Abstract
We describe an importance-sampling method for approximating likelihoods of population parameters based on multiple summary statistics. In this first application, we address the demographic history of closely related members of the Drosophila pseudoobscura group. We base the maximum-likelihood estimation of the time since speciation and the effective population sizes of the extant and ancestral populations on the pattern of nucleotide variation at DPS2002, a noncoding region tightly linked to a paracentric inversion that strongly contributes to reproductive isolation. Consideration of summary statistics rather than entire nucleotide sequences permits a compact description of the genealogy of the sample. We use importance sampling first to propose a genealogical and mutational history consistent with the observed array of summary statistics and then to correct the likelihood with the exact probability of the history determined from a system of recursions. Analysis of a subset of the data, for which recursive computation of the exact likelihood was feasible, indicated close agreement between the approximate and exact likelihoods. Our results for the complete data set also compare well with those obtained through Metropolis-Hastings sampling of fully resolved genealogies of entire nucleotide sequences.
ESTIMATION of population parameters in classical population genetics has traditionally proceeded through a moments approach, based on closed-form expressions for the expectation and variance of summary statistics (reviewed by Hey and Machado 2003; Beaumont 2004). For the purposes of this discussion, we regard as a summary statistic any measure of genetic variation that can be determined directly from numbers of derived mutations observed in a sample without explicit reference to the genealogy of the sampled genes.
Felsenstein (1992) proposed a maximum-likelihood (ML) approach to the estimation of population parameters, treating the intervals separating the nodes of the sample genealogy as missing data. He showed that ML methods that use knowledge of the node intervals scaled to the mutation rate have greater statistical power than classical moments-based methods that use summary statistics. Metropolis-Hastings (MH) sampling to simulate the posterior distribution of the gene genealogy based on entire nucleotide sequences now forms the basis of the dominant ML and Bayesian approaches to both population parameter estimation and phylogeny reconstruction (Kuhner et al. 1995; Huelsenbeck and Ronquist 2001).
Among the appealing properties of estimation procedures based on summary statistics are greater simplicity and potential for customization to specific biological systems. While computation of summary statistics by definition requires no knowledge of the genealogy of the sample, the intimate relationship between the pattern of segregating variation and the sample genealogy lies at the core of coalescence theory and molecular population genetics (Ewens 1972; Watterson 1975). In deriving not only the first two moments but also entire probability distributions of summary statistics, those works laid the basis for parameter estimation through likelihoods as well as for the parametric tests for which they are well known. Our objective here is to develop a likelihood-based method for inferring demographic history from multiple summary statistics.
Classes of segregating mutations:
Wakeley and Hey (1997) introduced a set of summary statistics suitable for exploring the demographic history of two groups: numbers of sites polymorphic in both groups, polymorphic in only one group, or showing fixed differences. Our classification expands theirs by using an outgroup to distinguish between ancestral and derived bases (see materials and methods). With respect to the genes sampled from a given group, we describe each segregating derived mutation as absent (a), segregating (s; present in at least one but not all), or fixed (f; present in all). A joint classification with respect to two groups comprises only seven types because segregating mutations are neither absent nor fixed in both groups.
Distinguishing between the effects of recent divergence and introgression after divergence on the sharing of polymorphisms between closely related species can be difficult, particularly in single-locus analyses (Wang et al. 1997). To address the time since divergence alone, we chose to study variation at DPS2002, a noncoding region for which tight linkage to a paracentric inversion strongly associated with multiple reproductive isolating mechanisms precludes its introgression between the closely related Drosophila species Drosophila pseudoobscura and D. persimilis (Noor and Smith 2000; Noor et al. 2001a,b) (see materials and methods).
Table 1 shows the array of mutations observed in DPS2002 haplotypes sampled from D. persimilis and two subspecies of D. pseudoobscura, D. p. pseudoobscura and D. p. bogotana (Machado et al. 2002). Consistent with the absence of introgression at DPS2002, the D. persimilis sample shares no mutations with the D. p. bogotana sample and only one (f/s) with the D. p. pseudoobscura sample. In contrast, the two subspecies of D. pseudoobscura share 10 mutations, suggesting recent divergence or ongoing gene flow. In the interspecific comparison involving D. p. bogotana (per/bog), the observation of both reciprocal arrangements of fixed differences (f/a and a/f) indicates that the most recent common ancestor (MRCA) of genes sampled from each species postdates any between-species coalescence events: the DPS2002 gene genealogy shows reciprocal monophyly. While the reciprocal numbers of DPS2002 polymorphisms restricted to one species (s/a and a/s) appear similar between D. p. bogotana and D. persimilis, D. p. pseudoobscura shows fourfold more polymorphisms than D. persimilis, perhaps reflecting larger effective population size. These observations suggest that the numbers of the various kinds of mutations segregating in the sample contain information about genealogical and demographic history.
TABLE 1.
Segregating mutations
| Type | Group 1 | Group 2 | pera/pseb | per/bogc | bog/pse |
|---|---|---|---|---|---|
| 1 | Segregating | Absent | 16 | 16 | 9 |
| 2 | Fixed | Absent | 5 | 6 | 0 |
| 3 | Absent | Segregating | 65 | 18 | 57 |
| 4 | Absent | Fixed | 0 | 2 | 0 |
| 5 | Segregating | Segregating | 0 | 0 | 9 |
| 6 | Fixed | Segregating | 1 | 0 | 1 |
| 7 | Segregating | Fixed | 0 | 0 | 0 |
D. persimilis, 13 sequences.
D. p. pseudoobscura, 19 sequences.
D. p. bogotana, 13 sequences.
Models of speciation:
Under the Dobzhansky-Muller scenario for the origin of species (reviewed by Turelli et al. 2001), genetic factors contributing to reproductive isolation arise during an initial phase of allopatry between the incipient species. We identify species divergence with the onset of the allopatric phase and not in particular with the origin of genetic isolating mechanisms. Speciation corresponds to a change in coalescence structure, the most recent point at which ancestral lineages with descendants in different species can have coalesced.
We assume that the paracentric inversion that precludes introgression at DPS2002 between D. pseudoobscura and D. persimilis arose as a neutral mutation and became associated with isolating barriers during the allopatric phase. That this second chromosome region is homosequential in D. pseudoobscura and outgroup D. miranda (Dobzhansky and Tan 1936) indicates that it is the D. persimilis arrangement that is derived. This inversion appears to correspond to a fixed difference, not only in our sample but also between the species (Dobzhansky and Powell 1975). For simplicity, we assume instantaneous fixation of the alternative gene orders in the two descendant species, with the inversion having arisen immediately before the MRCA of the sampled inverted chromosomes.
Likelihoods from summary statistics:
Marjoram et al. (2003) introduced a Markov chain Monte Carlo (MCMC) method that directly approximates the posterior distribution of the model, obviating the need to determine the probability of the data. Numerically generating genealogical and mutational histories that match the data may require extensive simulation.
We address methods that base inferences about demographic history and population parameters (M) on a set of summary statistics (D). Implementation of likelihood-based approaches entails the development of a practical means of determining or approximating probability distributions that depend on the unknown genealogical history (G) of the sampled genes and the number and location (U) of mutations that occurred on it,
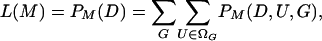 |
(1) |
for ΩG, the set of all possible placements of mutations on a given G. We adopt the infinite-sites model, under which neutral base substitutions occur independently, with the number in a given time interval following a Poisson distribution. Many likelihood-based methods proceed from the prior distribution of the genealogy, dependent on the population parameters alone, while others, including fully Bayesian approaches (Wilson and Balding 1998), incorporate the posterior distribution of histories given D (reviewed by Stephens and Donnelly 2000).
With respect to time since divergence of populations, exact analytical expressions are available for certain simple demographic histories (Watterson 1985; Takahata et al. 1995); however, the derivation of closed-form solutions is intractable for most systems of biological interest. Uyenoyama and Takebayashi (2004) described a method for the recursive determination of exact likelihoods from joint probability-generating functions of correlated summary statistics (compare Griffiths and Tavaré 1996). Takebayashi et al. (2004) used this approach to obtain a maximum-likelihood estimate (MLE) of the rate of recombination between a determinant of mating type in a flowering plant and a closely linked marker locus from the numbers of segregating sites at the two loci. While this method can in principle accommodate multiple summary statistics and general evolutionary models, the computation time makes its application impractical.
Likelihoods have been approximated by simulating complete genealogical and mutational histories under a coalescent model and computing summary statistics for each realization (for example, Fu and Li 1997; Weiss and von Haeseler 1998; Pritchard et al. 1999). While full simulation can accommodate a wide range of evolutionary questions, analysis of even small data sets under simple models may impose a considerable computational burden (Wall et al. 2002; Wall 2003), reflecting the exceedingly low probability of any particular realization (Stephens and Donnelly 2000; Marjoram et al. 2003).
Tavaré et al. (1997) introduced an acceptance-rejection algorithm for the estimation of the age of the MRCA of a sample of genes that entails simulating a gene genealogy and accepting the value for this random variable on the basis of the probability for the tree of the observed total number of segregating mutations. Beaumont et al. (2002) used a local regression method to approximate the likelihood of the actual data from realizations within a certain tolerance (Estoup et al. 2004; Tallmon et al. 2004; Hamilton et al. 2005).
Alternatively, the “fixed-S” approach (e.g., Hudson 1993; Depaulis and Veuille 1998) entails generating a genealogy under a standard coalescent model, randomly placing the observed total number of segregating sites (S) on the genealogy, and determining the fraction of outcomes consistent with the remaining observed summary statistics (H, for D = {S, H}). Markovtsova et al. (2001) showed that the distribution generated by this procedure does not in fact approximate PM(H|S), the conditional distribution of H given S. In particular, the genealogy should be sampled, not from a prior distribution determined by the standard coalescent, but from a conditional distribution, given S and the model parameters M. This discrepancy can become problematic under large departures of the observed value of S from its expectation (Depaulis et al. 2001; Markovtsova et al. 2001; Wall and Hudson 2001).
Approach through importance sampling:
We develop an importance-sampling (IS) approximation (see Liu 2001) to the likelihood (1). We use a proposal distribution to sample a genealogical and mutational history consistent with the observed array of seven types of segregating sites and then correct the bias by determining the exact probability of the history.
Here, a genealogical path (G) corresponds to an ordered list of the states assumed by the process at the nodes of the full genealogy, without specification of the mutations. Observation of the segregating sites present in a sample (D) provides multiple kinds of information,
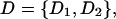 |
for D1 the types and D2 the numbers of base substitutions observed. We rewrite (1) as
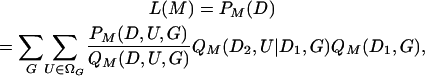 |
(2) |
for QM(D1, G), the stationary distribution of genealogies compatible with D1, and QM(D2, U|D1, G), a heuristic distribution of placements of mutations on G compatible with D2. We approximate this average (2) by
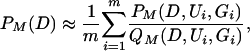 |
(3) |
for (Ui, Gi) independent and identically distributed (i.i.d.) samples from the proposal density QM(D2, U|D1, G)QM(D1, G).
Likelihoods approximated through this procedure may serve as the basis of either Bayesian or maximum-likelihood analyses. Here, we use IS to determine MLEs of the time since divergence between closely related species of Drosophila and the effective population sizes of the extant and ancestral species.
MATERIALS AND METHODS
Sequence information:
We studied the pattern of nucleotide variation segregating among DPS2002 sequences obtained by Machado et al. (2002) from the D. pseudoobscura species group, including 19 D. p. pseudoobscura, 13 D. p. bogotana, and 13 D. persimilis sequences (GenBank nos. AF450689–AF450734). This region, ∼940 bp in length, shows numerous single-nucleotide polymorphisms and variable oligonucleotide repetitive motif tracts, but no detectable open-reading frames. For each site segregating within the ingroup, we assumed that the base present in the single D. miranda sequence represented the ancestral base.
Although Machado et al. (2002) localized DPS2002 within a fixed paracentric inversion that distinguishes D. persimilis from D. pseudoobscura, the Noor group has recently determined that it lies ∼1.5 Mb outside this inversion, on the telomeric side (M. A. F. Noor, unpublished data). In general, recombination in inversion heterozygotes appears to be suppressed beyond the bounds of inversion breakpoints, perhaps reflecting disruption of chromosome pairing or production of unbalanced gametes upon crossing over (see Navarro et al. 1997 and references therein). In particular, DPS2002 has been shown to be tightly linked to the inverted region in inversion heterozygotes (0/357 recombinants) (Noor and Smith 2000).
In the present study, we assume complete linkage of sites within DPS2002 and of DPS2002 to the inversion. Association of the inversion with multiple reproductive isolating mechanisms, including hybrid male sterility, hybrid inviability, hybrid male courtship dysfunction, and behavioral isolation, prevents introgression of linked regions, including DPS2002 (Noor and Smith 2000; Noor et al. 2001a,b).
Classification of mutations:
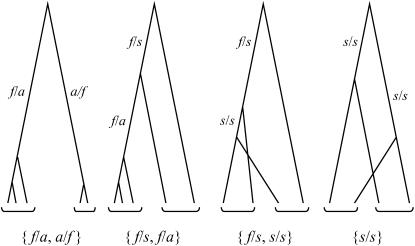
Our assumption of the absence of intragenic recombination entails that all sites within a locus share a single genealogical history. This common history constrains the observed array of neutral, independent mutations, 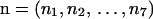 , for ni the number of mutations of type i (Table 2). For example, the observation of a mutation fixed in group 1 and absent from group 2 implies a topology in which the MRCA of the haplotypes sampled from group 1 postdates all between-group coalescence events. This topology excludes the presence in group 2 of mutations segregating in group 1 (s/s and s/f). With the exception of types 1 and 3, the presence of each mutational type excludes the presence of two other types, implying observation of a maximum of four distinct types of mutations. Figure 1 illustrates the four possible distinguishable topologies (modulo reciprocals) assumed by the sample genealogy: {f/a, a/f}, {f/s, f/a}, {f/s, s/s}, and {s/s}.
, for ni the number of mutations of type i (Table 2). For example, the observation of a mutation fixed in group 1 and absent from group 2 implies a topology in which the MRCA of the haplotypes sampled from group 1 postdates all between-group coalescence events. This topology excludes the presence in group 2 of mutations segregating in group 1 (s/s and s/f). With the exception of types 1 and 3, the presence of each mutational type excludes the presence of two other types, implying observation of a maximum of four distinct types of mutations. Figure 1 illustrates the four possible distinguishable topologies (modulo reciprocals) assumed by the sample genealogy: {f/a, a/f}, {f/s, f/a}, {f/s, s/s}, and {s/s}.
TABLE 2.
Incompatibilities
| Type | Group 1/group 2 | Incompatible | Compatible |
|---|---|---|---|
| 1 | s/a | None | All |
| 2 | f/a | s/s, s/f | a/f or f/s |
| 3 | a/s | None | All |
| 4 | a/f | s/s, f/s | f/a or s/f |
| 5 | s/s | f/a, a/f | f/s or s/f |
| 6 | f/s | a/f, s/f | f/a or s/s |
| 7 | s/f | f/a, f/s | a/f or s/s |
Figure 1.—
Possible topologies of gene genealogies of a nonrecombining region. Brackets below the trees indicate sequences sampled from the same group. Branch labels indicate branch type (Table 2).
Summary statistics:
We used CLUSTAL-W (Thompson et al. 1994), with minor manual modifications, to generate a multiple alignment of the 46 DPS2002 sequences. After elimination of sites with gaps in any of the sequences, 892 homologous sites remained. For each of three triads (outgroup D. miranda and a pair of ingroup taxa), we restricted attention to sites at which more than one base segregated within the ingroup taxa under consideration. In accordance with an infinite-sites model of mutation, we designated the base in the outgroup sequence as ancestral and any other base as a mutation. After removing sites at which the ancestral base was absent from both ingroup taxa, we counted the number of mutations in each category shown in Table 1. For sites at which more than two bases segregated, both derived bases contributed to our mutation counts: for example, at a site at which C was designated ancestral, C and G segregated in the sample from one ingroup taxon, and C and T in the other, we counted each mutation (G and T) as segregating in one group and absent in the other. A Bioperl (Stajich et al. 2002) script for counting the various types of mutations is available from its author on request (J.E.S.: jason.stajich@duke.edu).
METHODS OF ESTIMATION
We present an evolutionary model for the demographic history of a sample of haplotypes from two species. We then construct a recursion in the exact probability of the observed array of the seven kinds of segregating sites under this model and describe its importance-sampling approximation (3).
Evolutionary model:
We assume that the time since divergence of species 1 and 2 from ancestral species 0 follows an exponential distribution with parameter λ, treating λ as the rate of species fusion. During the interval spanned by the gene genealogy, species i (i = 0, 1, 2) maintains a constant effective size of Ni genes (for autosomal regions, twice the effective number of individuals).
At any point within the gene genealogy of a sample, we classify each lineage according to its species membership and the distribution of its descendants between the two groups of the initial sample. A type 1 lineage has descendants among the genes sampled from species 1 but not from species 2, and type 2 has those from species 2 but not from species 1. Type 3 lineages have descendants in both groups. On level l of the gene genealogy (the interval in which a total of l ancestral lineages remain), the state or configuration of the process corresponds to (l01, l02, l03, l11, l22), for lij, the number of ancestral lineages of type j (j = 1, 2, 3) in species i (i = 0, 1, 2).
Speciation corresponds to a change in population structure: transition from two isolated groups to a single panmictic group. Our assumption of the instantaneous fixation of the alternative chromosomal types upon speciation entails that postspeciation coalescence events occur at rate
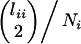 |
(i = 1, 2), in which
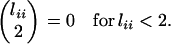 |
Our speciation scenario stipulates the origin of the inversion immediately before the MRCA of the D. persimilis subsample. For cases in which its origin predated the speciation event, we assume that it segregated in the ancestral species at frequency p. During the interval between the speciation event and the MRCA of the inverted chromosomes, coalescence among type 1 lineages occurs at rate
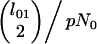 |
and among type 2 lineages at rate
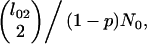 |
with no between-type coalescence events. Upon the origin of the inversion, all pairs of lineages coalesce at the same rate (1/N0), irrespective of type.
A genealogical path G corresponds to an ordered list of descriptions of the nodes of the full genealogy, without specification of mutations or within-level transitions,
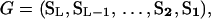 |
(4) |
for 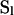 , the entry (most recent) state on level l, and L total sample size. For a sample comprising L1 haplotypes from species 1 and L2 from species 2 (L = L1 + L2), the initial state
, the entry (most recent) state on level l, and L total sample size. For a sample comprising L1 haplotypes from species 1 and L2 from species 2 (L = L1 + L2), the initial state 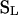 corresponds to (0, 0, 0, L1, L2) and the MRCA of the entire sample
corresponds to (0, 0, 0, L1, L2) and the MRCA of the entire sample  to (0, 0, 1, 0, 0). Characterization of the stationary distribution of genealogical paths requires only determination of Markov matrices of within- and between-level transition rates (appendix a). We extend the procedure introduced by Wiuf and Donnelly (1999) to condition the gene genealogies proposed in the IS procedure to have a topology compatible with the observed combination of mutational types (appendix b). Restricting sampling to compatible genealogies affords a considerable increase in efficiency for a modest computational cost.
to (0, 0, 1, 0, 0). Characterization of the stationary distribution of genealogical paths requires only determination of Markov matrices of within- and between-level transition rates (appendix a). We extend the procedure introduced by Wiuf and Donnelly (1999) to condition the gene genealogies proposed in the IS procedure to have a topology compatible with the observed combination of mutational types (appendix b). Restricting sampling to compatible genealogies affords a considerable increase in efficiency for a modest computational cost.
Exact recursion in likelihoods:
We derive a recursion in the joint probability-generating function (PGF) for the array of summary statistics.
Array of mutations in the sample:
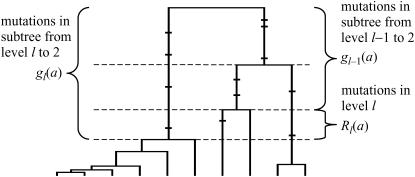
For each configuration on level l, we determine a PGF for the array of mutations accumulated in the subtree extending from level l to the MRCA. Let 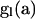 denote the vector of these PGFs, for
denote the vector of these PGFs, for 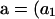 , a2, a3, a4, a5, a6, a7) comprising PGF parameters corresponding to the seven types of segregating mutations (Table 1). Figure 2 illustrates that the total number of mutations accumulated within the subtree beginning on level l corresponds to the sum of numbers accumulated on level l and in the subtree beginning on level l − 1:
, a2, a3, a4, a5, a6, a7) comprising PGF parameters corresponding to the seven types of segregating mutations (Table 1). Figure 2 illustrates that the total number of mutations accumulated within the subtree beginning on level l corresponds to the sum of numbers accumulated on level l and in the subtree beginning on level l − 1:
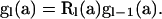 |
(5) |
appendix a presents the derivation of 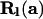 , the PGF of mutations accumulated within level l. Recursion (5) has initial condition
, the PGF of mutations accumulated within level l. Recursion (5) has initial condition
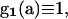 |
reflecting that mutations in the MRCA do not segregate in the sample. The PGF of the numbers of the seven types of mutations observed in a sample of size L corresponds to
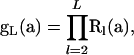 |
in which the matrix product begins on the left with the largest index value.
Figure 2.—
Recursion in probability-generating functions. Mutations in the subtree extending from the MRCA to level l comprise those occurring within level l and those in the subtree extending to level l − 1. Independence of the mutational process in these disjunct time periods implies that the PGFs of mutation numbers to level l correspond to the product of PGFs of mutations to level l − 1 and within level l (5).
Recursive computation of exact likelihoods:
We determine likelihoods from a recursion in probabilities rather than in the PGFs themselves. Taking derivatives of (5), we obtain an expression for the probability of observing the array 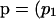 , p2, … , p7) of mutations in the subtree extending from level l to the MRCA,
, p2, … , p7) of mutations in the subtree extending from level l to the MRCA,
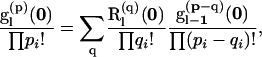 |
(6) |
in which q denotes the array of mutations that arose within level l and p − q the remaining mutations; the summation in q runs over all possible subsets of the total array p; and superscripts of the form (p) indicate the order of derivatives with respect to the parameters representing the mutational types (the pith derivative with respect to ai, i = 1, 2, … , 7).
We initialize the recursion in probabilities (6) by considering all possible assignments of mutations to level 2,
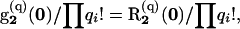 |
for q, a subset of the observed mutations n. We then determine  for all possible mutational arrays p that can occur in the subtree comprising levels 2 and 3. This recursion ends with
for all possible mutational arrays p that can occur in the subtree comprising levels 2 and 3. This recursion ends with 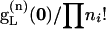 , the probability of the array of mutations observed in the initial sample.
, the probability of the array of mutations observed in the initial sample.
Importance-sampling approximation:
To approximate (2), we first sample from an analytical stationary distribution a genealogical path G (4) consistent with the types of mutations observed in the sample (D1) as well as the speciation scenario, place the observed numbers of mutations n along G according to a heuristic distribution, and then correct the bias introduced by the proposal, using the exact probability of the genealogical and mutational history.
Genealogical path:
Given entry state Sl on level l, the stationary distribution of the level l − 1 entry state Sl−1 is given by the corresponding row of
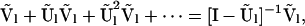 |
(7) |
for Ũl and Ṽl matrices of rates of within- and between-level transition probabilities (appendix a), conditional on the observed types of mutations (D1; appendix b). Beginning with the state of the initial sample (0, 0, 0, L1, L2), we construct a genealogical path Gi by sampling, for each successive level until termination in the MRCA, a path segment from the row of (7) that corresponds to the entry state for the level.
Proposal distribution:
Given a genealogical path Gi, we propose placements of the observed mutations according to a multinomial distribution,
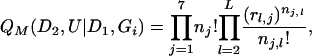 |
in which nj,l represents the number of type j mutations on level l (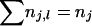 ) and rl,j the probability that a lineage on level l receives a type j mutation. For el,j the number of lineages on level l that are eligible to receive mutations of type j,
) and rl,j the probability that a lineage on level l receives a type j mutation. For el,j the number of lineages on level l that are eligible to receive mutations of type j,
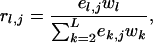 |
(8) |
in which wl represents the relative weight assigned to level l. Weight wl reflects the expected duration of level l, which has an exponential distribution with parameter corresponding to the rate of coalescence within the level (appendix c).
For the path segment corresponding to level l, we obtain the true probability PM(D, Ui,l, Gi,l) from the element of 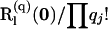 (A8) in the row and column associated with the entry (most recent) states on levels l and l − 1, respectively, for q, the array of mutations assigned to level l.
(A8) in the row and column associated with the entry (most recent) states on levels l and l − 1, respectively, for q, the array of mutations assigned to level l.
Likelihood function:
Griffiths and Tavaré (1994) described an importance-sampling procedure for generating likelihoods of arbitrary models (M) from those obtained under a particular driving model (M0). This approach entails first intensively sampling genealogical paths and placements of mutations under M0 and then characterizing the entire likelihood function by rescaling the probabilities (see Griffiths and Tavaré 1994; Kuhner et al. 1995; Felsenstein et al. 1999; Stephens and Donnelly 2000). For (Ui, Gi) i.i.d. samples from 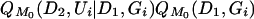 , we approximate the likelihood by
, we approximate the likelihood by
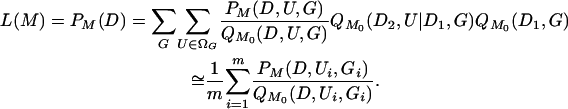 |
Because the choice of M0 affects the reliability of the approximation (Kuhner et al. 1995, 1998; Stephens and Donnelly 2000), we first estimate the MLE through a two-tier search (appendix d) and then sample intensively under this driving model.
APPLICATION
We began our exploration with a comparison between D. p. bogotana and D. persimilis. For this smaller data set (Table 1, per/bog), determination of both the exact likelihoods by recursive computation (6) and their importance-sampling approximations was in fact feasible. Having established a basis for confidence in our IS implementation, we then addressed the estimation of population parameters from the comparison between D. p. pseudoobscura and D. persimilis (Table 1, per/pse).
That the inversion difference appears to be fixed between the species as well as in our sample (Dobzhansky and Powell 1975) suggests the absence or segregation in very low frequency (p) of the derived gene order in the ancestral species. In accordance with this expectation, preliminary results (not shown) indicated higher likelihoods of lower values of p. As the data set contains little information about this aspect, we arbitrarily assigned p as 0.0001 in estimating the remaining parameters.
Comparison of D. p. bogotana and D. persimilis:
We used our two-phase search procedure (appendix d) to locate the maximum-likelihood values of the four parameters of the system (λ/u, uN0, uN1, uN2). Our IS approximations compare well to the exact likelihood computed using (6).
Maximum-likelihood estimates:
Table 3 provides the MLE array (“Unconstrained” column), with probabilities estimated using (3), based on a sample of 4 × 106 genealogical paths. A likelihood-ratio test detected no significant differences in effective population size among the species (comparison to “N0-N1-N2”).
TABLE 3.
D. p. bogotana/D. persimilis divergence
| Parameter | Unconstrained | N0 = N1 = N2 | |
|---|---|---|---|
| λ/u | 0.17 | 0.18 | |
| uN0 | 2.31 | 3.21 | |
| uN1 | 2.91 | 3.21 | |
| uN2 | 3.51 | 3.21 | |
| Likelihood | 1.05 × 10−5 | 8.78 × 10−6 | |
| P-value | 0.83 |
Comparison of exact and IS likelihood curves:
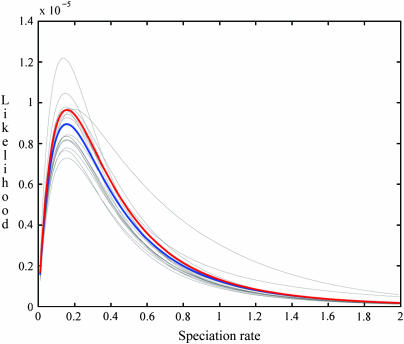
To assess the accuracy of our IS approximation, we constructed conditional-likelihood curves for the speciation rate (λ/u) with the remaining parameters assigned to their MLE values (Table 3) under both the exact recursion (6) and our IS method. Figure 3 presents the exact-likelihood function and 18 IS curves, each based on 500,000 sampled genealogies generated using the MLEs as the driving values. The average of the IS curves (blue line), based on a total of 9 × 106 samples, corresponds well to the exact conditional likelihood (red line), although it somewhat underestimates the absolute value of the likelihood.
Figure 3.—
Exact and IS-approximated conditional-likelihood function of the instantaneous rate of speciation (λ/u), with uN0, uN1, and uN2 assigned to their MLE values. Using the unconstrained MLEs (Table 3) as the driving values, we generated 18 IS curves, each based on 500,000 sampled genealogies. Red indicates the exact likelihood surface of λ/u, computed using (6) under the same assignments of the other parameters, and blue shows the average of the IS curves (9 × 106 total samples).
Computation using (6) of a single point of the exact-likelihood function required ∼4 hr on a Macintosh PowerPC G5 (2.5-GHz processor, 3.5 GB DDR SDRAM). Construction of the 200 points constituting the entire conditional-likelihood curve in Figure 3 required ∼800 hr using the exact recursion, compared to ∼30 min using our IS method based on 500,000 sampled genealogical histories. While computation time under the exact recursion increases roughly with the product of the numbers of mutations observed (elements of n), observation of more mutations has virtually undetectable effect on computation time under IS.
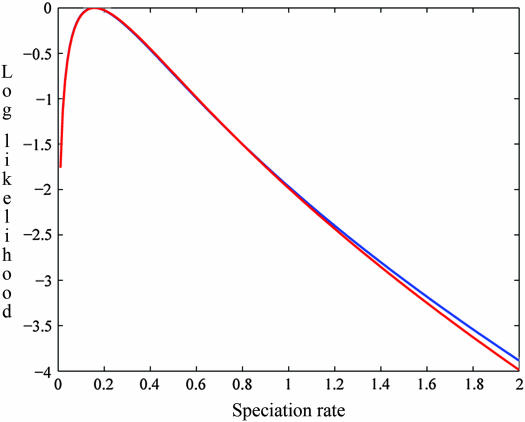
Figure 4 compares the exact log-likelihood function for the speciation rate (λ/u) computed under (6) to the IS approximation. Our IS curve provides an excellent indication of both the MLE and the breadth of the likelihood function. As noted by Stephens and Donnelly (2000), error of the IS approximation generally increases farther from the driving value. In the case studied here, however, detectable discrepancies arise only in regions quite distant from the MLE, well beyond the ∼95% confidence range spanned by 2 log-likelihood units.
Figure 4.—
Exact and IS-approximated log-likelihood functions of speciation rate (λ/u) under the same conditions as indicated for Figure 3. The red line represents the log of the exact-likelihood function, scaled to its maximum value, and the blue line shows our IS approximation, based on 9 × 106 sampled genealogies.
Comparison of D. p. pseudoobscura and D. persimilis:
The interspecific comparison involving D. p. pseudoobscura (Table 1) comprises more sequences (longer genealogies) and many more mutations (87 compared to 42). Both factors, but especially the increase in mutation number, render computation of the exact-likelihood function impractical.
Maximum-likelihood estimates:
Table 4 provides maximum-likelihood estimates of the population parameters, determined by our IS method using 4 × 106 sampled genealogies. Likelihood-ratio tests suggest that the effective size of D. p. pseudoobscura (uN2) significantly exceeds those of both D. persimilis (uN1) and the ancestor (uN0).
TABLE 4.
D. p. pseudoobscura/D. persimilis divergence
| Parameter | Unconstrained | N0 = N1 | N0 = N2 | N1 = N2 | |
|---|---|---|---|---|---|
| λ/u | 0.12 | 0.12 | 0.12 | 0.09 | |
| uN0 | 0.81 | 2.51 | 11.51 | 0.91 | |
| uN1 | 2.71 | 2.51 | 2.71 | 12.61 | |
| uN2 | 18.21 | 15.61 | 11.51 | 12.61 | |
| Likelihood | 4.59 × 10−6 | 2.88 × 10−6 | 2.34 × 10−7 | 9.23 × 10−9 | |
| P-value | 0.33 | 1.5 × 10−2 | 4.3 × 10−4 |
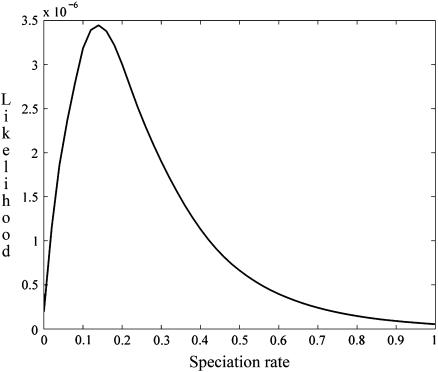
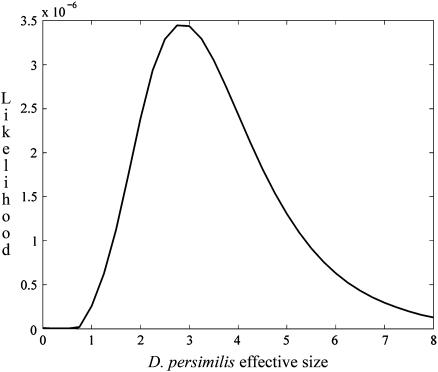
Profile-likelihood curves:
Various approaches exist for conveying a sense of the level of confidence in the estimate of a parameter in a multiple-parameter model (e.g., Berger et al. 1999). Within the maximum-likelihood framework adopted here, we have chosen to present our results in terms of the profile likelihood, under which the likelihood of a given value of a parameter corresponds to the maximum achieved over all assignments of the other parameters. appendix e describes our procedure for approximating the full four-dimensional likelihood surface using interpolating splines.
Figure 5 shows the profile-likelihood curve for the relative rate of speciation (λ/u), and Figure 6 shows the relative effective size of D. persimilis (uN1). Table 5 provides 90% confidence intervals for our MLEs, determined under the assumption of a χ2-distribution for the log-likelihood.
Figure 5.—
Profile-likelihood function for the scaled instantaneous rate of speciation (λ/u).
Figure 6.—
Profile-likelihood function for the scaled effective population size of D. persimilis (uN1).
TABLE 5.
MLEs and confidence intervals for D. p. pseudoobscura/D. persimilis divergence
| Parameter | MLEa | 90% confidence intervalsb |
|---|---|---|
| λ/u | 0.12 | (0.02, 0.46) |
| uN0 | 0.81 | (0.1, 5.0) |
| uN1 | 2.71 | (1.25, 5.75) |
| uN2 | 18.21 | (8, —)c |
Based on 4 × 106 sampled genealogies.
From profile likelihood.
Beyond bounds of approximated surface.
DISCUSSION
We have described a likelihood-based method for the estimation of population parameters. This first application addresses the time since divergence of closely related species in the D. pseudoobscura group. More importantly, it serves as a proof-of-concept demonstration of the speed and reliability of our approach, based on summary statistics rather than on entire nucleotide sequences.
Divergence between D. pseudoobscura and D. persimilis:
Our maximum-likelihood analysis indicates a significantly larger effective population size for D. pseudoobscura than for D. persimilis (Table 4) and a speciation event somewhat more ancient than some previous estimates.
Table 6 presents our estimates (Table 5) scaled to the rate of mutation per kilobase (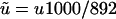 , for our 892-bp region). Numbers for effective population size correspond to
, for our 892-bp region). Numbers for effective population size correspond to 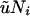 (i = 0, 1, 2) and those for divergence time to
(i = 0, 1, 2) and those for divergence time to 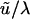 , the expectation of the exponentially distributed speciation-time variable.
, the expectation of the exponentially distributed speciation-time variable.
TABLE 6.
Maximum-likelihood estimates
| Absolute (×106)
|
|||
|---|---|---|---|
| Parameter | Scaleda | HNb | TSKc |
| Divergence time | 9.34 | 1.76 | 0.85 |
| Ancestral effective size | 0.91 | 0.34 | 0.17 |
| D. persimilis effective size | 3.04 | 1.15 | 0.55 |
| D. p. pseudoobscura effective size | 20.4 | 7.70 | 3.71 |
Mutations per kilobase.
5.3 × 10−6 mutations/kb/year.
1.1 × 10−5 mutations/kb/year.
Calibration of mutation rate:
Rescaling of our estimates into units of numbers of years or individuals requires determination of the rate of substitution of neutral mutations in noncoding regions. For the same Drosophila species studied here, Hey and Nielsen (2004) (HN) estimated 5.3 × 10−6 mutations per kilobase per year. This number, an average across 14 regions for which their analysis indicated substantial heterogeneity, reflects divergence in both coding and noncoding regions and both synonymous and nonsynonymous substitutions.
For mammals (humans and rodents), Bustamante et al. (2002) estimated a 70% reduction in the rate of substitution at silent sites in expressed genes compared to their homologous pseudogenes. This accelerated substitution in pseudogenes, particularly marked in genes with high GC content, reflects in part the release from selective constraints on hypermutable CpG dinucleotides (Sved and Bird 1990). In primates, Subramanian and Kumar (2003) found a significant overall excess in neutral substitution in exons compared to noncoding regions (pseudogenes, introns, and intergenic tracts), which disappeared upon exclusion of CpG sites.
While Drosophila genomes do not show significant CpG deficiency (Gentles and Karlin 2001), selective constraints on codon usage do influence the pattern of substitution at synonymous sites (Akashi 1995). Using the skew in base composition as an index of codon usage bias, Tamura et al. (2004) (TSK) inferred a neutral substitution rate of 1.1 × 10−5 mutations per kilobase per year for recently diverged (5.1 million years) Drosophila species.
Effective population size:
We identify Wright's “inbreeding effective number” of individuals (Crow and Denniston 1988) with half the inverse of the rate of coalescence (half the expected number of generations to coalescence between a random pair of genes; see Slatkin 1991). Our estimates of scaled population size (uNi), a ratio of rates, have units of mutations per coalescence. Division of these estimates by twice the rate of mutation per generation converts the scale to generations per coalescence, which we identify with effective numbers of individuals. Table 6 reports “absolute” effective population sizes (millions of individuals), obtained under the assumption of four generations per year (Schaeffer 1995) and HN and TSK mutation rates.
Under the TSK rate, our analysis suggests an effective population size for D. p. pseudoobscura of 3.7 × 106, with 1.4 × 106 the lower bound of the 90% confidence interval, comparable to Schaeffer's (1995) estimates (1.9 × 106, 4.5 × 106), based on the Adh region. Hey and Nielsen's (2004) estimate obtained from DPS2002 alone corresponds to 7.3 × 106 and 3.5 × 106 under the HN and TSK rates, respectively. Credible intervals, presumably obtainable from their method, were not reported.
For the effective population size of D. persimilis, we obtained an MLE of 0.55 × 106, with 90% confidence interval (0.25, 1.17). Hey and Nielsen's (2004) value based on DPS2002 alone (“θ2 × uΣ” in their Table 1) corresponds to 0.81 × 106 and 0.39 × 106 under the HN and TSK rates, respectively (credible intervals not provided).
Divergence time:
Under the TSK mutation rate, our MLE of the time since speciation between D. pseudoobscura and D. persimilis corresponds to ∼850 thousand years (KY), identical to the number obtained by Tamura et al. (2004) from their moments-based analysis of nuclear protein-coding genes located throughout the genome. Our estimate shows an insignificant excess over that of Aquadro et al. (1991) (500 KY), based on restriction site differences at the Amy region calibrated by DNA-DNA hybridization data.
Hey and Nielsen (2004) based their ML analysis on MCMC reconstruction of the gene genealogy of entire nucleotide sequences from 14 genomic regions. It permits variation among regions in rates of substitution and introgression, but does not accommodate the population substructuring induced by linkage of DPS2002 to the D. persimilis inversion, which prevents its introgression. Their estimates of divergence time scaled to substitution rate (t × uΣ, analogous to our 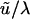 ) vary over a 13-fold range across the 14 regions, and those of absolute divergence time vary 238-fold. Their overall estimate of absolute divergence time (589 KY) corresponds to the average scaled divergence time divided by the average substitution rate.
) vary over a 13-fold range across the 14 regions, and those of absolute divergence time vary 238-fold. Their overall estimate of absolute divergence time (589 KY) corresponds to the average scaled divergence time divided by the average substitution rate.
Hey and Nielsen's (2004) estimate of divergence time based on DPS2002 alone exceeds the average across the 14 regions by >86%. This number corresponds to 1113 KY under the average substitution (HN) rate and to 536 KY under the TSK rate. Expressed on the same scale, our MLEs (λ/u = 0.12, uN0 = 0.81, uN1 = 2.71, uN2 = 18.21) show nonsignificant differences from theirs for DPS2002 alone (0.19, 0.9, 1.9, 17.2), in which we associate their number for divergence time with the inverse of the parameter of the exponentially distributed speciation-time variable.
Approaches to estimation:
Basing the estimation on summary statistics rather than on entire nucleotide sequences permits considerable simplification of the description of genealogical history. Streamlining of the evidentiary and computational basis affords greater computational and analytical freedom to address more realistic demographic scenarios and biological processes.
Genealogies as nuisance parameters:
Our primary interests lie in the characterization of the evolutionary process of speciation and genetic divergence. In this context, the genealogy of the sampled genes represents a nuisance parameter, an unknown aspect that influences the estimation of population parameters but that holds little interest in itself. More precisely, the gene genealogy is not a parameter but another manifestation of the evolutionary process under study (Donnelly and Tavaré 1995; Stephens 2001).
To accommodate some of the diversity of evolutionary processes, a number of methods entail estimation of a great many parameters. For example, full genealogical reconstruction in structured populations requires estimation of the ages of all nodes, mutations, migration events, changes in population size, and population divergences (Beerli and Felsenstein 2001; Nielsen and Wakeley 2001; Wilson et al. 2003; Hey and Nielsen 2004). A given biological system may correspond to a set of parameter assignments within the full model (for example, Yang 1996; Hey and Nielsen 2004). However, a model that accommodates the unique origin of the second chromosome inversion that prevents introgression at the DPS2002 region is not nested within available data analysis packages.
Griffiths and Tavaré (1996) showed that the variance of estimates of population parameters obtained from full genealogical reconstruction of entire nucleotide sequences tends to be smaller than that based on summary statistics. Even so, one may well prefer simpler methods based on summary statistics in cases for which they are nearly sufficient for the estimation of the population parameters of interest (Marjoram et al. 2003). The development and implementation of analyses capable of accommodating additional evolutionary processes can demand literally years of effort (Felsenstein et al. 1999), and even entire sequences may contain insufficient information to support the estimation of fully resolved gene genealogies as well as all parameters of a heavily parameterized model (Wiuf 2003). A related observation is that less detailed models can sometimes generate more accurate estimates (Takahashi and Nei 2000; Piontkivska 2004; Kosakovsky Pond and Frost 2005).
Reflecting our interest in population parameters rather than gene genealogies themselves, our method adopts a much-condensed genealogical description. A genealogical path in our analysis corresponds to an ordered list of lineage types associated with the nodes of the gene genealogy (4). It differs, in particular, from the genealogical history of Griffiths and Tavaré (1994), for which the state space includes the mutations.
Reduction of the computational burden invested in estimating genealogy may permit analysis of more realistic biological processes or demographic histories for which full genealogical reconstruction of entire nucleotide sequences may be altogether infeasible. For the application at hand, our model explicitly conditions the genealogical histories on the unique origin of the inversion that prevents introgression in the genomic region studied (appendix b). Incorporation of this biological information into the estimation procedure entailed only modification of Markov matrices of rates of within- and between-level transitions (appendix a). This structural flexibility may permit customization of the analysis to a wide variety of biological systems.
Estimation of divergence times:
Species divergence corresponds to a change in coalescence structure: the most recent point at which ancestral lineages with descendants in different species can have coalesced. Using the age of the most recent node with descendants sampled from different populations as a surrogate for divergence time generates negligible error only for ancient divergence events involving small ancestral population sizes and little interpopulation gene flow (see Nichols 2001). A number of recent reviews of moment- and likelihood-based approaches have addressed the estimation of population divergence apart from node age (Arbogast et al. 2002; Rosenberg and Feldman 2002; Takahata and Satta 2002).
Many likelihood-based methods related to ours treat time since speciation as a parameter. Takahata et al. (1995), Rannala and Yang (2003), and Wall (2003) based the estimation of divergence time and ancestral population size on numbers of segregating sites in a present-day sample comprising one sequence from each of two or more species. Nielsen and Wakeley (2001) and Hey and Nielsen (2004) used MH sampling to approximate the posterior distribution of fully resolved gene genealogies, including all node ages and time since speciation.
To incorporate time into the method of Griffiths and Tavaré (1994), for which the genealogical histories record only the relative order of events, Nielsen (1998) determined the probability distribution of the numbers of ancestral lineages remaining in each group at the divergence event. Our method characterizes the time since speciation as an exponentially distributed random variable and estimates the instantaneous rate of speciation (λ/u). This construction obviates the need to incorporate time into the backward construction of genealogical histories.
Unique evolutionary events:
Slatkin and Rannala (1997, 2000) based the estimation of the age of an advantageous or deleterious mutation on the relative magnitudes of neutral variation segregating within the affected and unaffected subsamples. The sample genealogy reflects coalescence of affected lineages only among themselves, with the number of ancestors from which they could have descended determined from a branching-process model. In contrast, Wiuf and Donnelly (1999) addressed the age of a neutral marker restricted to a subset 𝒟 of genes sampled from a population. Determination of the likelihood entails conditioning a random gene genealogy to contain a node from which all members of 𝒟 and none of the other sampled genes descend and requiring the occurrence of exactly one neutral mutation on the branch immediately ancestral to that node. These approaches differ with respect to more than statistical philosophy. In the latter case, the partitioning of the sample arises only after observation of the mutation of interest, while in the former, the distinction between affected and unaffected genes exists before observation of the sample.
We chose to study the DPS2002 region precisely because tight linkage to a second chromosome inversion precludes its introgression. We have imposed the simplifying assumption that the origin of the inversion occurred immediately before the MRCA of the inverted lineages. Further, for cases in which the MRCA of the inverted lineages predates the speciation event, our model assumes a constant frequency (p) of the inversion in the ancestral species and permits coalescence only within and not between gene orders. A more detailed analysis would incorporate a description of evolutionary change in p (e.g., Slatkin and Rannala 1997; De Iorio and Griffiths 2004).
Importance sampling:
The high complexity of virtually all biological systems of interest ensures that any particular realization of the evolutionary process occurs with extremely low probability, making approximation of likelihoods by “naive” Monte Carlo simulation impractical (Stephens and Donnelly 2000). Importance sampling offers a means of compensating for discrepancies generated by sampling from convenient but incorrect proposal distributions. As discussed in the introductory section, sprinkling the observed number of mutations over a random genealogy under the “fixed-S” procedure very rapidly generates genealogical and mutational histories consistent with the data, but approximates an incorrect distribution (Markovtsova et al. 2001).
Our method (2) proposes genealogical paths (4) by sampling, not from a posterior distribution given the full data (D = {D1, D2}), but from an analytically determined stationary distribution (7) of paths consistent with the types of segregating mutations observed (QM(D1, G)). It then sprinkles the observed numbers of mutations on the genealogical path (QM(D2, U|D1, G)) according to a heuristic weighting scheme (8). We then correct the bias introduced by the proposal distribution using the exact probability of the proposed genealogical and mutational history (appendix a).
Expansion of the evidentiary basis to include additional summary statistics would extract more information from the sampled sequences. Through straightforward redefinition of the state space, our method can incorporate counts of mutations of various kinds, including numbers of mutations classified according to their distribution among groups (Wakeley and Hey 1997; Wakeley et al. 2001), the number of haplotypes and their frequency spectrum (Ewens 1972), and the frequency spectrum of mutation numbers (Fu 1995). Our approach is less well suited to summary statistics defined as various moments, including average pairwise differences, variances, regressions, and correlations. However, the distribution of mutations among groups can replace pairwise FST values as the basis for the characterization of gene flow (Wakeley and Hey 1997), and the relative numbers of segregating sites at linked loci can replace pairwise linkage disequilibrium as the basis for the estimation of recombination rate (Takebayashi et al. 2004). The simplicity of our approach (appendix a) facilitates structural modification both to incorporate more information contained in the sampled sequences and to broaden the scope of evolutionary processes amenable to analysis.
Acknowledgments
We thank Beatrix Jones for perceiving the relevance of importance sampling to this problem, Michael Lavine for comments and suggestions that improved the analysis, Sudhir Kumar for insights into rate calibration and CpG avoidance, John Willis for reminding us of the unique evolutionary origin of inversions, and two anonymous reviewers and John Wakeley for comments. This research was supported in part by funding from National Science Foundation (NSF) [DMS-0203762 (Y.C.), NSF DEB-0314552 (M.A.F.N.), and an NSF predoctoral fellowship (J.E.S.)] and by the National Institutes of Health [GM 37841 (M.K.U.)].
APPENDIX A: PROBABILITY-GENERATING FUNCTIONS
For convenience, we summarize the recursive determination of a PGF of the array n of segregating sites observed in a sample of arbitrary size from the two species (see Uyenoyama and Takebayashi 2004).
Matrices Pl and and Ql, respectively, provide per-generation rates of within-level and between-level transitions for states on level l. For state α within level l, let Cl;α denote the total rate of transition to any other configuration, irrespective of level,
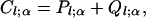 |
(A1) |
for Pl;α and Ql;α representing row sums of the within- and between-level transition rate matrices. Matrices Ul and Vl denote within- and between-level transition probabilities, given the occurrence of a transition,
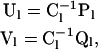 |
(A2) |
for Cl, a diagonal matrix in which the diagonal element in row α corresponds to Cl;α (A1).
Under a geometric distribution for the total number of mutations accumulated in the interval terminated by the first transition from state α and a multinomial distribution, given this total number, for the number of mutations arising on the three types of lineages, we obtain the joint PGF of mutation numbers,
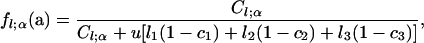 |
(A3) |
in which a = (a1, a2, a3, a4, a5, a6, a7) represents the array of seven PGF parameters corresponding to the seven types of observed segregating mutations (Table 1), and the assignment of these PGF parameters to c1, c2, and c3 depends on the configuration of state α:
 |
(A4) |
These expressions indicate that the conditional transition matrices (A2) and the joint distribution of mutation numbers (A3) depend only on the relative rates of transition and mutation: λ/u, uN0, uN1, and uN2.
To obtain an expression for 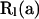 (5), the joint PGF of mutations occurring within level l of the sample genealogy, we consider the array of mutations that occurred before and after the most recent transition,
(5), the joint PGF of mutations occurring within level l of the sample genealogy, we consider the array of mutations that occurred before and after the most recent transition,
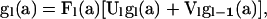 |
(A5) |
for 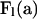 , a diagonal matrix with the PGFs of mutation numbers (A3) for states within level l arrayed along the diagonal. Rearrangement of (A5) completes the recursion (5),
, a diagonal matrix with the PGFs of mutation numbers (A3) for states within level l arrayed along the diagonal. Rearrangement of (A5) completes the recursion (5),
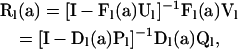 |
(A6) |
for
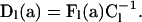 |
Because speciation alone induces within-level transitions and occurs exactly once in a genealogy,
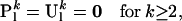 |
(A7) |
under which the matrix inverses in (7) and (A6) reduce to
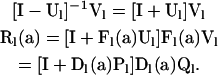 |
In determining the recursion in probabilities (6), we observe that the PGF parameters (a1, a2, … , a7) appear only in the 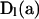 . Derivatives of these diagonal matrices with respect to the ai take the form
. Derivatives of these diagonal matrices with respect to the ai take the form
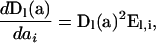 |
for 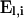 , a diagonal matrix of the absolute values of the coefficients of ai in the denominators of the elements of
, a diagonal matrix of the absolute values of the coefficients of ai in the denominators of the elements of 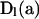 (A3). All configurations within a given block (l1, l2, l3) share the same coefficient of ai, implying that the corresponding submatrix of
(A3). All configurations within a given block (l1, l2, l3) share the same coefficient of ai, implying that the corresponding submatrix of 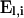 is proportional to the identity matrix
is proportional to the identity matrix
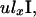 |
for x, the lineage type associated with ai in this block (A4). Using
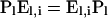 |
and (A7), we obtain
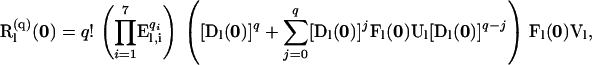 |
(A8) |
in which 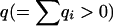 represents the total number of mutations arising on level l. The product of the
represents the total number of mutations arising on level l. The product of the 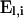 matrices in (A8) is nonzero only for arrays q that specify a combination of mutations that can occur on level l.
matrices in (A8) is nonzero only for arrays q that specify a combination of mutations that can occur on level l.
APPENDIX B: CONDITIONED GENEALOGIES
We extend the method of Wiuf and Donnelly (1999) to restrict the genealogical paths proposed by our IS procedure to those consistent with the kinds of mutations observed in the sample (D1). We describe the conditioning of genealogies on each of the four possible topologies ({f/a, a/f}, {f/a, f/s}, {s/s, f/s}, and {s/s}), both with and without the substructuring of the ancestral species induced by the presence of the chromosomal inversion that precludes introgression at DPS2002. In the system under study, the unique origin of this inversion entails the presence of an f/a branch (first two topologies), whether or not the data set includes an f/a mutation.
Let T0(x, y, z) represent the probability that a process presently in the prespeciation state (l01 = x, l02 = y, l03 = z, 0, 0) has a genealogy of the required kind, and let T1(x, y, 0) be the corresponding probability for a process in the postspeciation state (0, 0, 0, l11 = x, l22 = y). Conditioning genealogical topologies entails multiplying the within- and between-level transition rates (appendix a) by T0(x, y, z) or T1(x, y, z), for (x, y, z) the array of lineages of types 1, 2, and 3 at the destination (ancestral) state. For data sets containing fewer than two mutational types informative for topology, the probabilities are summed. For example, observation of an f/s mutation indicates two possible topologies, {f/a, f/s} or {s/s, f/s}, with the transition rates modified by the sum of the T0 and T1 probabilities associated with these topologies.
Wiuf and Donnelly (1999) derived a closed-form expression for the weightings to ensure a single branch of type f/a in the genealogy of a sample from a single, unstructured population. We obtain the T0 and T1 weights recursively, for each assignment of the parameters of the model. For the application at hand, this additional recursion (not simulation) step imposes a modest computational burden for each set of IS driving values, but permits a vast improvement in efficiency by guaranteeing that all genealogical histories proposed are compatible with the data.
{f/a, a/f}:
Because all lineages sampled from each group must coalesce among themselves, with the only between-group coalescence generating the MRCA, the genealogy cannot contain any type 3 lineages:
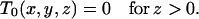 |
In the prespeciation phase without substructuring due to the chromosomal inversion, we obtain T0(x, y, 0) (x, y > 0) by iterating
 |
(B1) |
under boundary condition
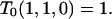 |
Under substructuring, we replace (B1) by
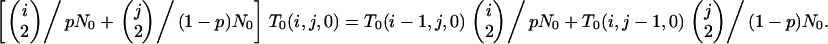 |
Similarly, in the postspeciation phase, we obtain T1(x, y, 0) from
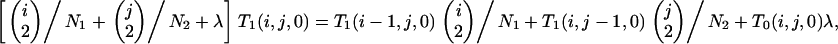 |
with boundary condition
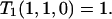 |
{f/a, f/s}:
On any given level of the gene genealogy, the presence of an f/s branch excludes the existence of all other branch types except a/s. Consequently, an f/s lineage retains its type back to the MRCA because any coalescence event that includes it generates an ancestral branch of type f/s. Because the f/s branch must coexist with at least one a/s branch,
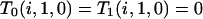 |
(B2) |
for positive i. The existence of an f/a branch excludes all type 3 branches (s/s or s/f) other than f/s, from which it must descend. In particular, branches of type 1 cannot occur on the same level with branches of type 3:
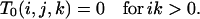 |
(B3) |
Because T0(0, j, k) corresponds to a state from which f/a branches cannot arise, we need consider only T0(x, y, 0). We first obtain T0(1, y, 0) from
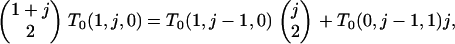 |
(B4) |
in which
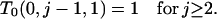 |
In the absence of substructuring due to the chromosomal inversion, we then generate T0(x, y, 0) from
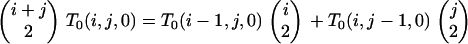 |
and, in its presence, from
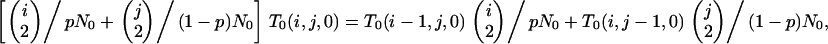 |
both viewed as recursions in j under (B2).
Similarly, in the postspeciation phase, we first obtain T1(1, y, 0) and then T1(x, y, 0) from
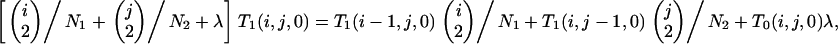 |
(B5) |
using (B2).
{f/s, s/s}:
We restrict consideration to cases without substructuring due to the chromosomal inversion because s/s branches cannot arise in its presence. We begin with prespeciation states that include a type 3 branch. Because at least one a/s branch must exist on any level containing the f/s branch,
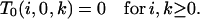 |
(B6) |
We first determine T0(0, y, z) from
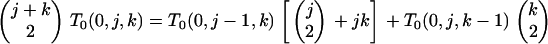 |
(B7) |
Because states on the right side of (B7) represent those reached from the state indicated on the left, which includes an s/s branch,
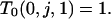 |
This expression together with (B6) for i = 0 implies
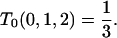 |
Beginning with this boundary value, we use (B7) to generate T0(0, j, 2) for all positive j. For successive values of k (k > 2), we obtain T0(0, j, k), given T0(0, j, k − 1) for all positive j, treating (B7) as a recursion in j.
Beginning with T0(0, y, z), we determine T0(x, y, z) given T0(x − 1, y, z) for arbitrary positive y and z from
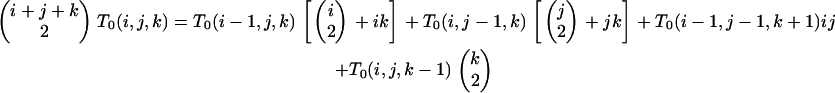 |
(B8) |
We first treat (B8) as a recursion in k under the assignment i = x and j = 1 and then as a recursion in j for arbitrary k.
For states that lack a type 3 branch, we determine T0(x, y, 0) from
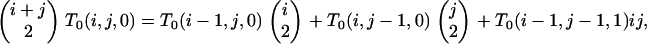 |
(B9) |
for T0(i − 1, j − 1, 1) obtained from (B8). In the postspeciation phase, we generate T1(x, y, 0) from (B5) under (B6).
{s/s}:
Because genealogies designated {s/s} contain no branches of types f/s, s/f, f/a, or a/f,
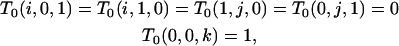 |
for positive i, j, and k. Using these boundary conditions, we obtain T0(x, y, z) with positive z from (B8), T0(x, y, 0) from (B9), and T1(x, y, 0) from (B5).
APPENDIX C: PROPOSED PLACEMENT OF MUTATIONS
Our proposal density reflects the placement of mutations on levels of a given genealogical path according to the expected length of the levels (8). In the prespeciation phase, the most recent event entails either a within-type coalescence or, in the absence of more than one type 1 lineage, a between-type coalescence. Waiting times to these various transitions correspond to exponentially distributed random variables. Because the minimum of two or more independent exponentially distributed random variables also has an exponential distribution, with parameter equal to the sum of the parameters of the base variables, the expected time to the most recent event corresponds to the inverse of the sum of their parameters:
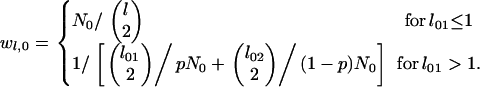 |
(C1) |
In the postspeciation phase, the most recent event involves coalescence in species 1, coalescence in species 2, or speciation, with expected time to the most recent event given by
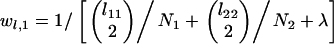 |
Speciation implies transfer of the lineages to the ancestral population without termination of the level, with some additional time required for coalescence. However, because our experimentation with the weights suggests that increasing the weight of the level by wl,0 (C1) for levels that include the speciation event tends to generate more error, our present implementation weights such levels by the expected time to the most recent event rather than to the most recent coalescence.
APPENDIX D: APPROXIMATION OF LIKELIHOODS
Our method for locating the mode of the likelihood function 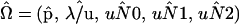 relies on a two-phase search procedure. The first phase characterizes the major features of likelihood surface across the five-dimensional parameter space and determines a preliminary estimate of
relies on a two-phase search procedure. The first phase characterizes the major features of likelihood surface across the five-dimensional parameter space and determines a preliminary estimate of 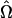 . This point is then used to seed a more refined, steepest-descent search for
. This point is then used to seed a more refined, steepest-descent search for  .
.
Random search:
Likelihoods of a large number of points randomly chosen in the five-dimensional parameter space are estimated. The parameter space is subdivided into bins and the likelihoods of points falling within the same bin are averaged to obtain an estimate of the likelihood of the point at the center of the bin. In the study described here, preliminary exploration of the likelihood surface suggested a trust region spanning the zero point up to a limit for each of the five parameters (p, 1; λ/u, 1.5; uN0, 10; uN1, 10; uN2, 40) within which the likelihoods of ∼200,000 random points were estimated, each from 10,000 sampled histories. The size of the bins corresponded to 0.01 for the p and λ/u dimensions and 0.1 for uN0, uN1, and uN2.
For each parameter, we generate an approximate Bayesian posterior marginal distribution under uniform prior distributions for all parameters. Our preliminary estimate of the maximum-likelihood parameter set corresponds to the modes for the five parameters. For each parameter, a conditional-likelihood curve is estimated using 500,000 sampled histories under a driving value corresponding to the preliminary ML parameter values. The seed passed to the second phase of the mode search corresponds to these preliminary MLEs, subject to small modifications to improve the correspondence between the driving values of the conditional-likelihood curves and their modes.
Steepest-descent search:
Beginning with the assignment of the seed point as the driving model, the steepest-ascent code determines a succession of driving models. For iteration i with driving model Ω(i), a local estimate of the direction of higher likelihoods is determined by comparing likelihoods at points on a lattice around Ω(i). For the jth parameter 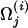 , we consider three values,
, we consider three values,
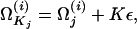 |
for K ∈ {−1, 0, 1} and ɛ a small step size. The lattice point with greatest IS-estimated likelihood determines the search direction Δ. Restricting consideration to search direction Δ, we then determine for each parameter the best number (up to a specified maximum) of steps of size ɛ by estimating likelihoods at all combinations of step numbers for all parameters. We propose a move to 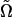 , corresponding to the point with the highest likelihood.
, corresponding to the point with the highest likelihood.
We accept the proposed point 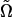 only if its likelihood estimated using itself as the driving model exceeds that estimated using the present Ω(i) as the driving model. Upon acceptance, the algorithm sets
only if its likelihood estimated using itself as the driving model exceeds that estimated using the present Ω(i) as the driving model. Upon acceptance, the algorithm sets
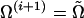 |
and initiates another cycle. Upon rejection, it either terminates, after a specified number of consecutive rejections, or initiates another cycle from Ω(i).
To guard against settling on local modes, we repeat this procedure several times, beginning from perturbations of the seed value.
APPENDIX E: PROFILE-LIKELIHOOD SURFACE
Having assigned p as 0.0001, we used IS with the MLEs (Table 4) as driving values to approximate the likelihood surface over the remaining four dimensions at 20,000 grid points (λ/u, 10 values beginning at 0.0 with a step size of 0.1; uN0 and uN1, 10 values from 0.01, step size 0.75; uN2, 20 values from 0.01, step size 2.0), each based on 300,000 sampled genealogical histories. Invoking the csapi function of the MatLab spline toolbox, we approximated the full four-dimensional likelihood surface using multivariate cubic splines (de Boor 2001). We then discretized the interpolated surface on a finer grid, beginning at zero for each of the four parameters with smaller step sizes (λ/u, 0.02; uN0 and uN1, 0.25; uN2, 1.0). From these >1.7 × 106 estimated and interpolated points, we generated the profile-likelihood curves by determining for each value of a given parameter the values of the remaining three parameters that gave the highest likelihood.
References
- Akashi, H., 1995. Inferring weak selection from patterns of polymorphism and divergence at “silent” sites in Drosophila DNA. Genetics 139: 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquadro, C. F., A. L. Weaver, S. W. Schaeffer and W. W. Anderson, 1991. Molecular evolution of inversions in Drosophila pseudoobscura: the amylase region. Proc. Natl. Acad. Sci. USA 88: 305–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbogast, B. S., S. V. Edwards, J. Wakeley, P. Beerli and J. B. Slowinski, 2002. Estimating divergence times from molecular data on phylogenetic and population genetic timescales. Annu. Rev. Ecol. Syst. 33: 707–740. [Google Scholar]
- Beaumont, M. A., 2004. Recent developments in genetic data analysis: What can they tell us about human demographic history? Heredity 92: 365–379. [DOI] [PubMed] [Google Scholar]
- Beaumont, M. A., W. Zhang and D. J. Balding, 2002. Approximate Bayesian computation in population genetics. Genetics 162: 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli, P., and J. Felsenstein, 2001. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc. Natl. Acad. Sci. USA 98: 4563–4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, J. O., B. Liseo and R. L. Wolpert, 1999. Integrating likelihood methods for eliminating nuisance parameters. Stat. Sci. 14: 1–28. [Google Scholar]
- Bustamante, C. D., R. Nielsen and D. L. Hartl, 2002. A maximum likelihood method for analyzing pseudogene evolution: implications for silent site evolution in humans and rodents. Mol. Biol. Evol. 19: 110–117. [DOI] [PubMed] [Google Scholar]
- Crow, J. F., and C. Denniston, 1988. Inbreeding and variance effective population numbers. Evolution 42: 482–495. [DOI] [PubMed] [Google Scholar]
- de Boor, C., 2001. A Practical Guide to Splines. Springer-Verlag, New York.
- De Iorio, M., and R. C. Griffiths, 2004. Importance sampling on coalescent histories. I. Adv. Appl. Probab. 36: 417–433. [Google Scholar]
- Depaulis, F., and M. Veuille, 1998. Neutrality tests based on the distribution of haplotypes under an infinite-site model. Mol. Biol. Evol. 15: 1788–1790. [DOI] [PubMed] [Google Scholar]
- Depaulis, F., S. Mousset and M. Veuille, 2001. Haplotype tests using coalescent simulations conditional on the number of segregating sites. Mol. Biol. Evol. 18: 1136–1138. [DOI] [PubMed] [Google Scholar]
- Dobzhansky, T., and J. R. Powell, 1975. Drosophila pseudoobscura and its American relatives, Drosophila persimilis and Drosophila miranda, pp. 537–587 in Invertebrates of Genetic Interest, edited by R. C. King. Plenum Press, New York.
- Dobzhansky, T., and C. C. Tan, 1936. Studies on hybrid sterility III. A comparison of the chromosome structure in two related species, Drosophila pseudoobscura and Drosophila miranda. Z. Indukt. Abstammungs-Vererbungsl. 72: 88–113. [Google Scholar]
- Donnelly, P., and S. Tavaré, 1995. Coalescents and genealogical structure under neutrality. Annu. Rev. Genet. 29: 401–421. [DOI] [PubMed] [Google Scholar]
- Estoup, A., M. A. Beaumont, F. Sennedot, C. Moritz and J.-M. Cornuet, 2004. Genetic analysis of complex demographic scenarios: spatially expanding populations for the cane toad, Bufo marinus. Evolution 58: 2021–2036. [DOI] [PubMed] [Google Scholar]
- Ewens, W. J., 1972. The sampling theory of selectively neutral alleles. Theor. Popul. Biol. 3: 87–112. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 1992. Estimating effective population size from samples of sequences: inefficiency of pairwise and segregation sites as compared to phylogenetic estimates. Genet. Res. 59: 139–147. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., M. K. Kuhner, J. Yamato and P. Beerli, 1999. Likelihoods on coalescents: a Monte Carlo sampling approach to inferring parameters from population samples of molecular data, pp. 163–185 in Statistics in Molecular Biology and Genetics, edited by F. Seillier-Moiseiwitsch. Institute of Mathematical Statistics and American Mathematics Society, Haywood, CA.
- Fu, Y.-X., 1995. Statistical properties of segregating sites. Theor. Popul. Biol. 48: 172–197. [DOI] [PubMed] [Google Scholar]
- Fu, Y.-X., and W.-H. Li, 1997. Estimating the age of the common ancestor of a sample of DNA sequences. Mol. Biol. Evol. 14: 195–199. [DOI] [PubMed] [Google Scholar]
- Gentles, A. J., and S. Karlin, 2001. Genome-scale compositional comparisons in eukaryotes. Genome Res. 11: 540–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, R. C., and S. Tavaré, 1994. Simulating probability distributions in the coalescent. Theor. Popul. Biol. 46: 131–159. [Google Scholar]
- Griffiths, R. C., and S. Tavaré, 1996. Monte Carlo inference methods in population genetics. Math. Comput. Model. 23: 141–158. [Google Scholar]
- Hamilton, G., M. Currat, N. Ray, G. Heckel, M. A. Beaumont et al., 2005. Bayesian estimation of recent migration rates after a spatial expansion. Genetics 170: 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey, J., and C. A. Machado, 2003. The study of structured populations—new hope for a difficult and divided science. Nat. Rev. Genet. 4: 535–543. [DOI] [PubMed] [Google Scholar]
- Hey, J., and R. Nielsen, 2004. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167: 747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., 1993. The how and why of generating gene genealogies, pp. 23–36 in Mechanisms of Molecular Evolution, edited by N. Takahata and A. G. Clark. Sinauer, Sunderland, MA.
- Huelsenbeck, J. P., and F. Ronquist, 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond, S. L., and S. D. W. Frost, 2005. A simple hierarchical approach to modeling distributions of substitution rates. Mol. Biol. Evol. 22: 223–234. [DOI] [PubMed] [Google Scholar]
- Kuhner, M. K., J. Yamato and J. Felsenstein, 1995. Estimating effective population size and mutation rate from sequence data using Metropolis-Hastings sampling. Genetics 140: 1421–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhner, M. K., J. Yamato and J. Felsenstein, 1998. Maximum-likelihood estimation of population growth rates based on the coalescent. Genetics 149: 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. S., 2001. Monte Carlo Strategies in Scientific Computing. Springer-Verlag, New York.
- Machado, C. A., R. M. Kliman, J. A. Markert and J. Hey, 2002. Inferring the history of speciation from multilocus DNA sequence data: the case of Drosophila pseudoobscura and close relatives. Mol. Biol. Evol. 19: 472–488. [DOI] [PubMed] [Google Scholar]
- Marjoram, P., J. Molitor, V. Plagnol and S. Tavaré, 2003. Markov chain Monte Carlo without likelihoods. Proc. Natl. Acad. Sci. USA 100: 15324–15328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovtsova, L., P. Marjoram and S. Tavaré, 2001. On a test by Depaulis and Veuille. Mol. Biol. Evol. 18: 1132–1133. [DOI] [PubMed] [Google Scholar]
- Navarro, A., E. Betrán, A. Barbadilla and A. Ruiz, 1997. Recombination and gene flux caused by gene conversion and crossing over in inversion heterozygotes. Genetics 146: 695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols, R., 2001. Gene trees and species trees are not the same. Trends Ecol. Evol. 16: 358–364. [DOI] [PubMed] [Google Scholar]
- Nielsen, R., 1998. Maximum likelihood estimation of population divergence times and population phylogenies under the infinite sites model. Theor. Popul. Biol. 53: 143–151. [DOI] [PubMed] [Google Scholar]
- Nielsen, R., and J. Wakeley, 2001. Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics 158: 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor, M. A. F., and K. R. Smith, 2000. Recombination, statistical power, and genetic studies of sexual isolation in Drosophila. J. Hered. 91: 99–103. [DOI] [PubMed] [Google Scholar]
- Noor, M. A. F., K. L. Grams, L. A. Bertucci, Y. Almendarez, J. Reiland et al., 2001. a The genetics of reproductive isolation and the potential for gene exchange between Drosophila pseudoobscura and D. persimilis via backcross hybrid males. Evolution 55: 512–521. [DOI] [PubMed] [Google Scholar]
- Noor, M. A. F., K. L. Grams, L. A. Bertucci and J. Reiland, 2001. b Chromosomal inversions and reproductive isolation of species. Proc. Natl. Acad. Sci. USA 98: 12084–12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piontkivska, H., 2004. Efficiencies of maximum likelihood methods of phylogenetic inferences when different substitution models are used. Mol. Phylogenet. Evol. 31: 865–873. [DOI] [PubMed] [Google Scholar]
- Pritchard, J. K., M. T. Seielstad, A. Perez-Lezaun and M. W. Feldman, 1999. Population growth of human Y chromosomes: a study of Y chromosome microsatellites. Mol. Biol. Evol. 16: 1791–1798. [DOI] [PubMed] [Google Scholar]
- Rannala, B., and Z. Yang, 2003. Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics 164: 1645–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, N. A., and M. W. Feldman, 2002. The relationship between coalescence times and population divergence times, pp. 130–164 in Modern Developments in Theoretical Population Genetics—The Legacy of Gustave Malécot, edited by M. Slatkin and M. Veuille. Oxford University Press, Oxford.
- Schaeffer, S. W., 1995. Population genetics in Drosophila pseudoobscura: a synthesis based on nucleotide sequence data for the Adh gene, pp. 329–352 in Genetics of Natural Populations: The Continuing Importance of Theodosius Dobzhansky, edited by L. Levine. Columbia University Press, New York.
- Slatkin, M., 1991. Inbreeding coefficients and coalescence times. Genet. Res. 58: 167–175. [DOI] [PubMed] [Google Scholar]
- Slatkin, M., and B. Rannala, 1997. Estimating the age of alleles by use of intraallelic variability. Am. J. Hum. Genet. 60: 447–458. [PMC free article] [PubMed] [Google Scholar]
- Slatkin, M., and B. Rannala, 2000. Estimating allele age. Annu. Rev. Genomics 1: 225–249. [DOI] [PubMed] [Google Scholar]
- Stajich, J. E., D. Block, K. Boulez, S. E. Brenner, S. A. Chervitz et al., 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12: 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, M., 2001. Inference under the coalescent, pp. 213–238 in Handbook of Statistical Genetics, edited by D. J. Balding, M. J. Bishop and C. Cannings. John Wiley & Sons, Chichester, UK.
- Stephens, M., and P. Donnelly, 2000. Inference in molecular population genetics. J. R. Stat. Soc. B 62: 605–635. [Google Scholar]
- Subramanian, S., and S. Kumar, 2003. Neutral substitutions occur at a faster rate in exons than in noncoding DNA in primate genomes. Genome Res. 13: 838–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved, J., and A. Bird, 1990. The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc. Natl. Acad. Sci. USA 87: 4692–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, K., and M. Nei, 2000. Efficiencies of fast algorithms of phylogenetic inference under the criteria of maximum parsimony, minimum evolution, and maximum likelihood when a large number of sequences are used. Mol. Biol. Evol. 17: 1251–1258. [DOI] [PubMed] [Google Scholar]
- Takahata, N., and Y. Satta, 2002. Pre-speciation coalescence and the effective size of ancestral populations, pp. 52–71 in Modern Developments in Theoretical Population Genetics—The Legacy of Gustave Malécot, edited by M. Slatkin and M. Veuille. Oxford University Press, Oxford.
- Takahata, N., Y. Satta and J. Klein, 1995. Divergence time and population size in the lineage leading to modern humans. Theor. Popul. Biol. 48: 198–221. [DOI] [PubMed] [Google Scholar]
- Takebayashi, N., E. Newbigin and M. K. Uyenoyama, 2004. Maximum-likelihood estimation of rates of recombination within mating-type regions. Genetics 167: 2097–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallmon, D. A., G. Luikart and M. A. Beaumont, 2004. Quantitative evaluation of a new effective population size estimator based on approximate Bayesian computation. Genetics 167: 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., S. Subramanian and S. Kumar, 2004. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 21: 36–44. [DOI] [PubMed] [Google Scholar]
- Tavaré, S., D. J. Balding, R. C. Griffiths and P. Donnelly, 1997. Inferring coalescence times from DNA sequence data. Genetics 145: 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli, M., N. H. Barton and J. A. Coyne, 2001. Theory and speciation. Trends Ecol. Evol. 16: 330–343. [DOI] [PubMed] [Google Scholar]
- Uyenoyama, M. K., and N. Takebayashi, 2004. A simple method for computing exact probabilities of mutation numbers. Theor. Popul. Biol. 65: 271–284. [DOI] [PubMed] [Google Scholar]
- Wakeley, J., and J. Hey, 1997. Estimating ancestral population parameters. Genetics 145: 847–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley, J., R. Nielsen, S. N. Liu-Cordero and K. Ardlie, 2001. The discovery of single-nucleotide polymorphisms—and inferences about human demographic history. Am. J. Hum. Genet. 69: 1332–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, J. D., 2003. Estimating ancestral population sizes and divergence times. Genetics 163: 395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, J. D., and R. R. Hudson, 2001. Coalescent simulations and statistical tests of neutrality. Mol. Biol. Evol. 18: 1134–1135. [DOI] [PubMed] [Google Scholar]
- Wall, J. D., P. Andolfatto and M. Przeworski, 2002. Testing models of selection and demography in Drosophila simulans. Genetics 162: 203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. L., J. Wakeley and J. Hey, 1997. Gene flow and natural selection in the origin of Drosophila pseudoobscura and close relatives. Genetics 147: 1091–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson, G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Watterson, G. A., 1985. The genetic divergence of two populations. Theor. Popul. Biol. 27: 298–317. [Google Scholar]
- Weiss, G., and A. von Haeseler, 1998. Inference of population history using a likelihood approach. Genetics 149: 1539–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, I. J., and D. J. Balding, 1998. Genealogical inference from microsatellite data. Genetics 150: 499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, I. J., M. E. Weale and D. J. Balding, 2003. Inferences from DNA data: population histories, evolutionary processes and forensic match probabilities. J. R. Stat. Soc. A 166: 155–201. [Google Scholar]
- Wiuf, C., 2003. Inferring population history from genealogical trees. J. Math. Biol. 46: 241–264. [DOI] [PubMed] [Google Scholar]
- Wiuf, C., and P. Donnelly, 1999. Conditional genealogies and the age of a neutral mutant. Theor. Popul. Biol. 56: 183–201. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1996. Maximum-likelihood models for combined analyses of multiple sequence data. J. Mol. Evol. 42: 587–596. [DOI] [PubMed] [Google Scholar]