Abstract
Linkage disequilibrium (LD) mapping using natural populations results in higher resolution of marker-trait associations compared to family-based quantitative trait locus (QTL) studies. Depending on the extent of LD, it is possible to identify alleles within candidate genes associated with a trait. Analysis of a natural mutant in Arabidopsis has shown that mutations in cinnamoyl CoA reductase (CCR), a key lignin gene, affect physical properties of the secondary cell wall such as stiffness and strength. Using this gene, we tested whether LD mapping could identify alleles associated with microfibril angle (MFA), a wood quality trait affecting stiffness and strength of wood. We identified 25 common single-nucleotide polymorphism (SNP) markers in the CCR gene in Eucalyptus nitens. Using single-marker and haplotype analyses in 290 trees from a E. nitens natural population, two haplotypes significantly associated with MFA were found. These results were confirmed in two full-sib families of E. nitens and Eucalyptus globulus. In an effort to understand the functional significance of the SNP markers, we sequenced the cDNA clones and identified an alternatively spliced variant from the significant haplotype region. This study demonstrates that LD mapping can be used to identify alleles associated with wood quality traits in natural populations of trees.
LINKAGE disequilibrium (LD) mapping or association mapping is useful in identifying phenotype and genotype associations at higher resolution than that of linkage mapping. Quantitative trait locus (QTL) mapping uses only the recombinations found in the progeny of pedigrees, which are typically two to three generations. Therefore, linkage analysis can identify only chromosomal regions associated with a trait, but to increase the resolution of the QTL mapping, a large number of individuals per generation and/or special populations such as recombinant inbred lines are required (Darvasi et al. 1993). However, even by using advanced crosses such as recombinant inbred lines, which may take several years to generate depending on the species, QTL could be mapped only to 5- to 10-cM regions. (Long et al. 1995; Alpert and Tanksley 1996). LD mapping provides an alternative approach to increase the resolution of marker trait associations. Unlike traditional linkage analysis where phenotype and genotype associations are analyzed in progeny from a cross between two parents, LD mapping is done in natural populations or in breeding populations of unrelated individuals. Many recombination events accumulated over the history of a population remove any long-range disequilibria between the loci except for the closely linked loci. Thus any marker-trait associations found using LD mapping will be of higher resolution (Mackay 2001). Moreover, using LD mapping, variation in the species as a whole can be accessed whereas QTL studies are limited to the variation present between the parents of a cross (Neale and Savolainen 2004).
LD mapping has been successfully applied in human genetics to identify and clone many disease genes (Kerem et al. 1989; Harley et al. 1991; Hästbacka et al. 1992; Fallin et al. 2001). In humans, LD extends over 50 kb (Reich et al. 2001), making genome-wide LD mapping feasible whereas in plant species there is a large variation in the extent of LD. In inbreeding species such as Arabidopsis, LD extends up to 250 kb (Nordborg et al. 2002) and up to 10 cM in barley (Kraakman et al. 2004). In contrast, in maize, an outcrossing species, LD extends to only a few kilobases (Remington et al. 2001). Similarly in loblolly pine, an outcrossing tree, LD declined within several kilobases (Brown et al. 2004). This suggests that in outbreeding species genome-wide LD mapping may not be feasible but candidate-gene-based LD mapping should be possible. However, for such studies, criteria to identify correct candidate genes will be critical. In addition, it is not clear what density of markers within genes is required to detect any associations.
The presence of population structure, which may lead to spurious associations, is one of the potential limitations to the wide usage of LD mapping in plants (Buckler and Thornsberry 2002). Genetic diversity studies in plants have found generally much less genetic population structure in outbreeding species compared to inbreeding species. Inbreeding species usually have less variation within populations but greater genetic differentiation between populations (Hamrick and Godt 1996). Therefore, the confounding effect of population structure on LD mapping may not be a serious problem in outcrossing species. However, even when there is a population structure, statistical methods are now available to identify and control for the population structure (Pritchard et al. 2000). Recently a few studies in plants have used LD mapping to identify candidate gene alleles associated with traits (Thornsberry et al. 2001; Hagenblad et al. 2004; Olsen et al. 2004). However, as yet there are no such studies in forest trees. Candidate-gene-based LD mapping could be particularly useful in breeding programs of forest trees since domestication is only a few generations old and not long enough to create significant linkage disequilibrium.
We examined whether LD mapping can be used to identify alleles associated with microfibril angle (MFA). Microfibril angle is a major determinant of timber strength (stiffness) in trees (Cave and Walker 1994; Evans and Ilic 2001). Secondary xylem (wood) is composed of cellulose, lignin, and hemi-cellulose. Cellulose microfibrils are embedded in lignin and a hemi-cellulose matrix, which gives strength to the wood tissue (Plomion et al. 2001). The angle at which microfibrils are arranged with respect to the longitudinal axis of the cell determines the stiffness of the wood. If the angle is low, stiffness and strength of the wood will be high, and as the angle increases, wood becomes weaker (Evans and Ilic 2001). MFA is under genetic control (Donaldson 1993; Nakada et al. 2003; Hannrup et al. 2004; Lima et al. 2004), but it can be measured directly only in samples from mature trees. Early selection based on molecular markers is therefore an attractive option in programs seeking to improve this trait.
In the biosynthetic pathway of lignins, enzymes belonging to the common phenylpropanoid pathway catalyze the reactions, starting with deamination of phenylalanine and leading to the synthesis of hydroxycinnamoyl CoA esters. Hydroxycinnamoyl CoA esters are directed toward lignin synthesis through two enzymes. Cinnamoyl CoA reductase (CCR) converts hydroxycinnamoyl CoA esters into cinnamaldehydes, and cinnamoyl alcohol dehydrogenase (CAD) catalyzes the reduction of cinnamaldehydes into hydroxycinnamoyl alcohols, the precursors of lignin (Hahlbrock and Scheel 1989; Lacombe et al. 1997; Lauvergeat et al. 2002). A natural lignin mutant (irx4) of Arabidopsis thaliana was shown to affect physical properties of the secondary cell wall such as stiffness and strength. Further analysis of the mutant has shown that alternative splicing in the CCR gene is responsible for the changes in physical properties (Jones et al. 2001). As mutations in CCR in A. thaliana have been shown to affect stiffness and strength, we wanted to determine if polymorphisms in this gene would affect the MFA of woody trees.
Here we tested single-marker- and haplotype-based LD mapping methods to identify alleles associated with wood quality traits in a natural population of Eucalyptus nitens, an outcrossing species. We identified two common haplotypes associated with MFA, demonstrating the potential of LD mapping in identifying alleles and haplotypes within the candidate genes associated with quantitative traits. This is the first LD mapping study in a forest tree species and it demonstrates the potential of LD mapping in identifying useful alleles.
MATERIALS AND METHODS
Populations:
We used 290 trees from different open-pollinated families of E. nitens (an association population) for initial SNP marker analyses. These were from the Victoria (Australia) central highlands region of the species' natural distribution and were grown in a field trial in northwestern Tasmania. To confirm the results from LD mapping, we genotyped the SNP markers in 287 first-generation trees of a full-sib family of E. nitens and 148 trees from a full-sib family of E. globulus, a species closely related to E. nitens. SNP 120, a nonsynonymous polymorphism in exon 3, was variable between the E. globulus parents and was used to genotype the E. globulus family. Wood cores were collected from each tree at 1.3 m from ground level, and wood properties, including MFA, were measured using SilviScan 2 (Evans et al. 2000; Evans and Ilic 2001).
SNP discovery and genotyping:
Primers were designed for sequencing the CCR gene in E. nitens and E. globulus on the basis of the E. gunnii sequence of CCR (Lacombe et al. 1997, 2000). Five trees from an E. nitens association population were used to sequence the CCR gene to identify common SNPs. A total of 3.3 kb of the gene, including the promoter region, was sequenced. Identified SNPs were genotyped by the single-nucleotide primer extension method using a Beckman Coulter sequencing system. We also genotyped the E. nitens association population with 14 microsatellite markers to identify any population structure using the model-based program STRUCTURE (Pritchard et al. 2000). FST statistics were estimated using the GDA program (Weir 1996) to identify the differentiation between subpopulations on the basis of geographic area distribution.
Detection of alternatively spliced mRNA:
RNA isolated from the cambial tissue of six trees of the E. nitens full-sib family was used to make cDNA. DNA contamination from RNA was removed by DNAse treatment (DNA free), and amplification of genomic DNA was further reduced by designing the primers spanning exon-exon borders. Complementary DNA was synthesized with oligo(dt)15 primers using the (Promega, Madison, WI) reverse transcription system. RT-PCR was performed using primers spanning exon-exon borders and the amplified products were cloned into pGEM-T Easy (Promega) vector. An alternatively spliced variant was detected by electrophoresis and by sequencing the amplified clones.
Real-time PCR:
Real-time PCR was done using cDNA samples from 30 trees and a Corbett 3000 machine. We analyzed both normally and abnormally spliced transcripts. Normal and splice variant expression levels were normalized by the housekeeping gene, SEC13. Standard curves were used to estimate the transcript levels using three replications for each tree.
Statistical analysis:
LD and Hardy-Weinberg equilibrium (HWE) tests were done using SAS/Genetics software (Czika et al. 2002). Two SNPs, SNP 20 and SNP 23, had shown significant departures from HWE with lower heterozygosity after applying the Bonferroni correction for multiple testing. Composite linkage disequilibrium coefficient (CLD; Weir 1979, 1996) was used to test the linkage disequilibrium between pairs of markers. The composite linkage disequilibrium (CLD) coefficient does not require a HWE assumption as it measures both intra- and intergametic LD to account for the ambiguity associated with double heterozygotes. LD-measure r2 values are calculated by dividing the CLD by the product of four allele frequencies at the two loci. Departures from linkage equilibrium were tested by the permutation version of the exact test.
Single-marker analysis:
Single-marker analysis based on the analysis of variance (ANOVA) was used to identify markers associated with the trait. ANOVA was done by fitting the model y = μ +mi + eij, where y is the trait value, μ is the mean, mi is genotype of ith marker, and eij is the residual associated with the jth individual in the ith genotypic class. Smoothing measures that implement Fisher's method for multiple hypothesis testing were used. The Bonferroni correction was used to correct for multiple testing of smoothed P-values. These tests were done using PROC PSMOOTH procedures in SAS/Genetics.
Haplotype analysis:
Haplotype frequencies were estimated and the association of haplotypes with the trait values were tested using haplotype trend regression (HTR; Zaykin et al. 2002). This method is based on an overlapping sliding window of markers approach and provides an overall association test and tests for individual haplotypes. HTR estimates haplotype frequencies using the expectation maximization (EM) algorithm. EM-inferred haplotype frequencies are fairly accurate even when some of the loci are not in HWE (Fallin and Schork 2000). This was confirmed using another program, PHASE (Stephens et al. 2001), which is not affected by departures from HWE and with which we obtained exactly the same haplotype frequencies as from HTR. The significance of the haplotype associations were tested using a permutation test.
We also compared the haplotype frequencies among three regions of an E. nitens association population. The association population of E. nitens was divided into three regions—Macalister, Rubicon, and Toorongo—based on geographic area distribution of the species. We used the program PHASE to estimate and assign haplotypes to individual trees. Haplotype differences among three regions were tested using χ2 statistics for contingency tables.
RESULTS AND DISCUSSION
Population structure:
In LD mapping,the presence of population structure may lead to spurious associations. To identify the population structure, we genotyped the sampled trees of the E. nitens association population with 14 microsatellite markers and the data were analyzed with the STRUCTURE program. This model-based approach using multilocus genotype information identifies clusters of populations that have distinctive allele frequencies and assigns individuals to each cluster. Using this program with an admixed model and without using the geographic area information, we found that the proportion of individuals assigned to each subpopulation (K) remained symmetric, with varying subpopulation or cluster values (Table 1). With population structure, a proportion of the individuals will be strongly assigned to one or another group and the proportions assigned to each subpopulation will be asymmetric (Pritchard et al. 2000). Under each K value tested, most of the individuals remained admixed. Therefore, results from this analysis suggest that there is no significant substructure in this material. We analyzed genetic differentiation among three geographic regions of central Victoria distribution of this population. Pairwise comparison of FST estimates showed low levels of differentiation among the population regions (Table 2). These results confirm that there was no significant structure within the sampled population. Previous studies using isozyme and RFLP markers have also shown low levels of differentiation among the populations from this region (Byrne et al. 1998). A low level of differentiation is expected in forest trees because most tree species are outcrossing with large effective populations and show higher levels of gene flow among populations (Neale and Savolainen 2004).
TABLE 1.
Cluster analysis of subpopulations of E. nitens
| K | Proportion of the individuals assigned to inferred clusters |
|---|---|
| 2 | 0.50, 0.50 |
| 3 | 0.33, 0.33, 0.34 |
| 4 | 0.25, 0.25, 0.25, 0.25 |
| 5 | 0.20, 0.20, 0.21, 0.20, 0.19, 0.19 |
| 6 | 0.17, 0.17, 0.17, 0.16, 0.17, 0.17 |
The number of clusters (K) was predicted by the STRUCTURE program.
TABLE 2.
Overall and pairwise comparison of FST estimates across population regions
| MA | RU | Overall | |
|---|---|---|---|
| RU | 0.018 | — | — |
| TO | 0.006 | 0.015 | — |
| Total | — | — | 0.011 |
MA, Macalister; RU, Rubicon; TO, Toorongo.
SNP detection and pairwise linkage disequilibrium:
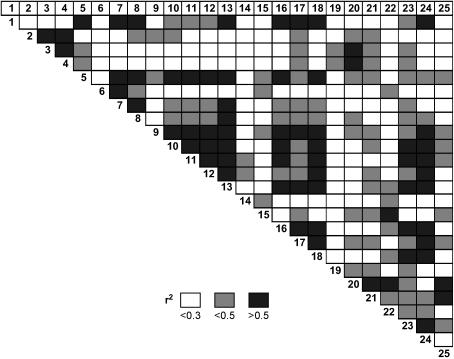
CCR is a well-characterized gene, and its full-length sequence including the promoter (4.3 kb) is available in public databases (Lacombe et al. 1997, 2000). We sequenced 3.3 kb of the gene, including part of the promoter in five unrelated trees from an open-pollinated population of E. nitens. In total, 35 SNPs were identified, including 7 from the promoter region. We designed SNP primers for some singletons as well as other polymorphisms and ran them in 96 samples. From these, 25 common SNPs (minor allele frequency >0.10) have been genotyped in 290 trees from an open-pollinated population. Most of the SNPs are from intron regions, one each from an exon (synonymous) and the 3′-UTR, and four are from the promoter region. One SNP (SNP 4) from the promoter is in the cis-regulatory element (Table 3). LD analysis has shown that LD does not extend over the entire gene and that there are a number of markers that are in linkage equilibrium (r2 < 0.3; Figure 1). Similar results of limited LD were reported in candidate genes of outcrossed species such as maize (Remington et al. 2001) and Pinus (Dvornyk et al. 2002; Brown et al. 2004; Neale and Savolainen 2004). However, there are not many studies on genome-wide LD in plants. Genome-wide analysis of LD in Arabidopsis has indicated that LD extends over 250 kb (Nordborg et al. 2002). A consequence of low LD observed in this study is that the resolution of associations between the marker and the trait will be high. However, if this limited LD extends to the whole genome, genome-wide LD mapping may not be possible, as a large number of markers are required to cover the whole genome.
TABLE 3.
SNP positions, allele frequencies, and associations with MFA
| Marker (SNP) | Region | Position | Frequency | Pa |
|---|---|---|---|---|
| 1 | Promoter | −806 | 0.36 | NS |
| 2 | Promoter | −782 | 0.21 | NS |
| 3 | Promoter | −196 | 0.12 | NS |
| 4 | Promoter | −94 | 0.12 | NS |
| 5 | Intron 2 | 666 | 0.44 | NS |
| 6 | Intron 2 | 694 | 0.13 | NS |
| 7 | Intron 2 | 707 | 0.38 | NS |
| 8 | Intron 2 | 713 | 0.36 | NS |
| 9 | Intron 2 | 976 | 0.46 | NS |
| 10 | Intron 2 | 998 | 0.43 | NS |
| 11 | Intron 2 | 1021 | 0.43 | NS |
| 12 | Intron 2 | 1023 | 0.46 | NS |
| 13 | Intron 2 | 1091 | 0.34 | NS |
| 14 | Intron 2 | 1103 | 0.14 | NS |
| 15 | Intron 2 | 1104 | 0.49 | NS |
| 16 | Intron 2 | 1132 | 0.33 | NS |
| 17 | Intron 2 | 1174 | 0.47 | NS |
| 18 | Intron 2 | 1202 | 0.29 | NS |
| 19 | Intron 3 | 1502 | 0.48 | NS |
| 20 | Intron 3 | 1573 | 0.35 | 0.0018 |
| 21 | Intron 3 | 1600 | 0.31 | 0.0002 |
| 22 | Exon 4 | 1921 | 0.27 | 0.05 |
| 23 | Intron 4 | 2051 | 0.34 | NS |
| 24 | Intron 4 | 2064 | 0.28 | NS |
| 25 | 3′-UTR | 3114 | 0.28 | NS |
NS, not significant.
P-values of markers associated with MFA. Fisher's smoothing method for multiple hypothesis testing and Bonferroni correction for multiple testing have been used.
Figure 1.
Pairwise linkage disequilibrium (r2) between the SNP markers.
LD mapping in association population:
MFA values ranged from 16.2° to 29.0° with a mean of 21° and a standard deviation (SD) of 2.2° in an association population of E. nitens. To identify SNP markers associated with MFA, we used single-marker analysis (ANOVA). With this analysis we found two SNP markers, SNP20 and SNP21, significantly associated with MFA (Table 3). Both of these markers are located in the 3′-end of intron 3 and are in significant linkage disequilibrium with each other. Each of these markers explained 4.6% of total variation in MFA. We used a HTR test (Zaykin et al. 2002) to identify the significant haplotypes associated with MFA. Results from this method with a three-marker sliding window were essentially similar to those from single-marker analysis. We found two common haplotypes surrounding SNP 20 and SNP 21, with a significant effect on MFA. These haplotypes span exon 3, intron 3, and exon 4, a total length of 850 bp (Table 4). Haplotype 1 is made up of a A-A-A-C-A-G sequence while haplotype 2 is made up of A-A-C-T-G-G. The portion of haplotype 1 with a significant effect on trait (A-A-C from SNP 19 to SNP 21; Table 4) explained 5.9% of total variation in MFA while the significant portion of haplotype 2 (C-T-G from SNP 20 to SNP 22) explained 3.4% of total variation in MFA.
TABLE 4.
Haplotypes associated with MFA in an association population of E. nitens
| Haplotype | Frequency | P(overall) | Mean(deg) | P(ind) |
|---|---|---|---|---|
| SNPs 18–20 | 0.03 | |||
| A-A-A | 0.17 | 21.6 | 0.001 | |
| A-A-C | 0.23 | 20.6 | 0.04 | |
| SNPs 19–21 | 0.009 | |||
| A-A-C | 0.24 | 21.6 | 0.0001 | |
| A-C-T | 0.23 | 20.6 | 0.01 | |
| SNPs 20–22 | 0.04 | |||
| A-C-A | 0.26 | 21.3 | 0.01 | |
| C-T-G | 0.28 | 20.5 | 0.003 | |
| SNPs 21–23 | 0.05 | |||
| C-A-G | 0.26 | 21.3 | 0.01 | |
| T-G-G | 0.27 | 20.5 | 0.004 |
P(overall) is the test of significance for overall association with all haplotypes; P(ind) is the test of significance for individual haplotypes.
Comparison of haplotype frequencies among three regions of an E. nitens association population:
We tested whether the haplotype frequencies differ among three geographic regions of an E. nitens association population. To estimate and assign haplotypes to individual trees, we used the program PHASE because its accuracy in comparison with other methods for inferring and reconstructing haplotypes was found to be high (Adkins 2004). We further tested the accuracy of the haplotype assignment of PHASE by comparing it with the sequence data of five individuals. We found haplotype assignment to be accurate in the samples that we tested (data not shown). We used this program to estimate haplotypes from the region (SNP 18 to SNP 23) that showed significant effect on MFA using HTR analysis. We used the three-marker sliding-window method similar to HTR analysis to estimate and assign haplotypes to individual trees. From three SNP markers, a total of eight haplotypes were estimated and assigned to individual trees. Haplotype frequencies from three common haplotypes from two regions, i.e., SNPs 18–20 and SNPs 19–21, are presented in Table 5. There is a twofold difference in haplotype frequencies between Toorongo and the other two regions for haplotypes A-A-A from the SNP 18–20 region and A-A-C from the SNP 19–21 region. These differences are found to be significant on the basis of chi-square tests (Table 5). These two haplotypes are also the two most significant haplotypes detected with HTR analysis (Table 4). However, two other haplotypes that were significant in HTR analysis (Table 4) from the same SNP marker regions were not significant in this analysis.
TABLE 5.
Haplotype frequencies among the three geographic regions of the E. nitens association population
| Haplotype | MA (n = 78) | RU (n = 46) | TO (n = 158) | Overall (n = 282) | Pa |
|---|---|---|---|---|---|
| SNPs 18–20 | |||||
| AAA | 0.25 | 0.23 | 0.12 | 0.17 | 0.02 |
| AAC | 0.22 | 0.18 | 0.28 | 0.25 | NS |
| AGA | 0.19 | 0.18 | 0.29 | 0.24 | NS |
| SNPs 19–21 | |||||
| AAC | 0.33 | 0.33 | 0.16 | 0.23 | 0.003 |
| ACT | 0.20 | 0.21 | 0.28 | 0.24 | NS |
| GAC | 0.33 | 0.37 | 0.43 | 0.39 | NS |
MA, Macalister; RU, Rubicon; TO, Toorongo. NS, not significant.
P is the probability level from χ2 contingency tests.
We then tested for variation in MFA among three geographic regions. Significant difference in trait values among three geographic regions would suggest the association of haplotypes that showed significant frequency differences with the trait. ANOVA showed a significant difference in MFA values among three regions. Therefore, this analysis identified the same haplotypes identified by HTR analysis. Association studies based on haplotype frequency differences are common in human genetics where differences in case and control groups are used for the identification markers associated with a disease. Results from this analysis suggest a significant potential for selecting populations with high frequencies of useful haplotypes. Such selection could be particularly useful in forest tree breeding programs where domestication is in early stages and breeding material is still being sourced from natural populations.
Confirmation of association studies in two full-sib families:
MFA values in the full-sib family of E. nitens ranged from 15.6° to 29.6° with a mean of 22.6° and an SD of 2.4°; in E. globulus, the range of MFA was 12.3° to 28° with a mean of 17.5° and an SD of 2.7°. To confirm the association results from the E. nitens association population in full-sib families of E. nitens and E. globulus, we sequenced the parents of these families. One of the E. nitens parents was homozygous for haplotype 2, identified in the association population, while the other parent had only part of haplotype 1 in a heterozygous condition. Similarly, one of the parents of E. globulus had part of haplotype 1 in a homozygous condition (Table 6). The significant markers SNP 20 and SNP 21 were variable between the parents, but homozygous within both parents of the E. nitens family (Table 6). However, SNP 18, SNP 22, and SNP 23 from the significant haplotype region are variable between the parents with the maternal parent heterozygous for all three loci. In a full-sib family any markers within a small genomic region will be in complete linkage disequilibrium with each other and so we used SNP 18 alone to genotype 287 trees of the E. nitens family and to separate parental haplotypes. Single-marker analysis in this family showed that SNP 18 is significantly associated with MFA, indicating that haplotypes separated by this marker are associated with MFA. Similarly, by genotyping 148 trees from the E. globulus family with SNP 120 from exon 3, which is part of the significant haplotype in this species, a significant association was observed between the parental haplotype separated by the marker and MFA (Table 6). Therefore, results from the E. nitens association population were confirmed in full-sib families from two species showing a clear association between variation in the CCR gene and the MFA. By using the natural population we were able to identify the subgenic region in CCR affecting the trait, a result that would not have been possible with family-based studies alone. The two significant haplotypes extend to ∼850 bp. The high resolution of marker and trait association found in this study is in contrast with A. thaliana CRY2 flowering-time association in which haplogroups extend over 65 kb (Olsen et al. 2004). This is expected because Eucalyptus is predominantly an outcrossing species (Moran 1992) while A. thaliana is predominantly a selfing species with low rates of recombination (Borevitz and Nordborg 2003). Because of low recombination rates in selfing species, haplotype blocks extend over a long distance. Therefore, identifying the causal variant within the extensive haplotype block is difficult as all the variants within the haplotype block are linked.
TABLE 6.
Parental haplotypes of E. nitens and E. globulus full-sib families from SNPs 18–23 and associations with MFA
| Family | Haplotype | Marker | Pa |
|---|---|---|---|
| E. nitens | SNP 18 | ||
| Maternal | A-G-A-C-A-G/C-G-A-C-G-A | AC | 0.02 |
| Paternal | A-A-C-T-G-G/A-A-C-T-G-G | AA | |
| E. globulus | SNP 120 | ||
| Maternal | C-A-G-C-C-A-G/C-G-G-C-C-A-G | AG | 0.04 |
| Paternal | C-G-G-A-C-A-G/C-G-G-A-C-A-G | GG |
Underlined alleles in haplotypes represent the alleles that are common to the significant haplotypes from the association population of E. nitens. In E. gobulus parental haplotypes, the position of SNP 120 between SNP 18 and SNP 19 is in italics.
P-values of the association between SNP markers and MFA.
Discovery of alternative splicing:
Having established the association with MFA, we then sought to determine the functional significance of these haplotypes as all the markers from the significant haplotypes are from intron regions. We sequenced the intron 2 to exon 5 region in the parental trees of the E. nitens family to identify polymorphisms that may change amino acids, but did not detect any nonsynonymous polymorphism. However, we did detect a C → T polymorphism in intron 3 at position 1587 that lies between SNP 20 and SNP 21 and is heterozygous in the female parent, as is SNP 18. Mutations in exon-splicing enhancer (ESE) elements found in introns have been shown to disrupt normal splicing (Kanopka et al. 1996; Mine et al. 2003). Using an ESE-finding program (http://exon.cshl.edu/ESE/index.html), several ESEs, which were disrupted by haplotype polymorphisms associated with MFA, were detected in intron 3.
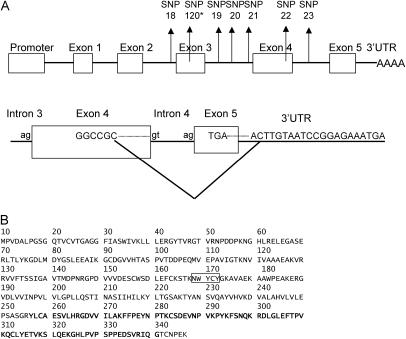
We wanted to determine if the haplotypes containing these polymorphisms would cause alternative splicing due to their location in ESE. We cloned the RT-PCR products from six trees of the E. nitens full-sib family. Three of the six trees had the AA genotype and the remaining trees had the AC genotype of SNP 18, which was used to genotype this family. Analysis of the inserts by gel electrophoresis identified an insert of smaller than expected size in one tree. By sequencing this insert, we detected an alternatively spliced variant with a 292-bp in-frame deletion. In the splice variant, part of exon 4 and the entire terminal exon 5 were skipped. An alternative termination codon in 3′-UTR, which is 40 bp away from the original stop codon, was included in the splice variant (Figure 2A). To determine if this variant was present in other trees, we designed primers spanning part of exon 4 and the 3′-UTR. RT-PCR using these primers showed amplified products in both groups of trees with AA and AC genotypes. Cloning and sequencing RT-PCR products further confirmed identification of the alternative splicing in these trees. Alternative splicing was not specific to any one allele or haplotype. We observed alternative splicing in clones with both A and C alleles; i.e., both parental haplotypes are associated with alternative splicing. It therefore appears that although the haplotype polymorphisms associated with MFA may disrupt ESE elements, they are not sufficient to cause alternative splicing.
Figure 2.
Expression of CCR. (A) Schematic of the CCR gene showing the positions of exons and introns. Exons are shown as boxes and introns as lines. Positions of SNP markers from the significant haplotype region are shown. Alternative splicing is caused by skipping part of exon 4 and all of exon 5. Alternative stop codon in 3′-UTR has been included in the splice variant. (B) Amino acid sequence of the CCR gene. The boxed motif NWYCY (bottom) is conserved in all plant CCR genes and is thought be the catalytic site of the gene. Amino acid sequences (boldface type) are deleted and a new sequence of six amino acids is added to the splice variant. “*SNP 120” is from the E. globulus full-sib family.
In a number of studies intron mutations were associated with a quantitative trait. In Drosophila melanogaster many noncoding mutations from several loci were found to be associated with bristle number (Long et al. 1998; Lyman et al. 1999; Robin et al. 2002). Extensive analysis of the Delta gene in D. melanogaster did not identify any nonsynonymous mutations while two intron mutations were associated with bristle number and it was suggested that cis-acting regulatory and/or splicing variants may be contributing to the variation in bristle number (Genissel et al. 2004). Results from this study also suggest that intron mutations rather than coding region mutations underlie the variation in MFA. These results show the importance of using coding as well as noncoding SNPs in LD mapping to identify significant associations.
Relative expression analysis of the splice variant:
Having established that the splice variant is produced by both groups of trees (AA and AC), we were interested to know if the expression level of alternatively spliced mRNA varied between the two groups of trees. A significant difference between AA and AC groups would indicate that the splice variant is associated with MFA. We used real-time PCR to quantify the mRNA levels in 30 trees, but did not observe a significant difference between the two groups. The reason for this result could be that all the progeny trees were heterozygous and still had one copy of the haplotype associated with MFA. Further work in the natural population, comparing splice variant expression level in trees with the haplotype associated with MFA and trees without the haplotype, may clarify the role of the splice variant in controlling the trait.
CCR is part of a group of related genes showing high homology not only with CCR genes from other plant species, but also with other genes such as dihydroflavonol-4-reductase (DFR) in plants and the mammalian gene 3B-hydroxysteroid dehydrogenase and bacterial UDP-galactose-4-epimirase. The N-terminal region of CCR between positions 15 and 35 is extremely conserved among all the enzyme families (Figure 2B). This conserved N-terminal region has been proposed as the putative cofactor binding site (Lacombe et al. 1997). Within the CCR sequences from different plant species there is an extremely conserved motif, NWYCY (position 168–173) at the beginning of exon 4 (Figure 2B). This motif was suggested to be the catalytic site of the CCR gene (Pichon et al. 1998). All of these critical sequences are retained in the splice variant.
Conclusion and implications:
No significant population structure was observed in the E. nitens population used in this study. Limited levels of LD were observed within the CCR gene. From LD mapping using single-marker, haplotype analyses and family-based studies, we found specific polymorphisms in CCR associated with variation in MFA. We have identified an alternatively spliced mRNA from the haplotype associated with MFA but the role of the variant in controlling the trait needs further testing.
This study demonstrates that LD mapping can be used to identify alleles associated with a trait at higher resolution and that the variation at the whole-population level can be analyzed. The latter advantage is particularly useful in characterizing the variation of breeding lines. The limited information available on the extent of LD in outcrossing plant species suggests that LD does not extend over a long distance (Remington et al. 2001; Dvornyk et al. 2002; Brown et al. 2004; Neale and Savolainen 2004). This implies that genome-wide LD mapping may not be possible in outcrossing species. Therefore, linkage mapping may be used to identify the chromosomal regions and candidate genes associated with a trait and then LD mapping may be used for fine-scale mapping of identified regions or candidate genes.
Results from this study show that the success of LD mapping of candidate genes using natural populations is dependent upon using a number of markers across the gene. This requires complete characterization of candidate genes in terms of the position of introns, exons, and, if possible, promoter regions before using them in LD mapping. Careful selection of candidate genes through different approaches such as microarray analysis, EST database searches, and QTL mapping is very important as a large amount of effort is needed for LD mapping. Success of LD mapping in outcrossing plants therefore depends upon careful selection of candidate genes, complete characterization of the identified genes, and discovering sufficient markers to cover the whole gene. Once sufficient markers have been discovered, depending on the patterns of LD, a subset of markers from these can be used in LD mapping.
LD mapping should have a major impact on selection procedures in the breeding of tree species. Much of the natural variation in tree species is contained in breeding populations since at most only a few breeding generations have occurred since domestication. This study shows that LD mapping in natural populations can locate useful alleles and haplotypes, such that selection for quantitative traits is feasible without a highly structured breeding population.
Acknowledgments
We thank M. E. Devey, S. Nambiar, A. Brown, and W. Spielmeyer for their critical comments on the manuscript and S. Southerton for providing RNA samples. We thank Forestry Tasmania for access to the field trial of E. nitens. This research was supported in part by grants from the Co-operative Research Centre for Sustainable Production Forestry.
References
- Adkins, R. M., 2004. Comparison of the accuracy of methods of computational haplotype inference using a large empirical dataset. BMC Genet. 5: 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpert, K. B., and S. D. Tanksley, 1996. High-resolution mapping and isolation of a yeast artificial chromosome contig containing fw2.2: a major fruit weight quantitative trait locus in tomato. Proc. Natl. Acad. Sci. USA 24: 15503–15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz, J. O., and N. Nordborg, 2003. The impact of genomics on the study of natural variation in Arabidopsis. Plant Physiol. 132: 718–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, G. R., G. P. Gill, R. J. Kuntz, C. H. Langley and D. B. Neale, 2004. Nucleotide diversity and linkage disequilibrium in loblolly pine. Proc. Natl. Acad. Sci. USA 101: 15255–15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler, E. S., and J. M. Thornsberry, 2002. Plant molecular diversity and applications to genomics. Curr. Opin. Plant Biol. 5: 107–111. [DOI] [PubMed] [Google Scholar]
- Byrne, M., T. L. Parrish and G. F. Moran, 1998. Nuclear RFLP diversity in Eucalyptus nitens. Heredity 81: 225–233. [Google Scholar]
- Cave, I. D., and J. C. F. Walker, 1994. Stiffness of wood in fast grown plantation softwoods: the influence of microfibril angle. For. Prod. J. 44: 43–48. [Google Scholar]
- Czika, W., X. Yu and R. D. Wolfinger, 2002. An Introduction to Genetic Data Analysis Using SAS/Genetics. SAS Institute, Cary, NC.
- Darvasi, A., A. Weinreb, V. Minke, J. I. Weller and M. Soller, 1993. Detecting marker-QTL linkage and estimating QTL gene effect and map location using a saturated genetic map. Genetics 134: 943–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson, L. A., 1993. Variation in microfibril angle among three genetic groups of Pinus radiata trees. New Zealand J. For. Sci. 23: 90–100. [Google Scholar]
- Dvornyk, V., A. Sirvio, M. Mikkonene and O. Savolainen, 2002. Low nucleotide diversity at two phytochrome loci along a latitudinal cline in Pinus sylvestris. Mol. Biol. Evol. 19: 179–199. [DOI] [PubMed] [Google Scholar]
- Evans, R., and J. Ilic, 2001. Rapid prediction of wood stiffness from microfibril angle and density. For. Prod. J. 51: 53–57. [Google Scholar]
- Evans, R., S. Stringer and P. Kibblewhite, 2000. Variation of microfibril angle, density and fibre orientation in twenty-nine Eucalyptus nitens trees. Appita J. 53: 450–457. [Google Scholar]
- Fallin, D., and N. J. Schork, 2000. Accuracy of haplotype frequency estimation for biallelic loci, via the expectation-maximization algorithm for unphased diploid genotype data. Am. J. Hum. Genet. 67: 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallin, D., A. Cohen, L. Essioux, I. Chumakov, M. Blumenfeld et al., 2001. Genetic analysis of case/control data using estimated haplotype frequencies: application to APOE locus variation and Alzheimer's disease. Genome Res. 11: 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genissel, A., T. Pastinen, A. Dowell and T. F. C. Mackay, 2004. No evidence for an association between common nonsynonymous polymorphisms in Delta and bristle number variation in natural and laboratory populations of Drosophila melanogaster. Genetics 166: 291–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenblad, J., C. Tang, J. Molitor, J. Werner, K. Zho et al., 2004. Haplotype structure and phenotypic associations in the chromosomal regions surrounding two Arabidopsis thaliana flowering time loci. Genetics 168: 1627–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahlbrock, K., and D. Scheel, 1989. Physiology and molecular biology of phenylpropanoid metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40: 347–369. [Google Scholar]
- Hamrick, J. L., and M. J. W. Godt, 1996. Effects of life history traits on genetic diversity in plant species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 351: 1291–1298. [Google Scholar]
- Hannrup, B., C. Cahalan, G. Chantre, M. Grabner, B. Karlsson et al., 2004. Genetic parameters of growth and wood quality traits in Picea abies. Scand. J. For. Res. 19: 14–29. [Google Scholar]
- Harley, H. G., J. D. Brook, J. Floyd, S. Crow, M. C. Thibault et al., 1991. Detection of linkage disequilibrium between the myotonic dystrophy locus and a new polymorphic DNA marker. Am. J. Hum. Genet. 49: 68–75. [PMC free article] [PubMed] [Google Scholar]
- Hästbacka, J., A. De La Chapelle, I. Kaitil, P. Sistonen, A. Weaver et al., 1992. Linkage disequilibrium mapping in isolated founder populations: diastrophic dysplasia in Finland. Nat. Genet. 2: 204–211. [DOI] [PubMed] [Google Scholar]
- Jones, L., A. R. Ennos and S. R. Turner, 2001. Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J. 26: 205–216. [DOI] [PubMed] [Google Scholar]
- Kanopka, A., O. Muhlemann and G. Akusjarvi, 1996. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature 381: 535–538. [DOI] [PubMed] [Google Scholar]
- Kerem, B. S., J. M. Rommens, J. A. Buchanan, D. Markiewicz, T. K. Cox et al., 1989. Identification of the cystic fibrosis gene: genetic analysis. Science 245: 1073–1080. [DOI] [PubMed] [Google Scholar]
- Kraakman, A. T. W., R. E. Niks, P. M. M. M. Van Den Berg, P. Stam and F. A. Van Eeuwijk, 2004. Linkage disequilibrium mapping of yield and yield stability in modern spring barley cultivars. Genetics 168: 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe, E., S. Hawkins, J. V. Doorsselaere, J. Piquemal, D. Goffner et al., 1997. Cinnamoyl CoA reductase, the first committed enzyme of the lignin branch biosynthetic pathway: cloning, expression and phylogenetic relationships. Plant J. 11: 429–441. [DOI] [PubMed] [Google Scholar]
- Lacombe, E., J. Doorsselaere, W. Boerjan, A. Boudet and J. Grima-pettenati, 2000. Characterization of cis-elements required for vascular expression of the Cinnamoyl CoA Reductase gene and for protein-DNA complex formation. Plant J. 23: 663–676. [DOI] [PubMed] [Google Scholar]
- Lauvergeat, V., P. Rech, A. Jauneau, C. Guez, P. Coutos-Thevenot et al., 2002. The vascular expression pattern directed by the Eucalyptus gunnii cinnamyl alcohol dehydrogenase Eg CAD2 promoter is conserved among woody and herbaceous plant species. Plant Mol. Biol. 50: 497–509. [DOI] [PubMed] [Google Scholar]
- Lima, J. T., M. C. Breese and C. M. Cahalan, 2004. Variation in microfibril angle in Eucalyptus clones. Holzforschung 58: 160–166. [Google Scholar]
- Long, A. D., S. L. Mullaney, L. A. Reid, J. D. Fry, C. H. Langley et al., 1995. High resolution mapping of genetic factors affecting abdominal bristle number in Drosophila melanogaster. Genetics 139: 1273–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, A. D., R. F. Lyman, C. H. Langley and T. F. C. Mackay, 1998. Two sites in the Delta gene region contribute to naturally occurring variation in bristle number in Drosophila melanogaster. Genetics 149: 999–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman, R. F., C. Lai and T. F. C. Mackay, 1999. Linkage disequilibrium mapping of molecular polymorphisms at the scabrous locus associated with naturally occurring variation in bristle number in Drosophila. Genet. Res. 74: 303–311. [DOI] [PubMed] [Google Scholar]
- Mackay, T. F. C., 2001. The genetic architecture of quantitative traits. Annu. Rev. Genet. 35: 303–339. [DOI] [PubMed] [Google Scholar]
- Mine, M., M. Brivet, G. Touati, P. Grabowski, M. Abitbol et al., 2003. Splicing error in E1 α pyruvate dehydrogenase mRNA caused by novel intronic mutation responsible for lactic acidosis and mental retardation. J. Biol. Chem. 278: 11768–11772. [DOI] [PubMed] [Google Scholar]
- Moran, G. F., 1992. Patterns of genetic diversity in Australian tree species. New Forests 6: 49–66. [Google Scholar]
- Nakada, R., Y. Fujisawa and Y. Hirakawa, 2003. Effects of clonal selection by microfibril angle on the genetic improvement of stiffness in Cryptomeria japonica D. Don. Holzforschung 57: 553–560. [Google Scholar]
- Neale, D. B., and O. Savolainen, 2004. Association genetics of complex traits in conifers. Trends Plant Sci. 9: 325–330. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., J. O. Borevitz, J. Bergelson, C. C. Berry, J. Chory et al., 2002. The extent of linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 30: 190–193. [DOI] [PubMed] [Google Scholar]
- Olsen, K. M., S. S. Halldorsdottir, J. R. Stinchcombe, C. Weinig, J. Schmitt et al., 2004. Linkage disequilibrium mapping of Arabidopsis CRY2 flowering time alleles. Genetics 167: 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon, M., I. Courbou, M. Beckert, A. Boudet and J. Grima-Peteenati, 1998. Cloning and characterization of two maize cDNA encoding Cinnamoyl-CoA Reductase (CCR) and differential expression of the corresponding genes. Plant Mol. Biol. 38: 671–676. [DOI] [PubMed] [Google Scholar]
- Plomion, C., G. Leprovost and A. Stokes, 2001. Wood formation in trees. Plant Physiol. 127: 1513–1523. [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K., M. Stephens and P. Donnelly, 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich, D. E., M. Cargill, S. Bolk, J. Ireland, P. C. Sabeti et al., 2001. Linkage disequilibrium in the human genome. Nature 411: 199–204. [DOI] [PubMed] [Google Scholar]
- Remington, D. L., J. M. Thornsberry, Y. Matsuoka, L. M. Wilson, S. R. Whitt et al., 2001. Structure of linkage disequilibrium and phenotype associations in the maize genome. Proc. Natl. Acad. Sci. USA 20: 11479–11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin, C., R. F. Lyman, A. D. Long, C. H. Langley and T. F. C. Mackay, 2002. hairy: a quantitative trait locus for Drosophila sensory bristle number. Genetics 162: 155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, M., N. J. Smith and P. Donnelly, 2001. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68: 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry, J. M., M. M. Goodman, J. Doebley, S. Kresovich and D. Nieksen, 2001. Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28: 286–289. [DOI] [PubMed] [Google Scholar]
- Weir, B. S., 1979. Inferences about linkage disequilibrium. Biometrics 35: 235–254. [PubMed] [Google Scholar]
- Weir, B. S., 1996. Genetic Data Analysis II: Methods for Discrete Population Genetic Data. Sinauer Associates, Sunderland, MA.
- Zaykin, D. V., P. H. Westfall, S. S. Young, M. A. Karnoub, M. J. Wagner et al., 2002. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum. Hered. 53: 79–91. [DOI] [PubMed] [Google Scholar]