Abstract
The concept of selective (or bin) mapping is used here for the first time, using as an example the Prunus reference map constructed with an almond × peach F2 population. On the basis of this map, a set of six plants that jointly defined 65 possible different genotypes for the codominant markers mapped on it was selected. Sixty-three of these joint genotypes corresponded to a single chromosomal region (a bin) of the Prunus genome, and the two remaining corresponded to two bins each. The 67 bins defined by these six plants had a 7.8-cM average length and a maximum individual length of 24.7 cM. Using a unit of analysis composed of these six plants, their F1 hybrid parent, and one of the parents of the hybrid, we mapped 264 microsatellite (or simple-sequence repeat, SSR) markers from 401 different microsatellite primer pairs. Bin mapping proved to be a fast and economic strategy that could be used for further map saturation, the addition of valuable markers (such as those based on microsatellites or ESTs), and giving a wider scope to, and a more efficient use of, reference mapping populations.
LINKAGE map construction typically requires cosegregation analyses of hundreds of Mendelian loci, most of them molecular markers, using a relatively large number of plants from a population in linkage disequilibrium (usually F2, BC1, or similar progenies). The mapping effort is considerable, particularly when the objective is to obtain a high-density map or to incorporate a large number of functionally meaningful markers, such as those based on expressed sequence tag (EST) sequences, or markers that are particularly suitable for breeding applications, such as simple-sequence repeat (SSR) markers, into an existing map.
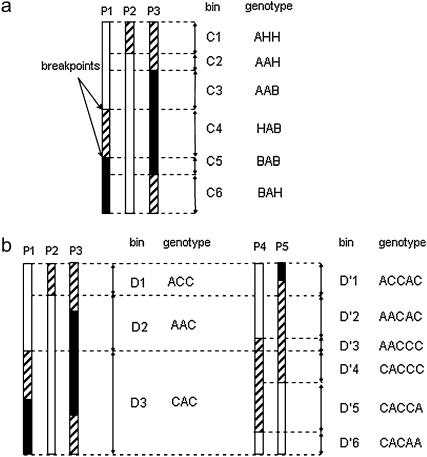
A strategy to improve the efficiency of mapping, named selective mapping, was proposed by Vision et al. (2000). It consists of a two-step process in which, first, a mapping population of usual size (N = 60–250) is used to construct a saturated framework map with markers placed on it with high precision, and second, new markers are added to this map with lower precision using a selected subset of highly informative plants. The final objective is to lower the cost of genotyping new markers with a minimal loss of mapping precision. The selection of this subset of plants is based on the number and position of recombinational crossover sites (or breakpoints) detected with the framework marker data in each plant. The breakpoints identified by the ensemble of the selected plants define a set of bins, i.e., chromosome fragments bounded by two adjacent breakpoints or by a distal breakpoint and the telomere, characteristic of each subset (Figure 1). For a given marker, the joint genotype of the selected subset of plants ideally identifies a unique small bin in the genome. The optimal subset of a given size would have the maximum possible number of breakpoints evenly spaced throughout the genome, resulting in a high number of small bins of uniform size. Vision et al. (2000) developed methods and designed a software program (MapPop) to facilitate the selection of optimal (or nearly optimal) subsets from mapping populations.
Figure 1.
Graphical genotype of one chromosome with indication of the bins detected in a selected set of three plants (P1–P3) of a F2 progeny. Open fragments are homozygous for the allele of the female parent (A), solid fragments are homozygous for the allele of the male parent (B), and stripped fragments are heterozygous (H). The boundaries of the bins are indicated with broken horizontal lines. (a) Codominant loci: six bins (C1–C6) with different genotypes can be identified. (b) Bins detected by the same three plants in the case of dominant loci for the allele of the female parent, where only two genotypes can be identified: A and C (B or H). Fewer breakpoints are detected, and none of the transitions from B to H or vice versa can be identified, resulting in only three bins (D1–D3). With two additional plants (P4 and P5), the number of bins increases again to six (D′1–D′6), but only one of them (D′1) is identical to a bin (C1) of the codominant case.
We have applied this concept using as a framework population the F2 progeny of almond (Prunus dulcis) × peach (P. persica) used to construct the Prunus map (Joobeur et al. 1998). The genus Prunus includes all stone fruit species (peach, cherry, apricot, and plum) and almond, which share a common genome (Dirlewanger et al. 2004a). A map in this highly polymorphic almond × peach progeny is available containing currently 562 loci (Dirlewanger et al. 2004a), all of them highly transferable (isozymes, RFLPs, SSRs, and other STSs) across species of the genus, and can be considered a high-density map (<1 marker/cM on average). This map is currently accepted as the reference or general map for the genus. Many SSRs have been obtained in Prunus since the first set was developed for peach by Cipriani et al. (1999). Some of them (185) have already been mapped in the reference map (Aranzana et al. 2003; Dirlewanger et al. 2004a), but a higher SSR map density is needed for complete coverage of the Prunus genome and to have a high probability of finding a polymorphic SSR in any map region of 10–15 cM of any progeny, particularly of peach, the most economically important and least variable species of the genus (Byrne 1990). A reference map densely populated with SSRs would be useful for gene/QTL tagging, whole-genome selection, and other plant breeding applications (Tanksley et al. 1989). At the time of starting this research, several hundred new SSR markers, developed by the authors, other research groups, or obtained from the increasingly important set of Prunus ESTs held at the Genome Database for the Rosaceae (GDR; http://www.genome.clemson.edu/gdr/) were available with an unknown map position. In this article we have applied the bin mapping approach for the first time and used it to place 264 newly developed microsatellite-derived markers on the almond × peach reference map.
MATERIALS AND METHODS
The population used was the F2 obtained from selfing a single plant (MB1-73) of the cross between “Texas” almond and “Earlygold” peach. Marker data are available for 88 plants of this population (referred to as the T × E population), and the marker data set used was that of the most recent map (Dirlewanger et al. 2004a).
Our main criterion for selecting the set of plants for bin mapping (the bin set) was for the number of plants included in this set to be minimal. Additional criteria were a good combination of the following: the minimal number of joint genotypes that each correspond to more than one bin (“duplicate bins”), the smallest maximum bin length, and the highest number of bins (minimal average bin length). By visual inspection, we found that fewer than six plants would generate a high number of duplicate bins. Six was considered a desirable size, because a set of eight individuals (six plants of the F2 plus the two parents or one parent and the F1 hybrid) would be enough for bin mapping. Eight is a suitable unit of analysis as the plates used for PCR reactions are usually of 8 × 12 wells or multiples of this number. Two approaches were followed to find this set of six plants: (a) the Mappop v.1.0 software (Vision et al. 2000) and (b) selection by visual inspection. The algorithm used for selecting the bin set of MapPop, based on minimizing the expected and maximum bin lengths (Vision et al. 2000), is more efficient in finding optimal plant subsets than visual inspection, which lies essentially in finding a good combination of plants among those that have high numbers of breakpoints. In contrast, visual inspection allowed us a better control of the genotypically identical bins.
For these analyses we considered only those plants and codominant markers with at least 70% of the data points, which reduced the data set from 88 plants and 562 markers to 60 plants and 388 markers. Once a set of plants was selected, the final number of bins and their genotype were determined using all 562 loci.
For Mappop we used the 60 × 388 data set with the same notation as that of the Mapmaker mapping software (Lander et al. 1987): A and B for homozygotes for female and male alleles, respectively, and H for heterozygotes. The commands used for the selection of the set of plants were “loadframe” with the typestring AB-CDH and “samplemax” for six plants. For visual inspection we selected 3 of the 14 plants with 11 or more breakpoints from the set of 60, which together detected a high number of bins. The other 3 were found by adding, to the first set, 13 additional plants with 9 or 10 recombination events and looking for combinations that complemented the first 3 and were within the selection criteria mentioned previously.
Given that a proportion of the markers is expected to be dominant and that these markers are less informative for bin mapping (Figure 1b), additional plants need to be included in the bin set to obtain a level of resolution similar to that found for codominant markers with the six plants of the codominant set (the AHB set). For that purpose, after selection of the AHB set, the set was complemented with two sets of six plants, one for markers where the dominant allele was that of the almond parent (the DB set) and the other for markers dominant for the peach allele (the AC set). The ensemble of these 12 plants (6 of the AHB set plus 6 more of the DB or AC sets) allowed us to map dominant loci with the required precision. Selection of these new plants was done visually and with criteria similar to those defined previously.
To minimize the size of the experiment, we limited the parental information to Earlygold and the MB1-73 hybrid plant. With two inbred parental lines, there is a simple interpretation of the results, but peach and almond cultivars are heterozygous at many loci. The Earlygold parent was chosen as we expected a higher level of homozygosis in peach than in almond (Byrne 1990). If Earlygold has a heterozygous genotype for alleles of the same size as the hybrid, the assignment of the A and B genotypes to the sample of six plants is ambiguous, because two reciprocal interpretations are possible (i.e., HHBHAB or HHAHBA). We found this for three markers, but in all of them only one of the two possible interpretations corresponded to a bin, which was accepted as the correct one. A more common event, including bins covering 73 cM (14% of the T × E distance), occurs if the Earlygold allele is dominant, and the bin has no homozygotes for the almond allele. Here, the marker would be taken as monomorphic. When looking for markers for these specific regions, in the monomorphic cases it may be advisable to run Texas or other plants of the F2, known to have the A genotype.
Bins of the AHB set were coded with the linkage group number of the bin location, followed by a colon, and then a two-digit number, corresponding to the position of the last marker included in the bin according to the map of Dirlewanger et al. (2004a). For example, bin 7:48 ends with a marker 48 cM from the top of linkage group 7. Some dominant markers, or codominant markers with missing data, were in two contiguous bins. These markers were not considered when determining the position of each bin.
In total, 401 microsatellite primer pairs were assayed: 68 (with the letters M and MA) come from a peach (cv. Akatsuki) cDNA library (Yamamoto et al. 2001); 7, MD201a–MD207a, were obtained from microsatellites within the peach gene sequences of GenBank accession nos. AF414988, AF317062, AJ271438, X96856, AF129074, AF129073, and X77231, respectively; 63 (the UDAp series) from an apricot (cv. Portici) genomic library, enriched for AG/CT repeats (Messina et al. 2004); 42 (the UDA series) from an almond (cv. Ferragnès) genomic DNA library enriched for AC/TG repeats (Testolin et al. 2004); 14 (the CPSCT series) were obtained from an enriched (AG/CT repeats) genomic DNA library of Japanese plum cv. Santa Rosa (Mnejja et al. 2004); and 15 (the UCD series) from a genomic DNA library of sweet cherry cv. Valerij Tschkalov (Struss et al. 2003). A total of 180 microsatellites were found in EST collections, 153 (the EPPCU series) obtained from Clemson University and from the GDR (http://www.genome.clemson.edu/gdr/), and 27 from the collection of ESTs of INRA-Bordeaux (the EPPB series). The four-digit number given to the EST-derived microsatellites corresponds to the last four numbers of the accession number of the sequences from which they were obtained. We started with 220 EST sequences containing a microsatellite, but 40 of them were duplicates of other sequences already included. Using the methodology described by Georgi et al. (2002), nine SSRs (pchgms48, 49, 51, 55, 56, 57, 59, 60, and 61) were obtained by searching for microsatellite sequences in peach BAC clones, which contained RFLP probes detecting markers located in different genome regions (AG37, Pru1, AG2, AG12, AG44, AC43, AG56, AC55, and B4A9, respectively). An additional SSR, pchgms58, in the BAC “Nemared” clone 39B10, was also studied. Finally, the DREa microsatellite was found in the sequence of a dehydration-responsive element-binding protein homolog of Prunus. Sequences of all markers reported in this article are recorded in GenBank.
DNA was extracted in Cabrils as previously described (Viruel et al. 1995) and transferred to Udine, Tsukuba, and Bordeaux for analysis. Methods for PCR amplification, electrophoretic separation, and labeling were those currently used in the laboratories of the authors (Yamamoto et al. 2001; Aranzana et al. 2003; Dirlewanger et al. 2004b; Testolin et al. 2004). Data of Bordeaux, Tsukuba, and Udine were double checked at Cabrils, and those of Cabrils were checked independently by two of the authors. The joint genotype of each marker was used to map each marker by visually matching the joint genotype with that of the set of bins obtained in the framework map.
RESULTS
The best AHB set found by visual inspection included plants 5, 12, 23, 30, 34, and 83 of the T × E population. This set detected a total of 67 bins, including 65 found with the previous T × E data and 2 more detected in this work, with two pairs (2:45/3:04 and 5:41/8:30) having an identical joint genotype. In total, 63 bins could be identified by a single joint genotype and two bin pairs each corresponded to only one joint genotype. The average bin length was 7.8 cM and the longest bin (7:25) spanned 24.7 cM. The AHB set found with the Mappop software included plants 5, 27, 56, 63, 67, and 108, with a shorter maximum bin length (18.7 cM). This set identified 56 bins (average length of 9.4 cM/bin), but three groups with the same joint genotype involved seven bins (each group including bins from two chromosomes), resulting in 52 different joint genotypes, 49 of which were able to identify a single bin. We considered the first AHB set more suitable for bin mapping and selected it for analysis of the new SSRs. One additional plant (no. 27) distinguished the two pairs of redundant bins and was analyzed with the markers that fell in either of them.
The final unit of analysis was Earlygold, the MB1-73 hybrid plant and the F2 plants 5, 12, 23, 30, 34, and 83 (Figure 2). When analyzed for the 401 SSR primer pairs, 253 (63%) were polymorphic, giving 264 loci. For 243 primer pairs, we found a single polymorphic locus, and 10 segregated for more than one locus. Nine of these resulted in two polymorphic loci and one resulted in three. From the 148 SSR primer pairs (37% of all SSR primer pairs used) that did not yield any scorable polymorphism, 97 (24%) produced a monomorphic band in the progeny studied, 8 (2%) produced a multi-banded pattern difficult to score, and 43 (11%) did not amplify. Of the 350 primer pairs that produced scorable patterns (97 monomorphic plus 253 polymorphic), 72% segregated in T × E. The characteristics of the bins identified and the positions of the markers added to this map are listed in Table S1 at http://www.genetics.org/supplemental/.
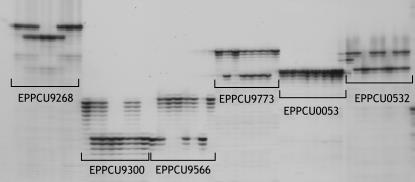
Figure 2.
Acrylamide sequencing gel showing the segregation of the AHB bin set for six SSR markers. Plants of this set, from left to right, are: Earlygold, Texas × Earlygold F1 hybrid, and plants 5, 12, 23, 30, 34, and 83 of the F2. Genotypes for the different SSRs (only for the F2 plants) and corresponding bins, from left to right, are: EPPCU9268, HAAHBB, bin 4:46; EPPCU9300, HAAHHA, bin 6:25; EPPCU9566, AAHABA, bin 5:08; EPPCU9773, BHHHHB, bins 2:45 or 3:04; EPPCU0053, monomorphic; and EPPCU0532, AHHAHH, bin 3:49.
The majority of the polymorphic loci detected (229 of 264 or 87%) were codominant, and only 35 (13%) were dominant. A total of 204 of the codominant loci mapped to a single bin and 21 (9%) fell in duplicate bins, corresponding to two map positions, and required the analysis of an additional plant for assignment to one bin (11 were placed in 2:45, 1 in 3:04, 8 in 5:41, and 1 in 8:30). Only three markers had a genotype that did not correspond to any of the bins established with the T × E map. These were MA059a, EPPCU9168, and UDA-042. The first detected a new bin in the middle of G4, and the other two were located in the same bin, on the top of G5. These three SSRs were assayed in all plants of the T × E population, proving them to be in new bins and allowing us to establish their map position more precisely. With the two markers of G5, this group increased its length in 5 cM compared to the map of Dirlewanger et al. (2004a), bringing the total map length to 524 cM.
For the SSRs where the peach allele was dominant we selected the AC set, consisting of six more plants (15, 27, 57, 74, 102, and 117). The combined information of sets AHB and AC resulted in 66 bins, all corresponding to a different joint genotype, with a maximum bin size of 25.0 cM. The same was done with the almond allele dominant (BD set), and we found that plants 3, 7, 17, 64, 91 and 95, plus the AHB set, detected 59 different bins, the longest being 20.1 cM. The bins detected with these new sets did not always match perfectly the bins found with the AHB set (data not shown).
From the 35 dominant markers found, 18 were dominant for the almond allele and 17 for the peach allele. Fourteen dominant markers (40%) could be assigned either to a single bin (31%) or to two contiguous bins on the same linkage group (9%), using only the AHB set. With the six additional plants for 18 of the 21 dominant markers assigned to more than one linkage group, we found that all of them were in one or two contiguous bins of the AHB set.
New markers were found in 60 of the 67 bins. The bins without new markers were small, including only two to six markers of the previous T × E map. Those with the largest number of markers coincided with some of the longest and more populated bins in T × E. The bin where most new SSRs were located was 1:50, with 15 SSRs and an interval of 14.1 cM (30 markers in the previous T × E map), followed by bin 3:37 with 13 SSRs with an interval of 11.7 cM (27 markers), and bin 7:25, with 12 markers and the longest interval, of 24.7 cM (24 markers).
DISCUSSION
The bin mapping approach has proven to be successful and efficient when used in Prunus, and it can be used in any other plant or animal species, provided that it has a sufficiently saturated framework map. The main achievement of our work was to minimize the effort involved in placing new markers on a map by selecting as small a set of plants as possible. The use of an F2 population, with three possible genotypes per data point, is more efficient for the selection of a bin set of small size, compared to the examples of backcross or radiation progeny given by Vision et al. (2000). Finding a set of 6–10 plants with a resolution for bin mapping similar to ours should be possible in F2 progeny used for the construction of framework maps of any species. We used a relatively compact map (524 cM) in our example, but similar results can be expected in longer maps, as the total genetic distance of a map is proportional to the number of breakpoints of the average gamete, and therefore the information provided by the average individual would be higher in species or populations with longer maps.
When using bin sets of small size, several bins (i.e., duplicate bins) may correspond to the same joint genotype. This is more likely to occur in maps with longer total distances when using the same bin set size, because more bins are expected, and the number of possible joint genotypes remains the same. This can be solved by increasing the bin set size proportionally to the map length. Duplicate bins were found in both the set of plants selected visually and that obtained with the Mappop software, but in the first case their number was lower. We adopted the visual approach, but it is time consuming and it cannot be discounted that a more efficient set of six plants exists in T × E. Improving the Mappop software to detect possible duplicate bins and to provide different sets of plants ordered by their efficiency in different respects (number of duplicate bins, maximum bin size, and average bin size), allowing the users to choose the set more appropriate for their needs, would be a solution.
A total of 264 new SSRs, obtained from 401 microsatellite primer pairs, could be placed on the T × E map with an average accuracy of 7.8 cM, using only 6 plants of the population instead of 88, i.e., with less than one-tenth of the effort. The current number of markers on this map is now 826, which, considering that the total distance is 524 cM (Dirlewanger et al. 2004a; this work), corresponds to an average map density of 0.63 cM/marker. The number of SSRs of T × E has increased from 185 to 449 (average map density of 1.2 cM/SSR). We found only two new bins, indicating that the coverage of the T × E map is almost complete.
The analysis of a reduced number of plants implies that any scoring errors could lead to gross mistakes in the assignment of the marker position. In our case, the number of possible bins obtained with a codominant marker in the set of six plants is 36 = 729. We found only 67 bins, and predicted 5 more, considering the five cases where two contiguous bins are separated by two recombination events instead of one, as in the other cases. Additional bins may be found in the extremes of each linkage group, although we considered this an event with low probability, given the high level of saturation of the T × E map. Thus, considering the 72 predicted and the 729 possible bins, erroneous interpretation would give a new bin 90% of the time. Submitting markers that detect new bins to a more exhaustive analysis, i.e., confirmation with a larger set of plants or the analysis of the whole population, would make bin mapping a very robust method against errors.
Some of the map positions of the markers were expected, such as those of seven mapped SSRs (the pchgms series) developed from BACs that contained another mapped marker, or for EPPCU2288 and EPPCU6309, that correspond to different SSRs located on the same gene. In all these cases, the pairs of markers expected to be in the same physical region were also located in the same bin. Thirty-three more SSRs have already been placed on other maps, 26 in an intraspecific peach F2 (Yamamoto et al. 2001; T. Yamamoto, unpublished data), and 17 in a three-way interspecific progeny, involving almond, peach, and myrobolan plum (Dirlewanger et al. 2004b). All but one of them fell in bins located in the expected linkage group and in a similar position to that found in these maps. The exception was MA040a on one of the ends of G3 of the myrobolan plum map (Dirlewanger et al. 2004b) while it was expected to be in bin 6:74. The same marker was located on the expected G6 region by T. Yamamoto (unpublished data) in the “Akame” × “Juseitou” peach F2. This may be because the MA040a primers detect an additional locus in myrobolan plum, the locus is misplaced in this map, or there is a small genetic rearrangement in this species.
One area where bin mapping would be particularly efficient is in the candidate gene approach, where a large number of possible candidates must be tested for colocation with specific genes or QTL. The use of bin mapping combined with the high polymorphism of T × E (72% of the scorable microsatellite primer pairs segregated) would facilitate this task, allowing the selection of only those candidates that fall into the target regions, which could be later studied in more detail in the whole T × E population or in other populations. Another important advantage of the bin mapping approach is to facilitate the scientific community access to a reference mapping population and to cooperate in placing new markers or characters by exchanging a limited number of vegetatively propagated plants or DNA samples.
Acknowledgments
We thank T. J. Vision for advice about the use of the Mappop software and A. Caño-Delgado for critical review of the manuscript. Funds for this research have been partly provided by projects FIT-010000-2001-112 and AGL2003-04691 of the Spanish Ministry of Science and Technology.
References
- Aranzana, M. J., A. Pineda, P. Cosson, E. Dirlewanger, J. Ascasibar et al., 2003. A set of simple-sequence repeat (SSR) markers covering the Prunus genome. Theor. Appl. Genet. 106: 819–825. [DOI] [PubMed] [Google Scholar]
- Byrne, D. H., 1990. Isozyme variability in four diploid stone fruits compared with other woody perennial plants. J. Hered. 81: 68–71. [Google Scholar]
- Cipriani, G., G. Lot, W. G. Huang, M. T. Marrazzo, E. Peterlunger et al., 1999. AC/GT and AG/CT microsatellite repeats in peach (Prunus persica (L) Batsch): isolation, characterisation and cross-species amplification in Prunus. Theor. Appl. Genet. 99: 65–72. [Google Scholar]
- Dirlewanger, E., E. Graziano, T. Joobeur, F. Garriga-Calderé, P. Cosson et al., 2004. a Comparative mapping and marker assisted selection in Rosaceae fruit crops. Proc. Natl. Acad. Sci. USA 101: 9891–9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirlewanger, E., P. Cosson, W. Howad, G. Capdeville, N. Bosselu et al., 2004. b Microsatellite genetic linkage maps of Myrobalan plum and an almond-peach hybrid—location of root-knot nematode resistance genes. Theor. Appl. Genet. 109: 827–832. [DOI] [PubMed] [Google Scholar]
- Georgi, L. L., Y. Wang, D. Yvergniaux, T. Ormsbee, M. Iñigo et al., 2002. Construction of a BAC library and its application to the identification of simple sequence repeats in peach (Prunus persica (L.) Batsch). Theor. Appl. Genet. 105: 1151–1158. [DOI] [PubMed] [Google Scholar]
- Joobeur, T., M. A. Viruel, M. C. de Vicente, B. Jáuregui, J. Ballester et al., 1998. Construction of a saturated linkage map for Prunus using an almond × peach F2 progeny. Theor. Appl. Genet. 97: 1034–1041. [Google Scholar]
- Lander, E. S., P. Green, J. Abrahamson, A. Barlow, M. J. Daly et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Messina, R., O. Lain, M. T. Marrazzo, G. Cipriani and R. Testolin, 2004. New set of microsatellite loci isolated in apricot. Mol. Ecol. Notes 4: 432–434. [Google Scholar]
- Mnejja, M., J. Garcia-Mas, W. Howad, M. L. Badenes and P. Arús, 2004. Simple-sequence repeat (SSR) markers of Japanese plum (Prunus salicina Lindl.) are highly polymorphic and transferable to peach and almond. Mol. Ecol. Notes 4: 163–165. [Google Scholar]
- Struss, D., R. Ahmad, S. M. Southwick and M. Boritzki, 2003. Analysis of sweet cherry (Prunus avium L.) cultivars using SSR and AFLP markers. J. Am. Soc. Hortic. Sci. 128: 904–909. [Google Scholar]
- Tanksley, S. D., N. D. Young, A. H. Paterson and M. W. Bonierbale, 1989. RFLP mapping in plant breeding: new tools for an old science. BioTechnology 7: 257–263. [Google Scholar]
- Testolin, R., R. Messina, O. Lain, M. T. Marrazzo, W. H. Huang et al., 2004. Microsatellites isolated in almond from an AC-repeat enriched library. Mol. Ecol. Notes 4: 459–461. [Google Scholar]
- Viruel, M. A., R. Messeguer, M. C. de Vicente, J. Garcia-Mas, P. Puigdomènech et al., 1995. A linkage map with RFLP and isozyme markers for almond. Theor. Appl. Genet. 91: 964–971. [DOI] [PubMed] [Google Scholar]
- Vision, T. J., D. G. Brown, D. B. Shmoys, R. T. Durrett and S. D. Tanksley, 2000. Selective mapping: a strategy for optimizing the construction of high-density linkage maps. Genetics 155: 407–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, T., T. Shimada, T. Imai, H. Yaegaki, T. Haji et al., 2001. Characterization of morphological traits based on a genetic linkage map in peach. Breed. Sci. 51: 271–278. [Google Scholar]