Abstract
Circadian clocks maintain robust and accurate timing over a broad range of physiological temperatures, a characteristic termed temperature compensation. In Arabidopsis thaliana, ambient temperature affects the rhythmic accumulation of transcripts encoding the clock components TIMING OF CAB EXPRESSION1 (TOC1), GIGANTEA (GI), and the partially redundant genes CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY). The amplitude and peak levels increase for TOC1 and GI RNA rhythms as the temperature increases (from 17 to 27°C), whereas they decrease for LHY. However, as temperatures decrease (from 17 to 12°C), CCA1 and LHY RNA rhythms increase in amplitude and peak expression level. At 27°C, a dynamic balance between GI and LHY allows temperature compensation in wild-type plants, but circadian function is impaired in lhy and gi mutant plants. However, at 12°C, CCA1 has more effect on the buffering mechanism than LHY, as the cca1 and gi mutations impair circadian rhythms more than lhy at the lower temperature. At 17°C, GI is apparently dispensable for free-running circadian rhythms, although partial GI function can affect circadian period. Numerical simulations using the interlocking-loop model show that balancing LHY/CCA1 function against GI and other evening-expressed genes can largely account for temperature compensation in wild-type plants and the temperature-specific phenotypes of gi mutants.
INTRODUCTION
The circadian clock is an endogenous 24-h timer found in most eukaryotes and in photosynthetic bacteria. The clock drives rhythms in the physiology, biochemistry, and metabolism of the organisms (reviewed in Hall and McWatters, 2006). Circadian rhythms are defined as rhythms that persist in constant conditions with a periodicity of ∼24 h, that can be entrained to local time, and that are capable of maintaining a constant periodicity over a broad range of physiological temperatures. Intriguingly, although these characteristics of the clock are shared across taxonomic groups, there is little conservation of the molecular components used (reviewed in Harmer et al., 2001).
In Arabidopsis thaliana, a transcription feedback loop model has been proposed for the clock, similar to the architecture of the clock in other organisms. The loop comprises two myb transcription factors, LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK ASSOCIATED1 (CCA1), which are proposed to function by repressing the expression of TIMING OF CAB EXPRESSION1 (TOC1), which encodes a pseudoresponse regulator of unknown biological function. TOC1, in turn, activates LHY/CCA1 (Alabadi et al., 2001). It is clear that this model is not sufficient to explain current experimental data, as abnormal rhythms persist in plants lacking both LHY and CCA1 or in those lacking TOC1 (Alabadi et al., 2002; Mizoguchi et al., 2002; Locke et al., 2005). Mathematical simulations incorporating current experimental data have led to the proposal of a model comprising two interlocking feedback loops, with GIGANTEA (GI) and TOC1 as candidates for components of a second loop (Locke et al., 2005).
One of the key characteristics of all circadian rhythms is that they are able to maintain robust rhythms with a period close to 24 h over a broad range of physiological temperatures; this property is termed temperature compensation. In Drosophila, studies identified a role for the key clock component PERIOD (PER) in temperature responses, including temperature-dependent variation in Per protein properties (Sawyer et al., 1997) and differential splicing of the Per RNA (Majercak et al., 1999). In Neurospora, two isoforms of the central clock component Frequency (FRQ) have been implicated in maintaining rhythm robustness over a physiological range of temperatures: mutants lacking either form are rhythmic in only a limited temperature range (Liu et al., 1997). Both studies suggest the hypothesis that temperature compensation is an intrinsic feature of the central clock components, rather than a result of additional regulators.
To identify components of the temperature compensation mechanism in the model plant Arabidopsis, a quantitative genetic approach has been used. This exploits accession-specific variations in the pattern of temperature compensation for rhythms of leaf movement between Columbia (Col) and Landsberg erecta (Ler) and between Ler and Cape Verde Islands (Cvi). This strategy uncovered a number of quantitative trait loci (QTL) across the genome. The genes underlying some of these, such as the flowering regulator FLOWERING LOCUS C (Edwards et al., 2006), had no known function in the Arabidopsis clock mechanism. By contrast, further characterization of one of these QTL, PerCv1b, identified GI as a candidate for the PerCv1b trait (Edwards et al., 2005).
The gi alleles were originally identified as supervital mutants having increased fitness as measured by high fecundity, resulting in increased seed yield per plant. The basis for this was the large rosettes and late flowering of the mutants (Rédei, 1962). More recently, GI has been identified as acting upstream of CONSTANS in the photoperiodic regulation of flowering (Suarez-Lopez et al., 2001). The GI gene encodes a 127-kD nuclear protein of 1173 amino acids (Fowler et al., 1999; Park et al., 1999; Huq et al., 2000). GI is conserved in plants; however, GI homologs appear to be absent from the genomes of the microalga Chlamydomonas (Mittag et al., 2005) and those of human, mouse, and Drosophila. GI is expressed throughout the plant and is regulated by the circadian clock, with a maximal level of expression 12 h after dawn (Fowler et al., 1999). Mutations in the clock-affecting genes LHY, CCA1, and TIME FOR COFFEE alter GI RNA expression levels (Mizoguchi et al., 2002; Hall et al., 2003). In addition to its role in flowering, GI has also been phenotypically characterized as affecting the circadian clock, as the gi-1 mutant causes a short-period phenotype for both leaf movement and CAB:LUCIFERASE (LUC) expression. Another allele, gi-2, causes a similar short-period phenotype for leaf movement, although CAB:LUC expression oscillates with a long period (Park et al., 1999). The gi mutants cause a reduction in the expression of CCA1 and LHY (Fowler et al., 1999). Recent investigations (Mizoguchi et al., 2005), together with this study, suggest that the flowering and circadian functions of GI are distinct.
In this study, we demonstrate that the gene GI plays a critical role in increasing the temperature range permissive for rhythmicity, extending the temperature range over which both the robust rhythms of CAB expression and leaf movement rhythms can be maintained. We go on to investigate, first, how key components of the wild-type circadian mechanism (GI, LHY, CCA1, and TOC1) change in response to altered temperature. Second, we describe how this temperature-specific regulation is altered in the gi mutant, identifying a mechanism for the temperature-specific phenotype of GI. We conclude that a dynamic balance between LHY and GI is key for the effective temperature compensation of the circadian clock at high temperatures, whereas at low temperature LHY appears to be substituted by CCA1. In addition to this role in temperature compensation, GI also plays a critical role in extending the temperature range over which robust and accurate rhythmicity can be maintained.
RESULTS
GI Plays a Critical Role in Maintaining Rhythmicity in Leaf Movement at Higher Temperatures
The QTL PerCv1b was identified as a locus involved in temperature compensation of the circadian clock from a screen of Cvi × Ler recombinant inbred lines. Further mapping of this QTL using near isogenic lines localized the QTL to an ∼900-kb region of chromosome 1. Within this region was the flowering-time gene GI. Sequencing GI from Ler and Cvi revealed a number of polymorphisms, two of which resulted in amino acid substitutions (Edwards et al., 2005). This, together with the period effects of the gi mutant (Park et al., 1999), confirmed GI as a strong candidate for a gene involved in the temperature compensation mechanism.
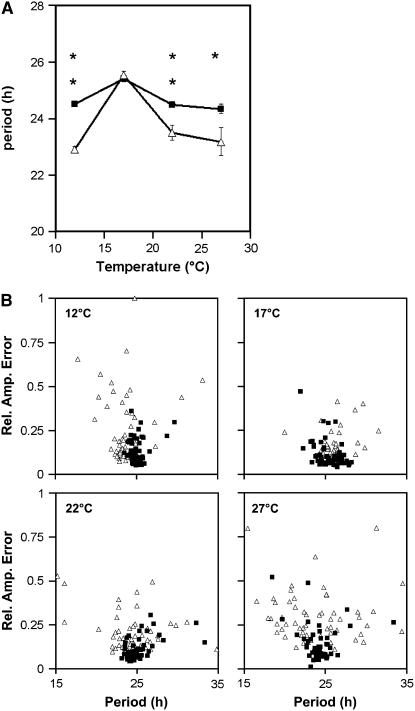
To identify a role for GI in the temperature buffering of the circadian clock, circadian rhythms of leaf movement in the gi null mutant gi-11 over a range of temperatures (12, 17, 22, and 27°C) were characterized. The gi-11 null mutant is a T-DNA insertion mutation and has been described previously (Richardson et al., 1998; Fowler et al., 1999); furthermore, the late-flowering phenotype of the gi-11 mutation could be complemented with a wild-type GI cDNA (see Supplemental Figure 1 online). Both gi-11 and wild-type plants were grown under 12-h-light/12-h-dark cycles (12L:12D) at 22°C to entrain the clock and were then transferred to constant light at 12, 17, 22, or 27°C. The free-running periods of the leaf movement rhythms were measured. Figure 1A shows that at 17°C, the gi mutant (open triangles) and the wild type (closed squares) had no significant period difference. However, as the temperature increased, the period of gi became significantly shorter than that of the wild type. A significant period shortening was also observed in response to a decrease in temperature. This temperature-dependent effect on the free-running period is consistent with GI playing a function in the temperature compensation mechanism of the Arabidopsis clock.
Figure 1.
Temperature Compensation of Leaf Movement Rhythms for Wassilewskija (Ws) Wild Type and Ws gi-11.
Seedlings were entrained under 12L:12D cycles for 7 d, after which they were transferred to constant light at 12, 17, 22, or 27°C, at which rhythms of leaf movement were assayed. For each temperature, 25 to 30 wild-type plants and 25 to 30 gi mutant plants were assayed, corresponding to 50 to 60 leaf movement rhythms.
(A) Variable-weighted mean of period estimates plotted against temperature. Closed squares, wild type; open triangles, gi-11. Error bars represent variance-weighted se. Asterisks indicate t test P values: * P < 0.05, ** P < 0.01.
(B) Period estimates for individual leaves plotted against their relative amplitude errors (Rel. Amp. Error). Closed squares, wild type (n = 50 to 60); open triangles, gi-11 (n = 50 to 60). This experiment was performed independently three times at each of the four temperatures; the results shown are representative.
In addition to a shortening of the period, a temperature-dependent increase in the variance of period was seen for the gi mutant, with the variance increasing from 3.1 at 17°C to 18.0 at 27°C (F test, P = 1.8 × 10−9). Although an increase in the variance of period with temperature was also observed for the wild type, this was not of the same magnitude (from 1.9 at 17°C to 3.8 at 27°C; F test, P = 4.1 × 10−3). The huge increase in the variance of period in the gi mutant at 27°C demonstrates a breakdown in the precision of the clock. This reduced precision could clearly be seen when period estimates for each leaf movement rhythm were plotted against the relative amplitude error (Figure 1B), a measure of rhythm robustness varying from 0 (a perfect fit to the cosine wave) to 1 (not statistically significant). For wild-type period estimates, the associated relative amplitude errors remained low, with all 60 points clustering tightly at each temperature (Figure 1B). However, for the gi mutant, as temperature increased, both the relative amplitude error and the range of period estimates increased. Similarly, as temperature decreased to <17°C, the variability and relative amplitude errors increased (Figure 1B). Similar results were seen for the gi-3 allele in the Ler background, with temperature-dependent effects on both period and rhythm robustness observed (see Supplemental Figure 2 online). However, unlike the null mutant, the period of gi-3 was not wild type at 17°C. The gi-3 mutation introduces a stop codon at the 3′ end of the gene; therefore, a truncated GI protein can be produced. Furthermore, the expression of GI mRNA in the gi-3 mutant had only been reduced by half (Fowler et al., 1999). Similarly, another gi allele isolated in our laboratory, gi-611, which lacked the characteristic flowering phenotype of the standard gi mutants, also had a temperature-dependent period phenotype, although it did not have a reduced robustness phenotype (see Supplemental Figure 3 online). Together, the leaf movement analysis suggests a role for GI not only in buffering the period of the clock against temperature change but also in extending the range of temperature over which rhythmicity can be maintained.
GI Plays a Comparable Role Maintaining the Rhythmicity of Expression of CAB:LUC at High and Low Temperatures
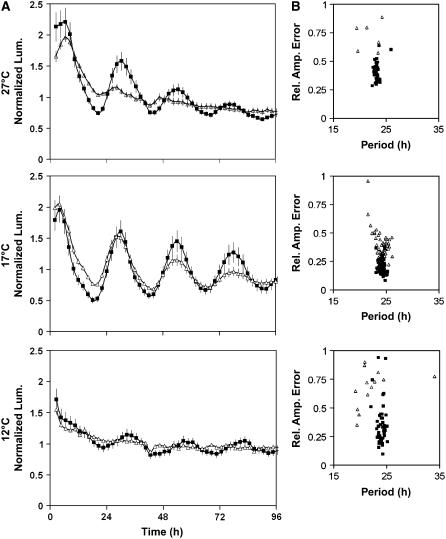
It is clear that GI functions to extend the temperature range at which accurate leaf movement rhythms can be maintained. To investigate whether this extended to other rhythms, the CAB:LUC marker was used to assay the rhythmic expression of CAB in the gi-11 background in plants entrained at 22°C and then free run in constant light at 12, 17, or 27°C. After transfer to 17°C, the rhythmic expression patterns of CAB in the wild type and gi-11 were almost identical (Figure 2A), with no significant difference in period and >98% of gi-11 seedlings producing periods within the circadian range (15 to 35 h). The only difference in the circadian phenotype between gi-11 and the wild type at 17°C was the low amplitude of CAB expression in gi-11 and an increase in the variance of period (period variance of 0.16 for the wild type and 1.14 for gi; F test, P = 3 × 10−12). Thus, at 17°C, the circadian clock is able to sustain rhythms of CAB expression even in the absence of GI. However, upon transfer of gi-11 seedlings from 22 to 27°C, CAB expression rapidly dampened, and by 48 h expression was arrhythmic (Figure 2A). This was reflected by the fact that only 30% (8 of 26) of gi-11 seedlings produced periods for CAB expression within the circadian range, all with relative amplitude errors of >0.5, indicative of weak rhythms. By contrast, 100% (31 of 31) of wild-type seedlings were scored as producing robust rhythms in CAB expression at 27°C. This response was mirrored when gi-11 plants were transferred from 22 to 12°C, with CAB expression rapidly becoming arrhythmic (Figure 2A) and only 35% (15 of 43) of seedlings producing rhythms within the circadian range. These observations are consistent with the leaf movement analysis in that GI functions to extend the temperature range permissive for rhythmicity. Furthermore, its similar effect on these two outputs is consistent with GI acting on the central clock.
Figure 2.
GI Is Required for the Rhythmicity of CAB at 12 and 27°C.
Transgenic seedlings carrying the CAB:LUC reporter gene were entrained under 12L:12D cycles for 7 d, after which the seedlings were transferred to 12, 17, or 27°C and constant red and blue light. Luminescence was monitored in the wild-type Ws (closed squares) and gi-11 (open triangles). For both the wild type and gi-11, expression was monitored in at least three independently transformed lines.
(A) Plots represent the normalized average expression of 8 to 27 single seedlings of one representative transformed line. Error bars indicate se.
(B) Summary of the mathematical analysis of the expression rhythms of the seedlings, represented by plots of period estimates plotted against the respective relative amplitude errors (Rel. Amp. Error) for wild-type Ws and gi-11. At 27°C, wild-type n = 31, gi-11 n = 26; at 17°C, wild-type n = 116, gi-11 n = 51; at 12°C, wild-type n = 52, gi-11 n = 43.
Temperature Regulation of Clock-Related Gene Expression
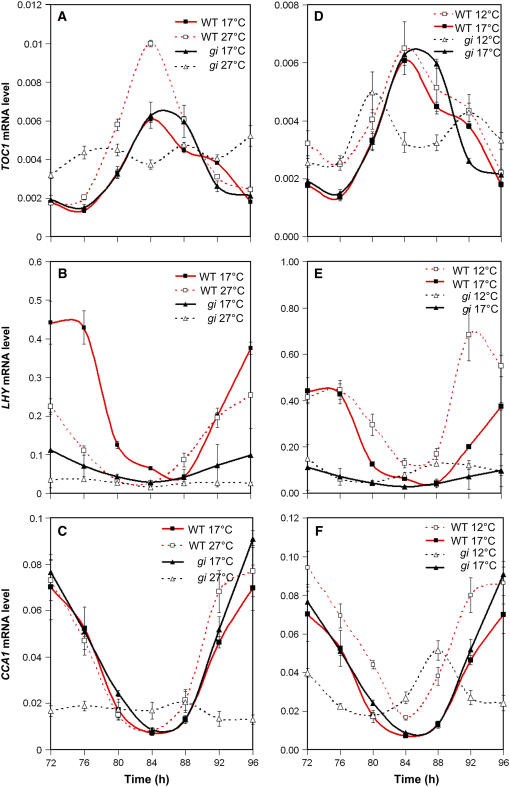
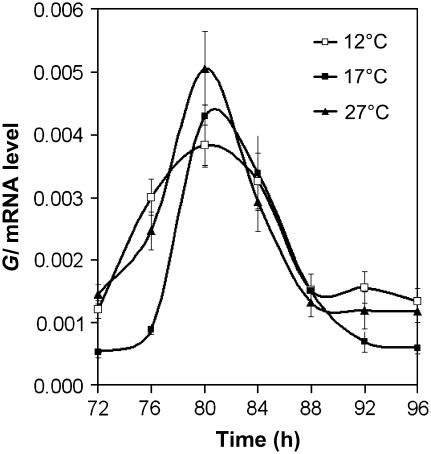
To investigate how key molecular clock components respond to ambient temperature in wild-type plants, the accumulation of transcripts that encoded CCA1, LHY, and TOC1 was measured. Plants were entrained for 7 d with 12L:12D cycles before transfer to 12, 17, or 27°C and constant light. Transcript abundance was then assayed, starting at 72 h after transfer, every 4 h over a 24-h period. Figures 3A to 3C compare transcript abundance and expression patterns of plants free-running at 17 and 27°C. At 27°C, the amplitude of the TOC1 expression rhythm was almost double that at 17°C, with the trough level of expression remaining constant but the peak level increased. Also, the shoulder seen after the peak of expression was lost at 27°C. A temperature-dependent increase in expression was also observed for GI (Figure 4). The reciprocal was observed for LHY levels, with the amplitude of the oscillation of LHY decreasing at 27°C. CCA1 expression showed no significant change with increased temperature. The difference in response of LHY and CCA1 was consistent with the two genes not playing completely redundant roles. A temperature-dependent increase in clock components, similar to that seen for TOC1, has also been observed in Neurospora, in which the amount/amplitude of FRQ protein increased with temperature (Liu et al., 1998). A comparison of the free-running clock at 17 and 12°C revealed no significant differences in the expression of TOC1. However, although less pronounced than the effect of high temperature, peak levels of CCA1 and LHY did increase at 12°C compared with 17°C, and the peak level of GI decreased (Figure 4). Similarly, a reciprocal decrease in GI was also seen (Figures 3D to 3F).
Figure 3.
The Clock Components TOC1, LHY, and CCA1 Have Altered Amplitudes in Response to Temperature and GI Function.
Seedlings were grown at 22°C in 12L:12D conditions for 7 d; the seedlings were then transferred to constant light and 12, 17, or 27°C. They were harvested after 72 h and every 4 h thereafter for the next 24 h. Total RNA was assayed by real-time quantitative PCR, and the accumulation of TOC1 ([A] and [D]), LHY ([B] and [E]), and CCA1 ([C] and [F]) was measured relative to an internal UBIQUITIN (UBQ) control. The plots compare the accumulation of mRNAs at 27°C (open symbols) with that at 17°C (closed symbols) ([A] to [C]) or at 17°C (closed symbols) with that at 12°C (open symbols) ([D] to [F]). Expression in the gi-11 background is represented by triangles, and that in the wild type is represented by squares. Error bars indicate se. Each point represents the average of three biological repeats, with each biological repeat made up of three technical repeats.
Figure 4.
GI mRNA Expression Is Weakly Temperature-Regulated.
Wild-type seedlings were grown at 22°C in 12L:12D conditions for 7 d, and the seedlings were then transferred to constant light and 12°C (open squares), 17°C (closed squares), or 27°C (closed triangles). They were harvested after 72 h and every 4 h thereafter for the next 24 h. Total RNA was assayed by real-time quantitative PCR, and the accumulation of GI was measured relative to an internal UBQ control. Error bars indicate se. Each point represents the average of three biological repeats, with each biological repeat made up of three technical repeats.
Although only subtle changes in the regulation of clock components were observed between 17 and 12°C, in a microarray experiment comparing seedlings grown at 22 and 4°C, transcripts encoding many of the known clock components rapidly dampened to a high level upon transfer to 4°C (NASCArrays experiment reference number NASCARRAYS-138; see Supplemental Figure 4 online). Although this microarray analysis is consistent with a loss of clock function at 4°C, a longer time course will be required to confirm this finding. However, this apparent loss of rhythmicity has also been observed in chestnut (Castanea sativa), in which the levels of Cs LHY and Cs TOC1 dampened to a high level upon transfer to 4°C, with LHY failing to repress TOC1 levels (Ramos et al., 2005).
Temperature-Dependent GI Function at the Molecular Level
Both our leaf movement experiments and CAB:LUC expression data suggest that GI functions to maintain accurate and robust rhythmicity at high and low temperatures. Moreover, the effect on multiple outputs would support the hypothesis that GI acts directly on the central clock at higher and lower temperatures. Previous studies investigating GI's role in the clock found that CCA1 and LHY levels are reduced in plants grown under long days at 22°C in the gi-3 mutant. This finding supports a function for GI in the regulation of CCA1 and LHY (Fowler et al., 1999). To determine whether GI's effects on the key clock components are temperature-dependent, we compared the expression profiles of TOC1, CCA1, and LHY in wild-type and gi-11 seedlings free-running in constant light at 27 and 17°C, as described above (Figure 3). At 17°C, TOC1 expression levels were identical in both the gi-11 and wild-type plants. At 27°C, TOC1 levels failed to increase in the gi-11 background and were expressed at an almost constant level. At 17°C, the expression of LHY was lower in the gi mutant, and this may contribute to the reduction in amplitude of the CAB:LUC oscillations observed in gi-11 at 17°C. At 27°C, the expression level and amplitude of LHY were further significantly reduced. For CCA1 expression, in contrast with LHY, the level, amplitude, and expression profile were identical in the gi mutant and the wild type at 17°C, demonstrating a clear difference in the regulation of the LHY and CCA1 genes by GI. At 27°C in the wild type, CCA1 expression was temperature-independent, whereas in the gi mutant, expression became temperature-dependent, with levels and amplitude of CCA1 decreasing at 27°C. Together, our expression analysis of key clock components suggests a mechanism whereby GI maintains rhythmicity and accuracy at higher temperatures by the temperature-dependent regulation of TOC1, thereby maintaining sufficient levels/amplitude of CCA1 and LHY to sustain accurate and robust clock function.
We also tested changes in gene expression in CCA1, LHY, and TOC1 in the wild type and the gi mutant at 12°C (Figure 3). Little change in the expression of clock components in response to decreases in temperature was observed in the wild type; however, in the gi mutant, the expression of both CCA1 and TOC1 decreased, with the expression of TOC1 becoming biphasic, with two peaks of expression only 12 h apart. This was not observed for CCA1 or LHY, consistent with a breakdown of the feedback regulatory loop composed of CCA1/LHY and TOC1 described by Alabadi et al. (2001). The expression of LHY remained at a similar level at both 17 and 12°C, although both CCA1 and LHY expression had an altered phase, suggesting that the period length of CCA1 and LHY is shorter at 12°C, which is consistent with the short-period phenotype of rhythms of leaf movement (Figure 1A).
The Interlocking Loop Model Supports GI Function in Temperature Compensation
GI is proposed to contribute to a key component, Y, of the interlocking loop model for the Arabidopsis clock mechanism (Locke et al., 2005). Therefore, we tested whether the observed temperature-dependent effects of GI were consistent with the function of Y in this model. Without explicit modeling, it would be difficult to determine whether this was the case or whether our experimental data reflected additional temperature-dependent functions of GI. mRNA expression profiles for TOC1 and LHY/CCA1 at 27, 17, and 12°C for both wild-type and gi genotypes were simulated (see Methods; see also Supplemental Methods and Supplemental Figures 5 and 6 online). As temperature effects were not explicitly included in the model, a qualitative fit was obtained by varying only the transcription rates of the model components LHY and Y. LHY in the model represents both LHY and CCA1. Y represents GI, although unidentified genes with functions that overlap with GI may also contribute to Y.
The wild type at 27°C was modeled as having reduced LHY expression (Figure 3), which would shorten the circadian period if this were the only effect of temperature. Higher GI expression in the model (following the trend in Figure 4) compensated for the reduced LHY levels, substantially restoring the period length and also recapitulating several features of the molecular data in Figure 3 (see Supplemental Methods and Supplemental Figure 5 online). In particular, a gi mutation in the model had a more severe low-amplitude phenotype at 27°C compared with the standard temperature (see Supplemental Figure 5 online), consistent with the experimental data (Figures 1 and 2). The wild type at 12°C was modeled by increasing the transcription rate of LHY, as LHY RNA levels are increased at lower temperatures (see Supplemental Figure 3C online). GI RNA levels were little reduced at 12°C (Figure 4), but reducing Y transcription rates in the model again compensated the circadian period. The simulated gi mutation again recapitulated the observed effects of the gi mutant at 12°C (see Supplemental Figure 6 online). We propose that the reduced Y function that was required to simulate the wild type at 12°C might represent downregulation of another gene that contributes to Y, overlapping with GI function (see Supplemental Methods online). Altering only the LHY and Y transcription rates was sufficient to recreate the major effects of temperature on clock gene expression, showing that the temperature-dependent effects of GI proposed here are consistent with GI's location as a component of Y in the two-feedback-loop model.
LHY and CCA1 Contribute Differentially to Temperature Compensation
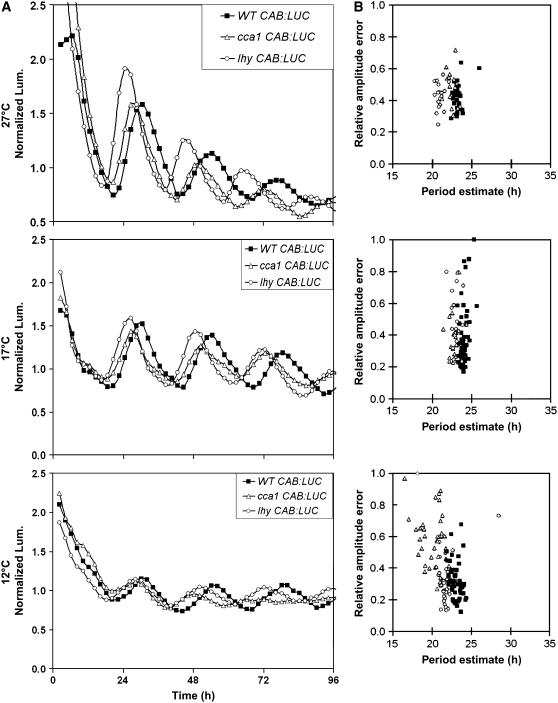
Our data suggest that a dynamic balance between LHY and GI functions maintains robust and accurate rhythmicity at higher temperatures and that LHY function is distinct from CCA1 in this respect. To confirm the function of LHY in this mechanism, we tested temperature compensation in the lhy and cca1 loss-of-function mutants. Both lhy and cca1 maintained robust rhythms of CAB:LUC expression at 17 and 27°C (Figure 5). Both mutants had the same short-period phenotype at 17°C (wild type, 23.9 h; cca1, 22.8 h; lhy, 22.8 h). At 27°C, a period difference of ∼1 h was maintained between the wild type (23.0 h) and cca1 (22.2 h), whereas in the lhy mutant, the period difference increased to nearly 2 h (wild type, 23.0 h; lhy, 21.1 h) (Figure 5). The lhy mutant, therefore, has a shorter period than cca1 (Student's t test, P < 0.01) at 27°C, supporting a role for LHY in the temperature compensation mechanism. This was suggested previously by analysis of natural genetic variation (Edwards et al., 2005), as LHY was noted as a candidate gene for the temperature-specific QTL PerCv1a and PerCola. At 12°C, the period of the wild type (23.3 h) differed from that of lhy (21.6 h) by 1.7 h. In cca1, however, the rhythm of CAB:LUC expression was dampened (Figure 5), and the period difference between the wild type (23.3 h) and cca1 (20.0 h) was 3.3 h. This temperature-dependent shortening of the circadian period and the reduction in rhythm robustness almost phenocopied the effect of gi-11 at 12°C (Figures 1 and 2). At lower temperatures, our data indicate that CCA1 plays a greater role than LHY in temperature compensation and the maintenance of rhythm robustness, confirming that CCA1 and LHY functions are not completely overlapping.
Figure 5.
The lhy Mutant Is Compromised in Its Ability to Buffer the Clock against Increases in Temperature, and the cca1 Mutant Is Compromised in Its Buffering Capacity at Lower Temperatures.
Transgenic seedlings carrying the CAB:LUC reporter gene were entrained under 12L:12D cycles for 7 d, after which the seedlings were transferred to 12, 17, or 27°C and constant red and blue light. Luminescence was monitored over the subsequent 96 h.
(A) Plots represent average normalized expression of CAB in wild-type Ws (closed squares), the cca1 mutant (open triangles), and the lhy mutant (open circles) at 12, 17, or 27°C. Mutant and wild-type expression was monitored in at least four independently transformed lines, and the plots show average expression in representative CAB:LUC lines.
(B) Summary of the mathematical analysis of the expression rhythms of the seedlings, represented by plots of period estimates plotted against the respective relative amplitude errors for wild-type Ws (closed squares), cca1 (open triangles), and lhy (open circles).
DISCUSSION
Our characterization of key components of the circadian clock over a range of temperatures has uncovered temperature-dependent changes in their gene expression. We found that levels of LHY mRNA decrease with temperature increases and that this was counterbalanced by increases in TOC1 and GI. By contrast, CCA1 levels changed little between 27 and 17°C; however, we did see an increase at lower temperatures (Figures 3 and 4). This balance between clock components is consistent with the antagonistic balance model of temperature compensation put forward by Ruoff et al. (1997). We tested whether these new data could fit within the constraints of a proposed interlocking-loop model of the circadian clock, incorporating GI (Locke et al., 2005).
First, we considered compensation of the clock in response to increasing temperature. The effects of increases in temperature could be simulated by reducing LHY transcription rates, causing a shortening of the period; however, this could be balanced by increasing GI transcription levels. Although the experimental increases in GI expression were not pronounced, there was a gradual increase in peak expression with temperature (Figure 4). It has been shown that the GI protein oscillates and that this is regulated at the posttranscriptional level (David et al., 2006). Therefore, it is possible that levels of GI could also be modulated by temperature. This is consistent with the temperature compensation mechanisms of other organisms (Rutila et al., 1996; Liu et al., 1997; Lahiri et al., 2005). By altering the transcription parameters of GI and LHY alone, the period of the model clock system was balanced and there was little difference in the phase of either LHY or TOC1 mRNA with temperature (see Supplemental Figure 5 online). Together, the simulation and experimental data are supportive of the conclusion that LHY and GI form a dynamic counterbalance to temperature-compensate the clock at high temperature.
In support of this dynamic counterbalance hypothesis, we show that a null gi mutant (gi-11) has a temperature-dependent circadian phenotype, affecting both robustness and period (Figures 1 and 2). In gi-11, the counterbalance between GI and LHY is lost and LHY RNA levels decrease at 27°C. A similar temperature compensation phenotype was also observed in the gi-611 and gi-3 alleles; however, the partial gi-611 mutation does not display the loss of rhythm robustness with temperature (see Supplemental Figure 3 online). The lhy loss-of-function mutant likewise shows a strong period alteration at high temperature (Figure 5).
This proposal can account for temperature compensation at 27°C, but our experimental data show that different processes are involved at low temperature. Specifically, between 17 and 12°C, GI expression is little altered in the wild type, and the gi mutation reduces the expression of CCA1 rather than LHY. Furthermore, at 12°C, the cca1 loss-of-function mutant alters period more severely than the lhy mutation and also reduces rhythmic robustness, the reverse of the situation at 27°C. At low temperature, our data are consistent with CCA1 replacing LHY in temperature compensation of the clock. The switch in the apparent importance of these two transcription factors may reflect unknown differences in temperature-dependent biochemical properties of the two proteins. GI participates by regulating the relative expression levels of CCA1 and LHY in a temperature-dependent manner, as demonstrated by the effects of the gi mutation, thus contributing to the stable period over a wide range of temperatures. The qualitative effect of decreases in temperature could be simulated by increasing LHY transcription rates, causing an increase in the period, which could be balanced by decreasing the transcription rates of components of Y redundant to GI (see Supplemental Methods Figure 6 online). In addition to its effect on period, the gi null mutant affects the robustness of circadian rhythms, an effect not shared by the partial gi-611 allele. The lhy mutant at 27°C affects only the circadian period, not rhythmic robustness, but cca1 at 12°C affects both, phenocopying the gi mutant. This difference between the gi and lhy mutants suggests that the effect of GI on rhythm robustness at high temperature functions via a mechanism that is less dependent on LHY than the effect on circadian period.
Simulating the effects of temperature by altering LHY and Y expression showed that these counterbalancing effects were largely sufficient to replicate the observed effects of temperature on clock gene expression in the wild type. The simulations also confirmed that the effects of the gi mutation on the clock at all temperatures are consistent with the identification of GI as part of Y. Temperature, in fact, will affect many processes in addition to LHY and Y transcription rates (potentially, all 61 kinetic parameters in the model). Obtaining simulations that exactly fit the data would likely require all of the parameters to be estimated afresh for each temperature. To constrain the estimation, most of the experimental results that were used in developing the original model would also have to be replicated at 12 and 27°C. The current clock model must also be extended to account for our temperature-specific data. For example, LHY and CCA1 were modeled as a single gene and must be separated to explore the differences in their functions at high and low temperature.
This role of GI in extending the temperature range over which robust rhythms can be maintained is analogous to that of the two alternatively spliced forms of the Neurospora clock gene FRQ, FRQ1-989 and FRQ100-989 (Garceau et al., 1997). Both the long and short forms of FRQ are capable of restoring rhythmicity in a frq null strain; however, the long form is unable to sustain rhythmicity at low temperatures, and the short form is unable to sustain rhythmicity at high temperatures (Liu et al., 1997). Thus, the two spliced forms together extend the range of temperatures over which rhythms can be maintained. From our data, it is not clear how GI extends the range of rhythmicity. However, it is possible that the GI/TOC1 loop of the interlocking-loop model proposed by Locke et al. (2005) plays an important function in stabilizing the clock at higher temperatures.
Previously, it was demonstrated that the flowering phenotype of gi mutants is temperature-independent, with the exception of gi-2 (Araki and Komeda, 1993). Here, we demonstrate that the circadian phenotype of gi alleles is temperature-dependent. Together, these two observations suggest that the circadian and flowering phenotypes of gi mutants result from distinct functions of GI. This conclusion is further supported by the isolation of two new gi alleles, gi-611 and gi-596 (see Supplemental Figure 1 online). These have short- and long-period circadian phenotypes, respectively, but neither shows the striking late-flowering phenotype of other gi alleles under long photoperiods. Indeed, gi-611 mutants flowered early in short photoperiods, which might be predicted from their short-period phenotype (Yanovsky and Kay, 2002). A similar conclusion has been drawn by Mizoguchi et al. (2005) based on the characterization of a GI overexpressor and gi-3. They demonstrated a disproportionate effect of both GI overexpression and the gi-3 mutation on the control of circadian clock–regulated flowering genes compared with other circadian clock–regulated genes.
Here, we have shown that GI plays a critical role in temperature compensation of the clock and in extending the range of temperatures at which rhythmicity can be maintained. We recently demonstrated that having a robust and accurate clock increases photosynthesis and productivity in Arabidopsis (Dodd et al., 2005); thus, our new insight into how the clock maintains robustness and accuracy at high temperatures is likely to have implications for enhancing the performance of plants at higher and lower temperatures. It is feasible that this could be used to extend the geographical range of crops and potentially allow the development of new varieties able to cope better with global climate change.
METHODS
Plant Material
Arabidopsis thaliana gi-11 was isolated in a screen of T-DNA insertion lines described by Richardson et al. (1998) and Fowler et al. (1999). The CAB:LUC+ transgene, in the Ws background, was as described by Hall et al. (2002). The cca1-11 and lhy-21 mutants were isolated from the Arabidopsis Functional Genomics Consortium population (Krysan et al., 1999). Single mutants have been described (Hall et al., 2003). The mutants gi-611 and gi-596 were isolated in a screen for mutants with altered temporal expression of CAB from an ethyl methanesulfonate population of the Ws CAB:LUC transgenic line 6A.
Growth Conditions
For luminescence, RNA extraction, and leaf movement analysis, seedlings were first surface-sterilized in 70% ethanol for 2 min, immediately followed by 50% bleach for 10 min. They were then rinsed twice in sterile distilled water and resuspended in 0.15% agar. The seedlings were then sown on Murashige and Skoog medium containing 3% sucrose and 1.5% agar. Seeds were kept at 4°C for 2 d and then grown in 12L:12D cycles of 80 μmol·m−2·s−1 in a Sanyo MLR350 plant growth chamber. Temperatures both during entrainment and during experiments were logged using Hobo temperature loggers (Onset Computer).
Rhythm Analysis
Leaf movement rhythms were measured using a time-lapse video imaging system as described by Edwards et al. (2005). However, instead of Ultra-track cameras, Sony Exwave HAD cameras (Sovereign International) were used, having high resolution and allowing the number of plants assayed on one plate to be increased from 12 to 15. Luminescence levels were analyzed using an ORCA-II-BT 1024 16-bit camera cooled to −80°C (Hamamatsu Photonics). The camera was housed on top of a Sanyo MIR-553 cooled incubator maintaining a uniform temperature ±0.5°C (Sanyo Gallenkamp). Illumination was provided by four red/blue light-emitting diode arrays (MD Electronics). Image acquisition and light control were driven by WASABI imaging software (Hamamatsu Photonics). The images were processed using Metamorph 6.0 image-analysis software (Molecular Devices). Individual period estimates were generated by importing data into BRASS (available from www.amillar.org) and using BRASS to run fast Fourier transform nonlinear least-squares analysis programs (Plautz et al., 1997) on each data trace to generate period estimates and relative amplitude errors.
RNA Analysis
Plants entrained in 12L:12D for 7 d at 22°C were transferred to constant light at 12, 17, 22, and 27°C. After 72 h of free-running conditions, ∼100 seedlings were harvested every 4 h for 24 h and frozen in liquid nitrogen. Total RNA was extracted and treated with DNase using RNeasy plant mini kits (Qiagen) according to the manufacturer's instructions. A 2-μg portion was reverse-transcribed using the Advantage-for-PCR kit (BD Bioscience) with random hexamer primers according to the manufacturer's instructions. TOC1, LHY, CCA1, and GI transcript abundance was measured in each sample relative to UBQ10 using quantitative real-time PCR in a Rotor-Gene 3000 real-time PCR machine (Corbett Research). Each reaction contained 2 μL of cDNA product and 6 μL of QuantiTect SYBR Green PCR mix (Qiagen) together with gene-specific primers: UBQ10 forward primer, 5′-CACACTCCACTTGGTCTTGCGT-3′; UBQ10 reverse primer, 5′-TGGTCTTTCCGGTGAGAGAGTCTT-3′; TOC1 forward primer, 5′-TCTTCGCAGAATCCCTGTGAT-3′; TOC1 reverse primer, 5′-GCTGCACCTAGCTTCAAGCA-3′; LHY forward primer, 5′-ACGAAACAGGTAAGTGGCGACA-3′; LHY reverse primer, 5′-TGGGAACATCTTGAACCGCGTT-3′; CCA1 forward primer, 5′-GATGATGTTGAGGCGGATG-3′; CCA1 reverse primer, 5′-TGGTGTTAACTGAGCTGTGAAG-3′; GI forward primer, 5′-GGTCGACGGTTTCTCCAATCTA-3′; GI reverse primer, 5′-CGGACTATTCATTCCGTTCTTC-3′.
The UBQ10 and LHY primers have been described previously (Czechowski et al., 2004), as have the CCA1 and GI primers (Hall et al., 2003) and the TOC1 primers (Farre et al., 2005). The efficiency value of amplification for each set of primers was determined beforehand by measuring the abundance of transcripts from a cDNA dilution series. Efficiency values were computed for each primer set using REST (http://www.wzw.tum.de/gene-quantification/; Pfaffl et al., 2002). Each RNA sample was assayed in triplicate, and RNAs were assayed from three biological repeats. The TOC1, LHY, CCA1, and GI transcript abundance levels were normalized to UBQ using Q-gene software (http://www.biotechniques.com/softlib/qgene.html; Muller et al., 2002).
Computational Methods
Simulations were performed in MATLAB and Circadian Modeling, a user-friendly simulation interface that is freely available from www.amillar.org/downloads.html. Starting parameter values and model equations were as described (Locke et al., 2005). Parameter values were modified to replicate experimental data at high and low temperatures, as described in the Supplemental Methods online.
Accession Numbers
Sequence data for GI, LHY, CCA1, and TOC1 can be found in the GenBank/EMBL data libraries under the following accession numbers: At1g22770 (GI), At1g01060 (LHY), At2g46830 (CCA1), and At5g61380 (TOC1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Late-Flowering Phenotype of the gi-11 Mutation Can Be Complemented with a Wild-Type GI cDNA.
Supplemental Figure 2. Temperature Compensation of Leaf Movement Rhythms for Ler Wild Type and Ler gi-3.
Supplemental Figure 3. Amino Acid Substitutions in the N-Terminal Half of the GI Protein Can Lengthen or Shorten Circadian Period but Do Not Result in Late Flowering.
Supplemental Figure 4. At 4°C, the Oscillations of Key Clock Genes Dampen Rapidly.
Supplemental Figure 5. Modeling the Temperature Compensation Mechanism at High Temperatures.
Supplemental Figure 6. Modeling the Temperature Compensation Mechanism at Low Temperatures.
Supplemental Methods.
Supplementary Material
Acknowledgments
This research was initiated in A.J.M.'s group at the University of Warwick and pursued in A.H.'s group at the University of Liverpool. A.H. and C.L. carried out the leaf movement analysis in Liverpool, A.H. in Warwick. A.H. and P.D.G. carried out the luciferase analysis of the gi mutants and cca1 and lhy mutants in Liverpool, A.H. and M.M.S. in Warwick. Real-time PCR was performed by P.D.G. Modeling work was carried out by J.C.W.L. The gi-611 and gi-596 mutants were isolated in a screen performed by M.M.S., S.H., A.H., and S.J.D., and the flowering-time measurement was carried out by J.P. We are grateful to Victoria Hibberd and Nazir Sharrif for expert technical assistance in Warwick, to Ferenc Nagy and László Kozma-Bognár for sequencing the gi alleles, and to N. Bueno del Carpio for reading the manuscript. M.M.S. was supported by a PhD studentship from the Biotechnology and Biological Science Research Council (BBSRC), S.J.D. by a Department of Energy Fellowship of the Life Sciences Research Foundation, and S.H. in part by a postdoctoral fellowship from the Japan Society for the Promotion of Science. Research at Liverpool was funded by BBSRC Grant BBS/B/11125 and Royal Society Grant R4917/1 to A.H. Research at Warwick was funded by BBSRC Awards G08667, G13967, and G15231 to A.J.M.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Andrew J. Millar (andrew.millar@ed.ac.uk) and Anthony Hall (anthony.hall@liverpool.ac.uk).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.039990.
References
- Alabadi, D., Oyama, T., Yanovsky, M.J., Harmon, F.G., Mas, P., and Kay, S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1. Science 293 880–883. [DOI] [PubMed] [Google Scholar]
- Alabadi, D., Yanovsky, M.J., Mas, P., Harmer, S.L., and Kay, S.A. (2002). Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol. 12 757–761. [DOI] [PubMed] [Google Scholar]
- Araki, T., and Komeda, Y. (1993). Analysis of the role of the late-flowering locus, GI, in the flowering of Arabidopsis thaliana. Plant J. 3 231–239. [Google Scholar]
- Czechowski, T., Bari, R.P., Stitt, M., Scheible, W.R., and Udvardi, M.K. (2004). Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: Unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J. 38 366–379. [DOI] [PubMed] [Google Scholar]
- David, K.M., Armbruster, U., Tama, N., and Putterill, J. (2006). Arabidopsis GIGANTEA protein is post-transcriptionally regulated by light and dark. FEBS Lett. 580 1193–1197. [DOI] [PubMed] [Google Scholar]
- Dodd, A.N., Salathia, N., Hall, A., Kevei, E., Toth, R., Nagy, F., Hibberd, J.M., Millar, A.J., and Webb, A.A. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309 630–633. [DOI] [PubMed] [Google Scholar]
- Edwards, K.D., Anderson, P.E., Hall, A., Salathia, N.S., Locke, J.C., Lynn, J.R., Straume, M., Smith, J.Q., and Millar, A.J. (2006). FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, K.D., Lynn, J.R., Gyula, P., Nagy, F., and Millar, A.J. (2005). Natural allelic variation in the temperature-compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics 170 387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre, E.M., Harmer, S.L., Harmon, F.G., Yanovsky, M.J., and Kay, S.A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15 47–54. [DOI] [PubMed] [Google Scholar]
- Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Coupland, G., and Putterill, J. (1999). GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garceau, N.Y., Liu, Y., Loros, J.J., and Dunlap, J.C. (1997). Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell 89 469–476. [DOI] [PubMed] [Google Scholar]
- Hall, A., Bastow, R.M., Davis, S.J., Hanano, S., McWatters, H.G., Hibberd, V., Doyle, M.R., Sung, S., Halliday, K.J., Amasino, R.M., and Millar, A.J. (2003). The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15 2719–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, A., Kozma-Bognar, L., Bastow, R.M., Nagy, F., and Millar, A.J. (2002). Distinct regulation of CAB and PHYB gene expression by similar circadian clocks. Plant J. 32 529–537. [DOI] [PubMed] [Google Scholar]
- Hall, A., and McWatters, HG. (2006). Endogenous Plant Rhythms. (Oxford, UK: Blackwell Publishing).
- Harmer, S.L., Panda, S., and Kay, S.A. (2001). Molecular bases of circadian rhythms. Annu. Rev. Cell Dev. Biol. 17 215–253. [DOI] [PubMed] [Google Scholar]
- Huq, E., Tepperman, J.M., and Quail, P.H. (2000). GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 97 9789–9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri, K., Vallone, D., Gondi, S.B., Santoriello, C., Dickmeis, T., and Foulkes, N.S. (2005). Temperature regulates transcription in the zebrafish circadian clock. PLoS Biol. 3 e351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Garceau, N.Y., Loros, J.J., and Dunlap, J.C. (1997). Thermally regulated translational control of FRQ mediates aspects of temperature responses in the Neurospora circadian clock. Cell 89 477–486. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Merrow, M., Loros, J.J., and Dunlap, J.C. (1998). How temperature changes reset a circadian oscillator. Science 281 825–829. [DOI] [PubMed] [Google Scholar]
- Locke, J.C.W., Southern, M.M., Kozma-Bognar, L., Hibberd, V., Brown, P.E., Turner, M.S., and Millar, A.J. (2005). Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol. Syst. Biol. http://dx.doi.org/10.1038/msb4100018. [DOI] [PMC free article] [PubMed]
- Majercak, J., Sidote, D., Hardin, P.E., and Edery, I. (1999). How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24 219–230. [DOI] [PubMed] [Google Scholar]
- Mittag, M., Kiaulehn, S., and Johnson, C.H. (2005). The circadian clock in Chlamydomonas reinhardtii. What is it for? What is it similar to? Plant Physiol. 137 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi, T., Wheatley, K., Hanzawa, Y., Wright, L., Mizoguchi, M., Song, H.R., Carre, I.A., and Coupland, G. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2 629–641. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Wright, L., Fujiwara, S., Cremer, F., Lee, K., Onouchi, H., Mouradov, A., Fowler, S., Kamada, H., Putterill, J., and Coupland, G. (2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17 2255–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, P.Y., Janovjak, H., Miserez, A.R., and Dobbie, Z. (2002). Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32 1372–1374, 1376, 1378–1379. [PubMed] [Google Scholar]
- Park, D.H., Somers, D.E., Kim, Y.S., Choy, Y.H., Lim, H.K., Soh, M.S., Kim, H.J., Kay, S.A., and Nam, H.G. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285 1579–1582. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W., Horgan, G.W., and Dempfle, L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30 e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz, J.D., Straume, M., Stanewsky, R., Jamison, C.F., Brandes, C., Dowse, H.B., Hall, J.C., and Kay, S.A. (1997). Quantitative analysis of Drosophila period gene transcription in living animals. J. Biol. Rhythms 12 204–217. [DOI] [PubMed] [Google Scholar]
- Ramos, A., Perez-Solis, E., Ibanez, C., Casado, R., Collada, C., Gomez, L., Aragoncillo, C., and Allona, I. (2005). Winter disruption of the circadian clock in chestnut. Proc. Natl. Acad. Sci. USA 102 7037–7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rédei, G.P. (1962). Supervital mutants of Arabidopsis. Genetics 47 443–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, K., Fowler, S., Pullen, C., Skelton, C., Morris, B., and Putterill, J. (1998). T-DNA lagging of a flowering-time gene and improved gene transfer by in planta transformation of Arabidopsis. Aust. J. Plant Physiol. 25 125–130. [Google Scholar]
- Ruoff, P., Rensing, L., Kommedal, R., and Mohsenzadeh, S. (1997). Modeling temperature compensation in chemical and biological oscillators. Chronobiol. Int. 14 499–510. [DOI] [PubMed] [Google Scholar]
- Rutila, J.E., Zeng, H.K., Le, M., Curtin, K.D., Hall, J.C., and Rosbash, M. (1996). The tim(sl) mutant of the Drosophila rhythm gene timeless manifests allele-specific interactions with period gene mutants. Neuron 17 921–929. [DOI] [PubMed] [Google Scholar]
- Sawyer, L.A., Hennessy, J.M., Peixoto, A.A., Rosato, E., Parkinson, H., Costa, R., and Kyriacou, C.P. (1997). Natural variation in a Drosophila clock gene and temperature compensation. Science 278 2117–2120. [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410 1116–1120. [DOI] [PubMed] [Google Scholar]
- Yanovsky, M.J., and Kay, S.A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419 308–312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.