Abstract
Differential expression of mRNA among animal strains is one of the mechanisms for their diversity. cDNA microarray analysis of the prostates of BUF/Nac (BUF) and ACI/N (ACI) rats, which show different susceptibility to prostate cancers, found 195 differentially expressed genes. To identify loci that control differential expression of 13 genes with diverse expression levels, their expression levels were measured by quantitative RT-PCR in 89 backcross rats, and expression quantitative trait locus (eQTL) analysis was performed. Nine genes [Aldh1a1, Aldr1, Bmp6, Cdkn1a (p21), Cntn6, Ghr, Jund, Nupr1, and RT1-M3] were controlled by cis-acting loci. Cdkn1a, a cell cycle regulator and a candidate for a prostate cancer susceptibility gene, was mapped to its own locus and had polymorphisms, including a 119-bp insertion in the 5′ upstream region in BUF rats. Four genes (Kclr, Pbsn, Psat1, and Ptn) were controlled by trans-acting loci. Pbsn, a prostate-specific gene on chromosome X, was controlled by a QTL on chromosome 8. Depending upon which gene that we selected from the genes widely used for normalization (Actb, Gapd, or Ppia), different QTL were mapped for Kclr, Psat1, and Ptn. Normalization using Actb most appropriately explained the expression levels in a congenic strain for chromosome 3. eQTL analysis with precise measurement of expression levels and appropriate normalization was shown to be effective for mapping loci that control gene expression in vivo.
THE diversity in gene expression is one of the mechanisms for diversity among individuals, such as susceptibility to common diseases. A differential gene expression could be due to a cis-acting polymorphism at the gene itself (Baier et al. 2000; Kuramoto et al. 2001) or due to a polymorphism of a gene upstream. Therefore, analysis of determinants of differential gene expression is important not only for studying the architecture of transcriptional regulation in vivo, but also for identifying causative genes for various phenotypes. However, it is often difficult to determine whether a differential gene expression is a cause or a consequence, in other words, whether it is due to a cis-acting or a trans-acting polymorphism.
A new approach to this issue is to treat mRNA expression levels as quantitative traits and to map quantitative trait loci (QTL) that control the expression levels in vivo. Using cDNA microarray technology, several studies performed comprehensive analysis of mRNA expression and QTL controlling the expression in budding yeast (Brem et al. 2002), eucalyptus (Kirst et al. 2005), mice (Schadt et al. 2003), rats (Hubner et al. 2005), and humans (Cheung et al. 2003; Schadt et al. 2003; Monks et al. 2004; Morley et al. 2004). The studies indicated that such an approach, expression QTL (eQTL) analysis, is effective for identifying the cause of the expression differences in vivo and can be applied to various genes. For generalized use of eQTL analysis, however, validation regarding whether a causative polymorphism is present in a mapped QTL and whether eQTL analysis is effective for genes with even low expression levels is necessary. The genes with low expression are not amenable to analysis by microarray. Expression analysis using only cDNA microarrays can mistake a sequence polymorphism in a probe sequence as an expression difference (Yamashita et al. 2003).
Here, we identified genes differentially expressed in the prostates of BUF/Nac (BUF) and ACI/N (ACI) rats that show different susceptibilities to prostate cancers (Isaacs 1984; Inaguma et al. 2003). Loci responsible for different prostate cancer susceptibility in rats were recently mapped (Yamashita et al. 2005), and if differentially expressed genes or their controlling genes are present in the mapped loci, they are considered to be good candidates. eQTL analysis was performed for 13 genes with diverse expression levels, and a putative causative polymorphism was identified for one of the cis-controlled genes, Cdkn1a. Effects of genes used for normalization (referred to as “control genes” in other studies) were analyzed at the same time using congenic rats.
MATERIALS AND METHODS
Animals, prostate RNA, and genomic DNA samples:
BUF/NacJcl (BUF), ACI/NJcl (ACI), and F344/Jcl (F344) rats were purchased from CLEA Japan (Tokyo). BN/Crj (BN) rats were purchased from Charles River Japan (Yokohama, Japan). F1 rats were produced by mating female ACI rats with male BUF rats, and backcross rats were produced by mating female ACI rats with male F1 rats. Two congenic strains, BUF.ACI-Gcr2 and BUF.ACI-Gcr3, which had homozygous ACI chromosome 3 (D3Rat56–D3Rat83) and chromosome 16 (D16Rat31–D16Arb1), respectively, in the BUF background, were developed by the speed congenic method (Serreze et al. 1996). Specifically, in each generation of backcrossing, the male rat that had the most substituted loci after analysis of 171 loci was used to produce 30–100 progeny rats. At N5F12 and at N6F8, respectively, complete substitution of the ACI background by the BUF background was confirmed using the 171 genome-wide genetic markers. The congenic strains are deposited in The National Bio Resource Project for the Rat in Japan (http://www.anim.med.kyoto-u.ac.jp/NBR/home.htm).
Total RNA was extracted from the entire prostate, including the ventral and lateral lobes, of male rats at 10 weeks of age using ISOGEN (NIPPON GENE, Tokyo). All the rats had been given 83 mg/liter N-methyl-N′-nitro-N-nitrosoguanidine (MNNG, Aldrich Chemical, Milwaukee) in drinking water for 2 weeks. RNA was purified using an RNeasy mini kit (QIAGEN, Valencia, CA), and the quality was examined by the ratio of S28 and S18 after running in a 1% agarose gel containing formalin. Genomic DNA was extracted from the tails of rats by an automated DNA extractor, GENEXTRACTOR TA-100 (Takara Shuzo, Kyoto, Japan).
Oligonucleotide microarray analysis:
Equal amounts of RNA were pooled from three BUF and three ACI rats. cDNA microarray analysis was performed using GeneChip Rat Genome U34A (Affymetrix, Santa Clara, CA) as in our previous studies (Kuramoto et al. 2002; Abe et al. 2003; Yamashita et al. 2003). The signal intensities were normalized so that the average of all the genes on a GeneChip would be 500, and the data were processed using Affymetrix Microarray Suite version 5.0. Differentially expressed genes were selected by their 2-fold increase or a 0.5-fold decrease.
Quantitative RT-PCR:
cDNA was synthesized from 2 μg of total RNA using SuperScript II reverse transcriptase (Invitrogen, Groningen, The Netherlands) and oligo(dT)12-18 primer (Invitrogen). Real-time PCR was performed using the iCycler iQ detection system (Bio-Rad Laboratories, Hercules, CA) with SYBR green PCR core reagents (Applied Biosystems, Foster City, CA). Samples of 89 backcross rats were simultaneously analyzed in a 96-well plate. The primers used are listed in supplementary Table S1 at http://www.genetics.org/supplemental/. By monitoring amplification curves of a test sample and samples that contained 101–106 molecules of the gene of interest, the number of target molecules in the test sample was analyzed. The number was normalized to that of Actb, Gapd, and Ppia, which are widely used as internal controls (Weisinger et al. 1999; Feroze-Merzoug et al. 2002; Lee et al. 2002).
Genotyping:
A total of 146 microsatellite markers that distributed all the autosomes and spanned 1512 cM were used (supplementary Table S2 at http://www.genetics.org/supplemental/). The mean and median of intermarker distances were 12.0 and 12.3 cM, respectively. PCR was carried out using 20 ng of genomic DNA, and the products were electrophoresed in a 3 or 4% NuSieve GTG agarose gel.
The genotype of the Cdkn1a 5′ upstream region among rat strains was determined using an upper primer (5′-GCGCTGTTATTAGACATGA-3′) and a lower primer (5′-AGAGCCACGCACATCTATG-3′) that amplified −501 to −264 (transcription start site, 0). The genotype of Cdkn1a 3′ downstream was determined using an upper primer (5′-ATGTAGAACCATTATTTAAGTCC-3′) and a lower primer (5′-GCGAGATGCGAGATGCAGATG-3′) (6292–6415).
Linkage analysis:
A linkage map was constructed by MAPMAKER/EXP (version 3.0b) software (Lander et al. 1987) that was modified by the Fink project (http://fink.sourceforge.net/) and installed on Mac OSX. Interval mapping of QTL was performed using MAPMAKER/QTL (version 1.1b) software that was similarly installed. To obtain improved normality, expression levels measured by quantitative RT-PCR were logarithm transformed and treated as quantitative traits. Logarithm of odds (LOD) score values of 1.9 and 3.3 were considered as thresholds for “suggestive” and “significant” linkage, respectively (Lander and Kruglyak 1995). With these values, linkage was expected to occur 1 and 0.05 times, respectively, at random in a genome scan.
Sequencing analysis:
Genomic DNAs of ACI and BUF rats were amplified by PCR using primers listed in supplementary Table S1 at http://www.genetics.org/supplemental/ and inserted into pGEM-T Easy Vector (Promega, Madison, WI). Cycle sequencing was performed using a DYEnamic ET terminator cycle sequencing kit (Amersham Biosciences, Piscataway, NJ) and an ABI310 DNA sequencer (Applied Biosystems).
Luciferase reporter assay:
5′ regions of ACI (without insertion) and BUF Cdkn1a (with insertion) were amplified using an upper primer (5′-GCTGTCAAAAGAAGCTTGAACTCC-3′) and a lower primer (5′-ACACACGAAAGCTTGTGGGGACAC-3′). The PCR fragments were digested with HindIII and cloned in the pGL3-Basic vector (Promega). Along with a control for transfection efficiency (33 ng phRL-TK, Promega), 330 ng of pGL3-Basic, pGL3-Control (SV40 promoter and enhancer), pGL3-Cdkn1a-ACI, and pGL3-Cdkn1a-BUF was transiently transfected to rat fibroblast cell line 3Y1 cl-3 (Ushijima et al. 1994) and rat prostate cancer cell line AT6.1 (Dong et al. 1995). The cells were seeded at 5 × 104 cells per well (12-well plate) 24 hr ahead, and transfection was performed using 1 μl of FuGENE 6 transfection reagent (Roshe Diagnostics, Hague Road, IN). At 24 hr after transfection, cells were harvested, and luciferase activity was measured with a dual-luciferase reporter assay system (Promega) in Lumat LB 9507 (Berthold Technologies, Bad Wildbad, Germany). Each transfection was performed in triplicate and experiments were repeated twice.
RESULTS
Genes differentially expressed between BUF and ACI prostates:
We first analyzed expression levels of 8800 probe sets on GeneChip rat genome U34A in the prostate of BUF and ACI rats. A total of 153 probes (134 genes) showed higher expression levels at twofold or more in the BUF prostates, and 72 probes (61 genes) showed higher expression levels in the ACI prostates (supplementary Table S3 at http://www.genetics.org/supplemental/). From these 195 genes, we selected 12 genes with high and low expression levels (Aldh1a1, Aldr1, Bmp6, Cntn6, Ghr, Jund, Nupr1, Pbsn, Psat1, Ptn, RT1-M3, and Sv2b), including Pbsn. We also added Cdkn1a (p21) and Kclr, which we had previously identified as differentially expressed, although these were classified as “no change” by GeneChip analysis due to their low expression levels. The expression levels of the 14 genes in the prostate were not altered by MNNG administration (data not shown).
By quantitative RT-PCR analysis, differential expression at 1.5-fold or more was confirmed for 13 of the 14 genes (Sv2b was excluded) (Table 1). Since sequence polymorphisms in PCR products could produce apparent expression differences, we further confirmed the absence of sequence polymorphisms. For all the 13 genes, no differences in the melting temperatures of the PCR products were observed between BUF and ACI rats. Further, for Cdkn1a and Jund, quantitative RT-PCR was performed using different regions of the genes, and results were in good accordance (data not shown).
TABLE 1.
GeneChip and real-time RT-PCR analyses in the prostate of rats
| GeneChip result
|
Real-time result
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BUF vs. ACI: | Signal
|
Copy number/β-actin (×104, mean ±SD)
|
||||||||
| Probe | Signal log ratio change | BUF | ACI | BUF (n = 3) | ACI (n = 3) | Backcross (n = 89) | Gene symbol | Chromosomal location | UniGene ID | Gene title |
| AF001898_at | 3.5 I | 5520 | 470 | 885.4 ± 235.5 | 97.0 ± 11.2 | 120.7 ± 79.4 | Aldh1a1 | Chr:1q51 | Rn.6132 | Aldehyde dehydrogenase family 1, member A1 |
| M60322_g_at | 2.4 I | 144 | 21 | 32.4 ± 10.4 | 10.2 ± 4.1 | 17.4 ± 6.0 | Aldr1 | Chr:4q22 | Rn.107801 | Aldehyde reductase 1 (low Km aldose reductase) |
| U66298_at | 6.8 I | 996 | 5 | 448.4 ± 216.8 | 2.7 ± 0.1 | 102.7 ± 109.6 | Bmp6 | Chr:17p12 | Rn.40476 | Bone morphogenetic protein 6 |
| X58830_at | 4.1 I | 2856 | 140 | |||||||
| L41275cds_s_at | 1.5 NC | 50 | 18 | 17.9 ± 2.1 | 5.8 ± 2.3 | 8.9 ± 9.6 | Cdkn1a | Chr:20p12 | Rn.10089 | Cyclin-dependent kinase inhibitor 1A (p21) |
| D87248_at | 2.9 I | 515 | 65 | 177.4 ± 44.9 | 35.2 ± 10.0 | 47.9 ± 26.6 | Cntn6 | Chr:4q41 | Rn.10644 | Contactin 6 |
| Z83757mRNA_at | 3.9 I | 73 | 4 | 25.5 ± 11.8 | 5.2 ± 1.8 | 8.9 ± 4.1 | Ghr | Chr:2q16 | Rn.2178 | Growth hormone receptor |
| Z83757mRNA_g_at | 4.1 I | 58 | 2 | |||||||
| D26307cds_at | −3.5 D | 23 | 296 | 6.0 ± 3.1 | 39.1 ± 5.3 | 23.6 ± 13.0 | Jund | Chr:16p14 | Rn.46225 | Jun D proto-oncogene |
| M55532_at | 4.1 NC | 66 | 3 | 0.94 ± 0.58 | 0.30 ± 0.05 | 0.61 ± 0.25 | Kclr | Chr:4q34 | Rn.9886 | Kupffer cell receptor |
| AF014503_at | 1.9 I | 3061 | 903 | 2280 ± 1394 | 442 ± 123 | 584 ± 303 | Nupr1 | Chr:1q36 | Rn.11182 | Nuclear protein 1 (p8) |
| M27156_at | 2.2 I | 16461 | 2990 | 19580 ± 10480 | 1310 ± 580 | 5440 ± 6140 | Pbsn | Chr:Xq22 | Rn.9862 | Probasin |
| M27156_g_at | 2.5 I | 10249 | 1563 | |||||||
| rc_AI102868_g_at | 2.1 I | 969 | 156 | 227.7 ± 94.1 | 28.9 ± 5.1 | 74.8 ± 42.2 | Psat1 | Chr:1q43 | Rn.100813 | Phosphoserine aminotransferase 1 |
| rc_AI230228_at | 1.8 I | 474 | 130 | |||||||
| rc_AI102795_at | 1.8 I | 324 | 134 | 391.5 ± 220.1 | 95.7 ± 11.0 | 141.1 ± 55.7 | Ptn | Chr:4q22 | Rn.1653 | Pleiotrophin |
| U16025_g_at | 2.4 I | 50 | 12 | 35.9 ± 20.8 | 20.8 ± 2.8 | 23.5 ± 14.0 | RT1-M3 | Chr:20p12 | Rn.92606 | RT1 class Ib, locus M3 |
| L10362_at | 2.9 I | 47 | 7 | 0.49 ± 0.21 | 0.50 ± 0.22 | Sv2b | Chr:1q31 | Rn.58137 | Synaptic vesicle glycoprotein 2 b | |
eQTL analyses:
Expression levels of the 13 genes were measured by quantitative RT-PCR in 89 ACI × (ACI × BUF)F1 backcross rats and normalized to that of Actb (β-actin). By linkage analysis, expression levels of 9 genes (Aldh1a1, Aldr1, Bmp6, Cdkn1a (p21), Cntn6, Ghr, Jund, Nupr1, RT1-M3) were mapped to their own loci with relatively high LOD scores (4.0–78.0) (Table 2). In contrast, expression levels of 4 genes (Kclr, Pbsn, Psat1, Ptn) were mapped to loci different from their own with LOD scores between 2.2 and 5.0 (Table 2). The expression level of even Cdkn1a, which had very low expression, was mapped to its own locus (chromosome 20) with a LOD score of 37.5. The expression level of Pbsn, a prostate-specific gene on chromosome X with abundant expression, was mapped to a locus on chromosome 8 with a LOD score of 4.0. By quantitative RT-PCR using different aliquots of the same samples, reproducibility of mapping was confirmed for Jund, which had very low expression, and Pbsn.
TABLE 2.
LOD score of linkage analyses using gene expression as QTL (LOD >1.9)
| Gene/ normalization | Peak 1 chromosome (cM) | Closest markers | LOD score | Variance explained (%) | Peak 2 chromosome (cM) | Closest markers | LOD score | Variance explained (%) | Peak 3 chromosome (cM) | Closest markers | LOD score | Variance explained (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aldh1a1 (Chr:1q51) | ||||||||||||
| Actb | 1 (115) | D1Rat70 | 23.2 | 77.7 | 3 (76) | D3Rat10 | 2.6 | 14.1 | 19 (59) | D19Rat4 | 2.1 | 10.5 |
| Gapd | 1 (117) | D1Rat298 | 22.4 | 75.8 | 3 (80) | D3Rat10 | 2.9 | 15.3 | 19 (57) | D19Rat4 | 2.0 | 10.9 |
| Ppia | 1 (117) | D1Rat298 | 24.1 | 76.0 | 3 (85) | D3Rat10 | 2.7 | 15.6 | 19 (59) | D19Rat4 | 2.4 | 11.8 |
| Aldr1 (Chr:4q22) | ||||||||||||
| Actb | 4 (25) | D4Rat15 | 16.5 | 60.6 | ||||||||
| Gapd | 4 (27) | D4Rat30 | 16.5 | 60.4 | ||||||||
| Ppia | 4 (31) | D4Rat30 | 9.8 | 40.1 | ||||||||
| Bmp6 (Chr:17p12) | ||||||||||||
| Actb | 17 (6) | D17Rat15 | 65.9 | 97.1 | ||||||||
| Gapd | 17 (6) | D17Rat15 | 69.9 | 97.6 | ||||||||
| Ppia | 17 (6) | D17Rat15 | 78.0 | 98.4 | ||||||||
| Cdkn1a (Chr:20p12) | ||||||||||||
| Actb | 20 (10) | D20Rat3 | 37.5 | 87.6 | ||||||||
| Gapd | 20 (10) | D20Rat3 | 39.4 | 88.8 | ||||||||
| Ppia | 20 (10) | D20Rat3 | 42.2 | 90.3 | 10 (6) | D10Rat44 | 2.1 | 11.0 | ||||
| Cntn6 (Chr:4q41) | ||||||||||||
| Actb | 4 (62) | Ampp | 6.5 | 31.3 | 3 (110) | D3Rat3 | 2.4 | 12.0 | 12 (0) | D12Rat58 | 2.0 | 9.7 |
| Gapd | 4 (62) | Ampp | 7.2 | 34.4 | 3 (110) | D3Rat3 | 3.2 | 15.5 | ||||
| Ppia | 4 (60) | Ampp | 7.6 | 33.6 | 3 (107) | D3Rat3 | 4.6 | 24.6 | ||||
| Ghr (Chr:2q16) | ||||||||||||
| Actb | 2 (22) | D2Rat75 | 4.4 | 26.3 | ||||||||
| Gapd | 2 (20) | D2Rat75 | 6.9 | 39.2 | ||||||||
| Ppia | 2 (18) | D2Rat298 | 6.2 | 33.2 | ||||||||
| Jund (Chr:16p14) | ||||||||||||
| Actb | 16 (9) | D16Rat18 | 6.0 | 29.3 | ||||||||
| Gapd | 16 (15) | D16Rat17 | 6.6 | 31.6 | ||||||||
| Ppia | 16 (15) | D16Rat17 | 5.4 | 27.5 | ||||||||
| (trial2) | ||||||||||||
| Actb | 16 (18) | D16Rat17 | 4.2 | 20.4 | ||||||||
| Gapd | 16 (18) | D16Rat17 | 5.1 | 24.7 | 12 (4) | D12Rat58 | 2.2 | 12.2 | ||||
| Ppia | 16 (15) | D16Rat17 | 3.6 | 19.1 | ||||||||
| Kclr (Chr:4q34) | ||||||||||||
| Actb | 3 (27) | D3Rat98 | 3.8 | 17.8 | 8 (20) | D8Rat47 | 2.3 | 12.3 | 18 (18) | Adrb | 2.3 | 11.2 |
| Gapd | 8 (19) | D8Rat49 | 2.8 | 13.7 | 3 (29) | D3Rat98 | 2.8 | 14.7 | 5 (110) | D5Rat50 | 2.2 | 11.7 |
| Ppia | 1 (33) | D1Rat10 | 2.3 | 12.7 | 8 (18) | D8Rat49 | 2.2 | 13.3 | 18 (16) | Adrb | 2.1 | 12.6 |
| Nupr1 (Chr:1q36) | ||||||||||||
| Actb | 1 (104) | D1Rat57 | 7.8 | 38.5 | 16 (42) | D16Rat15 | 2.1 | 10.6 | 3 (27) | D3Rat98 | 2.1 | 10.3 |
| Gapd | 1 (104) | D1Rat57 | 9.0 | 42.8 | 2 (100) | D2Rat65 | 2.0 | 11.4 | 5 (110) | D5Rat50 | 1.9 | 9.6 |
| Ppia | 1 (104) | D1Rat57 | 10.4 | 47.2 | 16 (42) | D16Rat15 | 2.4 | 11.9 | 2 (92) | D2Rat65 | 2.1 | 12.9 |
| Pbsn (Chr:Xq22) | ||||||||||||
| Actb | 8 (42) | D8Rat44 | 4.0 | 21.0 | 11 (38) | D11Mit8 | 2.8 | 14.2 | ||||
| Gapd | 8 (44) | D8Rat44 | 4.7 | 24.7 | 11 (38) | D11Mit8 | 2.4 | 11.9 | ||||
| Ppia | 8 (42) | D8Rat44 | 5.0 | 26.2 | 11 (38) | D11Mit8 | 2.3 | 11.5 | ||||
| (trial2) | ||||||||||||
| Actb | 8 (42) | D8Rat44 | 4.5 | 25.6 | 11 (25) | D11Rat63 | 2.9 | 21.4 | 15 (0) | D15Rat69 | 2.1 | 11.0 |
| Gapd | 8 (44) | D8Rat44 | 5.2 | 29.3 | 11 (38) | D11Mit8 | 2.2 | 12.1 | 15 (0) | D15Rat69 | 2.1 | 11.0 |
| Ppia | 8 (44) | D8Rat44 | 5.1 | 28.9 | 14 (41) | D14Rat18 | 2.2 | 11.7 | 11 (38) | D11Mit8 | 2.1 | 11.7 |
| Psat1 (Chr:1q43) | ||||||||||||
| Actb | 3 (27) | D3Rat98 | 3.1 | 15.1 | 2 (52) | D2Rat183 | 2.5 | 14.4 | 5 (110) | D5Mgh9 | 2.0 | 9.9 |
| Gapd | 5 (110) | D5Mgh9 | 2.8 | 13.6 | 2 (52) | D2Rat183 | 2.7 | 15.2 | 3 (27) | D3Rat98 | 2.3 | 11.5 |
| Ppia | 12 (30) | Planh | 2.2 | 12.0 | 1 (115) | D1Rat70 | 2.1 | 11.6 | 2 (56) | D2Rat183 | 2.1 | 13.6 |
| Ptn (Chr:4q22) | ||||||||||||
| Actb | 3 (43) | D3Rat40 | 3.1 | 16.8 | 8 (77) | D8Rat10 | 2.0 | 11.4 | ||||
| Gapd | 3 (45) | D3Rat40 | 2.3 | 13.1 | 10 (49) | D10Rat195 | 2.2 | 11.3 | 8 (81) | D8Rat10 | 1.9 | 10.5 |
| Ppia | 20 (43) | D20Rat29 | 2.6 | 12.5 | ||||||||
| RT1-M3 (Chr:20p12) | ||||||||||||
| Actb | 20 (16) | D20Rat5 | 2.3 | 11.3 | ||||||||
| Gapd | 20 (16) | D20Rat5 | 2.3 | 11.1 | ||||||||
| Ppia | 20 (16) | D20Rat5 | 2.9 | 13.9 |
Numbers in parentheses are the distances in centimorgans from the first marker used on the chromosome. Underlining indicates a significant result (LOD score >4.3)
Identification of Cdkn1a polymorphisms and their effects in mRNA expression:
Cdkn1a (p21) was expressed at a higher level in BUF rats than in ACI rats, and classified as a cis-controlled gene. To identify a causative polymorphism for the differential expression, we sequenced the 5′ upstream region, exon 1, exon 2, intron 2, exon 3, and the 3′ downstream region (−3374–181 and 5378–6400, transcription start site, 1). A 119-bp insertion in the 5′ upstream region (at −477) in BUF rats (accession no. AB194279) and a 14-bp repeat number difference (ACI = 2, BUF = 3) in the 3′ downstream region (6345, accession no. AB218281∼3) were found. Six other polymorphisms were found in the 5′ upstream region and in intron 2 (supplementary Table S4 at http://www.genetics.org/supplemental/).
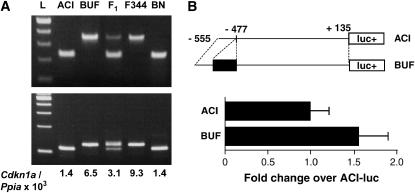
The 119-bp insertion in the 5′ upstream region consisted of a 15-bp duplication, a 85-bp rat identifier (ID) sequence (one of short interspersed nuclear elements in the rat) (Daniels and Deininger 1985), and a 19-bp poly(A) in an antisense direction with Cdkn1a. A putative p53-binding site was present in the 15-bp duplication. Among five rat strains [BUF, ACI, F344, BN, and (ACI × BUF)F1], the presence of these polymorphisms and high Cdkn1a expression in the prostate were associated (Figure 1A). BUF and F344, which showed higher Cdkn1a expressions among five strains of rats, had the same sequences.
Figure 1.
(A) Genotypes of Cdkn1a among five rat strains and correlation between genotypes and expression levels. (Top) The genotype 5′ upstream of the Cdkn1a gene (amplify −501 to −264, transcription start site, 0). The presence of insertions consisting of rat ID sequence were detected. (Bottom) The genotype 3′ downstream of the Cdkn1a gene (amplify 6292–6415). The PCR products were run in a 3.0% agarose gel. F1, (ACI × BUF)F1; L, 100-bp DNA ladder. Expression levels of the Cdkn1a gene were analyzed by quantitative RT-PCR. Concordance between the genotypes and expression level was observed. (B) Effect of the insertion 5′ upstream of the Cdkn1a gene for the regulation of gene expression in rat 3Y1 cl-3 evaluated by luciferase reporter assay. DNA fragments of the rat Cdkn1a 5′ region in ACI (without insertion) and in BUF (with insertion) were cloned in the pGL3-basic vector. The values of promoter activity were calculated on the basis of the activity observed upon cotransfection with the phRL-TK vector and expressed as the ratio to the promoter activity of ACI. Bars represent means +SD.
The effect of this insertion was further analyzed by a luciferase reporter assay using DNA fragments between −555 and +135 with and without the insertion. In the rat fibroblast cell line 3Y1 cl-3, the DNA fragment without the insertion showed a 35-fold increase of luciferase activity compared with the control vector without the DNA fragment, demonstrating its promoter activity. The 119-bp insertion resulted in a significant increase (1.6-fold) of the transcriptional activity (Figure 1B). A similar result (1.5-fold increase) was obtained using a rat prostate cancer cell line, AT6.1.
Effect of a gene used for normalization and usefulness of Actb:
Expression levels of each gene were normalized using three genes, Actb, Gapd, and Ppia, to correct variations in RNA quality among individual rats. Although all three genes are widely used for normalization by many researchers, different loci were mapped for Kclr, Psat1, and Ptn, depending upon the gene used for normalization (Table 2). For example, the main QTL that controlled the expression of Kclr was mapped on chromosome 3 by normalization with Actb (LOD score: 3.8) and on chromosome 8 with either Gapd (2.8) or Ppia (2.5). The main QTL that controlled expression of Psat1 was mapped on chromosome 3 with Actb (3.1), on chromosome 5 with Gapd (2.8), and on chromosome 12 near Planh with Ppia (2.2). Expression levels of Actb, Gapd, and Ppia were quantified twice. Each gene, whose expression level was quantified once, was normalized using the two values, and eQTL analysis was performed (supplementary Table S5 at http://www.genetics.org/supplemental/). Between the two quantifications, QTL mapped with LOD scores >3.0 were always reproducible, while those mapped with LOD scores <3.0 were occasionally not.
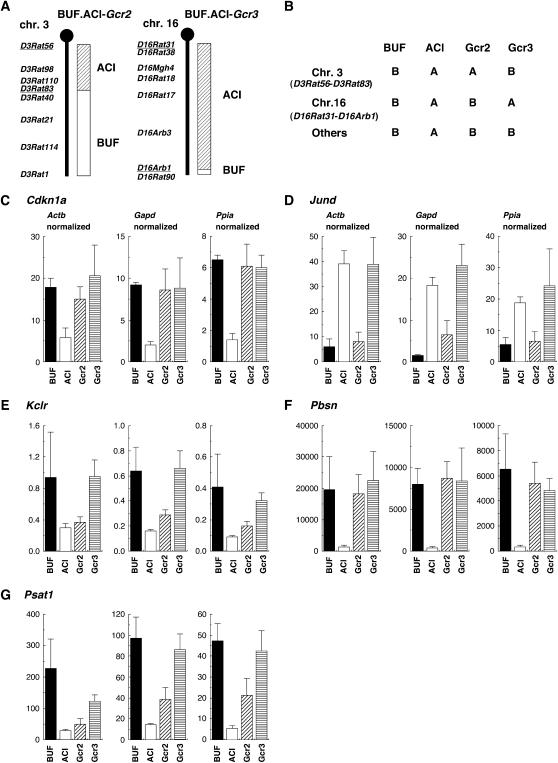
To clarify an appropriate gene for normalization among rat strains, two congenic rat strains were used. BUF.ACI-Gcr2 (Gcr2) had an ACI-derived genome around D3Rat98 in the BUF background, and BUF.ACI-Gcr3 (Gcr3) had an ACI-derived genome around D16Rat17 and D16Rat18 in the BUF background (Figure 2, A and B). Expression levels of five genes (Cdkn1a, Jund, Kclr, Pbsn, and Psat1) were analyzed in BUF, ACI, Gcr2, and Gcr3. If no controlling genes were on chromosome 3 and chromosome 16, Gcr2 and Gcr3 were expected to show expression levels equivalent to BUF rats. This was observed for Cdkn1a and Pbsn (Figure 2, C and F). In the case of Jund, whose QTL was mapped on chromosome 16, Gcr3 showed an expression level equivalent to that in ACI rats (Figure 2D). This confirmed that a locus on chromosome 16 controlled the expression of Jund. In the case of Kclr and Psat1, for which different controlling QTL were mapped, depending upon the gene used for normalization, Gcr2 showed expression levels close to those in ACI rats using any of the three genes for normalization (Figure 2, E and G). This showed that loci on chromosome 3 controlled the expression levels of Kclr and Psat1 and that the effect was most clearly detected using Actb for normalization (Table 2).
Figure 2.
(A) Genetic map of chromosome 3 of BUF.ACI-Gcr2 (Gcr2) and chromosome 16 of BUF.ACI-Gcr3 (Gcr3). Gcr2 and Gcr3 had homozygous ACI chromosome 3 (D3Rat56–D3Rat83) and chromosome 16 (D16Rat31–D16Arb1), respectively, in the BUF background. (B) Summary of genetic backgrounds of BUF, ACI, Gcr2, and Gcr3 in various chromosomes. (C–G) mRNA expression levels of Cdkn1a, Jund, Kclr, Pbsn, and Psat1 in BUF, ACI, Gcr2, and Gcr3. For each strain, three rats were analyzed, and the number of molecules was normalized to each of Actb, Gapd, and Ppia (means +SD; Actb normalized, ×104; Gapd and Ppia normalized, ×103).
DISCUSSION
Among the 195 genes differentially expressed in the prostates of ACI and BUF rats, eQTL analyses were performed for 13 genes selected for their wide range of expression levels and possible involvement in prostate carcinogenesis. Nine genes were cis-controlled, and 4 genes were trans-controlled.
Among the cis-controlled genes, a tumor-suppressor gene, Cdkn1a, was expressed 3.1-fold higher in BUF rats. We identified eight polymorphisms, including a 119-bp insertion in the 5′ upstream region, but not in the p53-binding tetramer (el-Deiry et al. 1995) or in a repressor in its 3′ untranslated region (Rishi et al. 1997). The 119-bp insertion contained the rat ID, which can act as a cis-acting positive regulator or enhancer (McKinnon et al. 1986; Osbourn et al. 1995), and a putative p53-binding sequence. However, our transient reporter gene assays demonstrated that the insertion upregulates the Cdkn1a expression only at 1.5- to 1.6-fold. The luciferase activities of the Cdkn1a promoter of both ACI and BUF rats exceeded even that of pGL3-Control, which has a strong promoter activity of SV40 promoter and enhancer. The Cdkn1a promoter activity is known to be induced by cellular stress (Park et al. 2002), which is caused by transfection itself, and the difference between ACI and BUF rats could have been attenuated. That a polymorphism(s) outside the regions that we sequenced is also responsible for the differential expression still remains a possibility.
ACI is susceptible to prostate carcinogenesis, and Wister, the original strain of BUF, is resistant (Isaacs 1984; Inaguma et al. 2003). By linkage mapping with prostate cancers, we have recently mapped Pcr1 (chromosome 2), Pcr2 (chromosome 1), Pcs1 (chromosome 19), and Pcs2 (chromosome 20) (Yamashita et al. 2005). Among the 195 differentially expressed genes, 7 were located on these loci, and, if cis-controlled, they are good candidates for prostate cancer susceptibility genes. Especially, Cdkn1a was on Pcs2 and cis-controlled and was a good candidate for it. An oncogenic transcription factor, Jund (5.1-fold higher in ACI rats), and a putative transcription factor, Nupr1 (p8 in human; 5.2-fold higher in BUF rats), were also cis-controlled, but were not within the four prostate cancer susceptibility loci.
Among the trans-controlled genes, Pbsn was expressed 15-fold higher in BUF rats and was shown to be controlled mainly by a locus on chromosome 8. Pbsn is known to be specifically expressed in the prostate and is preferred as a promoter for prostate-specific expression of transgenes (Greenberg et al. 1995; Asamoto et al. 2001). A polymorphic prostate-specific transcription factor is expected to be present on rat chromosome 8. Both Kclr and Psat1 were trans-controlled by QTL(s) in the same region on chromosome 3, and possibility that both genes are controlled by the same polymorphism was suggested. The trans-linkages show lower significance and explain less expression variance than do the cis-linkages. This pattern has been observed in another eQTL study (Schadt et al. 2003).
eQTL analysis was effective for genes with various expression levels when quantitative RT-PCR was used. Even for Cdkn1a, whose difference in expression level was difficult to detect using a microarray, the controlling locus was mapped with high LOD scores. To achieve precise analysis, melting temperatures of PCR products were confirmed to be the same between BUF and ACI rats since a polymorphism in a PCR product can potentially affect PCR efficiency. The expression levels were highly reproducible between two independent measurements, giving correlation coefficients of 0.78 and 0.92 for Jund and Pbsn, respectively.
Selection of a gene for normalization was important for quantitative RT-PCR since different QTL were mapped depending upon the gene, especially for trans-controlled genes with low expression. To determine an appropriate gene for normalization, we used congenic rats that had ACI-derived chromosome 3 in the BUF background (Gcr2). Expression levels of Kclr and Psat1 in Gcr2 were similar to that in ACI using any of the three genes for normalization, showing that their expression levels were controlled by a locus on chromosome 3. However, this effect of chromosome 3 was detected only when Actb was used for normalization. This indicated that expression levels of even genes for normalization had variations among rat strains. Although it was suggested that the variation of Actb was smaller than those of Gapd and Ppia among different strains of rats, analysis of more trans-controlled loci seems necessary. In contrast, for comparison between different tissues or different conditions in the same strain, Ppia was reported to give the most reproducible results (Weisinger et al. 1999; Feroze-Merzoug et al. 2002; Yamashita et al. 2004). Careful selection of a gene for normalization seems important.
In conclusion, we showed eQTL analysis for the rat prostate, the effectiveness of eQTL analysis for genes with a broad range of expression levels using quantitative RT-PCR, and an appropriate gene for normalization.
Acknowledgments
This work was supported by grants-in-aid for the third-term Comprehensive Cancer Control Strategy and for Cancer Research from the Ministry of Health, Labor and Welfare and by the Program for Promotion of Fundamental Studies in Health Sciences of the Organization for Pharmaceutical Safety and Research of Japan.
References
- Abe, M., S. Yamashita, T. Kuramoto, Y. Hirayama, T. Tsukamoto et al., 2003. Global expression analysis of N-methyl-N′-nitro-N-nitrosoguanidine-induced rat stomach carcinomas using oligonucleotide microarrays. Carcinogenesis 24: 861–867. [DOI] [PubMed] [Google Scholar]
- Asamoto, M., N. Hokaiwado, Y. M. Cho, S. Takahashi, Y. Ikeda et al., 2001. Prostate carcinomas developing in transgenic rats with SV40 T antigen expression under probasin promoter control are strictly androgen dependent. Cancer Res. 61: 4693–4700. [PubMed] [Google Scholar]
- Baier, L. J., P. A. Permana, X. Yang, R. E. Pratley, R. L. Hanson et al., 2000. A calpain-10 gene polymorphism is associated with reduced muscle mRNA levels and insulin resistance. J. Clin. Invest. 106: R69–R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem, R. B., G. Yvert, R. Clinton and L. Kruglyak, 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755. [DOI] [PubMed] [Google Scholar]
- Cheung, V. G., L. K. Conlin, T. M. Weber, M. Arcaro, K. Y. Jen et al., 2003. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 33: 422–425. [DOI] [PubMed] [Google Scholar]
- Daniels, G. R., and P. L. Deininger, 1985. Repeat sequence families derived from mammalian tRNA genes. Nature 317: 819–822. [DOI] [PubMed] [Google Scholar]
- Dong, J. T., P. W. Lamb, C. W. Rinker-Schaeffer, J. Vukanovic, T. Ichikawa et al., 1995. KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science 268: 884–886. [DOI] [PubMed] [Google Scholar]
- el-Deiry, W. S., T. Tokino, T. Waldman, J. D. Oliner, V. E. Velculescu et al., 1995. Topological control of p21WAF1/CIP1 expression in normal and neoplastic tissues. Cancer Res 55: 2910–2919. [PubMed] [Google Scholar]
- Feroze-Merzoug, F., I. M. Berquin, J. Dey and Y. Q. Chen, 2002. Peptidylprolyl isomerase A (PPIA) as a preferred internal control over GAPDH and beta-actin in quantitative RNA analyses. BioTechniques 32: 776–778, 780, 782. [DOI] [PubMed] [Google Scholar]
- Greenberg, N. M., F. DeMayo, M. J. Finegold, D. Medina, W. D. Tilley et al., 1995. Prostate cancer in a transgenic mouse. Proc. Natl. Acad. Sci. USA 92: 3439–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner, N., C. A. Wallace, H. Zimdahl, E. Petretto, H. Schulz et al., 2005. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat. Genet. 37: 243–253. [DOI] [PubMed] [Google Scholar]
- Inaguma, S., S. Takahashi, H. Ohnishi, S. Suzuki, Y. M. Cho et al., 2003. High susceptibility of the ACI and spontaneously hypertensive rat (SHR) strains to 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) prostate carcinogenesis. Cancer Sci. 94: 974–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs, J. T., 1984. The aging ACI/Seg versus Copenhagen male rat as a model system for the study of prostatic carcinogenesis. Cancer Res. 44: 5785–5796. [PubMed] [Google Scholar]
- Kirst, M., C. J. Basten, A. A. Myburg, Z-B. Zeng and R. R. Sederoff, 2005. Genetic architecture of transcript-level variation in differentiating xylem of a eucalyptus hybrid. Genetics 169: 2295–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramoto, T., K. Kitada, T. Inui, Y. Sasaki, K. Ito et al., 2001. Attractin/mahogany/zitter plays a critical role in myelination of the central nervous system. Proc. Natl. Acad. Sci. USA 98: 559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramoto, T., K. Morimura, S. Yamashita, E. Okochi, N. Watanabe et al., 2002. Etiology-specific gene expression profiles in rat mammary carcinomas. Cancer Res. 62: 3592–3597. [PubMed] [Google Scholar]
- Lander, E., and L. Kruglyak, 1995. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 11: 241–247. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., P. Green, J. Abrahamson, A. Barlow, M. J. Daly et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Lee, P. D., R. Sladek, C. M. Greenwood and T. J. Hudson, 2002. Control genes and variability: absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genome Res. 12: 292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon, R. D., T. M. Shinnick and J. G. Sutcliffe, 1986. The neuronal identifier element is a cis-acting positive regulator of gene expression. Proc. Natl. Acad. Sci. USA 83: 3751–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks, S. A., A. Leonardson, H. Zhu, P. Cundiff, P. Pietrusiak et al., 2004. Genetic inheritance of gene expression in human cell lines. Am. J. Hum. Genet. 75: 1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley, M., C. M. Molony, T. M. Weber, J. L. Devlin, K. G. Ewens et al., 2004. Genetic analysis of genome-wide variation in human gene expression. Nature 430: 743–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn, J. K., P. L. Weissberg and C. M. Shanahan, 1995. A regulatory element downstream of the rat SM22 alpha gene transcription start point enhances reporter gene expression in vascular smooth muscle cells. Gene 154: 249–253. [DOI] [PubMed] [Google Scholar]
- Park, S. H., Y. S. Lee, Y. Osawa, M. Hachiya and M. Akashi, 2002. Hsp25 regulates the expression of p21(Waf1/Cip1/Sdi1) through multiple mechanisms. J. Biochem. 131: 869–875. [DOI] [PubMed] [Google Scholar]
- Rishi, A. K., C. K. Hsu, X. S. Li, A. Hussain, T. M. Gerald et al., 1997. Transcriptional repression of the cyclin-dependent kinase inhibitor p21WAF1/CIP1 gene mediated by cis elements present in the 3′-untranslated region. Cancer Res. 57: 5129–5136. [PubMed] [Google Scholar]
- Schadt, E. E., S. A. Monks, T. A. Drake, A. J. Lusis, N. Che et al., 2003. Genetics of gene expression surveyed in maize, mouse and man. Nature 422: 297–302. [DOI] [PubMed] [Google Scholar]
- Serreze, D. V., H. D. Chapman, D. S. Varnum, M. S. Hanson, P. C. Reifsnyder et al., 1996. B lymphocytes are essential for the initiation of T cell-mediated autoimmune diabetes: analysis of a new “speed congenic” stock of NOD.Ig mu null mice. J. Exp. Med. 184: 2049–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima, T., H. Makino, M. Nakayasu, S. Aonuma, M. Takeuchi et al., 1994. Presence of p53 mutations in 3Y1-B clone 1–6: a rat cell line widely used as a normal immortalized fibroblast. Jpn. J. Cancer Res. 85: 455–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisinger, G., M. Gavish, C. Mazurika and O. Zinder, 1999. Transcription of actin, cyclophilin and glyceraldehyde phosphate dehydrogenase genes: tissue- and treatment-specificity. Biochim. Biophys. Acta 1446: 225–232. [DOI] [PubMed] [Google Scholar]
- Yamashita, S., T. Nomoto, T. Ohta, M. Ohki, T. Sugimura et al., 2003. Differential expression of genes related to levels of mucosal cell proliferation among multiple rat strains by using oligonucleotide microarrays. Mamm. Genome 14: 845–852. [DOI] [PubMed] [Google Scholar]
- Yamashita, S., T. Nomoto, M. Abe, M. Tatematsu, T. Sugimura et al., 2004. Persistence of gene expression changes in stomach mucosae induced by short-term N-methyl-N′-nitro-N-nitrosoguanidine treatment and their presence in stomach cancers. Mutat. Res. 549: 185–193. [DOI] [PubMed] [Google Scholar]
- Yamashita, S., S. Suzuki, T. Nomoto, Y. Kondo, K. Wakazono et al., 2005. Linkage and microarray analyses of susceptibility genes in ACI/Seg rats: a model for prostate cancers in the aged. Cancer Res. 65: 2610–2616. [DOI] [PubMed] [Google Scholar]