Abstract
TAF9 is a TATA-binding protein associated factor (TAF) conserved from yeast to humans and shared by two transcription coactivator complexes, TFIID and SAGA. The essentiality of the TAFs has made it difficult to ascertain their roles in TFIID and SAGA function. Here we performed a genomic synthetic genetic array analysis using a temperature-sensitive allele of TAF9 as a query. Results from this experiment showed that TAF9 interacts genetically with: (1) genes for multiple transcription factor complexes predominantly involving Mediator, chromatin modification/remodeling complexes, and regulators of transcription elongation; (2) virtually all nonessential genes encoding subunits of the SWR-C chromatin-remodeling complex and both TAF9 and SWR-C required for expressing the essential housekeeping gene RPS5; and (3) key genes for cell cycle control at the G1/S transition, as well as genes involved in cell polarity, cell integrity, and protein synthesis, suggesting a link between TAF9 function and cell growth control. We also showed that disruption of SAGA by deletion of SPT20 alters histone-DNA contacts and phosphorylated forms of RNA polymerase II at coding sequences. Our results raise the possibility of an unappreciated role for TAF9 in transcription elongation, perhaps in the context of SAGA, and provide further support for TAF9 involvement in cell cycle progression and growth control.
TRANSCRIPTION initiation by RNA polymerase II involves the assembly of general transcription factors on the core promoter to form a preinitiation complex (PIC). Transcriptional activators bind to specific cis-acting promoter elements within upstream activating sequences (UASs)/enhancers and stimulate PIC assembly through a mechanism thought to involve direct interactions with one or more components of the transcription machinery (Taatjes et al. 2004). The first step in PIC assembly is binding of the TATA-box-binding protein (TBP) or TFIID to the TATA box. TFIID is a multi-subunit complex consisting of TBP and a set of TBP-associated factors (TAFs) (Albright and Tjian 2000). TAFs are highly conserved from yeast to humans. In yeast, 14 TAFs have been identified, 13 of which are required for viability (Green 2000).
Several TAFs are associated with transcription factor complexes other than TFIID. In Saccharomyces cerevisiae, for example, TAF5, TAF6, TAF9, TAF10, and TAF12 (formerly known as Taf90, Taf60, Taf17, Taf 25, and Taf61/68, respectively) (Tora 2002) are also integral components of the Spt-Ada-Gcn5-acetyltransferase (SAGA) complex (Grant et al. 1998) and SAGA-like/SAGA altered, Spt8 absent complex (Pray-Grant et al. 2002; Sterner et al. 2002), which are involved in transcription of a subset of RNA polymerase II-dependent genes. Genetic mutations in (Durso et al. 2001; Michel et al. 1998) or experimental depletion of (Moqtaderi et al. 1998) shared TAFs have been shown to disrupt the integrity of both the TFIID and SAGA complexes.
Previous systematic analysis of the function of yeast TAFs was achieved by isolating temperature-sensitive mutants of each TAF followed by genome-wide expression profiling. These experiments revealed a selective requirement of TAFs for genome-wide transcription (Apone et al. 1998; Holstege et al. 1998; Lee et al. 2000; Shen et al. 2003). That is, none of the 13 essential TAFs is globally required for transcription, but rather each TAF appears to regulate transcription of a subset of genes in the yeast genome (Shen et al. 2003). The selective requirement of TAFs in genomic transcription raised the possibility that each TAF has a specific role in transcriptional regulation. Conceivably, selective requirement of TAFs in transcription may arise from specific biochemical and/or genetic interactions involving each TAF. Supporting this possibility, it has been shown by immunopurification and multi-dimensional mass spectrometry that each TAF associates with a distinct set of transcription factors (Sanders et al. 2002). Despite extensive biochemical characterization of TAFs from different organisms, systematic genetic analysis has not been performed due to their essential nature.
Large-scale genetic interaction analyses have been employed in various model organisms to dissect complex biochemical pathways by identifying mutations in genes that either enhance (synthetic lethality or sickness growth defects) or suppress (extragenic suppressors) the phenotype of a mutation in a known gene of that pathway. Recent development of yeast synthetic genetic array (SGA) analysis has facilitated the screening process for synthetic lethality/sickness by utilizing a comprehensive collection of yeast strains, each of which is devoid of one nonessential gene (∼4700 of ∼6200 total genes; Winzeler et al. 1999). The automation of pinning cells onto plates has streamlined the identification of synthetic genetic interactions for cells bearing double mutations (Tong et al. 2001). Synthetic interactions usually imply a functional connection between the genes involved. For instance, the secretory pathway of yeast has been divided into 10 different biochemical steps such as translocation to Golgi, maturation in ER, etc., and synthetic lethal interactions have been found for many genes in the pathway (Finger and Novick 2000). On the basis of this study, at least 75% of all interactions were subsequently shown to involve genes acting at either identical or separate steps within the same pathway (Hartman et al. 2001).
To further understand how TAFs function, we sought to identify genes that genetically interact with TAF9. TAF9 is an essential component of both TFIID and SAGA (Grant et al. 1998) and it was shown by microarray analysis that TAF9 is required for the expression of ∼60% of the yeast genome. This percentage is the highest among all TAF genes analyzed (Shen et al. 2003). We report here the results of synthetic lethality/sickness screens using a taf9-ts2 temperature-sensitive allele as a query. Both a conventional ade2/ade3 sectoring phenotype (Bender and Pringle 1991)-based screen and a SGA-based genome-wide screen were performed in this study. To our knowledge, this is the first systematic genome-wide genetic analysis of an essential transcription factor. The genetic interactions of TAF9 revealed a significant functional relationship with regulators of transcription elongation as well as initiation. Genetic interactions between TAF9 and genes for other specific cellular processes, including cell cycle progression and growth control, were also uncovered.
MATERIALS AND METHODS
Plasmids, yeast strains, and genetic methods:
Growth of yeast cells in rich (YPD) or synthetic media (SC, synthetic complete) was performed according to standard procedures (Sherman 1991). The construction of a starting strain for a conventional synthetic lethality screen for taf9-ts2 (Apone et al. 1998) was as follows. A 0.9-kb EcoRI fragment containing taf9-ts2 isolated from plasmid Lp35 (Apone et al. 1998) was cloned into pBSKS(+) to generate pWCS204. Plasmid pWCS204 was cut at the BglII site, which is located 176 bp downstream of the TAF9 stop codon, blunt ended by T4 DNA polymerase, and ligated to a filled-in 932-bp StuI-EcoRI fragment containing TRP1 isolated from pJJ246/280 (Jones and Prakash 1990). The resulting plasmid, pWCS206, contains a TRP1-marked taf9-ts2 allele on an EcoRI fragment. The 1.83-kb EcoRI fragment from pWCS206 was used to transform YPH500 (MATα, generating WCS123). WCS123 harboring the TRP1-marked taf9-ts2 allele was mated to PSY137 (MATa ade2 ade3 strain; Koepp et al. 1996), and the resulting diploid strain was sporulated and dissected to isolate haploid strain WCS129. WCS129 (MATα ade2 ade3 leu2 ura3 his3 lys2 taf9-ts2-TRP1) transformed with a plasmid pRS416-TAF9-ADE3 (a URA3-marked, centromere-containing plasmid) was used as the starting strain for mutagenesis and for screening for synthetic lethality by monitoring red/white sectoring of colonies.
The same cloning approach for creating a genetically tractable taf9-ts2 allele as described above was used to generate a query strain suitable for a genome-wide SGA screen, except that NAT1 (a nourseothricin-resistant gene) instead of TRP1 was used to mark the taf9-ts2 allele. The taf9-ts2-NAT1 allele was introduced into strain Y3656 (Tong et al. 2001) to replace the endogenous TAF9 gene. The genotype of the resulting query strain, WCS444, is MATα can1Δ∷MFA1pr-HIS3-MFα1pr-LEU2 ura3 leu3Δ his3Δ lys2Δ met15Δ taf9-ts2-NAT1. BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) is the parental strain of SGA deletion strains. The strain bearing a C-terminal Myc-tagged endogenous TAF9 (WCS311) used for chromatin immunoprecipitation analyses was generated as previously described (Longtine et al. 1998; Gao et al. 2004).
A genome-wide screen of TAF9 genetic interactions by SGA:
The SGA screen was performed essentially as previously described (Tong et al. 2001) except that a temperature-sensitive taf9-ts2 mutation was used in the query strain instead of the null allele of a nonessential gene. We performed the screen three times, and all candidates that scored reproducibly positive were confirmed by tetrad analysis. The confirmatory spot test comparing growth of taf9-ts2 xxxΔ double-mutant strains vs. taf9-ts2 and xxxΔ strains was performed at four different temperatures, 25°, 30°, 34°, and 37°, on SC and/or YPD media. In the course of these studies, we determined that the majority of false negatives that arise by SGA either interact weakly with taf9-ts2 or cause growth defects themselves, thereby hindering their identification. Genetic interactions were scored as strong when growth defects were evident at both 30° and 34° (Figure 1, red), and scored as conditional when growth defects were evident at 34° but not at 30° (Figure 1, blue).
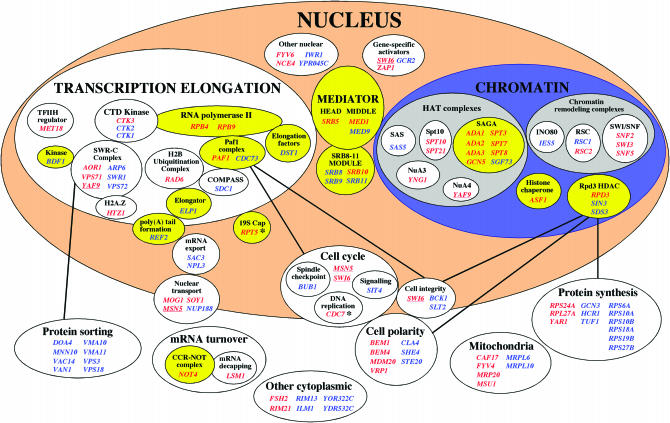
Figure 1.
Genome-wide synthetic interactions involving TAF9. Genes for transcription factors are classified into three major groups: Mediator, chromatin-modifying complexes, and regulators of transcription elongation. Yellow ovals denote sets of genes that encode transcription factors that have been demonstrated to biochemically associate with TAFs (see text). Genes marked in red indicate strong genetic interactions; those marked in blue indicate conditional synthetic interactions. Underlined genes are present in multiple categories. Asterisks denote two essential genes identified by a conventional synthetic lethality screen. Solid lines denote functional connections involving specific transcription factors associated with particular cellular processes (see text).
A synthetic lethality screen of taf9-ts2 allele by ade2/ade3-mediated red/white colony sectoring:
Ethyl methanesulfonate (EMS) was utilized to treat strain WCS129. EMS concentrations at 20 or 30 μl/ml cell culture were employed, which rendered cell survival at 25 or 15%, respectively. We screened ∼2.2 × 104 viable cells, and 443 mutant strains (∼2%) passed the first criterion of the screen and exhibited a constitutive red-color colony (nonsectoring), indicating failure to lose plasmid pRS416-TAF9-ADE3. Among 443 candidate strains, 127 strains passed the second criterion of 5-FOA sensitivity, an alternative assay for failure to lose plasmid pRS416-TAF9-ADE3 (a URA3-marked plasmid). As a third criterion, when pRS413-TAF9 (a HIS3-marked low-copy plasmid) is present, 82 strains of 127 candidates could grow on 5-FOA plates, which selected for Ura− cells. Those 82 strains were mated back to WCS128 (isogenic to WCS129 except opposite mating type) for further analyses of the dominant/recessive nature of EMS-introduced mutations. If the mutations were recessive, the resulting diploid strains were expected to be resistant to 5-FOA and able to form a red/white sectoring phenotype. We recovered 33 diploid strains, and among these candidate strains 12 were confirmed as recessive and 1 as dominant. Standard tetrad analysis on the 12 candidates was performed to determine which candidate(s) harbored a single recessive gene that rendered lethality in combination with the taf9-ts2 mutation. All spores derived from tetrad analyses were temperature sensitive due to the presence of the taf9-ts2 allele. Sets of four haploid strains derived from each tetrad that exhibited a 2:2 (5-FOAr and sectoring:5-FOAs and nonsectoring) segregation pattern for both 5-FOA sensitivity and colony-sectoring assays were an indication of the presence of a single recessive mutation.
To clone the corresponding wild-type genes from four synthetic lethal strains, sl54/WCS151, sl72/WCS148, sl284/WCS154, and sl336/WCS157, a YEp13-based genomic DNA library was transformed into the strains and the resulting transformants were selected for restoration of red/white colony sectoring and 5-FOA-resistant phenotypes. Since the YEp13 vector is a high-copy-number plasmid, the restoration of sectoring and 5-FOA-resistant phenotypes may reflect dosage dependency. To exclude this possibility, we subcloned the genomic insert into the low-copy plasmid pRS415 and repeated the phenotypic analysis. Sequencing of the respective genomic DNA fragments revealed the identities of the genes to be CDC7, RPT5, SAC3, and VPS72, corresponding to the mutations in synthetic lethal strains sl54, sl336, sl284, and sl72, respectively.
Northern blot and primer extension analyses:
Preparation of total RNA and Northern blot analysis were performed as described (Shen and Green 1997). Probes for GLN1, RPS5, and TRX1 transcripts were obtained by PCR amplification from genomic DNA as described (Shen et al. 2003). Primer extension was performed as described (Shen et al. 2003); the sequence of the PDR5 primer was 5′-CGAAAGTTCCTAGTTGCCAAT-3′.
Chromatin immunoprecipitation:
The conditions for formaldehyde-based in vivo crosslinking and chromatin immunoprecipitation (ChIP) were performed essentially as described (Gao et al. 2004) except no radioactively labeled nucleotide was used in PCR, and quantification of precipitated DNA after PCR was measured by ethidium bromide staining. Strains bearing Myc-tagged histone H4 as the only source of histone H4, immunoprecipitations using anti-Myc antibody, and sets of PDR5 primers used for PCR have been described (Gao et al. 2004). Primers used for ACT1 were the following:
promoter: ACT1-1, 5′-CTTATCGGATCCTCAAAACCC-3′ and ACT1-2, 5′-GGAGAGAGAGAGGCGAGTTTG-3′; and
coding sequences: ACT1-3, 5′-GAAGTGTGATGTCGATGTCCG-3′ and ACT1-4, 5′-CACTTGTGGTGAACGATAGATG-3′.
Washing conditions for immunoprecipitations using anti-Ser5 C-terminal domain (CTD) antibody (H14 clone) and anti-Ser2 CTD antibody (H5 clone) were based on previous descriptions with minor modifications (Komarnitsky et al. 2000).
Genetic nomenclature:
Standard nomenclature for S. cerevisiae was used throughout the text except for TAF proteins (capitalized according to the unified nomenclature for TBP-associated factors (Tora 2002).
RESULTS AND DISCUSSION
Synthetic lethal screens using taf9-ts2 as a query:
We first performed a genome-wide SGA screen with a collection of ∼4700 viable deletion strains (Tong et al. 2001) using a taf9-ts2 allele (Apone et al. 1998) as query. The screen was performed three times, each time in duplicate, and reproducible candidates were further analyzed. A total of 106 genetic interactions that were subsequently confirmed by tetrad analysis using standard synthetic-complete media without drugs were identified. Approximately 50% of the TAF9-interacting genes encode transcription factors. In addition to the SGA screen, a conventional ade2/ade3 synthetic lethal screen was performed in hopes of identifying essential genes that interact with taf9-ts2 (see materials and methods). From the latter screen we obtained mutations in four genes, two essential (CDC7 and RPT5) and two nonessential (SAC3 and VPS72). Notably, neither sac3Δ nor vps72Δ was detected in the taf9-ts2 SGA screens, although their conditional synthetic interactions with taf9-ts2 at 34° were subsequently confirmed by tetrad analysis. This observation suggested that our SGA screen might have missed some conditional synthetic interactions, consistent with previous reports indicating the presence of false negatives in SGA screens (Sarin et al. 2004; Tong et al. 2004; see also materials and methods). Genes definitively found not to interact with TAF9, and studied during the course of these experiments, are provided in Table 1.
TABLE 1.
Genes that do not exhibit a synthetic growth phenotype with TAF9
| Category | Gene |
|---|---|
| Chromatin structure | HDA1, CHD1, IES3, SAP30, PHO23 |
| Mediator | NUT1 |
| Transcription elongation | SPT4 |
| Repression | TUP1, SSN6, SKO1 |
| Gene-specific activator | GLN3 |
| Other transcription | MBF1, INO2, RIM9, RIM20, RPI1 |
| Other gene expression | CAF40, CDC40, LOC1 |
| Cell cycle | PHO80 |
| Cell polarity | SLA1, SMY1 |
| Protein synthesis | GCN20, RPS7A, RPS12, RPL13B, RPL39 |
| Protein sorting | VPS4, VPS20, VPS24, VPS28 |
| Protein degradation | DOA1, UBR2, UMP1 |
| Mitochondria | FMC1, LIP2, POR1, POS5, YHM1 |
| Carbohydrate metabolism | GPM2, TPS2 |
| Other cytoplasmic | ICE2, LTV1, NEW1, YOR302W |
| Uncharacterized | FYV5, YMR247C, YPR044C, YDR442W |
Spot tests were used to compare growth defects of taf9-ts2, xxxΔ, and corresponding double-mutant strains at 30° and 34° on SC media.
The combined results of our synthetic lethal/sick screens are summarized in Figure 1. Most TAF9-interacting genes are nuclear, the majority of which include previously characterized transcription factors that fall into three predominant categories: (1) Mediator; (2) complexes having roles in histone modification or chromatin remodeling; and (3) factors involved in transcription elongation. While interactions with each of these categories of transcription factors might be expected, remarkably few genes with roles in gene expression downstream of transcription elongation were identified in our screens. Cellular processes other than gene expression that are overrepresented in our screens, however, include the integrated processes of cell cycle, cell polarity, and cell integrity as well as protein synthesis and protein sorting. Below, each class of TAF9-interacting gene is described in more detail.
Mediator:
Mediator is a general transcription factor (Kornberg 2005b) composed of core Mediator (21 subunits) plus a negative regulatory module designated Srb8-11 (four subunits) (Bjorklund and Gustafsson 2005; Kornberg 2005a). Core mediator itself is composed of three distinct modules called “head,” “middle,” and “tail” (Chadick and Asturias 2005). The head module contacts RNA polymerase II in the PIC, and not surprisingly, six of eight of the respective subunits are essential. One gene from the head core module and two from the middle were identified as TAF9 interactors, as were all four genes from the Srb8-11 module, suggesting a robust relationship between Mediator and TAF9 function. This is consistent with reported biochemical interactions between Mediator and TFIID (Sanders et al. 2002), as well as SAGA-dependent recruitment of Mediator to the GAL1 UAS (Larschan and Winston 2005). While the Srb8-11 module of Mediator is thought to play a predominantly negative role in transcription (Bjorklund and Gustafsson 2005), evidence suggests that it can play a positive role as well (see Larschan and Winston 2005 and references therein). Isolation in our screen of all four subunits of the Srb8-11 module as TAF9 interactors would seem to be most consistent with the latter possibility. Interestingly, none of the genes for subunits of the tail module, four of five of which are nonessential, was obtained in our SGA screen.
Chromatin modification (SAGA, other HAT complexes, chromatin-remodeling complexes, Rpd3-HDAC complex, Asf1):
SAGA is an ∼15-subunit histone acetyltransferase (HAT) complex (Grant et al. 1998) that harbors the HAT enzyme Gcn5, as well as TAF9 itself. Not surprisingly, gcn5 as well as mutations in multiple other components of SAGA, were isolated in our SGA screen. Other HAT complexes in S. cerevisiae include ADA, NuA3, NuA4, Spt10, SAS, TFIID, Hat1, and Elongator (Brown et al. 2000), and our SGA screen identified at least one gene in each of these complexes, except Hat1, suggesting a tight correspondence between the function of TAF9 and HAT enzymes in general.
Chromatin-remodeling complexes in S. cerevisiae include Swi/Snf, RSC, Ino80, SWR-C, Chd1, and ISWI (ISW1a, ISW1b, and ISW2) (Narlikar et al. 2002). Representative members of each of these complexes, except ISWI and Chd1, were obtained in our SGA screen. This suggests that, as for Mediator and HAT complexes, a robust functional association exists between TAF9 and chromatin-remodeling complexes. The absence of genetic interactions with Chd1 and ISWI may reflect unique functions of these specific chromatin-remodeling complexes.
Rpd3-HDAC is one of several histone deacetylase (HDAC) complexes that are found in S. cerevisiae, which are ordinarily associated with gene silencing (Kurdistani and Grunstein 2003). While most HDAC complexes were not identified as genetic interactors with TAF9, genes for three subunits of the Rpd3-HDAC complex, Rpd3, Sin3, and Sds3, were obtained. Two different possibilities might account for this result. First, Rpd3-HDAC was recently shown to be involved in specific instances of gene activation in addition to its role in silencing (De Nadal et al. 2004; Kurdistani et al. 2004). Second, a genetic relationship between Rpd3-HDAC and TAF9 could, in principle, involve either activation or repression or both mechanisms in regard to different genes. Identification of direct biochemical interactions between Rpd3-HDAC and TFIID (Sanders et al. 2002) is consistent with the genetic interactions reported here.
The histone chaperone Asf1 has been shown to activate PHO5 and PHO8 by virtue of its nucleosome disassembly function (Adkins et al. 2004). Genome-wide transcriptional analysis of yeast lacking Asf1 shows it is involved in both activation and repression of many genes (Zabaronick and Tyler 2005). Identification of ASF1 as a synthetic interactor with TAF9 is consistent with reported biochemical and genetic interactions between Asf1 and TFIID (Chimura et al. 2002) and suggests a close relationship between these factors in control of gene expression.
Transcription elongation (TFIIS, SWR-C, H2A.Z, Bdf1 kinase, Paf1 complex, Rad6, Set1/COMPASS, Elongator, CTD kinase, Rpb9 and Rpb4 PolII subunits, 19S cap):
A wide variety of factors that control transcription elongation by RNA polymerase II have recently been characterized (Sims et al. 2004 and references therein). It is intriguing that TAF9 showed either strong (at 30°) or conditional (at 34°) synthetic interactions with almost all these factors. They include TFIIS (Dst1), the PolII C-terminal domain (CTD) kinase CTDK-I (composed of Ctk1, Ctk2, and Ctk3), the Paf1 complex (including Paf1 and Cdc73), the Set1-containing histone methylation complex COMPASS, the elongation-specific HAT enzyme Elongator, the nucleosome-remodeling complex SWR-C (including Swr1, Bdf1 kinase, and H2A.Z), the E2-ubiquitinating enzyme Rad6, which ubiquitinates histone H2B, the nonessential PolII subunits Rpb9 and Rpb4, and the 19S proteosomal cap complex. Notwithstanding, SPT4, encoding one of two subunits (including SPT5, which is essential) of the elongation factor DSIF (Winston 2001), is one of the few nonessential genes in this class that does not show a synthetic relationship to TAF9 (Table 1). These results raise the possibility that in addition to its well-established role in transcription initiation, TAF9 may play an important and previously unappreciated role, directly or indirectly, in transcription elongation.
The SWR-C complex, containing the Snf2 ATPase family member Swr1, plays a role in exchanging the histone variant H2A.Z (Htz1) for conventional H2A (Mizuguchi et al. 2004). While this modification has been shown to promote gene expression near silent heterochromatin (Meneghini et al. 2003), SWR-C also interacts genetically with multiple transcription elongation factors, suggesting a specific role in transcription elongation (Krogan et al. 2003). Interestingly, Htz1 also has been proposed to play a role in transcriptional elongation (Santisteban et al. 2000). In this regard, the relationship between TAF9 and SWR-C is especially noteworthy, since six of seven nonessential subunits of the purified SWR-C complex (Figure 1; Krogan et al. 2003) were identified in our SGA screen, including Htz1.
The Paf1 complex physically and functionally associates with specific elongation factors, including DSIF and FACT (Spt16 and Pob3; Squazzo et al. 2002). Moreover, Paf1 is essential for histone H2B monoubiquitination by the Rad6-Bre1-Lge1 complex, which subsequently signals histone methylation at Lys4 by the Set1/COMPASS complex at an early stage of elongation (Wood et al. 2003). Results from Madhani and colleagues (Hwang et al. 2003) suggest that H2B ubiquitination and H2A.Z substitution are redundant functions in the formation of active chromatin. Strong or conditional synthetic interactions identified here involving members of the Paf1 complex, Rad6, and the Set1/COMPASS complex, in addition to SWR-C and H2A.Z, raise the possibility of a role for TAF9 in transcription elongation related to the redundant functions of H2B ubiquitination and H2A.Z substitution. Supporting this possibility is the fact that the Paf1 complex biochemically interacts with TFIID (Sanders et al. 2002).
Elongator, a transcription-elongation-specific HAT enzyme (Gilbert et al. 2004), interacts genetically with TAF9 via its Elp1 subunit (Figure 1). This is consistent with biochemical interactions between TAF9 and all six subunits of Elongator (Sanders et al. 2002).
CTDK-I is a kinase that phosphorylates the RNA polymerase II CTD, subsequently affecting both transcription elongation and pre-mRNA processing (Ahn et al. 2004). CTK1 encodes the catalytic subunit of this kinase, which hyperphosphorylates serine 2 of the PolII CTD (Lee and Greenleaf 1991), thereby affecting RNA polymerase II elongation efficiency. Deletion of CTK1 prevents Set2 recruitment, required for Lys36 methylation, which is necessary in addition to Lys4 methylation by Set1/COMPASS for transcription elongation (Krogan et al. 2002). All three subunits of this kinase complex (Ctk1, Ctk2, and Ctk3) were identified in our SGA screen as TAF9 interactors. Additionally, both nonessential subunits of PolII, Rbp9 and Rpb4, which are functionally related to TFIIS (Wery et al. 2004), also interacted with TAF9.
The 19S regulatory cap of the 26S proteasome has been shown to play important roles in transcription elongation (Ferdous et al. 2001) as well as in initiation (Gonzalez et al. 2002). Biochemical interactions between components of the 19S cap and general transcription factors such as TFIID have also been reported (Sun et al. 2002). RPT5 encodes one of six ATPase components located in the “base” module of the 19S cap. All of these subunits are essential; identification of RPT5 as an interactor with TAF9 was made using a conventional ade2/ade3 screen. While identification of RPT5 as an interactor with TAF9 is consistent with a proposed role of the 19S cap in transcription, involvement of the 26S proteasome in a wide range of cellular processes, however, raises the possibility that this interaction may likewise result from transcription-independent mechanisms.
Gene-specific activators (Swi6, Zap1, Gcr2):
On the basis of the original coactivator hypothesis of the function of TAFs (Dynlacht et al. 1991) and the fact that >200 gene-specific activators have been identified in S. cerevisiae, many of which are nonessential (Harbison et al. 2004), synthetic interactions with multiple gene-specific activators might have been anticipated. However, only three gene-specific activators were identified in this investigation. Identification of swi6 as one of them is consistent with results of a previous synthetic lethality screen in which swi6Δ was used as a query (Macpherson et al. 2000). It is likewise consistent with biochemical interaction between TAF9 and Swi6 (Sanders et al. 2002). Swi6 binds to both Swi4 and Mbp1 to form the heterodimeric sequence-specific transcription factors SBF and MBF, respectively, both key regulators of the transcription of genes involved in the progression from G1 to S phase (Macpherson et al. 2000). In this regard, further connections to cell cycle control identified in this study (see below) support a prevalence of mechanisms linking TAF9 to cell cycle control.
A genetic relationship between TAF9 and ZAP1 may be a consequence of the large number of zinc-regulated genes in S. cerevisiae (Eide 2000). Gcr2 is involved in expression of glycolytic genes, and essentiality of glycolysis may underlie its conditional synthetic relationship to TAF9. Interaction of TAF9 with far more global transcription factors than gene-specific factors is consistent with the notion that TAFs function other than as general coactivators (Shen and Green 1997).
Other stages of gene expression: poly(A) tail formation, mRNA export, nuclear transport, mRNA turnover:
In contrast to the above results, few hits involving factors with roles in gene expression downstream of transcription were obtained. The fact that at least some interactions were identified, however, suggests correspondence between the function of TAF9 and downstream stages of gene expression. For example, identification of REF2 by SGA, which encodes a protein involved in poly(A) tail formation (Russnak et al. 1995), implicates a link between transcription initiation and termination. Indeed, widespread evidence supports the coupling of these two processes (Proudfoot 2004), including the fact that Ref2 interacts biochemically with TFIID (Sanders et al. 2002). Both Npl3 (Lei et al. 2001) and Sac3 (Lei et al. 2003) were also obtained as interactors with TAF9, suggesting that transcription is coupled to mRNA processing and mRNA export, consistent with current notions of nuclear organization and function (Maniatis and Reed 2002). On the basis of the regulatory integration predicted (Maniatis and Reed 2002), and the large number of proteins known to be involved in both mRNA processing and mRNA export (Burckin et al. 2005), we might have imagined more hits between TAF9 and genes in these categories. Nevertheless, four hits were obtained in genes encoding proteins with functions involved in nuclear transport, including Mog1, Soy1, Msn5, and Nup188. Recent studies suggest that transcriptional activation in S. cerevisiae may occur often at the nuclear periphery and involve contributions by nuclear pore proteins (see Casolari et al. 2005; Menon et al. 2005; and references therein). In this regard, it is intriguing, in light of the above results, that TAF9 interacts physically with Nup57 (Uetz et al. 2000; Menon et al. 2005). Further support of the possible involvement of TAFs in nuclear transport derives from the finding that mutation of Schizosaccharomyces pombe TAF7 causes poly(A)+ mRNAs to accumulate in the nucleus (Shibuya et al. 1999).
In addition to identification of nuclear genes involved in gene expression, synthetic interactions were found between TAF9 and the genes NOT4 and LSM1, which encode components of the cytoplasmic mRNA turnover apparatus. mRNA turnover in yeast requires the highly conserved Ccr4-Not complex, the predominant cytoplasmic mRNA deadenylase (Parker and Song 2004), of which Not4 is a component. Synthetic genetic interaction between NOT4 and TAF9 may indicate that altered stoichiometry among the cellular mRNA population, resulting from mRNA stabilization due to not4 in combination with reduced mRNA abundance due to taf9-ts2, is detrimental to cell growth. Alternatively, given that the Ccr4/Not complex also has been reported to play specific roles in both transcription initiation (Badarinarayana et al. 2000) and elongation (Denis et al. 2001), and has been shown to interact both biochemically and functionally with TAF1 (Deluen et al. 2002), it is possible that synthetic interaction between NOT4 and TAF9 results from interactions at transcription.
Cell cycle:
Coordination between transcription and cell cycle control in S. cerevisiae is well established (see Breeden 2003 and references therein). In fact, previous studies supported the involvement of specific TAFs in cell cycle control (Apone et al. 1996; Walker et al. 1997; Reese and Green 2001), including the fact that TAF9 interacts with SWI6 genetically and biochemically (Figure 1; Macpherson et al. 2000; Sanders et al. 2002). Identification in our SGA screen of MSN5, encoding a karyopherin required for proper nucleocytoplasmic transport of Swi6 (Queralt and Igual 2003), provides additional support for this notion, further linking TAF9 to control of the G1/S transition via Swi6 localization. Synthetic genetic interaction with TAF9 may involve a requirement of TAF9, along with Swi6, for activating G1 cyclin transcription (Macpherson et al. 2000).
Other genes identified in this study potentially linking TAF9 to cell cycle control include CDC7, SIT4, and BUB1. CDC7, isolated with a conventional ade2/ade3 screen, encodes an essential Dbf4-dependent kinase required for the G1/S transition via its role in activating prereplication complexes (Lei and Tye 2001). Lethality of taf9-ts2 cdc7 double mutants is unlikely due to transcriptional downregulation of CDC7, since transcription of CDC7 is independent of TAF9 on the basis of microarray analysis (Holstege et al. 1998). SIT4 encodes a type 2A-related serine-threonine phosphatase that functions in the G1/S transition of the cell cycle (Sutton et al. 1991). In summary, synthetic interactions with SWI6, MSN5, CDC7, and SIT4 strongly support a specific role for TAF9 in regulating the G1/S transition.
Cell polarity/cell integrity:
Related to the above results was the identification of a large number of TAF9-interacting genes having roles in cell polarity, including BEM1, BEM4, CLA4, MDM20, SHE4, STE20, and VRP1. This is consistent with tight coupling of cell polarity with cell cycle control (Lew and Reed 1993) and may be explained, at least in part, by the requirement of G1 cyclins for normal cell morphogenesis (Moffat and Andrews 2004). Remarkably, four of these genes, BEM1, BEM4, CLA4, and STE20, encode proteins that interact during G1 with a key small GTPase, Cdc42, which triggers polarized assembly of the actin cytoskeleton (Chang and Peter 2003).
Coinciding with the processes of cell polarity and cell cycle control is the cell integrity pathway, a Pkc1-mediated MAP kinase signal transduction cascade induced during periods of morphogenetic growth and in response to environmental conditions that jeopardize cell wall stability (Harrison et al. 2004). SWI6 appears to be critical for the function of this pathway (Madden et al. 1997), along with two other genes, BCK1 and SLT2, that were isolated as conditional synthetic interactors with TAF9 (Figure 1). BCK1 and SLT2 encode two key protein kinases, a MAPKKK (MEK kinase) and a MAP kinase (Harrison et al. 2004), respectively, which function as part of a linear pathway downstream of Pkc1. The MAP kinase Slt2, in particular, is required for recruiting the heterodimeric sequence-specific transcription factor SBF (Swi6 and Swi4) to the promoters of G1 cyclin genes under conditions of cell stress, a process that may involve direct interactions between Slt2 and the DNA-binding component of SBF, Swi4 (Baetz et al. 2001). Collectively, synthetic interactions of TAF9 with key genes involved in cell cycle, cell polarity, and cell integrity pathways support a major role of TAF9 in coordinating transcription with cell growth.
Protein synthesis:
A significant number of TAF9-interacting genes that are involved in protein synthesis were identified by SGA, the majority encoding ribosomal proteins, seven of eight of which are 40S subunit specific (RPS24A, RPS6A, RPS10A, RPS10B, RPS18A, RPS19B, RPS27B, and RPL27A; Figure 1). While ribosomal protein genes are commonly obtained as hits in SGA screens, a higher-than-average number obtained with TAF9 raises the question of whether TAF9 may be specifically involved in a process that couples transcription to translation. In this regard, it is noteworthy that genes for 40S ribosomal proteins were among the first S. cerevisiae genes identified as TAF dependent, specifically requiring TAF1 (Shen and Green 1997) and, as shown below, TAF9. Moreover, TFIID is required for transcription of the ribosomal protein gene regulon (Mencia et al. 2002), involving 137 ribosomal protein genes, which accounts for 50% of total mRNA in vivo (Warner 1999). Since most ribosomal proteins are produced from duplicate genes in S. cerevisiae (Warner 1999), strong or conditional synthetic interactions conceivably may derive simply from insufficient abundance of these proteins in the absence of TAF9. Alternatively, since it has been shown that translation is coupled to cell cycle control through the ribosomal proteins Rpl11 and Rpl23 (Dez and Tollervey 2004), perhaps an analogous mechanism couples translation with transcription involving the same or other ribosomal proteins. In this regard, it is noteworthy that, in addition to RPS24A being isolated as a TAF9 interactor (Figure 1), Rps24A was shown by co-immunopurification and mass spectrometry analysis to interact biochemically with TFIID (Sanders et al. 2002).
Protein sorting:
We identified eight different genes interacting with TAF9 whose products play roles related to protein sorting, including DOA4, MNN10, VAC14, VAN1, VMA10, VMA11, VPS3, and VPS18 (Figure 1). Connections between transcription and protein sorting have been described previously. For example, five (Aor1, Vps71, Yaf9, Arp6, and Vps72) of seven nonessential subunits of SWR-C as well as Bdf1 were found in a large-scale screen to identify genes required for vacuolar protein sorting (vps genes; Bonangelino et al. 2002), suggesting that SWR-C is specifically required for the expression of vps genes (Krogan et al. 2003). The fact that TAF9 interacts genetically with eight protein-sorting genes as well as five of the seven nonessential SWR-C genes raises the possibility that TAF9 plays a special role in vps gene expression alongside SWR-C.
Other nuclear/other cytoplasmic:
Four genes whose products localize to the nucleus but whose functions are unclear include FYV6, IWR1, NCE4, and YPR045C. Genes whose products localize to the cytoplasm but whose functions also are unclear include FSH2, ILM1, RIM13, RIM21, YOR322C, and YDR532C and will not be discussed.
Mitochondrial genes interacting synthetically with TAF9 include MRP6, MRP10, MRP20, MSU1, and FYV4. MRP6, MRP10, and MRP20 encode subunits of the mitochondrial ribosome, and strong or conditional synthetic interactions with these three genes raise the question of whether a connection exists between nuclear transcription and mitochondrial protein synthesis. While FYV4 encodes a deduced protein of unknown function, originally isolated by virtue of the hypersensitivity of an fyv4 strain to K1 killer toxin (Page et al. 2003), it was recently shown that both dst1 (TFIIS) and rpb9 (PolII subunit) cause a synthetic phenotype in combination with fyv4 (Malagon et al. 2004). The fact that TAF9 genetically interacts with FYV4, DST1, and RPB4 (Figure 1), and that both DST1 and RPB9 are linked to transcription elongation, supports a role for FYV4 in the control of nuclear transcriptional elongation by RNA PolII.
Spt20 (a SAGA subunit) is required for transcriptional elongation through PDR5:
The large number of TAF9 interactors involving complexes responsible for transcription elongation (Figure 1) raised the question of whether TFIID or SAGA may play a role in transcription elongation in addition to initiation. Although genetic interaction of TAF9 with particular elongation factors in SGA screens could result from a combined block on initiation and elongation of one or more essential genes, it also remained possible that many represent interactions of a more direct nature. Indeed, Berger and colleagues (Ingvarsdottir et al. 2005) recently showed by SGA analysis that genes encoding two SAGA-specific components, Spt3 and Spt8, interact synthetically with genes for many of the same transcription elongation proteins identified here as interacting with TAF9, although not with Mediator or most of the chromatin modification/remodeling complexes (Figure 1). This raises the possibility that TAF9-interacting genes for regulators of transcription elongation mostly represent SAGA-specific interactions, while those for Mediator and most chromatin modification/remodeling complexes largely represent TFIID-specific interactions. Supporting this notion, several studies are consistent with a role of SAGA in elongation (Van Mullem et al. 2002; Rodriguez-Navarro et al. 2004; Wery et al. 2004), including the fact that SAGA physically and functionally interacts with both the transcription elongation factor TFIIS (Wery et al. 2004) and the PolII subunit Rpb9, which has been linked to elongation control (Van Mullem et al. 2002).
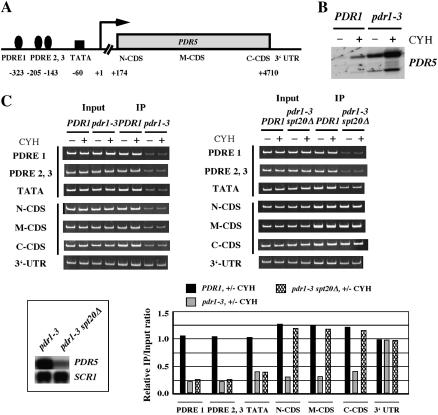
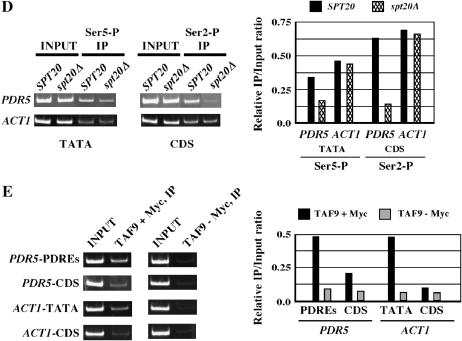
Hence, to further examine possible involvement of SAGA in transcriptional elongation, we used ChIP to analyze the integrity of nucleosomes within the coding sequence of a gene known to be controlled by SAGA, PDR5 (Gao et al. 2004). Previous studies have shown that nucleosomes partially disassemble via the loss of histone H3-H4 tetramers in genes undergoing transcription. This disruption is detected as a loss of signal when epitope-tagged versions of the respective histones, present in place of normal histones, are probed with the corresponding antibodies by ChIP (Kristjuhan and Svejstrup 2004; Lee et al. 2004; Schwabish and Struhl 2004). For our assay we used antibodies against Myc-tagged histone H4, the only source of cellular histone H4 (Gao et al. 2004; see materials and methods), to probe the integrity of nucleosomes in both PDR5 promoter and coding sequences and in a wild-type strain vs. a strain harboring a deletion of SPT20, which is required for the integrity of the SAGA complex (Sterner et al. 1999). Activation of PDR5 requires the binding of the activator protein encoded by PDR1 to three upstream sites called PDREs, shown in Figure 2A. Activation of PDR5 may be increased in two ways: first by the addition of a drug (such as cycloheximide) and second by mutation of the PDR1 activator gene to a constitutively active form, designated pdr1-3. As shown in Figure 2B in a pdr1-3 strain, which is characterized by resistance to drugs such as cycloheximide, PDR5 is expressed at a significantly higher degree than in the corresponding PDR1 strain (with or without drug treatment). This increased expression involves enhanced recruitment of specific transcription coactivators such as SAGA (Gao et al. 2004) and is further associated with the loss of contacts between histone H4 and DNA at both the PDR5 promoter region and coding sequences regardless of the presence of a drug, as shown in Figure 2C (left) (see also Gao et al. 2004). However, in a pdr1-3 spt20Δ strain, the PDR5-coding sequences appear to be associated with wild-type levels of Myc-histone H4, while the promoter region remains depleted (Figure 2C, right, and summary histogram at bottom right). This suggests that Spt20, and likely SAGA, bound at the PDR5 promoter (Gao et al. 2004), is required for the loss of histone-DNA contacts at PDR5-coding sequences, which would likely impact transcription elongation (Gao et al. 2004; Kristjuhan and Svejstrup 2004; Schwabish and Struhl 2004). Supporting this possibility, we found by ChIP analysis that at PDR5-coding sequences, RNA polymerase II containing phosphorylated serine 2 (Ser2) on the CTD of its large subunit is significantly less abundant in an spt20Δ strain than in the corresponding wild-type (SPT20) strain. Ser2 is phosphorylated predominantly during the elongation phase of transcription, while Ser5 is phosphorylated predominantly during the initiation phase (Komarnitsky et al. 2000). This effect of spt20Δ cannot be simply a consequence of defective transcription initiation, since the proportional loss of Ser5-phosphorylated CTD at PDR5 TATA sequences is substantially less than that of Ser2-phosphorylated CTD at PDR5-coding sequences in the corresponding strains (Figure 2D). By contrast, ACT1, which belongs to a class of TFIID-dependent genes (Huisinga and Pugh 2004), demonstrates comparable levels in spt20Δ and SPT20 strains of both Ser5-phosphorylated CTD at ACT1 TATA sequences and Ser2-phosphorylated CTD at ACT1-coding sequences (Figure 2D). We conclude that the function of Spt20, and presumably of SAGA, is not restricted to promoter regions of SAGA-dependent genes but extends to coding sequences as well.
Figure 2.
Spt20 is required for transcriptional elongation through PDR5. (A) Diagram of PDR5, showing the Pdr1 recognition elements (PDREs), TATA box, and coding sequences (CDS) relative to the transcription start site (+1). The promoter and CDS are drawn at different scales. (B) Primer extension analysis of PDR5 mRNA levels in wild type (PDR1) and drug-resistant (pdr1-3) strains with (+) and without (−) cycloheximide (CYH) induction. (C) ChIP of Myc-tagged histone H4 bound at PDR5 sequences in PDR1, pdr1-3, and pdr1-3 spt20Δ strains. Inset (below) shows PDR5 RNA levels in pdr1-3 vs. pdr1-3 spt20Δ strains; SCR1 served as a loading control. Relative immunoprecipitate/input ratio of ChIP experiments was quantified, with the results summarized in the histogram (bottom right). (D) ChIP of phosphorylated serine 5 (at TATA sequences) vs. serine 2 (at coding sequences) of the CTD of the RNA polymerase II large subunit in a wild-type PDR1 strain without CYH induction. PDR5 (a SAGA-dependent gene) and ACT1 (a TFIID-dependent gene) were analyzed. (E) ChIP of C-terminal Myc-tagged TAF9 on the promoters and coding sequences PDR5 and ACT1 in a wild-type PDR1 strain without CYH induction. Accompanying histograms in D and E show quantitation. Standard errors of ChIP experiments were <30%.
To determine how direct a role TAF9 might play in elongation, we compared its recruitment onto promoters vs. coding sequences at PDR5 and ACT1 (Figure 2E). As expected, and consistent with previous analyses of other SAGA subunits (Gao et al. 2004), Myc-tagged TAF9 is recruited more significantly to PDR5 UASs (PDREs) than to PDR5-coding sequences (Figure 2E) and to ACT1 TATA sequence than to ACT1-coding sequences (Figure 2E). Notwithstanding, the level of recruitment of Myc-tagged TAF9 to PDR5-coding sequences is measurably greater than that of the no-Myc tag control, raising the possibility that TAF9 in the context of SAGA contacts the coding sequences of PDR5 as well as promoter. While Myc-tagged TAF9 also showed higher-than-background recruitment to the ACT1-coding sequences, the difference between Myc-tagged TAF9 and the no-Myc tag control was within experimental error (∼30%; Figure 2 legend). This suggests that TAF9, presumably in the context of SAGA, may contact coding sequences as well as promoters, albeit with lower efficiency.
A role for SAGA in transcription elongation may not be unwarranted. For example, SAGA contains two distinct chromatin-modifying activities, histone acetylation involving Gcn5 and histone deubiqutination involving Ubp8, which may be required for transcription elongation of certain genes. In this regard, the Rad6-Bre1-Lge1 complex is required for ubiquitinating histone H2B, and a cycle of histone ubiquitination followed by deubiquitination is required for yeast GAL gene transcription, in which SAGA plays a role (Henry et al. 2003; Xiao et al. 2005). A putative role for SAGA and TAF9 in transcription elongation could involve direct or indirect interactions with coding sequences. Consider the following alternative possibilities: First, SAGA, bound to UASs or core promoters, may recruit elongation factors to the preinitiation complex. By such an indirect mechanism SAGA is thought to recruit, via its Sus1 subunit, the Sac3-Thp1 complex involved in mRNA export (Rodriguez-Navarro et al. 2004). Second, if transcription derives from DNA templates threading through PolII enzymes immobilized at transcription factories (Cook 1999), opportunities for direct cooperation between SAGA and traditional elongation factors may arise following transcription initiation. In this regard, recent identification of the chromodomain protein Chd1 as a subunit of SAGA (Pray-Grant et al. 2005) is of particular interest, since Chd1 has been shown to localize to coding sequences of transcribed genes (Simic et al. 2003) and, moreover, interacts both biochemically and genetically with the elongation factor complex Spt4-Spt5 (DSIF in mammals). Conceivably, TAF9 and Chd1, perhaps occupying neighboring domains of SAGA and bound to a nuclear transcription factory, may provide discrete functions that are temporally but not spatially distinct during the formation of an mRNA transcript. In this manner, TAF9 would be physically associated with Chd1 and functionally associated with various transcription elongation factors, although affiliated itself with promoter sequences. The observation that neither chd1Δ nor spt4Δ exhibits synthetic genetic interaction with taf9-ts2 (Table 1), coupled with the fact that TAF9 is recruited primarily to promoters (Figure 2E) and Chd1 to coding sequences (Simic et al. 2003), suggests that Chd1 and TAF9 play disparate roles in both SAGA function and transcription. Third, SAGA may act in conventional fashion as a coactivator at promoter sequences during preinitiation/initiation and then travel with the transcription machinery down the template during elongation, whereupon Chd1 participates directly and TAF9 mostly indirectly in elongation-related processes. Fourth, TAF9 acts independently of SAGA or TFIID in transcription elongation, perhaps as a component of another protein complex. The low molecular weight of TAF9 (17 kD), for example, may have caused it to elude detection in a protein complex previously affiliated with transcription elongation. Given the fact that the taf9-ts2 mutant used in this study is insensitive to the transcription elongation inhibitor 6-azauracil (data not shown), however, this fourth possibility seems unlikely. Regardless of the molecular mechanism, the culmination of our results, particularly in light of those of others (Ingvarsdottir et al. 2005; Kong et al. 2005), suggests that TAF9 and SAGA contribute to the control of transcription elongation in addition to transcription initiation.
TAF9 and SWR-C are required to express the essential housekeeping gene RPS5:
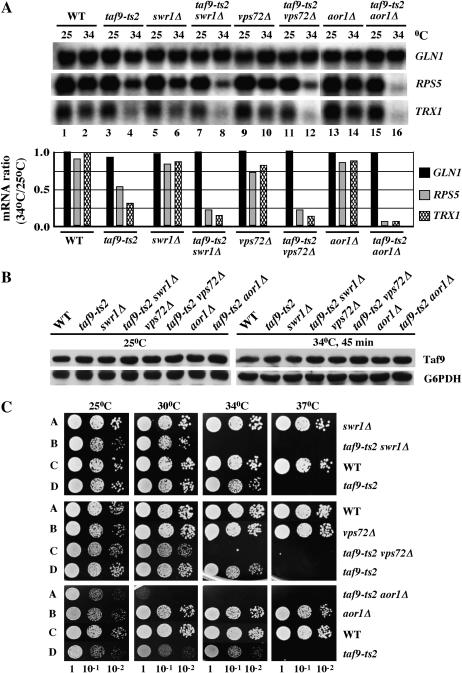
While this work was in progress the SWR-C complex was characterized by three different studies (Krogan et al. 2003; Kobor et al. 2004; Mizuguchi et al. 2004). Due to the large number of hits obtained in our SGA screen involving members of this complex (virtually all nonessential subunits of the 13-subunit complex; Krogan et al. 2003), we wanted to further study its relationship with TAF9. We asked whether the SWR-C complex was required for expressing genes known to require TAF9 for their expression. For this purpose, we analyzed the expression of RPS5 (Shen and Green 1997) and TRX1 (Shen et al. 2003). RPS5 encodes a small ribosomal protein subunit that is essential for cell growth, while TRX1 encodes thioredoxin, which is nonessential. GLN1 (Shen et al. 2003), whose transcription is insensitive to inactivation of TAFs, was used as a control for this experiment. Northern blot analysis was performed with total RNA collected from a taf9-ts2 strain and from strains that harbor null mutations in genes for three different members of the SWR-C complex, SWR1, VPS72, and AOR1. Expression in the corresponding mutant strains was examined before and after a shift in temperature from 25° to 34° (semipermissive for taf9-ts2). Figure 3A shows that RPS5 and TRX1 mRNA levels were only modestly reduced in swr1Δ, vps72Δ, aor1Δ strains or in a corresponding wild-type strain after a shift to the semipermissive temperature (Figure 3A, compare lanes 6, 10, and 14 to lane 2), and mRNA levels were more reduced in the taf9-ts2 strain (Figure 3A, lane 4). However, a synergistic decrease in mRNA levels was observed in double-mutant strains containing taf9-ts2 in combination with swr1Δ, vps72Δ, or aor1Δ (Figure 3A, lanes 8, 12, and 16). Interestingly, this synergistic effect was greater for aor1Δ taf9 than for swr1Δ taf9 or vps72Δ taf9 strains, corresponding with a greater growth defect of the former compared to the latter (Figure 3C). Less of a detectable effect of the double mutations on TRX1 than on RPS5 may be due to the fact that taf9-ts2 has a greater effect on TRX1 relative to RPS5 (Shen et al. 2003). No difference in transcription of GLN1, before or after the temperature shift, was detectable in any of these strains. To test the possibility that the expression of TAF9 may be dependent on SWR-C, we analyzed TAF9 protein levels in single and double mutants by Western blotting. Figure 3B shows that TAF9 protein levels were not affected by deletion of components of SWR-C. The combined results indicate that SWR-C becomes critical for transcription of the essential housekeeping gene RPS5 when the function of TAF9 is compromised. These results support a hypothesis in which one mechanism underlying a synthetic genetic relationship with TAF9 may entail the combined effect of a double block on the expression of a single essential gene. In light of results presented in the preceding section, and in conjunction with the possibility that SWR-C is involved in some aspect of transcription elongation (Santisteban et al. 2000; Krogan et al. 2003), a double block could arise due to defects in initiation plus elongation or defects in elongation only.
Figure 3.
Conditional synthetic genetic interactions between TAF9 and SWR-C. (A) Northern blot analysis showing RPS5 and TRX1 (TAF dependent) as well as GLN1 (TAF independent) RNA levels in single vs. double mutants. (Bottom) Histogram shows respective mRNA ratios (34°/25°). Cells were grown at permissive temperature (25°) with or without shifting to semipermissive temperature (34°) for 45 min. (B) Western blot analysis by rabbit polyclonal antibodies showing TAF9 protein levels for strains indicated in A. Glucose-6-phosphate dehydrogenase (G6PDH) served as a loading control. (C) Genetic interactions between taf9-ts2 and null alleles of SWR1, VPS72, or AOR1 assayed by a 10-fold dilution series spot test on SC media. Images were recorded after 2 days incubation at the indicated temperatures.
Concluding remarks:
Over one hundred genetic interactions with TAF9 were identified, approximately half involving previously characterized transcription factors. The higher-than-average number of genetic interactions, compared to ∼30 for an average nonessential gene (Tong et al. 2004), may reflect the fact that TAF9 is an essential gene required for transcription of ∼60% of the S. cerevisiae genome (Shen et al. 2003). The central role of TAF9 presumably derives not only from the fact that it is present in both TFIID and SAGA, but also from the fact that it is one of several histone-fold TAFs critical for maintaining the structural integrity of these complexes (Moqtaderi et al. 1998; Yatherajam et al. 2003 ; Wu et al. 2004; Timmers and Tora 2005). Considering an estimated 384 nonessential genes involved in transcriptional regulation in S. cerevisiae (Krogan et al. 2003), our identification of ∼50 of them as TAF9 interactors suggests the genetic interactions identified here are nevertheless quite selective (see also Table 1).
Data presented in this study expand the regulatory roles of TAF9 to include transcriptional elongation and general growth control and providing genetic support for specific biochemical interactions reported to occur between various transcription factors and TAF9 or TFIID (Figure 1, factors shown in yellow ovals; Sanders et al. 2002). Significantly, our genetic data in combination with those of Berger and colleagues (Ingvarsdottir et al. 2005) and others (Kong et al. 2005) suggest the possibility that TAF9 involvement in transcription elongation occurs specifically in the context of SAGA. This prediction is supported by the observation that the SAGA subunit Spt20 is required for proper histone-DNA contacts and elongating forms of RNA polymerase II in PDR5-coding region sequences. Extensive genetic interactions of TAF9 with components of SWR-C, whose function is affiliated with transcriptional elongation (Santisteban et al. 2000; Krogan et al. 2003), supports involvement of TAF9 in elongation control, as do physical interactions between TAF9 and all six subunits of Elongator (Sanders et al. 2002), as well as overlapping roles for the HAT activities of SAGA and Elongator (Wittschieben et al. 2000). During preparation of this article, Govind et al. (2005) published data supporting a role for SAGA in regulating transcription elongation.
Acknowledgments
The authors especially thank Dave Amberg for introducing the use of robotic SGA screens at SUNY Upstate Medical University. Constructive input from members of the yeast data club at SUNY Upstate Medical University and Syracuse University is highly appreciated. We also thank several anonymous reviewers for critical comments that improved the manuscript. A Faculty Development Fund and Hendrick's Fund provided by SUNY Upstate Medical University to W.-C.W.S supported this work.
References
- Adkins, M. W., S. R. Howar and J. K. Tyler, 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14: 657–666. [DOI] [PubMed] [Google Scholar]
- Ahn, S. H., M. Kim and S. Buratowski, 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13: 67–76. [DOI] [PubMed] [Google Scholar]
- Albright, S. R., and R. Tjian, 2000. TAFs revisited: more data reveal new twists and confirm old ideas. Gene 242: 1–13. [DOI] [PubMed] [Google Scholar]
- Apone, L. M., C. M. Virbasius, J. C. Reese and M. R. Green, 1996. Yeast TAF(II)90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 10: 2368–2380. [DOI] [PubMed] [Google Scholar]
- Apone, L. M., C. A. Virbasius, F. C. Holstege, J. Wang, R. A. Young et al., 1998. Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol. Cell 2: 653–661. [DOI] [PubMed] [Google Scholar]
- Badarinarayana, V., Y. C. Chiang and C. L. Denis, 2000. Functional interaction of CCR4-NOT proteins with TATAA-binding protein (TBP) and its associated factors in yeast. Genetics 155: 1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baetz, K., J. Moffat, J. Haynes, M. Chang and B. Andrews, 2001. Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4. Mol. Cell. Biol. 21: 6515–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, A., and J. R. Pringle, 1991. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorklund, S., and C. M. Gustafsson, 2005. The yeast Mediator complex and its regulation. Trends Biochem. Sci. 30: 240–244. [DOI] [PubMed] [Google Scholar]
- Bonangelino, C. J., E. M. Chavez and J. S. Bonifacino, 2002. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell 13: 2486–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden, L. L., 2003. Periodic transcription: a cycle within a cycle. Curr. Biol. 13: R31–R38. [DOI] [PubMed] [Google Scholar]
- Brown, C. E., T. Lechner, L. Howe and J. L. Workman, 2000. The many HATs of transcription coactivators. Trends Biochem. Sci. 25: 15–19. [DOI] [PubMed] [Google Scholar]
- Burckin, T., R. Nagel, Y. Mandel-Gutfreund, L. Shiue, T. A. Clark et al., 2005. Exploring functional relationships between components of the gene expression machinery. Nat. Struct. Mol. Biol. 12: 175–182. [DOI] [PubMed] [Google Scholar]
- Casolari, J. M., C. R. Brown, D. A. Drubin, O. J. Rando and P. A. Silver, 2005. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 19: 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadick, J. Z., and F. J. Asturias, 2005. Structure of eukaryotic Mediator complexes. Trends Biochem. Sci. 30: 264–271. [DOI] [PubMed] [Google Scholar]
- Chang, F., and M. Peter, 2003. Yeasts make their mark. Nat. Cell Biol. 5: 294–299. [DOI] [PubMed] [Google Scholar]
- Chimura, T., T. Kuzuhara and M. Horikoshi, 2002. Identification and characterization of CIA/ASF1 as an interactor of bromodomains associated with TFIID. Proc. Natl. Acad. Sci. USA 99: 9334–9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, P. R., 1999. The organization of replication and transcription. Science 284: 1790–1795. [DOI] [PubMed] [Google Scholar]
- Deluen, C., N. James, L. Maillet, M. Molinete, G. Theiler et al., 2002. The Ccr4-not complex and yTAF1 (yTaf(II)130p/yTaf(II)145p) show physical and functional interactions. Mol. Cell. Biol. 22: 6735–6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nadal, E., M. Zapater, P. M. Alepuz, L. Sumoy, G. Mas et al., 2004. The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 427: 370–374. [DOI] [PubMed] [Google Scholar]
- Denis, C. L., Y. C. Chiang, Y. Cui and J. Chen, 2001. Genetic evidence supports a role for the yeast CCR4-NOT complex in transcriptional elongation. Genetics 158: 627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez, C., and D. Tollervey, 2004. Ribosome synthesis meets the cell cycle. Curr. Opin. Microbiol. 7: 631–637. [DOI] [PubMed] [Google Scholar]
- Durso, R. J., A. K. Fisher, T. J. Albright-Frey and J. C. Reese, 2001. Analysis of TAF90 mutants displaying allele-specific and broad defects in transcription. Mol. Cell. Biol. 21: 7331–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynlacht, B. D., T. Hoey and R. Tjian, 1991. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell 66: 563–576. [DOI] [PubMed] [Google Scholar]
- Eide, D. J., 2000. Metal ion transport in eukaryotic microorganisms: insights from Saccharomyces cerevisiae. Adv. Microb. Physiol. 43: 1–38. [DOI] [PubMed] [Google Scholar]
- Ferdous, A., F. Gonzalez, L. Sun, T. Kodadek and S. A. Johnston, 2001. The 19S regulatory particle of the proteasome is required for efficient transcription elongation by RNA polymerase II. Mol. Cell 7: 981–991. [DOI] [PubMed] [Google Scholar]
- Finger, F. P., and P. Novick, 2000. Synthetic interactions of the post-Golgi sec mutations of Saccharomyces cerevisiae. Genetics 156: 943–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, C., L. Wang, E. Milgrom and W. C. Shen, 2004. On the mechanism of constitutive Pdr1 activator-mediated PDR5 transcription in Saccharomyces cerevisiae: evidence for enhanced recruitment of coactivators and altered nucleosome structures. J. Biol. Chem. 279: 42677–42686. [DOI] [PubMed] [Google Scholar]
- Gilbert, C., A. Kristjuhan, G. S. Winkler and J. Q. Svejstrup, 2004. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol. Cell 14: 457–464. [DOI] [PubMed] [Google Scholar]
- Gonzalez, F., A. Delahodde, T. Kodadek and S. A. Johnston, 2002. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science 296: 548–550. [DOI] [PubMed] [Google Scholar]
- Govind, C. K., S. Yoon, H. Qiu, S. Govind and A. G. Hinnebusch, 2005. Simultaneous recruitment of coactivators by Gcn4p stimulates multiple steps of transcription in vivo. Mol. Cell. Biol. 25: 5626–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, P. A., D. Schieltz, M. G. Pray-Grant, D. J. Steger, J. C. Reese et al., 1998. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94: 45–53. [DOI] [PubMed] [Google Scholar]
- Green, M. R., 2000. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. 25: 59–63. [DOI] [PubMed] [Google Scholar]
- Harbison, C. T., D. B. Gordon, T. I. Lee, N. J. Rinaldi, K. D. Macisaac et al., 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, J. C., T. R. Zyla, E. S. Bardes and D. J. Lew, 2004. Stress-specific activation mechanisms for the “cell integrity” MAPK pathway. J. Biol. Chem. 279: 2616–2622. [DOI] [PubMed] [Google Scholar]
- Hartman, J. L., IV, B. Garvik and L. Hartwell, 2001. Principles for the buffering of genetic variation. Science 291: 1001–1004. [DOI] [PubMed] [Google Scholar]
- Henry, K. W., A. Wyce, W. S. Lo, L. J. Duggan, N. C. Emre et al., 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17: 2648–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner et al., 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728. [DOI] [PubMed] [Google Scholar]
- Huisinga, K. L., and B. F. Pugh, 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13: 573–585. [DOI] [PubMed] [Google Scholar]
- Hwang, W. W., S. Venkatasubrahmanyam, A. G. Ianculescu, A. Tong, C. Boone et al., 2003. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11: 261–266. [DOI] [PubMed] [Google Scholar]
- Ingvarsdottir, K., N. J. Krogan, N. C. T. Emre, A. Wyce, N. J. Thompson et al., 2005. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol. Cell. Biol. 25: 1162–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J. S., and L. Prakash, 1990. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast 6: 363–366. [DOI] [PubMed] [Google Scholar]
- Kobor, M. S., S. Venkatasubrahmanyam, M. D. Meneghini, J. W. Gin, J. L. Jennings et al., 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2: E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp, D. M., D. H. Wong, A. H. Corbett and P. A. Silver, 1996. Dynamic localization of the nuclear import receptor and its interactions with transport factors. J. Cell Biol. 133: 1163–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky, P., E. J. Cho and S. Buratowski, 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14: 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, S. E., M. S. Kobor, N. J. Krogan, B. P. Somesh, T. M. Sogaard et al., 2005. Interaction of Fcp1 phosphatase with elongating RNA polymerase II holoenzyme, enzymatic mechanism of action, and genetic interaction with elongator. J. Biol. Chem. 280: 4299–4306. [DOI] [PubMed] [Google Scholar]
- Kornberg, R. D., 2005. a Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30: 235–239. [DOI] [PubMed] [Google Scholar]
- Kornberg, R. D., 2005. b Mediator comes of age. Trends Biochem. Sci. 30: 221. [DOI] [PubMed] [Google Scholar]
- Kristjuhan, A., and J. Q. Svejstrup, 2004. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 23: 4243–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N. J., J. Dover, S. Khorrami, J. F. Greenblatt, J. Schneider et al., 2002. COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 277: 10753–10755. [DOI] [PubMed] [Google Scholar]
- Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan et al., 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12: 1565–1576. [DOI] [PubMed] [Google Scholar]
- Kurdistani, S. K., and M. Grunstein, 2003. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol 4: 276–284. [DOI] [PubMed] [Google Scholar]
- Kurdistani, S. K., S. Tavazoie and M. Grunstein, 2004. Mapping global histone acetylation patterns to gene expression. Cell 117: 721–733. [DOI] [PubMed] [Google Scholar]
- Larschan, E., and F. Winston, 2005. The Saccharomyces cerevisiae Srb8-Srb11 complex functions with the SAGA complex during Gal4-activated transcription. Mol. Cell. Biol. 25: 114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl and J. D. Lieb, 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36: 900–905. [DOI] [PubMed] [Google Scholar]
- Lee, J. M., and A. L. Greenleaf, 1991. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1: 149–167. [PMC free article] [PubMed] [Google Scholar]
- Lee, T. I., H. C. Causton, F. C. Holstege, W. C. Shen, N. Hannett et al., 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405: 701–704. [DOI] [PubMed] [Google Scholar]
- Lei, E. P., H. Krebber and P. A. Silver, 2001. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 15: 1771–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, E. P., C. A. Stern, B. Fahrenkrog, H. Krebber, T. I. Moy et al., 2003. Sac3 is an mRNA export factor that localizes to cytoplasmic fibrils of nuclear pore complex. Mol. Biol. Cell 14: 836–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, M., and B. K. Tye, 2001. Initiating DNA synthesis: from recruiting to activating the MCM complex. J. Cell Sci 114: 1447–1454. [DOI] [PubMed] [Google Scholar]
- Lew, D. J., and S. I. Reed, 1993. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 120: 1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Macpherson, N., V. Measday, L. Moore and B. Andrews, 2000. A yeast taf17 mutant requires the Swi6 transcriptional activator for viability and shows defects in cell cycle-regulated transcription. Genetics 154: 1561–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden, K., Y. J. Sheu, K. Baetz, B. Andrews and M. Snyder, 1997. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science 275: 1781–1784. [DOI] [PubMed] [Google Scholar]
- Malagon, F., A. H. Tong, B. K. Shafer and J. N. Strathern, 2004. Genetic interactions of DST1 in Saccharomyces cerevisiae suggest a role of TFIIS in the initiation-elongation transition. Genetics 166: 1215–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T., and R. Reed, 2002. An extensive network of coupling among gene expression machines. Nature 416: 499–506. [DOI] [PubMed] [Google Scholar]
- Mencia, M., Z. Moqtaderi, J. V. Geisberg, L. Kuras and K. Struhl, 2002. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell 9: 823–833. [DOI] [PubMed] [Google Scholar]
- Meneghini, M. D., M. Wu and H. D. Madhani, 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112: 725–736. [DOI] [PubMed] [Google Scholar]
- Menon, B. B., N. J. Sarma, S. Pasula, S. J. Deminoff, K. A. Willis et al., 2005. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc. Natl. Acad. Sci. USA 102: 5749–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, B., P. Komarnitsky and S. Buratowski, 1998. Histone-like TAFs are essential for transcription in vivo. Mol. Cell 2: 663–673. [DOI] [PubMed] [Google Scholar]
- Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen et al., 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348. [DOI] [PubMed] [Google Scholar]
- Moffat, J., and B. Andrews, 2004. Late-G1 cyclin-CDK activity is essential for control of cell morphogenesis in budding yeast. Nat. Cell Biol. 6: 59–66. [DOI] [PubMed] [Google Scholar]
- Moqtaderi, Z., M. Keaveney and K. Struhl, 1998. The histone H3-like TAF is broadly required for transcription in yeast. Mol. Cell 2: 675–682. [DOI] [PubMed] [Google Scholar]
- Narlikar, G. J., H. Y. Fan and R. E. Kingston, 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475–487. [DOI] [PubMed] [Google Scholar]
- Page, N., M. Gerard-Vincent, P. Menard, M. Beaulieu, M. Azuma et al., 2003. A Saccharomyces cerevisiae genome-wide mutant screen for altered sensitivity to K1 killer toxin. Genetics 163: 875–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, R., and H. Song, 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11: 121–127. [DOI] [PubMed] [Google Scholar]
- Pray-Grant, M. G., D. Schieltz, S. J. McMahon, J. M. Wood, E. L. Kennedy et al., 2002. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol. Cell. Biol. 22: 8774–8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant, M. G., J. A. Daniel, D. Schieltz, J. R. Yates, III, and P. A. Grant, 2005. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433: 434–438. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N., 2004. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 16: 272–278. [DOI] [PubMed] [Google Scholar]
- Queralt, E., and J. C. Igual, 2003. Cell cycle activation of the Swi6p transcription factor is linked to nucleocytoplasmic shuttling. Mol. Cell. Biol. 23: 3126–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese, J. C., and M. R. Green, 2001. Genetic analysis of TAF68/61 reveals links to cell cycle regulators. Yeast 18: 1197–1205. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro, S., T. Fischer, M. J. Luo, O. Antunez, S. Brettschneider et al., 2004. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116: 75–86. [DOI] [PubMed] [Google Scholar]
- Russnak, R., K. W. Nehrke and T. Platt, 1995. REF2 encodes an RNA-binding protein directly involved in yeast mRNA 3′-end formation. Mol. Cell. Biol. 15: 1689–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, S. L., J. Jennings, A. Canutescu, A. J. Link and P. A. Weil, 2002. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22: 4723–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santisteban, M. S., T. Kalashnikova and M. M. Smith, 2000. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103: 411–422. [DOI] [PubMed] [Google Scholar]
- Sarin, S., K. E. Ross, L. Boucher, Y. Green, M. Tyers et al., 2004. Uncovering novel cell cycle players through the inactivation of securin in budding yeast. Genetics 168: 1763–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish, M. A., and K. Struhl, 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 24: 10111–10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W. C., and M. R. Green, 1997. Yeast TAF(II)145 functions as a core promoter selectivity factor, not a general coactivator. Cell 90: 615–624. [DOI] [PubMed] [Google Scholar]
- Shen, W. C., S. R. Bhaumik, H. C. Causton, I. Simon, X. Zhu et al., 2003. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J. 22: 3395–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., 1991. Getting started with yeast. Methods Enzymol. 194: 3–21. [DOI] [PubMed] [Google Scholar]
- Shibuya, T., S. Tsuneyoshi, A. K. Azad, S. Urushiyama, Y. Ohshima et al., 1999. Characterization of the ptr6(+) gene in fission yeast: a possible involvement of a transcriptional coactivator TAF in nucleocytoplasmic transport of mRNA. Genetics 152: 869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic, R., D. L. Lindstrom, H. G. Tran, K. L. Roinick, P. J. Costa et al., 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22: 1846–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims, R. J., III, R. Belotserkovskaya and D. Reinberg, 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18: 2437–2468. [DOI] [PubMed] [Google Scholar]
- Squazzo, S. L., P. J. Costa, D. L. Lindstrom, K. E. Kumer, R. Simic et al., 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21: 1764–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya et al., 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19: 86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner, D. E., R. Belotserkovskaya and S. L. Berger, 2002. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc. Natl. Acad. Sci. USA 99: 11622–11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L., S. A. Johnston and T. Kodadek, 2002. Physical association of the APIS complex and general transcription factors. Biochem. Biophys. Res. Commun. 296: 991–999. [DOI] [PubMed] [Google Scholar]
- Sutton, A., D. Immanuel and K. T. Arndt, 1991. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol. Cell. Biol. 11: 2133–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taatjes, D. J., M. T. Marr and R. Tjian, 2004. Regulatory diversity among metazoan co-activator complexes. Nat. Rev. Mol. Cell Biol. 5: 403–410. [DOI] [PubMed] [Google Scholar]
- Timmers, H. T., and L. Tora, 2005. SAGA unveiled. Trends Biochem. Sci. 30: 7–10. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader et al., 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu et al., 2004. Global mapping of the yeast genetic interaction network. Science 303: 808–813. [DOI] [PubMed] [Google Scholar]
- Tora, L., 2002. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 16: 673–675. [DOI] [PubMed] [Google Scholar]
- Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson et al., 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627. [DOI] [PubMed] [Google Scholar]
- Van Mullem, V., M. Wery, M. Werner, J. Vandenhaute and P. Thuriaux, 2002. The Rpb9 subunit of RNA polymerase II binds transcription factor TFIIE and interferes with the SAGA and elongator histone acetyltransferases. J. Biol. Chem. 277: 10220–10225. [DOI] [PubMed] [Google Scholar]
- Walker, S. S., W. C. Shen, J. C. Reese, L. M. Apone and M. R. Green, 1997. Yeast TAF(II)145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell 90: 607–614. [DOI] [PubMed] [Google Scholar]
- Warner, J. R., 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24: 437–440. [DOI] [PubMed] [Google Scholar]
- Wery, M., E. Shematorova, B. Van Driessche, J. Vandenhaute, P. Thuriaux et al., 2004. Members of the SAGA and Mediator complexes are partners of the transcription elongation factor TFIIS. EMBO J. 23: 4232–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston, F., 2001. Control of eukaryotic transcription elongation. Genome Biol. 2: REVIEWS1006. [DOI] [PMC free article] [PubMed]
- Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]
- Wittschieben, B. O., J. Fellows, W. Du, D. J. Stillman and J. Q. Svejstrup, 2000. Overlapping roles for the histone acetyltransferase activities of SAGA and elongator in vivo. EMBO J. 19: 3060–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, A., J. Schneider, J. Dover, M. Johnston and A. Shilatifard, 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278: 34739–34742. [DOI] [PubMed] [Google Scholar]
- Wu, P. Y., C. Ruhlmann, F. Winston and P. Schultz, 2004. Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell 15: 199–208. [DOI] [PubMed] [Google Scholar]
- Xiao, T., C. F. Kao, N. J. Krogan, Z. W. Sun, J. F. Greenblatt et al., 2005. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25: 637–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatherajam, G., L. Zhang, S. M. Kraemer and L. A. Stargell, 2003. Protein-protein interaction map for yeast TFIID. Nucleic Acids Res. 31: 1252–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabaronick, S. R., and J. K. Tyler, 2005. The histone chaperone anti-silencing function 1 is a global regulator of transcription independent of passage through S phase. Mol. Cell. Biol. 25: 652–660. [DOI] [PMC free article] [PubMed] [Google Scholar]