, this analysis revealed that not all factors that modified GMR>H likewise modified the effects of cell death inducers. In most cases, an effect was observed on GMR-p53, raising the possibility that H-mediated cell death feeds into this pathway (Table 2). For example, leo mutants rescued all but GMR-hid (Figure 5, C–F), whereas mutants of N-signaling components or overexpression of several different genes like wdb (Figure 3, K–N) had an influence on GMR-p53 and either GMR-grim or GMR-hid. Some mutants acted very specifically on GMR-p53, GMR-hid, or GMR-rpr (Table 2). Curiously, a mutation in defective proventriculus (dve) suppressed GMR-p53- and GMR>H-induced cell death alike, but enhanced apoptosis caused by GMR-hid and GMR-rpr. This differential influence on cell death inducers argues for rather specific roles for these factors in the regulation of apoptosis.
Altogether, 44 of the identified interactors of GMR>H modified the small rough eye phenotypes caused by different apoptotic stimuli in a similar way, thus strengthening our screening strategy for factors involved in the regulation of H-mediated apoptosis. It will be interesting to eludicate the molecular basis for these specific behaviors in the future.
DISCUSSION
Programmed cell death is used to remove damaged or supernumerary cells and serves as a substantial patterning mechanism during the development of complex animal structures. In Drosophila, apoptosis was shown to be required, e.g., for shaping of the nervous system, patterning of the pupal eye, metamorphosis, or proper development of germ cells. Crosstalk between different signaling pathways fuels differentiation and apoptosis alike. The N-signaling pathway is one example of a cell-cell communication pathway involved in a large number of cell fate decisions that is associated with apoptotic processes as well (Miller and Cagan 1998; Artavanis-Tsakonas et al. 1999; Wech and Nagel 2005).
In this study, we aimed at finding factors that modify apoptotic phenotypes resulting from overexpression of H in the eye. We performed a misexpression and a loss-of-function screen based on chromosomal deficiencies. This twofold approach allowed us to play the strengths of one off the weaknesses of the other. While a deficiency-based screen can quickly map loci interacting with H, it can be difficult to subsequently identify specific mutations that account for this interaction. In addition, as only a fraction of mutations results in visible phenotypes, modifiers may go unnoted especially in cases of gene duplication and redundancy. Therefore, a complementary overexpression screen may identify genes that are missed otherwise. In the past, gain-of-function genetics have been successful in identifying genes crucial to different developmental processes like oogenesis, tissue growth, sensory organ development, or thorax formation (Rørth et al. 1998; Abdelilah-Seyfried et al. 2000; Peña-Rangel et al. 2002; Tseng and Hariharan 2002).
The gain-of-function screen identified a total of 86 factors, including 57 enhancers and 29 suppressors. A potential drawback of this screen is the effects arising from the misexpression of these factors themselves: ∼40% of the enhancers and 60% of the suppressors displayed phenotypes on their own when overexpressed in the eye. However, >50% of them (44 of 86) showed the opposite effect on GMR>H when tested in the respective loss-of-function mutant background, arguing for a specific connection with H. Moreover, some of the genes identified in our screen may be involved in the expression of the glass gene itself. To check for this, we tested all identified suppressors for their ability to influence tissue loss and apoptosis caused by H during wing development (Figure 1J). Most of them (23 of 29) ameliorated the effects of H overexpression, arguing against an exclusive influence on the glass gene itself (data not shown).
In the loss-of-function screen we recovered 41 deficiencies and were able to subsequently map 36 different loci, 22 acting as suppressors and 14 as enhancers. Ten deficiencies were also recovered in the gain-of-function screen. One explanation might be that, altogether, the deficiencies uncovered just 75% of the genome, leaving a quarter uninspected. Moreover, the collection of EP lines that we used accounts for ∼10% of the genes in the entire genome (Rørth et al. 1998). These numbers illustrate the benefit of taking various genetic approaches and emphasize that no single screen will identify all or even most potential interactors.
Specification and subdivision of H interactors:
The N-signaling pathway regulates a plethora of developmental processes, including various differentiation steps and cell death during eye development. As H acts as a general antagonist of N, one might expect a variety of diverse factors to modify phenotypes caused by H overexpression. For this reason, the isolated modifiers were subjected to further analyses with regard to their own phenotypes and their general involvement in apoptosis.
EP enhancers primarily involved in N-dependent differentiation events:
A majority of the 57 enhancers (33 or 58%) caused no phenotype or even bigger eyes upon overexpression, indicating that they do not induce apoptosis on their own. Interestingly, 15 of them were also identified in screens conducted to find factors involved in thorax formation (Peña-Rangel et al. 2002), bristle development (Abdelilah-Seyfried et al. 2000), mesoderm development (Bidet et al. 2003), cell growth in the eye (Tseng and Hariharan 2002), or synapse formation (Kraut et al. 2001). As N signaling regulates various aspects in the development of these different tissues and organs, one might speculate that this group of enhancers affects N activity primarily during differentation processes. Although not identified in the aforementioned screens, the remaining 14 factors, belonging to functional categories as diverse as growth regulators, transcription factors, or protein kinases and enzymes, might be connected to the N-signaling pathway as well, thus reflecting the manyfold N-dependent processes in the development of Drosophila (Table 1 and Table S1 at http://www.genetics.org/supplemental/).
H interactors primarily involved in cell growth and proliferation:
A total of 24 factors showed no apparent effect in our cell death assays (Table 1; Tables S1 and S2 at http://www.genetics.org/supplemental/). These factors comprise several N pathway components (extramacrochaetae, scalloped, twin of m4), the novel factor CG8788, and also smr, which functions as a corepressor and also might mediate transcriptional repression of N target genes in Drosophila (Kao et al. 1998; Tsuda et al. 2002). However, most of the genes in this category have functions related to cell division and cell growth. For example, Dap overexpression reduces growth and proliferation in the eye-imaginal disc (Tseng and Hariharan 2002) and causes lethality upon combined misexpression with H. Consistent with the notion that levels of dMyc determine growth and cell proliferation, mutants in this gene were also lethal in trans with GMR>H. dMyc activity is regulated by several morphogens (Johnston et al. 1999; Moreno and Basler 2004) and our results suggest that N may also be involved for at least some aspects of dMyc regulation. Another group of genes, including bazooka, fat, if, or Rok, functions in cell adhesion and cell polarity. The incorrect establishment of epithelial polarity is accompanied by hyperplastic growth, which can be synergistically enlarged, followed by an ectopic N signal (Brumby and Richardson 2003). Our finding that mutations in any of these loci behave as suppressors of H overexpression raises the possibility of a rather direct connection to the N-signaling pathway. Thus, this screen uncovered several genes, which influence H and N activity during growth and proliferation, raising the question of their molecular role in the N-signaling pathway.
Regulators of cell death:
Exactly 50% of all our different modifiers (56 of 112) either were rescued by DIAP1 or influenced cell death inducers themselves. The recovery of factors known to be generally involved in apoptosis, such as rpr, th, and ban, or more specifically during eye development such as klu, not only was expected but also was demanded by our approach. Interestingly, two of these genes (Rac2 and Eip78C) were in a data set collected in the course of a genome-wide analysis of steroid-triggered cell death response in Drosophila (Lee et al. 2003). A further connection between N and the ecdysone regulatory network was recently established during metamorphosis of the midgut (Li and White 2003).
The regulatory input of EGFR signaling as well as crosstalk with Notch signaling in the control of cell death has been shown at different stages of Drosophila development, most notably in the eye (Wolff and Ready 1991; Bergmann et al. 1998; Kurada and White 1998; Miller and Cagan 1998; Wech and Nagel 2005). In agreement with these earlier findings, we identified several EGFR-pathway members as modifiers of H and cell death inducers alike (e.g., aop, DER, lilli, pnt, rho). More interestingly, we also identified several members of the JNK pathway (e.g., bsk, hep, msn, puc), which has been involved earlier in morphogenetic as well as stress-induced apoptosis (Stronach and Perrimon 1999; Adachi-Yamada and O'Connor 2002). Genetic analyses have demonstrated that JNK signaling is an effector of larval and pupal apoptosis (Adachi-Yamada et al. 1999; Moreno et al. 2002; Wech and Nagel 2005). During embryogenesis it was already described that N signaling has a negative effect on JNK signaling in the process of dorsal closure. However, this seems to involve noncanonical N signaling (Zecchini et al. 1999). In addition to this influence on patterning processes, our work points to an involvement of canonical N signaling in JNK-mediated morphological cell death. In this context it is interesting to note that our screen also identified a phosphatase subunit: overexpression of PP2A-β′ (B56) encoded by wdb strongly suppressed H-, p53-, and grim-induced cell death in the eye, whereas wdb mutants acted as an enhancer of H. In agreement, knockdown of B56 PP2A during embryogenesis resulted in caspase activation (Li et al. 2002; Van Hoof and Goris 2003). Genetically, it was placed in the p53-regulated path of apoptosis (Li et al. 2002 and our own observations). We find a strong correlation between H-induced cell death and p53-mediated apoptosis (see Table 2). For example, cell death induced by overexpression of p53 can be rescued by increasing N signals [N- or Su(H)-GOF or H-LOF; Table 2 and data not shown]. Further studies will determine the molecular mechanism underlying these genetic interactions.
Factors with novel functions in cell death:
Many of the identified interactors have been previously implicated in different aspects of development but not, at least to our knowledge, in apoptotic processes. One example is the IMP. IMP is ecdysone inducible and was suggested to be involved in the regulation of translation, maybe during metamorphosis (Garbe et al. 1993; Lasko 2000), arguing for a role of IMP in triggering cell death in this context.
Interestingly, we identified several genes involved in chromatin remodeling as strong interactors of H and the other tested cell death inducers. The Drosophila Brahma complex plays an important role during the G1 phase of the cell cycle (Papoulas et al. 1998; Brumby et al. 2002). In our hands, brm2 mutants behaved as an enhancer of H and the proapoptotic genes hid, rpr, and grim, but not of the stress-induced p53 apoptotic pathway (Table 2). This argues for an additional role of brm in the coordination of cell death, in addition to its well-defined function in the regulation of cell growth. Another example is DspI, which was identified in our screens as a general repressor of apoptosis. DspI encodes a transcriptional corepressor that binds to Dorsal and Relish (Rel) proteins (Lehming et al. 1998; Brickman et al. 1999). Like their mammalian counterparts, Rel proteins mediate the immune response via JNK signaling (Park et al. 2004). It is tempting to speculate that Rel proteins together with DspI might likewise protect against apoptosis by limiting the JNK signal. In this case, the effects of Dsp1 on H-mediated apoptosis can be easily explained and provide a further link for a crosstalk between JNK and Notch pathways. A third factor in this group is the Drosophila cAMP-response-element-binding protein, which is encoded by the nej locus and belongs to the CBP/p300 family (Goodman and Smolik 2000). nej is required at successive stages of eye development and overexpression caused severe retina degeneration (Ludlam et al. 2002; Kumar et al. 2004). As nej mutants have antiapoptotic effects on H and most cell death inducers alike, one might assume a “manager” function in the control of cellular homeostasis and apoptosis.
Finally, 7 of 15 interactors with a hitherto unknown function were shown to interfere with apoptosis. The future challenge will be to determine the molecular and functional relationship between these new genes and cell death induction by H.
Conclusions:
The screens provided us with a wealth of new information regarding cell death induction observed after overexpression of H. Our results are compatible with the notion that changes in N activity affect cell death as a response to abnormal or imbalanced developmental signals within a cell. In agreement, the identified modifiers include factors and signaling components like p53, JNK signaling, and hormone-triggered factors, all known to be involved in the coordination of a wide range of biological responses, including growth, differentiation, and programmed cell death. Apparently, N signaling is required for the correct interpretation of such developmental signals and for the crosstalk between different signaling pathways that is essential for cell survival and differentiation.
Acknowledgments
We thank the Bloomington Stock Center and Exelixis for providing mutant stocks and the EP collection and S. Clevers, S. Eaton, E. Kuranaga, and M. Muskavitch for fly stocks. We are indebted to T. Stoesser for his invaluable help in keeping the flies alive, V. Gesellchen for the initiation of the deficiency screen, and U. Walldorf for helpful comments on the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft grant NA 427/1-1 to A.C.N.
References
- Abdelilah-Seyfried, S., Y.-M. Chan, C. Zeng, N. J. Justice, S. Younger-Shepherd et al., 2000. A gain-of-function screen for genes that affect the development of the Drosophila adult external sensory organ. Genetics 155: 733–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi-Yamada, T., and M. O'Connor, 2002. Morphogenetic apoptosis: a mechanism for correcting discontinuities in morphogen gradients. Dev. Biol. 251: 74–90. [DOI] [PubMed] [Google Scholar]
- Adachi-Yamada, T., K. Fujimura-Kamada, Y. Nishida and K. Matsumoto, 1999. Distortion of proximodistal information causes JNK-dependent apoptosis in Drosophila wing. Nature 400: 166–169. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas, S., M. R. Rand and R. J. Lake, 1999. Notch signaling: cell fate control and signal integration in development. Science 284: 770–776. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., 1989. Drosophila: A Laboratory Handbook and Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Bangs, P., and K. White, 2000. Regulation and execution of apoptosis during Drosophila development. Dev. Dyn. 218: 68–79. [DOI] [PubMed] [Google Scholar]
- Barolo, S., T. Stone, A. G. Bang and J. W. Posakony, 2002. Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCTBP to Suppressor of Hairless. Genes Dev. 16: 1964–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann, A., J. Agapite, K. McCall and H. Steller, 1998. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell 95: 331–341. [DOI] [PubMed] [Google Scholar]
- Bidet, Y., T. Jagla, J.-P. Da Ponte, B. Dastugue and K. Jagla, 2003. Modifiers of muscle and heart cell fate identified by gain-of-function screen in Drosophila. Mech. Dev. 120: 991–1007. [DOI] [PubMed] [Google Scholar]
- Brickman, J. M., M. Adam and M. Ptashne, 1999. Interaction between an HMG-1 protein and members of the Rel family. Proc. Natl. Acad. Sci. USA 96: 10679–10683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, M. H., W. Nordstrom, G. Tsang, E. Kwan, G. M. Rubin et al., 2000. Drosophila p53 binds a damage response element at the reaper locus. Cell 101: 103–113. [DOI] [PubMed] [Google Scholar]
- Brumby, A. M., and H. E. Richardson, 2003. scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. EMBO J. 22: 5769–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumby, A. M., C. B. Zraly, J. A. Horsfield, J. Secombe, R. Saint et al., 2002. Drosophila cyclin E interacts with components of the Brahma complex. EMBO J. 21: 3377–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, J. L., Y. C. Tsai, S. J. Chiu and Y. H. Sun, 2004. Localized Notch signal acts through eyg and upd to promote global growth in Drosophila eye. Development 131: 3839–3847. [DOI] [PubMed] [Google Scholar]
- Chen, G., J. Fernandez, S. Mische and A. J. Courey, 1999. A functional interaction between the histone deacetylase rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 13: 2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P., H. Nordstrom, B. Gish and J. M. Abrams, 1996. grim, a novel cell death gene in Drosophila. Genes Dev. 10: 1773–1782. [DOI] [PubMed] [Google Scholar]
- de Celis, J. F., J. de Celis, P. Ligoxygakis, A. Preiss, C. Delidakis et al., 1996. Functional relationships between Notch, Su(H) and the bHLH genes of the E(spl) complex: the E(spl) genes mediate only a subset of Notch activities during imaginal development. Development 122: 2719–2728. [DOI] [PubMed] [Google Scholar]
- Frost, J. A., S. Xu, M. R. Hutchison, S. Marcus and M. H. Cobb, 1996. Actions of Rho family small G proteins and p21-activated protein kinases on mitogen-activated protein kinase family members. Mol. Cell. Biol. 16: 3707–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe, J. C., E. Yang and J. W. Fristrom, 1993. IMP-L2: an essential secreted immunoglobulin family member implicated in neural and ectodermal development in Drosophila. Development 119: 1237–1250. [DOI] [PubMed] [Google Scholar]
- Giraldez, A. J., and S. M. Cohen, 2003. Wingless and Notch signaling provide cell survival cues and control cell proliferation during wing development. Development 130: 6533–6543. [DOI] [PubMed] [Google Scholar]
- Go, M. J., D. S. Eastman and S. Artavanis-Tsakonas, 1998. Cell proliferation control by Notch signaling in Drosophila development. Development 125: 2031–2040. [DOI] [PubMed] [Google Scholar]
- Goodman, R. H., and S. Smolik, 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14: 1553–1577. [PubMed] [Google Scholar]
- Grether, M. E., J. M. Abrams, J. Agapite, K. White and H. Steller, 1995. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 9: 1694–1708. [DOI] [PubMed] [Google Scholar]
- Hannus, M., F. Feiguin, C. P. Heisenberg and S. Eaton, 2002. Planar cell polarization requires Widerborst, a B′ regulatory subunit of protein phosphatase 2A. Development 129: 3493–3503. [DOI] [PubMed] [Google Scholar]
- Hay, B. A., 2000. Understanding IAP function and regulation: a view from Drosophila. Cell Death Differ. 7: 1045–1056. [DOI] [PubMed] [Google Scholar]
- Hay, B. A., T. Wolff and G. M. Rubin, 1994. Expression of baculovirus p35 prevents cell death in Drosophila. Development 120: 2121–2129. [DOI] [PubMed] [Google Scholar]
- Hay, B. A., R. Maile and G. M. Rubin, 1997. P-element insertion-dependent gene activation in the Drosophila eye. Proc. Natl. Acad. Sci. USA 94: 5195–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner, M. O., 2000. The biochemistry of apoptosis. Nature 407: 685–687. [DOI] [PubMed] [Google Scholar]
- Jiang, C., E. Baehrcke and C. Thummel, 1997. Steroid regulated programmed cell death during Drosophila metamorphosis. Development 124: 4673–4683. [DOI] [PubMed] [Google Scholar]
- Jiang, C. A., A. F. J. Lamblin, H. Steller and C. Thummel, 2000. A steroid-triggered transcriptional hierarchy controls salivary gland cell death during Drosophila metamorphosis. Mol. Cell 5: 445–455. [DOI] [PubMed] [Google Scholar]
- Johnston, L. A., D. A. Prober, B. A. Edgar, R. N. Eisenman and P. Gallant, 1999. Drosophila myc regulates cellular growth during development. Cell 98: 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, H.-Y., P. Ordentlich, N. Koyano-Nakagawa, Z. Tang, M. Downes et al., 1998. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 12: 2269–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut, R., K. Menon and K. Zinn, 2001. A gain-of-function screen for genes controlling motor axon guidance and synaptogenesis in Drosophila. Curr. Biol. 11: 417–430. [DOI] [PubMed] [Google Scholar]
- Kumar, J. P., T. Jamal, A. Doetsch, F. R. Turner and J. B. Duffy, 2004. CREB binding protein functions during successive stages of eye development in Drosophila. Genetics 168: 877–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurada, P., and K. White, 1998. Ras promotes cell survival in Drosophila by downregulating hid expression. Cell 95: 319–329. [DOI] [PubMed] [Google Scholar]
- Lasko, P., 2000. The Drosophila melanogaster genome: translation factors and RNA binding proteins. J. Cell Biol. 150: F51–F56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit, T., W. J. Brook, M. Ng, M. Calleja, H. Sun et al., 1996. Two distinct mechanisms for long-range patterning by decapentaplegic in the Drosophila wing. Nature 381: 387–393. [DOI] [PubMed] [Google Scholar]
- Lee, C.-Y., E. A. Clough, P. Yellon, T. M. Teslovich, D. A. Stephan et al., 2003. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Curr. Biol. 13: 350–357. [DOI] [PubMed] [Google Scholar]
- Lehming, N., A. Le Saux, J. Schüller and M. Ptashne, 1998. Chromatin components as part of a putative transcriptional repressing complex. Proc. Natl. Acad. Sci. USA 95: 7322–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., A. Scuderi, A. Letsou and D. M. Virshup, 2002. B56-associated protein phosphatase 2A is required for survival and protects from apoptosis in Drosophila melanogaster. Mol. Cell. Biol. 22: 3674–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. R., and K. P. White, 2003. Tissue-specific gene expression and ecdysone-regulated genomic networks in Drosophila. Dev. Cell 5: 59–72. [DOI] [PubMed] [Google Scholar]
- Ludlam, W. H., M. H. Taylor, K. G. Tanner, J. M. Denu, R. H. Goodman et al., 2002. The acetyltransferase activity of CBP is required for wingless activation and H4 acetylation in Drosophila melanogaster. Mol. Cell. Biol. 22: 3832–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, D., A. C. Nagel, B. Johannes and A. Preiss, 1999. Subcellular localization of Hairless protein shows a major focus of activity within the nucleus. Mech. Dev. 89: 195–199. [DOI] [PubMed] [Google Scholar]
- Miller, D. T., and R. L. Cagan, 1998. Local induction of patterning and programmed cell death in the developing Drosophila retina. Development 125: 2327–2335. [DOI] [PubMed] [Google Scholar]
- Morel, V., V. Lecourtois, O. Massiani, D. Maier, A. Preiss et al., 2001. Transcriptional repression by Suppressor of Hairless involves binding of a Hairless-dCTBP complex in Drosophila. Curr. Biol. 11: 789–792. [DOI] [PubMed] [Google Scholar]
- Moreno, E., and K. Basler, 2004. dMYC transforms cells into super-competitors. Cell 117: 117–129. [DOI] [PubMed] [Google Scholar]
- Moreno, E., M. Yan and K. Basler, 2002. Evolution of TNF signaling mechanisms: JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Curr. Biol. 12: 1263–1268. [DOI] [PubMed] [Google Scholar]
- Mosrin-Huaman, C., L. Canaple, D. Locker and M. Decoville, 1998. DSP1 gene of Drosophila melanogaster encodes an HMG-domain protein that plays multiple roles in development. Dev. Genet. 23: 324–334. [DOI] [PubMed] [Google Scholar]
- Ollmann, M., L. M. Young, C. J. Di Como, F. Karim, M. Belvin et al., 2000. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell 101: 91–101. [DOI] [PubMed] [Google Scholar]
- Palaga, T., and B. Osborne, 2002. The 3 D's of apoptosis: death, degradation and DIAP's. Nat. Cell Biol. 4: E149–E151. [DOI] [PubMed] [Google Scholar]
- Papoulas, O., S. J. Moseley, C. M. Mccallum, M. Sarte, A. Shearn et al., 1998. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development 125: 3955–3966. [DOI] [PubMed] [Google Scholar]
- Park, J. M., H. Brady, M. G. Ruocco, H. Sun, D. Williams et al., 2004. Targeting of TAK1 by the NF-kappa B protein Relish regulates the JNK-mediated immune response in Drosophila. Genes Dev. 18: 584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazdera, T. M., P. Janardhan and J. S. Minden, 1998. Patterned epidermal cell death in wild type and segment polarity mutant Drosophila embryos. Development 125: 3427–3436. [DOI] [PubMed] [Google Scholar]
- Peña-Rangel, M. T., I. Rodriguez and J. R. Riesgo-Escovar, 2002. A misexpression study examining dorsal thorax formation in Drosophila melanogaster. Genetics 160: 1035–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow, S., T. A. Draizen and J. W. Truman, 1997. Genes that induce apoptosis: transcriptional regulation in identified, doomed neurons of the Drosophila CNS. Dev. Biol. 190: 206–213. [DOI] [PubMed] [Google Scholar]
- Rørth, P., 1996. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93: 12418–12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth, P., K. Szabo, A. Bailey, T. Laverty, J. Rehm et al., 1998. Systematic gain-of-function genetics in Drosophila. Development 125: 1049–1057. [DOI] [PubMed] [Google Scholar]
- Ryoo, H. D., T. Gorenc and H. Steller, 2004. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev. Cell 7: 491–501. [DOI] [PubMed] [Google Scholar]
- Schweisguth, F., 2004. Notch signaling activity. Curr. Biol. 14: R129–R138. [PubMed] [Google Scholar]
- Stronach, B. E., and N. Perrimon, 1999. Stress signaling in Drosophila. Oncogene 18: 6172–6182. [DOI] [PubMed] [Google Scholar]
- Thomas, B. J., 2005. Cell-cycle control during development: taking it up to Notch. Dev. Cell 8: 451–459. [DOI] [PubMed] [Google Scholar]
- Thompson, C. B., 1995. Apoptosis in the pathogenesis and treatment of disease. Science 267: 1456–1462. [DOI] [PubMed] [Google Scholar]
- Tseng, A.-S., and I. K. Hariharan, 2002. An overexpression screen in Drosophila for genes that restrict growth or cell-cycle progression in the developing eye. Genetics 162: 229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, L., R. Nagaraj, S. L. Zipursky and U. Banerjee, 2002. An EGFR/Ebi/Sno pathway promotes Delta expression by inactivating Su(H)/SMRTER repression during inductive Notch signaling. Cell 110: 625–637. [DOI] [PubMed] [Google Scholar]
- Van Hoof, C., and J. Goris, 2003. Phosphatases in apoptosis: to be or not to be, PP2A is in the heart of the question. Biochim. Biophys. Acta 1640: 97–104. [DOI] [PubMed] [Google Scholar]
- Vaux, D. L., and S. J. Korsmeyer, 1999. Cell death in development. Cell 96: 245–254. [DOI] [PubMed] [Google Scholar]
- Vernooy, S. Y., J. Copeland, N. Ghaboosi, E. E. Griffin, S. J. Yoo et al., 2000. Cell death regulation in Drosophila: conservation of mechanism and unique insights. J. Cell Biol. 150: F69–F76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voas, M. G., and I. Rebay, 2004. Signal integration during development: insights from the Drosophila eye. Dev. Dyn. 229: 162–175. [DOI] [PubMed] [Google Scholar]
- Watts, R. J., E. D. Hoopfer and L. Luo, 2003. Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron 38: 871–885. [DOI] [PubMed] [Google Scholar]
- Wech, I., and A. C. Nagel, 2005. Mutations in rugose promote cell type specific apoptosis in the Drosophila eye. Cell Death Differ. 12: 145–152. [DOI] [PubMed] [Google Scholar]
- White, K., E. Tagaoglu and H. Steller, 1996. Cell killing by the Drosophila gene reaper. Science 271: 805–807. [DOI] [PubMed] [Google Scholar]
- Wolff, T., and D. F. Ready, 1991. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development 113: 841–850. [DOI] [PubMed] [Google Scholar]
- Ye, Y., and M. E. Fortini, 1999. Apoptotic activities of the wild-type and Alzheimer's disease-related mutant presenilins in Drosophila melanogaster. J. Cell Biol. 146: 1351–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecchini, V., K. Brennan and A. Martinez-Arias, 1999. An activity of Notch regulates JNK signalling and affects dorsal closure in Drosophila. Curr. Biol. 9: 460–469. [DOI] [PubMed] [Google Scholar]