Abstract
Mutations in genes encoding components of the heterotrimeric G-protein complex were previously shown to confer altered sensitivity to increased levels of d-glucose. This suggests that G-protein coupling may be a novel sugar-signaling mechanism in Arabidopsis thaliana. THYLAKOID FORMATION1 (THF1) is here demonstrated in vivo as a Gα interaction partner that functions downstream of the plasma membrane–delimited heterotrimeric G-protein (GPA1) in a d-glucose signaling pathway. THF1 is a plastid protein localized to both the outer plastid membrane and the stroma. Contact between root plastidic THF1 and GPA1 at the plasma membrane occurs at sites where the plastid membrane abuts the plasma membrane, as demonstrated by Förster resonance energy transfer (FRET). A probable role for THF1 in sugar signaling is demonstrated by both biochemical and genetic evidence. Root growth in the thf1-1 null mutant is hypersensitive to exogenous d-glucose, and THF1-overexpressing roots are resistant to inhibition of growth rate by high d-glucose. Additionally, THF1 levels are rapidly degraded by d-glucose but not l-glucose. The interaction between THF1 and GPA1 has been confirmed by in vitro and in vivo coimmunoprecipitation, FRET analysis, and genetic epistasis and provides evidence of a sugar-signaling mechanism between plastids and the plasma membrane.
INTRODUCTION
d-Glucose, a vital cellular nutrient, serves as both a source of energy and a substrate. Not surprisingly given its paramount role, d-glucose also causes a number of hormone-like responses in a variety of eukaryotes, affecting the regulation of gene expression and developmental processes (Sheen et al., 1999; Rolland et al., 2001). Because all plants both produce and use d-glucose, they must be capable of specifically adapting to changing levels of this hexose. The mechanisms of d-glucose signaling in plants are largely unknown, but one involves at least in part a hexokinase-metabolizing d-glucose transported into cells (Sheen et al., 1999; Rolland et al., 2001; Moore et al., 2003).
There is emerging evidence that other sugar-signaling mechanisms exist in plants and yeast (Rolland et al., 2001; Eastmond and Graham, 2003; Tiessen et al., 2003; Kolbe et al., 2005), including a hexokinase-independent mechanism involving key components of the G-protein heterotrimer (Ullah et al., 2002; Chen et al., 2003; Chen and Jones, 2004). This is not without precedent, as a G-protein–coupled d-glucose signaling mechanism was recently identified in the yeast Saccharomyces cerevisiae, in which it has been shown that sugar agonists and antagonists bind with low affinity to Gpr1, a G-protein–coupled receptor (GPCR) (Lemaire et al., 2004). This, in turn, initiates cyclic AMP signaling in a pathway involving protein kinase A, a G-protein α subunit, and a regulator of G-protein signaling (RGS) protein (Colombo et al., 1998; Versele et al., 1999).
It is well established that the Arabidopsis thaliana genome encodes a single canonical Gα subunit (GPA1) (Ma et al., 1990; Ullah et al., 2001), a single Gβ subunit (AGB1) (Weiss et al., 1994), two Gγ subunits (AGG1 and AGG2) (Mason and Botella, 2000, 2001), and a single RGS protein (RGS1) (Chen et al., 2003). By contrast, there are an estimated 23 Gα, 6 Gβ, and 12 Gγ subunit genes in mammals (Vanderbeld and Kelly, 2000) and 37 RGS proteins (Siderovski and Willard, 2005), making Arabidopsis an advantageous model system for the study of G-protein–coupled signaling (Jones and Assmann, 2004). However, a plant GPCR, with its cognate ligand activating the plant G-protein complex, has not been identified, although candidate GPCRs have been proposed (Devoto et al., 1999; Pandey and Assmann, 2004). Furthermore, signaling elements activated by plant G-proteins (i.e., effectors) are few (Jones and Assmann, 2004). To date, only two candidate effectors have been shown to interact with GPA1 in vitro: one interaction suggests a role for heterotrimeric G-proteins in the regulation of germination and seedling development (Lapik and Kaufman, 2003), and the other demonstrates the regulation of phospholipase D activity, possibly via interaction at a DRY motif similar to that found in G-protein–coupled receptors (Zhao and Wang, 2004).
Despite the apparent lack of information about the components of G-protein signaling mechanisms, this linchpin element has been implicated in a wide array of signals in plants (Wang et al., 2001; Ullah et al., 2002, 2003; Booker et al., 2004; Joo et al., 2005). Thus, in plants, mutant analyses have shown that heterotrimeric G-proteins represent a critical nexus in the signal regulation of a variety of processes such as germination, cell division and elongation, stress responses, and plant morphology (Perfus-Barbeoch et al., 2004). As such, the field is wide open for the identification of novel interaction partners for Gα.
Because null mutations in GPA1 and RGS1 have been shown to confer altered sensitivities to d-glucose (Ullah et al., 2002; Chen et al., 2003; Chen and Jones, 2004), we explored the mechanism of G-protein–coupled d-glucose signaling in Arabidopsis. Because the main target of RGS1 is the activated GTP-bound form of GPA1 (GPA1GTP) (Chen et al., 2003) and because we have shown that the activated form of GPA1 can confer sugar resistance (this study), we sought physical interactors to GPA1GTP as potential effectors or modifiers in G-protein–coupled sugar signaling. We identified a GPA1-interacting protein designated THYLAKOID FORMATION1 (THF1) (Wang et al., 2004), and through both biochemical and genetic approaches, we provide evidence that THF1 functions as a downstream component of the Arabidopsis G-protein–coupled sugar-signaling pathway.
Here, we show that THF1 is ubiquitously expressed in Arabidopsis, with the highest THF1 promoter:β-glucuronidase (GUS) activity observed in root meristems. THF1 is localized to the outer plastid membrane and/or the plastid stroma and is also found in plastid stromules. Although the concept of extraplastidic signaling via stromules interacting with the plasma membrane was revisited recently (Kwok and Hanson, 2004a, 2004b, 2004c), this work demonstrates a specific signaling event between the plastid and the plasma membrane that represents a novel sugar-signaling mechanism in eukaryotic cells.
RESULTS
The Arabidopsis Heterotrimeric G-Protein Is Involved in Sugar Signaling in the Root
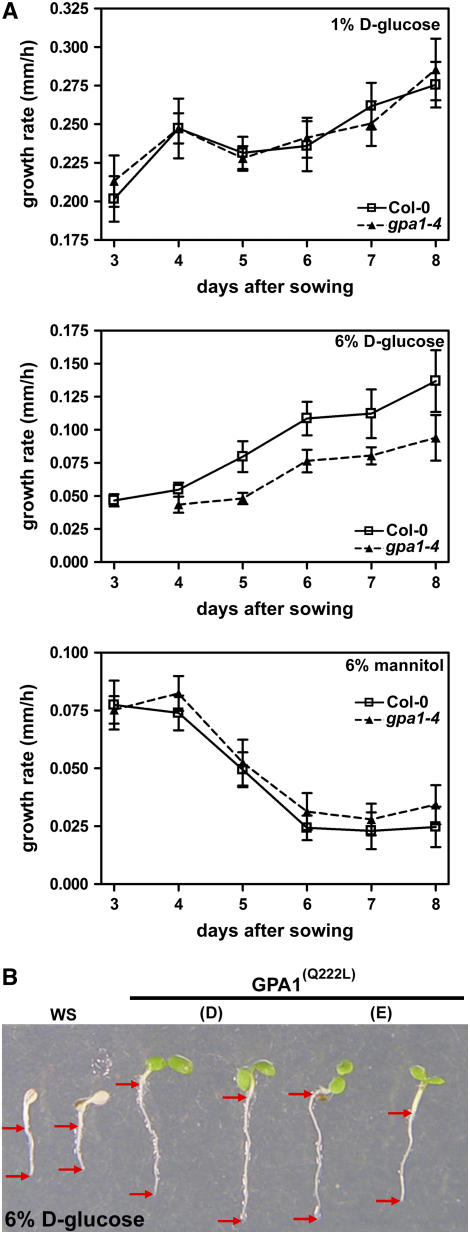
Previously, we and others have shown that gpa1 null seedlings were more sensitive to increased levels of d-glucose, but not mannitol, during germination (Ullah et al., 2002; Chen et al., 2006) and that gpa1 null seedlings were hypersensitive to d-glucose (Chen et al., 2003) in a green-seedling assay (Jang et al., 1997). As the highest levels of GPA1 expression are observed in roots, we investigated the effects of increased d-glucose during root development. Seeds of wild-type (Columbia) and gpa1-4 plants were germinated on medium containing either control (1%) or increased (6%) d-glucose or 6% mannitol as an osmotic control. The growth rate of the primary roots of these seedlings was assayed daily for 6 d, starting 48 h after the plates were placed in the light.
Consistent with previous findings using the green-seedling and germination assays (Ullah et al., 2002; Chen et al., 2003), plants lacking GPA1 were more sensitive to d-glucose. The growth rate of the primary root was reduced in gpa1-4 seedlings exposed to 6% d-glucose (Figure 1A, middle panel) in the days after germination but was not statistically different from the wild-type growth rate on the control level of 1% d-glucose. The retardation in root growth was relieved upon return to 1% d-glucose–containing plates (data not shown). Importantly, both wild-type and gpa1-4 seedlings were observed to be similarly sensitive to the 6% mannitol levels, confirming that the growth-arrest phenotype observed on increased d-glucose was not attributable to an osmotic effect.
Figure 1.
Heterotrimeric G-Protein α Subunit Is Involved in Sugar Signaling.
(A) Seedlings of wild-type Columbia (Col-0) and gpa1-4 genotypes were sown on normal (1%) and increased (6%) d-glucose and 6% mannitol as an osmotic control. The rate of growth of the primary root was assayed over a 6-d period, beginning 48 h after exposure to light. Shown are interval growth rates for each 24-h period expressed as millimeters of growth per hour. Error bars represent se. n > 10. As shown previously by Ullah et al. (2002), gpa1 mutants have delayed germination on high glucose. Student's t tests were performed with pairwise comparison between wild-type and mutant root growth rate on 6% d-glucose. For days 5 and 6, P < 0.1; for days 4, 7, and 8, P = ∼0.2.
(B) Seedlings overexpressing the constitutively active form of the Gα subunit [GPA1(Q222L)] are less sensitive to increased d-glucose than are wild-type seedlings. Images show 7-d-old seedlings. Arrows denote root tips and root/shoot junctions.
We confirmed a role for GPA1 in sugar signaling through an analysis of plants overexpressing a constitutively active form of GPA1 [GPA1(Q222L)]. Seedlings overexpressing GPA1(Q222L) have previously been shown to have other growth phenotypes, similar to rgs1-2 null seedlings (Chen et al., 2003). Root growth in seedlings from two lines overexpressing GPA1(Q222L) was compared with that in wild-type Wassilewskija (Ws). As shown in Figure 1B, the two GPA1(Q222L) transgenic lines (D and E) exhibited greater resistance to high d-glucose compared with wild-type Ws, indicating a role for the GTP-bound form of Gα (GPA1GTP) in overcoming increased d-glucose levels.
THF1 Is a GPA1 Interaction Partner
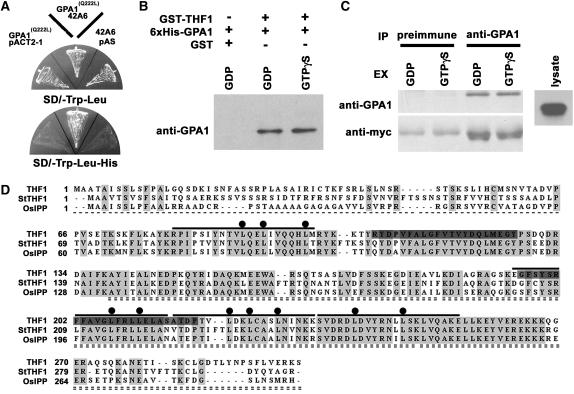
As shown in Figure 1B, overexpression of a constitutively active form of GPA1 conferred tolerance to increased levels of d-glucose. This demonstrates a role for G-protein–coupled signaling, and more specifically, GPA1GTP, in mediating Arabidopsis responses to high d-glucose. Therefore, to identify potential downstream components of this pathway, we sought effectors that interact with GPA1(Q222L) as bait in a yeast two-hybrid screen (Figure 2).
Figure 2.
Interaction between GPA1 and THF1.
(A) Isolation of THF1 as a GPA1(Q222L)-interacting protein in a yeast two-hybrid screen. GPA1(Q222L) is a GTPase-deficient form of GPA1. The triple-dropout plate indicates interaction between GPA1(Q222L) and prey 42A6 on the SD/−Leu/−Trp/−His plate. 42A6 is a partial clone corresponding to THF1.
(B) Both GDP- and GTPγS-bound forms of GPA1 interacted with THF1 in vitro. Purified 6xHis-GPA1 and full-length GST-THF1 were incubated in buffer with either GDP or GTPγS for 1 h, then precipitated with glutathione-agarose beads. The precipitates were probed with anti-GPA1 serum. GST alone did not bind to GPA1.
(C) Alignment of the coding sequences of THF1 homologues in higher plants using the Clustal method (BioEdit). Identical residues are highlighted in light gray, and dark gray denotes the two putative transmembrane domains (see Figure 4). Boldface lines above the sequences represent the matrix repeat sequences. Dots associated with the matrix repeats mark repeated Leu residues. The single dashed line shows the putative transit peptide. The C-terminal sequence indicated by the double dashed line was the identified prey protein from clone 42A6.
(D) GPA1 interaction with THF1 in vivo. Myc-tagged THF1 was expressed in Arabidopsis suspension cells and immunoprecipitated (IP) with preimmune or anti-GPA1 serum in the presence of GDP or GTPγS. The precipitates were probed with GPA1 or anti-myc antiserum.
We screened three yeast cDNA libraries constructed from different Arabidopsis tissues using both low- and high-copy versions of the bait and focused on a candidate clone that was subsequently confirmed to encode an interactor in the yeast two-hybrid configuration by recloning and retesting and by coimmunoprecipitation experiments (Figures 2A, 2B, and 2D). We demonstrated that the full-length protein interacts with GPA1 in the presence of both GDP and GTPγS. The prey encodes the 162–amino acid C-terminal sequence of a 300–amino acid protein (At2g20890), which we previously designated THF1 (Wang et al., 2004). As we reported previously (Wang et al., 2004), THF1 does not share significant sequence with any known protein, but similar proteins can be found in a number of other species, with potato (Solanum tuberosum) and rice (Oryza sativa) proteins shown for comparison (Figure 2C). Additional analyses predicted the three-dimensional folding structure to be weakly supported as an ENTH fold, as found in the human inositol triphosphate binding, clathrin assembly protein (clathrin assembly lymphoid myeloid leukemia, Q13492). However, no binding to a lipid profile (PIP Stips P-6001; Echelon Bioscience) was observed. Nonetheless, the predicted analogy is consistent with the observed altered membrane trafficking in our previous study (Wang et al., 2004).
As shown in Figure 2C, inspection revealed that THF1 has four stretches of low similarity to M repeats such as M6_STRPY from Streptococcus pyogenes (Fischetti, 1989), a motif found within protein interaction interfaces (bars). The interaction region between THF1 and GPA1, which lies within the C-terminal 162 amino acids of THF1, encompasses at least three of the four putative M repeats. The conserved Leu residues highlighted in Figure 2C (dots) are suggested to be important for coiled-coil secondary structure (Herwald et al., 2004).
We tested the interaction between GPA1 and full-length THF1 by an in vitro coprecipitation assay (Figure 2B). For this purpose, 6xHis-GPA1 and GST-THF1 were expressed and purified from Escherichia coli using nickel and glutathione S-transferase (GST) columns, respectively. To determine whether THF1 interacts preferentially with the GTP-bound form of GPA1, GPA1 was loaded with GDP or GTPγS before adding GST-THF1 to the binding assay buffer. We detected a qualitatively similar binding ability of THF1 to either the GDP- or GTP-bound form of GPA1 (Figure 2B), whereas GST alone was not able to coprecipitate GPA1. To test whether GPA1 interacts with THF1 in vivo, we transiently expressed a full-length C-myc–tagged THF1 clone in Arabidopsis suspension cells (Figure 2D). Total protein was extracted with buffer containing either GDP or GTPγS and then immunoprecipitated with anti-GPA1 serum. The GPA1 preimmune serum was unable to coprecipitate THF1. Samples were subjected to immunoblotting using anti-GPA1 or anti-myc serum. As shown in Figure 2D, GPA1 and THF1-myc interacted equally well both in vitro and in vivo in the presence of either GDP or GTPγS.
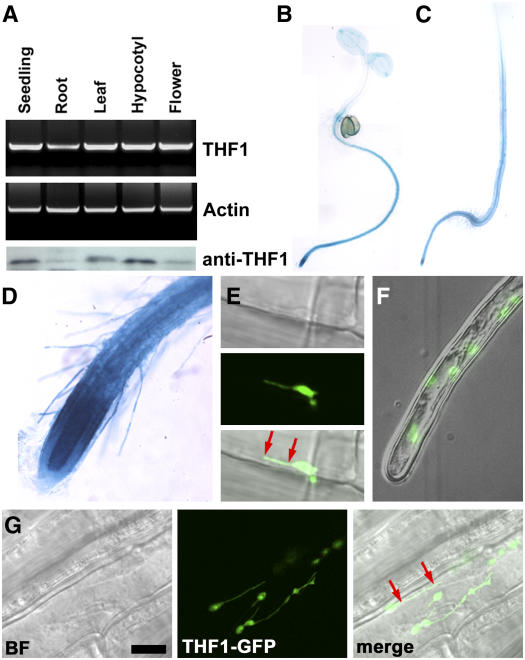
Ubiquitously Expressed THF1 Is Localized to Plastids and Plastid Stromules
Our initial report on THF1 described how its global gene expression could be regulated by light (Wang et al., 2004). Here, we show that it is expressed ubiquitously in all organs (Figure 3A), with the highest levels of THF1 promoter:uidA (GUS) expression observed in roots of both light-grown (Figure 3B) and dark-grown (Figure 3C) seedlings, showing that the gene is well expressed in roots regardless of light conditions. The highest THF1 promoter:uidA expression was observed in the root apical meristems (Figure 3D), similar to that observed for both GPA1 and RGS1 (Ullah et al., 2001; Chen et al., 2003). Immunoblot analysis of THF1 protein levels showed that the highest levels of THF1 protein were in hypocotyls (Figure 3A). The comparison between the immunoblot and GUS data suggests either a rapid turnover of THF1 protein or some form of posttranslational regulation in the root.
Figure 3.
Tissue Expression and Subcellular Localization of THF1.
(A) THF1 transcript and THF1 protein in different organs. Top, RT-PCR using RNA isolated from the indicated organs from 30-d-old light-grown plants and 10-d-old seedlings. Bottom, the same set of samples was used for immunoblot analysis probed with anti-THF1 serum (α-THF1). The lowest and highest signals were in the linear range of this assay.
(B) and (C) Transcriptional fusions between the THF1 promoter and the uidA gene encoding GUS were used to transform Arabidopsis. Seedlings were stained for GUS activity in 5-d-old seedlings grown either in constant light (B) or in darkness (C). Staining was stopped after 5 h to highlight the predominant expression locations, but overnight staining indicated ubiquitous expression.
(D) Root tip cells including hairs express THF1; the strongest expression was seen in the root meristem.
(E) to (G) A 35S-driven THF1-GFP fusion protein localizes to plastids and can be seen in plastid stromules that appear to tether the plastid to the plasma membrane (red arrows; see also Supplemental Movie 1 online). (E) and (G) show root epidermal cells; (F) shows a root hair cell. BF, bright field. Bar in (G) = 10 μm.
Confocal imaging of a 35S-driven THF1–green fluorescent protein (GFP) fusion protein in 5-d-old seedlings (Figures 3E to 3G) confirmed the earlier observation that THF1 is a plastid protein (Wang et al., 2004) but further demonstrated that THF1-GFP is present in root plastid stromules. Stromules are tubular extensions of plastids, enclosed by both the inner and outer plastid envelope (Gray et al., 2001), which are thought to increase the surface area of plastid membranes for an as yet undetermined function. It is important to note that the THF1-GFP–labeled stromules were most easily visualized in root tissues (Figures 3E and 3G), whereas none were found in leaf tissues. Time-lapse imaging of THF1-GFP in root epidermal cells and root hairs (Figure 3F; see Supplemental Movie 1 online) revealed the plastid compartments and stromules to be highly dynamic within cells, suggesting that the plastids may use their stromule extensions to interact and anchor to the plasma membrane (Figures 3E and 3G, red arrows). Tethering is a previously reported feature of stromules (Kwok and Hanson, 2004a) that may allow plastids to remain in proximity to cellular structures against the currents of cytoplasmic streaming (Gunning, 2005; Natesan et al., 2005).
THF1 Localizes to Both the Outer Plastid Membrane and the Stroma
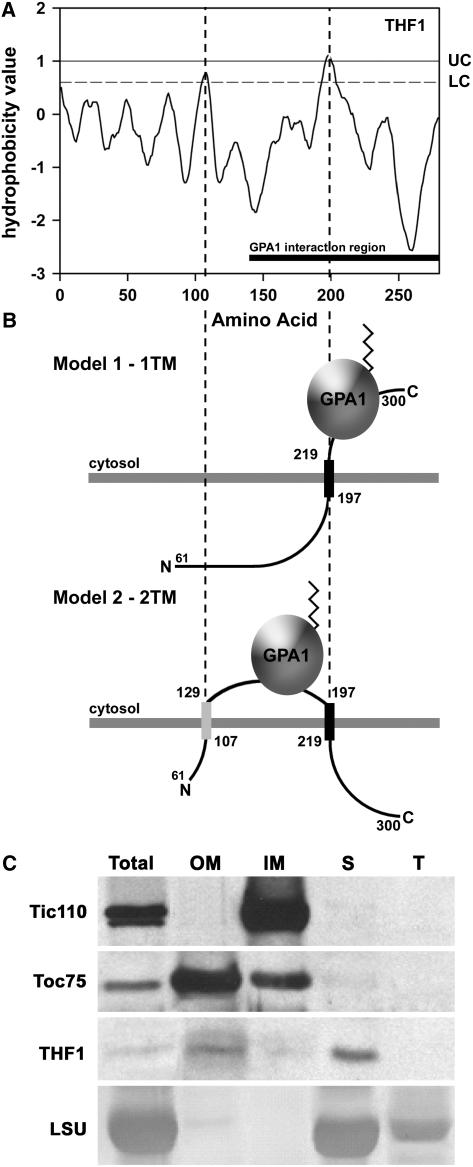
The data presented above describe a root plastidic interaction partner to a plasma membrane–delimited G-protein. Although the highly vacuolated nature of root cells means that plastids and other organelles are frequently pressed up against the plasma membrane, the concept that a specific protein–protein interaction was occurring prompted us to look in more detail at the physical possibility of this unusual situation. Kyte–Doolittle hydropathy analysis of full-length THF1 using the TOPRED algorithm (von Heijne, 1992; Claros and von Heijne, 1994) set to identify prokaryotic/plastidic proteins strongly predicts the presence of at least one and possibly two membrane-spanning domains, as shown by hydrophobicity values exceeding the upper (UC) and lower (LC) cutoff values (Figure 4A). This prediction is in contrast with that extracted from the Cornell Plastid Proteome Database (Friso et al., 2004), which suggests that THF1 does not posses membrane-spanning domains. However, this discrepancy is attributable to the different prediction algorithms used to determine the presence of eukaryotic membrane-spanning domains. Further analysis of these putative transmembrane domains revealed that both their presence and sequence are highly conserved among the highly divergent taxa that contain orthologs to THF1, including rice and potato (Figure 2C). We previously showed that THF1 contains a cleaved transit peptide that facilitates entry into the plastid (Wang et al., 2004), leading us to propose two possible topology models for the remaining 239 amino acids of the mature protein within the outer plastid membrane (Figure 4B).
Figure 4.
THF1 Is a Protein of the Outer Plastid Membrane and Stroma.
(A) Kyte–Doolittle hydropathy plot for THF1. UC and LC denote the upper and lower cutoffs for the prediction of membrane-spanning domains, respectively. The solid bar at bottom denotes the positions mapped by the yeast two-hybrid assay as the THF1–GPA1 interaction domain. Dashed lines denote positions of the two predicted membrane spans.
(B) Possible topologies for THF1 that enable interaction between the membrane-spanning pool of THF1 and the plasma membrane–delimited GPA1. THF1 contains at least one (model 1), and possibly two (model 2), membrane-spanning domains, which embed THF1 in plastid membranes. The angled line on GPA1 represents the N-terminal myristoyl group that anchors GPA1 to the plasma membrane (data not shown).
(C) Isolated plastids were fractionated and subjected to immunoblot analysis with the corresponding antisera to the indicated marker proteins or THF1. Fractions are represented as follows: Total, whole preparation; OM, outer plastid membrane; IM, inner plastid membrane; S, stromal fraction; T, thylakoid fraction. Controls used to determine THF1 localization were as follows: Tic110, a component of the inner plastid membrane; Toc75, a component of the outer plastid membrane; and the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (LSU), which is found in the stroma and thylakoid.
Both models allow for the THF1–GPA1 interaction on the external face of the plastid, consistent with our identification of the THF1-interaction region within the C-terminal 162 amino acids of the mature THF1 protein (denoted the GPA1 interaction region in Figure 4A). To confirm this in silico analysis, immunoblot analysis of fractionated plastids was performed. Intriguingly, as shown in Figure 4C, THF1 is enriched in both the outer membrane and stromal fractions. Although this is consistent with an outer membrane–spanning prediction for THF1 (Figure 4A), we applied our model for the THF1–GPA1 interaction only to the pool of THF1 that spans the outer membrane, as opposed to the stromal pool. The fact that a second pool of THF1 is found in the stromal fraction may suggest either multiple functions for THF1 in plastids or some requirement for the trafficking of THF1. Dual localization of THF1 in the fractionation data would also be consistent with a single localization in different plastid types. Although there are no known plastid-specific differences in the protein import machinery, this possibility should not be ruled out. Nonetheless, both modeling and biochemical fractionation of plastid membranes are consistent with our hypothesis of an outer membrane localization for THF1 on root plastids.
In Vivo Dynamics of the GPA1–THF1 Interaction
Förster resonance energy transfer (FRET) analysis between 35S-driven GPA1-CFP (for cyan fluorescent protein) and THF1-YFP (for yellow fluorescent protein) constructs in the gpa1-4 thf1-1 background provided spatial resolution of the GPA1–THF1 interaction in root epidermal cells. Two-filter channel FRET analysis confirmed our earlier observations that THF1 is plastidic (Figure 5A) and GPA1 is localized to the plasma membrane (Figure 5B). We observed that the ratio of YFP to CFP was high where a plastid was adjacent to the plasma membrane. The observed FRET signal, denoted nF/I (Figure 5), corresponds to the observed YFP-CFP ratio and represents an algorithmic output for two-filter channel FRET based on the equations of Gordon et al. (1998). Simply put, it is a mathematical representation of the YFP:CFP ratio, normalized to the donor intensity. FRET requires that the donor and receiver reside within 10 to 100 Å, strongly suggesting a physical interaction between GPA1 and THF1. FRET was never observed where root plastids did not abut the plasma membrane, consistent with our demonstration that a direct physical interaction between GPA1 and THF1 occurs in vivo. Additionally, the detection of FRET between GPA1 and THF1 is consistent with an outer membrane localization for the pool of THF1 relevant to G-protein signaling, as FRET cannot occur between two sequestered fluorophores, one on the plasma membrane and one on the plastid stroma, because of the extreme proximity required for energy transfer.
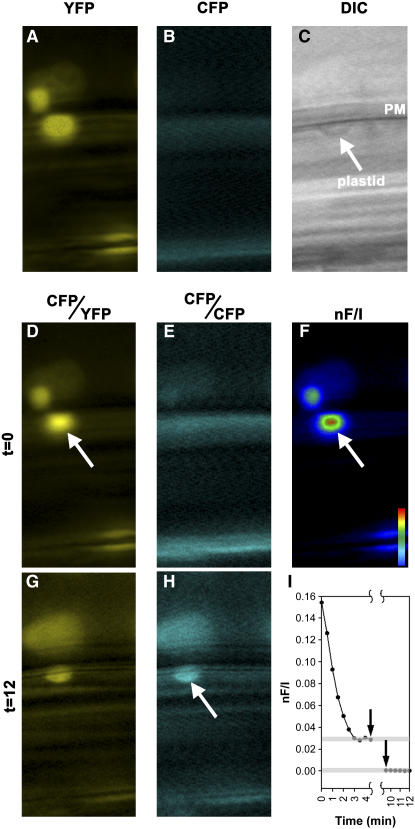
Figure 5.
FRET Analysis of the GPA1–THF1 Interaction.
FRET between GPA1-CFP and THF1-YFP was calculated using established algorithms for two-filter FRET with fluorescence microscopy.
(A) to (C) Single images of cells expressing THF1-YFP (A) and GPA1-CFP (B) and a corresponding differential interference contrast (DIC) image (C), showing plasma membrane (PM) and a plastid.
(D) to (H) FRET images were recorded before ([D] to [F]) and after ([G] and [H]) acceptor photobleaching. Arrows denote the localization of the observed FRET signal where a plastid abutted the plasma membrane ([D] and [F]) and the region of CFP dequenching after YFP photobleaching (H).
(I) Quantification of the observed FRET signals. The first arrow denotes the start of the 480-nm irradiation, and the second arrow indicates the end.
Although nF/I is a robust method to measure FRET, we further validated the observed FRET signal using an acceptor photobleaching method. This method is routinely used to confirm FRET between various fluorophore pairs (Bastiaens and Jovin, 1996; Bastiaens et al., 1996; Kenworthy, 2001; Vermeer et al., 2004). Its principle is that energy transfer is reduced or abolished when the acceptor fluorophore (in this case YFP) is photobleached, accompanied by dequenching of donor (CFP) fluorescence. This is a stringent diagnostic test for FRET, as in most circumstances fluorescence normally decreases after acceptor photobleaching (Karpova et al., 2003). After a 5-min irradiation at 480 nm, we found that CFP emission was dequenched in the same region where FRET was originally recorded (Figure 5H). A quantitative analysis of the nF/I recording is shown in Figure 5I. Before YFP photobleaching, the nF/I stabilized within 3 min (top gray line). After acceptor photobleaching, the FRET efficiency was reduced irreversibly to zero (bottom gray line).
thf1-1 Root Growth Is Hypersensitive to Increased d-Glucose
Our previous report described the phenotype of an insertion mutant in THF1. We demonstrated that a homozygous SALK insertion line (094925) is transcript null for THF1 and that the THF1 cDNA was capable of complementing the mutation (Wang et al., 2004). We found that the gene was necessary for correct development of the thylakoid membrane and that the null allele conferred altered plant morphology, specifically stunted growth and variegated leaves (Wang et al., 2004). To examine whether THF1 had any role in the G-protein–coupled sugar pathway, we tested the effect of increased d-glucose on the rate of thf1-1 primary root growth (Figure 6B). Additionally, although THF1 expression was observed in root hairs (Figure 3D), no obvious root hair phenotype was observed for this mutant (data not shown).
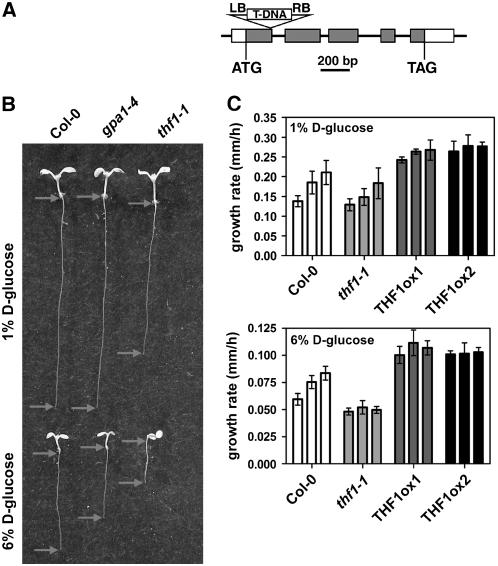
Figure 6.
THF1 Is Involved in d-Glucose Signaling.
(A) Scheme of the T-DNA insertion (SALK line 094925) in THF1, as described previously (Wang et al., 2004). LB, left border; RB, right border.
(B) Seedlings of Columbia, gpa1-4, and thf1-1germinated on both 1% and 6% d-glucose, showing the sensitivity of the thf1-1 mutant to increased d-glucose compared with both the wild type and gpa1-4. Images show randomly selected seedlings taken from the pool used for the determination of growth rate. Arrows mark the shoot/root transition and the root tip. The growth rate of roots on increased d-glucose is reduced in thf1-1 seedlings.
(C) Tolerance to high levels of d-glucose can be conferred by overexpression of THF1. The rate of growth of the primary root was assayed over a 3-d period (each set of three bars), beginning 48 h after exposure to light. The interval rate is expressed as millimeters of growth per hour, averaged over each 24-h period. Error bars represent se. n > 10. THF1ox, ectopically overexpressed THF1.
As shown previously, on 1% d-glucose, gpa1-4 roots were similar to wild-type roots, although thf1-1 roots were ∼25% shorter and thf1-1 cotyledons were pale green (Figure 6B). However, on 6% d-glucose, thf1-1 seedlings were ∼50% shorter than gpa1-4 seedlings. Because root length is a complex trait, one that can be a result of altered germination rates, we quantitated the growth rate of the primary roots over time. On 1% d-glucose, the thf1-1 root growth rate was ∼10% lower than the wild-type rate (statistically insignificant) (Figure 6C, top); however, thf1-1 roots were never able to recover from the growth-arresting effect of high d-glucose compared with wild-type roots (Figure 6C, bottom). This finding suggests that the loss of THF1 confers either increased sensitivity to, or the inability to adapt to, high d-glucose. To complement these data, the effects of THF1 overexpression were tested. Two independent THF1 overexpression lines were germinated on 1% and 6% d-glucose. Contrary to the loss of THF1, ectopic overexpression of THF1 caused roots to grow at faster rates on either low or high d-glucose compared with the wild type at all times.
This difference in phenotypes between gpa1-4 and thf1-1 enabled epistasis analysis between the gpa1-4 and thf1-1 alleles. The double mutant exhibited the same root-development phenotype as the thf1-1 null mutant on both normal and increased levels of d-glucose (see Supplemental Figure 1 online). This finding suggests a relationship between the two genes, which is consistent with our hypothesis that THF1 operates downstream of GPA1 in the G-protein–coupled sugar-signaling pathway.
THF1 Protein Levels Are Affected by d-Glucose
To test a mechanistic role for THF1 in sugar signaling, we quantitated the effect of d-glucose on the steady state level of THF1 protein by imaging and immunoblot analysis (Figure 7). As shown previously, 35S:THF1-GFP fluorescence was observed in plastids through the roots of 5-d-old seedlings (Figures 3E to 3G), including the root meristem. However, after 30 min of 6% d-glucose treatment, observable GFP fluorescence in roots was diminished. The four panels of Figure 7A are conventional fluorescence microscopy images of the same root and are representative of >20 roots assayed. No decrease in the levels of 35S:THF1-GFP fluorescence was observed after treatment with l-glucose (Figure 7A), NaCl, or mannitol. Sugar-induced changes in THF1-GFP fluorescence were not observed in the leaves.
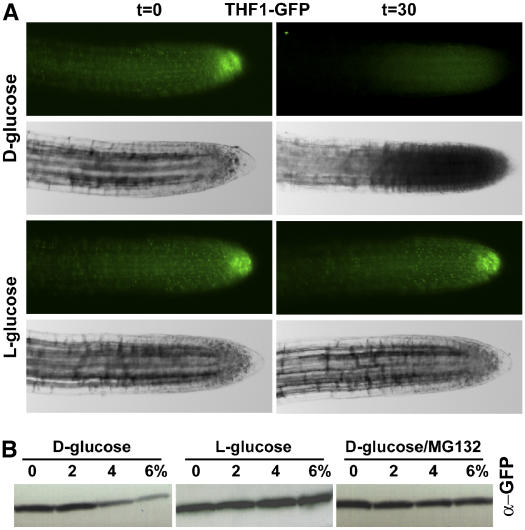
Figure 7.
d-Glucose Has a Direct Effect on THF1 Steady State Level.
(A) Green fluorescence in root cells expressing a 35S-driven THF1-GFP fusion protein disappeared 30 min after being treated with 6% d-glucose, but not l-glucose
(B) THF1-GFP degradation at the indicated applied glucose levels (%) for 30 min was confirmed by immunoblotting probed with anti-GFP antiserum. Degradation did not occur with l-glucose treatment and was inhibited by the general protease inhibitor MG132. Not shown are nontransformed roots, which did not display fluorescence under these excitation and digital capture conditions.
Immunoblot analysis was used to confirm that the observed loss of signal was specific to d-glucose and that it was attributable to protein degradation (Figure 7B). Protein samples from 35S:THF1-GFP seedlings were extracted from seedlings treated with 0, 2, 4, or 6% d-glucose for 30 min and subjected to SDS-PAGE and immunoblot analysis with anti-GFP antibodies. As seen in Figure 7B, the steady state level of THF1-GFP protein was reduced over 30 min in a dose-dependent manner. However, in samples treated with l-glucose, the steady state level of THF1-GFP remained constant, showing specificity for the d-stereoisomer. Samples treated with d-glucose in the presence of the protease inhibitor MG132 did not show changes over time in THF1 steady state levels, suggesting that THF1 half-life is influenced by high d-glucose levels.
DISCUSSION
THF1 Is an Interaction Partner for Gα in the Sugar-Signaling Pathway in Roots
Six lines of evidence indicate that THF1, a plastid protein, interacts with the plasma membrane–delimited G-protein, GPA1, and four lines of evidence indicate that THF1 has a role in sugar signaling.
The GPA1–THF1 interaction is demonstrated by the following findings. (1) In a yeast two-hybrid conformation, the C-terminal 162 residues of THF1 interact with the constitutively active form of GPA1. (2) This interaction in yeast was confirmed in vitro using recombinant full-length THF1 and GPA1. (3) In vivo interaction in plant cells was shown through coimmunoprecipitation of GPA1 with THF1. (4) Genetic epistasis analysis between the loss-of-function alleles gpa1-4 and thf1-1 is consistent with the two corresponding gene products operating in the same pathway. (5) THF1 is at least in part localized to the outer membrane of plastid stromules, which appear to associate with the plasma membrane. (6) FRET analysis between fluorophore-tagged GPA1 and THF1 confirms the interaction in vivo and provides spatial information.
The evidence that THF1 has a direct or indirect role in sugar signaling is based on both biochemical and genetic experiments. (1) thf1-1 loss-of-function mutants are hypersensitive to and unable to recover from exogenous d-glucose. (2) THF1-overexpressing plants have a higher rate of root growth on high d-glucose. (3) An epistatic relationship between the thf1-1 and gpa1-4 alleles places THF1 genetically on the G-protein–coupled sugar-signaling pathway. (4) Exogenous d-glucose, but not l-glucose, causes a rapid degradation of THF1 protein.
THF1 was shown to interact similarly with either the GDP- or GTP-bound form of GPA1 (Figure 2). THF1 may be a component of the heterotrimeric complex but does not preferentially interact with the activated form of the Gα subunit, suggesting a scaffolding or tethering role for THF1, perhaps to maximize signaling between the plasma membrane and plastids in response to d-glucose. This possibility implies that the primary function of THF1 is not signaling per se but to act as a maintenance or accessory protein. However, as in vitro and in vivo interaction shown by in vitro and in vivo coimmunoprecipitation is based on end-point analysis rather than kinetics, we do not exclude the possibility that THF1 affinity for GPA1 is affected by the GDP/GTP state and is not detected in our assay. Modeling based on data from kinetic analyses may reveal small affinity differences that have a large impact on the ratio of the GTP- and GDP-bound states as they are allosterically amplified through the coupled effector interactions, as is the case for human Gα subunits (Hatley et al., 2003). Detailed kinetics analyses are also needed to determine whether THF1 has GAP activity on GPA1GTP. Because the designation of THF1 as a Gα effector awaits evidence for either a demonstrated GAP activity or different binding affinities for the two nucleotide states of GPA1, other roles, such as structural or targeting, should not be ruled out.
It should be noted that it is likely that THF1 acts in pathways other than those coupled by the heterotrimeric G-protein, for three reasons. (1) The expression pattern of THF1 (Figure 3) extends outside the limited areas of GPA1. (2) The loss-of-function phenotypes of thf1-1 mutants are pleiotropic, beyond the narrowly defined traits of the gpa1 mutant. (3) Plastid fractionation data showed that THF1 was localized to both the outer plastid membrane and the stroma, suggesting that THF1 may perform an additional role in the stroma, where GPA1 should be excluded. Although the dual localization of plastid proteins is unusual, it is not unique to THF1, as other plastid proteins have dual localization and function (Tu et al., 1999). Thus, it should be considered that although GPA1 can be a regulator of THF1, it might not be the only one. Also, on this point, we do not exclude exclusive locations for THF1 of either the outer membrane or the stroma but in different pools of plastids, because our fractionation data do not distinguish between potentially different plastid types.
The initial observations of thf1-1, showing that it lacked properly formed thylakoid membranes, are consistent with a structural or trafficking role (Wang et al., 2004). Indeed, it could be the case that the phenotypes pertaining to improper thylakoid formation may relate to the lack of the stromal pool of THF1, whereas the d-glucose–sensitive phenotypes are related to a lack of the THF1–GPA1 interaction. If this interaction functions to either mediate or transduce signals between the plasma membrane and the plastids, a lack of this interaction may represent an aberration in the ability of root plastids to regulate such processes as starch synthesis.
Despite the broad expression pattern of THF1, the highest levels observed with the THF1 promoter:GUS fusion were in the root meristem, correlating with strong expression of GPA1 (Ullah et al., 2001). As thf1-1 meristems have normal histology and gravitropic responses (data not shown), this disfavors a developmental defect as the cause for the observed seedling hypersensitivity to d-glucose.
THF1 Mediates Signaling between the Plasma Membrane and the Plastids
How THF1, a plastid protein, is able to interact with the plasma membrane–delimited GPA1 is an intriguing question. Plastids are well known for their dynamic shape change and movement, but the diversity of plastids based on their function is essentially unknown beyond the classic three plastid types: chloroplasts, leukoplasts, and amyloplasts. Plastids can migrate freely in the cytoplasm or adhere to the plasma membrane through actin cages, depending on different cell types and growth conditions (Kwok and Hanson, 2004c; Gunning, 2005). The discovery of stromules, tube-like extensions of the plastid envelope and stroma, revealed a unique mechanism by which plastid molecules can communicate with other cell compartments and indeed other plastids (Kwok and Hanson, 2004a, 2004d). Others have reported that stromules are able to form bead-like structures that move around the cell as part of a stromule network or even as free vesicles (Pyke and Howells, 2002; Natesan et al., 2005). Such plastid-derived vesicles are highly reminiscent of vesicles trafficking along a filamentous network. Evidence for plastid communication with other subcellular compartments is scant but increasing (Chew et al., 2003; Strand et al., 2003). The observed FRET between GPA1 and THF1 indicates that the interaction occurs at the interface between the plastid and the plasma membrane, and because the interaction was monitored over several minutes (to allow for acceptor photobleaching), this may suggest that plastids that interact with the plasma membrane do not move as freely within the cytosol as untethered plastids. Our time-lapse acquisition of stromule movement in roots is consistent with this finding (see Supplemental Movie 1 online).
The prediction of membrane-spanning domains in THF1 and the identification of an outer membrane–localized pool of THF1 render the four interaction data sets interpretable and suggest a direct link between plastids and the plasma membrane. Of the two topological models proposed in Figure 4B, model 1 is the most likely because it fits the THF1-GFP degradation data shown in Figure 7E: in this model, the C terminus of THF1 is fully exposed to the cytosol for potential ubiquitination and subsequent degradation (Vierstra, 2003). However, although the inhibitor MG132 is typically associated with inhibition of the function of the cytosolic 26S proteasome (Yang et al., 2004), and considering that THF1 is both a plastidic and/or a membrane-spanning protein, we cannot rule out the possibility that the MG132 acts as an inhibitor of a protease not operating in the 26S proteasome pathway. Another possibility is that an as yet unidentified MG132-sensitive degradation machinery is present in plastids, which could be responsible for the degradation of both the membrane and soluble pools of THF1.
We expect that plant cells precisely coordinate biosynthetic activities with the export of molecules from plastids to their destinations in response to internal and external signals (Eastmond and Graham, 2003; Tiessen et al., 2003; Kolbe et al., 2005). Consideration of known vesicular trafficking mechanisms is a particularly suitable approach to such an issue, because trafficking machineries are highly conserved in eukaryotes and are responsible for delivering and recycling many kinds of molecules, including protein and lipid components of the membrane, to various compartments (Maxfield and McGraw, 2004). For example, in pancreatic β-cells, heterotrimeric G-proteins are important molecular switches that control insulin secretion via endocytosis and exocytosis to ensure the vital function of glucose homeostasis (Konrad et al., 1995; Lang, 1999; Skoglund et al., 1999; Luo et al., 2001). The colocalization of GPA1 and its physical interactor, THF1, at the plasma membrane suggests that plastid molecules such as THF1 communicate with plasma membrane–delimited molecules via two possible mechanisms. One possibility is to produce a lipid microdomain that can serve as a platform to assemble G-protein complexes at the plasma membrane. THF1 may establish the specificity of the G-protein signaling pathway or the regulation of receptor-mediated endocytosis in a GPA1-dependent manner. The second possibility is that THF1 together with GPA1 plays a role in exocytosis at stromule–plasma membrane sites.
Heterotrimeric G-Protein–Coupled Sugar Signaling
All of the data presented here and elsewhere are consistent with the prevailing notion that d-glucose acts as a hormone-like signal, although a physiological concentration range of extracellular d-glucose in signaling has yet to be defined. High applications of d-glucose (Arenas-Huertero et al., 2000) cause physiologically appropriate responses, and genetic screens for mutants resistant to high d-glucose have revealed known elements in sugar signaling (Xiao et al., 2000; Moore et al., 2003). Because cells develop in a wide range of d-glucose concentrations, a sugar-signaling mechanism operating from presumably 30 to 300 mM or higher d-glucose is expected. Molar levels of d-glucose are found in tissues such as fruit, but the levels of extracellular d-glucose in vegetative tissues such as root have yet to be successfully evaluated. Indirect measurements have been applied to address this problem, and estimates of apoplastic levels of d-glucose at 150 mM (3%) or higher have been proposed (McLaughlin and Boyer, 2004; Makela et al., 2005). For example, sorghum (Sorghum bicolor) embryos develop in as high as 6% apoplastic d-glucose (Maness and Mcbee, 1986).
What is the nature and mechanism of the high-glucose-induced root growth arrest? Little is known about how sugars control growth and development in plants. We found that root cells grown on high d-glucose become severely expanded (data not shown), suggesting a possible control of the cell division/expansion pathway. This is entirely consistent with the now firmly established role of the G-protein complex in the modulation of cell division (Ullah et al., 2001, 2003; Chen et al., 2003; Jones et al., 2004). Sugars control cyclin D expression in plant cells (Riou-Khamlichi et al., 2000), and a loss-of-function mutation in a moss cyclin D gene caused a d-glucose–resistant phenotype (Lorenz et al., 2003). In addition, sugars control the G1-to-S phase transition (Menges and Murray, 2002). Thus, it is becoming clear that sugars act as signals to modulate plant cell proliferation and do so by a complex set of signaling mechanisms. Hexokinase is needed for intracellular glucose detection, and now, with the introduction of both THF1 and RGS1 (Chen et al., 2003; Chen and Jones, 2004), a signaling network involving a heterotrimeric G-protein is added to our understanding of d-glucose signaling in Arabidopsis. Because these organelles either use or produce d-glucose or its catabolites, it is reasonable to assume that extracellular glucose availability would influence cellular glucose economy potentially as it occurs in yeast.
METHODS
Plant Growth Conditions, Plasmid Construction, Antiserum, and Transformation
The Arabidopsis thaliana ecotype used in this study was Columbia with the exception of the GPA1-overexpressing lines (Figure 1B), which were in Ws. Seeds were surface-sterilized and stratified for 4 to 5 d before germination at 22°C on half-strength Murashige and Skoog medium containing the indicated concentrations of d-glucose. Plants were grown under short-day conditions. Where required, plates were scored for root length daily and the data analyzed using ImageJ. All whole-seedling images were taken with a Sony DSC-F717 digital camera.
The various plasmids were constructed using the Gateway cloning system (Invitrogen). For clarity in gene, mutant, and protein designations, the gene corresponding to the locus of At2g20890 is referred to as THF1, its open reading frame is referred to as THF1, the null mutant is referred to as thf1-1, and the protein is referred to as THF1. THF1 was inserted into the entry vector pENTR/D-TOPO (Invitrogen) by topoisomerase-mediated ligation and then recombined into destination vectors pDEST15, pDEST24 (Invitrogen), pGWB5 (35S-driven C-terminal GFP fusion), pGWB6 (35S-driven overexpression), and pGWB41 (35S-driven C-terminal YFP fusion) (Research Institute of Molecular Genetics) to create the various expression vectors for Escherichia coli and plants. The GTPase-deficient form of GPA1(Q222L) is described by Chen et al. (2003). Arabidopsis plants were transformed by the flower-dip method (Bechtold et al., 1993). Transgenic T1 plants were identified by kanamycin resistance.
Antiserum to THF1 was raised in rabbits (Cocalico Biologicals) as described previously (Wang et al., 2004). Antisera to GPA1 (9271 and 9272) were generated in rabbits using as antigen the C-terminal peptide of GPA1 (DETLRRRNLLEAGLL) coupled to keyhole limpet hemocyanin via an N-terminal Cys thioester linkage.
Identification and Analysis of Mutants and Transgenic Plants
The homozygous T-DNA insertion mutant thf1-1 was generated from the Salk Institute sequence-indexed insertion mutant collection (http://signal.salk.edu/smission.html) as described previously (Wang et al., 2004). The THF1 transcript level was quantified using the ThermoScript RT-PCR system (Invitrogen) together with gene-specific primers and THF1 protein levels by immunoblot analysis with the anti-THF1 polyclonal antibody. The gpa1-4 thf1-1 double mutant was generated by genetic crosses, and homozygous lines were identified by genotyping and confirmed by PCR-based genotyping. The GPA1-CFP and THF1-YFP single transformants were generated by the floral-dipping method from gpa1-4 and thf1-1 plants, respectively, and screened by fluorescence microscopy and RT-PCR. GPA1-CFP and THF1-YFP double-transgene plants were created by genetic crosses of the transformed parents using lines selected to ensure that transgene expression was not higher than in the wild type. The THF1 overexpression lines were screened by immunoblot analysis (data not shown). GUS activity in situ was performed as described by Jefferson et al. (1987).
Yeast Two-Hybrid Screens
Yeast strains were grown at 30°C in standard rich medium (yeast peptone dextrose [YPD]) or synthetic medium (synthetic dextrose [SD]) supplemented with the appropriate amino acids. Yeast two-hybrid screens were performed using an interaction mating protocol as described previously (Soellick and Uhrig, 2001). A yeast mating procedure was performed for the verification of positive clones. Gap-repair cloning was used to recombine the candidate PCR products with the linearized vector pACT2-1 in yeast strain AH109. The bait vector pAS-GPA1(Q222L) was transformed into Y187 strain. For mating, one colony of each type was suspended in YPD medium and incubated at 30°C for 5 h. YPD medium was removed after centrifugation, and cells were resuspended in 200 μL of water. Half of the culture was spread on SD/−Leu/−Trp plates and half on SD/−Leu/−Trp/−His/+3-aminotriazole plates. The growth of yeast was checked 48 h after plating. Growth on the corresponding single-dropout plates indicated transformation (data not shown). The cDNAs used for the prey screens were generously provided by Csaba Koncz and Hans Sommer (Max-Planck Institüt für Züchtungsforschung).
Protein Coimmunoprecipitation Assay
For in vitro protein purification and interaction, 6xHis-GPA1 and GST-THF1 fusion proteins were purified from BL21(DE3) cells using affinity chromatography according to the manufacturer's instructions (6xHis-GPA1, Clontech; GST-THF1, Amersham). Poly-His-GPA1 was loaded with GDP or GTPγS before adding GST-THF1 to test the specific binding of GPA1 to THF1 in buffer A (50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 2 mM DTT, 5 mM MgSO4, protease inhibitor cocktail for bacterial cell extracts [Sigma-Aldrich], and 100 μM GDP or GTPγS). The samples were incubated at 25°C for 1 h in the presence of glutathione-agarose, washed five times with the buffer, and eluted with 2× SDS-PAGE sample buffer. Proteins were separated by SDS-PAGE and immunoblotted with anti-GPA1 (rabbit polyclonal antiserum raised against a peptide representing the last 15 amino acids of GPA1, designations 9271 and 9272) and anti-GST (Sigma-Aldrich) antibodies.
For in vivo coimmunoprecipitation, the myc epitope–tagged THF1 binary vector was transformed into Arabidopsis suspension cells (a gift from Csaba Koncz, Max-Planck Institüt für Züchtungsforschung) by Agrobacterium tumefaciens–mediated transformation. Cells were cultured and harvested as described by Ferrando et al. (2000). Total protein was extracted with buffer (as described above plus the addition of 1% Triton X-100). The expression level of the myc-THF1 fusion protein was confirmed by immunoblotting with an anti–c-myc antibody (mouse monoclonal 9E10; Santa Cruz Biotechnology) before coimmunoprecipitation was performed according to procedures described elsewhere (Booden et al., 2002). Immunoprecipitated proteins were then resolved by SDS-PAGE, transferred to HybondP polyvinylidene difluoride membranes (Amersham Biosciences), and probed with the indicated antisera and antibodies.
FRET Microscopy
Fluorescence images of GPA1-CFP and THF1-YFP seedlings were captured using an Olympus IX81 inverted microscope controlled by IPlab software version 3.6 (Scanalytics). Images of CFP, YFP, and the CFP:YFP ratio were observed through a ×60 water-immersion objective and simultaneously captured by a cooled charge-coupled device camera (Photometrics Cascade digital camera; Roper Scientific) equipped with an OI-05-EM CFP/YFP FRET emission filter in a dual-view mounting tube. Filter sets used were YFP (excitation, 500/20 nm; emission, 535/30 nm), CFP (excitation, 436/20 nm; emission, 480/40 nm), and FRET (505dcxr; excitation, 436/20 nm; emission, 480/30 and 535/40 nm). For the YFP photobleaching FRET analysis, the YFP acceptor was photobleached by a 5-min, 480-nm irradiation. Calculation of normalized net FRET (nF/I) was performed with IPlab version 3.6 software, which uses established algorithms for two-filter FRET with fluorescence microscopy (Gordon et al., 1998).
Plastid Fractionation
Plastids were isolated and fractionated as described previously (Inaba et al., 2005). Essentially, chloroplasts were isolated from 15- to 20-d-old seedlings grown on 0.5× Murashige and Skoog plates supplemented with sucrose as described previously (Smith et al., 2002). Isolated intact chloroplasts were separated into membrane and soluble fractions by lysis in 0.1 M Na2CO3, pH 11.5, followed by centrifugation at 200,000g for 20 min. The pellets (membrane fraction) were directly dissolved in SDS-PAGE sample buffer. The soluble proteins were recovered by precipitation with trichloroacetic acid and dissolved into SDS-PAGE sample buffer. Intact chloroplasts were treated with trypsin as described previously (Jackson et al., 1998).
Total protein extracts from Arabidopsis were obtained by directly homogenizing leaves in SDS-PAGE sample buffer, unless specified otherwise. To avoid proteolytic degradation, the extraction buffer was supplemented with 2000-fold diluted protease inhibitor cocktail for plant cell extracts (Sigma-Aldrich). Extraction of total proteins from different organs of soil-grown plants was done as described (Rensink et al., 1998).
Accession Numbers
Sequence data of the genes used in this article can be found in the GenBank/EMBL data libraries under accession numbers NM_127659 (THF1) and NM_128187 (GPA1). Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are At2g20890 (THF1) and At2g26300 (GPA1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Movie 1. Dynamics of Stromule Interactions with the Plasma Membrane.
Supplemental Figure 1. Epistasis Analysis between thf1 and gpa1 Loss-of-Function Alleles for Sugar Sensitivity.
Supplementary Material
Acknowledgments
We thank Peter Devreotes and Tian Jin of Johns Hopkins University for providing YFP and CFP cDNAs and advice on FRET microscopy, and Robert Bagnell, Jing Yang, Chris Breen, KwangChul Oh, and Tony Perdue, all of the University of North Carolina at Chapel Hill, for technical assistance. We thank Xin He (University of North Carolina at Chapel Hill) for help with the FRET normalization algorithm, and Brett Tyler (Virginia Bioinformatics Institute, Virginia Polytechnic Institute and State University) for proposing to us that THF1 is an integral membrane protein and for his supporting data. Work in A.M.J.'s laboratory on the Arabidopsis G-protein is supported by the National Institute of General Medical Sciences (Grant GM65989-01), the National Science Foundation (Grant MCB-0209711), and the U.S. Department of Energy (Grant DE-FG02-05ER15671).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Alan M. Jones (alan_jones@unc.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.037259.
References
- Arenas-Huertero, F., Arroyo, A., Zhou, L., Sheen, J., and Leon, P. (2000). Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 14 2085–2096. [PMC free article] [PubMed] [Google Scholar]
- Bastiaens, P.I., and Jovin, T.M. (1996). Microspectroscopic imaging tracks the intracellular processing of a signal transduction protein: Fluorescent-labeled protein kinase C beta I. Proc. Natl. Acad. Sci. USA 93 8407–8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiaens, P.I., Majoul, I.V., Verveer, P.J., Soling, H.D., and Jovin, T.M. (1996). Imaging the intracellular trafficking and state of the AB5 quaternary structure of cholera toxin. EMBO J. 15 4246–4253. [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316 1194–1199. [Google Scholar]
- Booden, M.A., Siderovski, D.P., and Der, C.J. (2002). Leukemia-associated Rho guanine nucleotide exchange factor promotes G alpha q-coupled activation of RhoA. Mol. Cell. Biol. 22 4053–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker, F.L., Burkey, K.O., Overmyer, K., and Jones, A.M. (2004). Differential responses of G-protein Arabidopsis thaliana mutants to ozone. New Phytol. 162 633–641. [DOI] [PubMed] [Google Scholar]
- Chen, J.G., and Jones, A.M. (2004). AtRGS1 Function in Arabidopsis thaliana. Methods Enzymol. 389 338–350. [DOI] [PubMed] [Google Scholar]
- Chen, J.G., Willard, F.S., Huang, J., Liang, J., Chasse, S.A., Jones, A.M., and Siderovski, D.P. (2003). A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301 1728–1731. [DOI] [PubMed] [Google Scholar]
- Chen, Y., Ji, F., Xie, H., Liang, J., and Zhang, J. (2006). The regulator of G-protein signaling proteins involved in sugar and abscisic acid signaling in Arabidopsis seed germination. Plant Physiol. 140 302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew, O., Rudhe, C., Glaser, E., and Whelan, J. (2003). Characterization of the targeting signal of dual-targeted pea glutathione reductase. Plant Mol. Biol. 53 341–356. [DOI] [PubMed] [Google Scholar]
- Claros, M.G., and von Heijne, G. (1994). TopPred II: An improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10 685–686. [DOI] [PubMed] [Google Scholar]
- Colombo, S., Ma, P., Cauwenberg, L., Winderickx, J., Crauwels, M., Teunissen, A., Nauwelaers, D., de Winde, J.H., Gorwa, M.F., Colavizza, D., and Thevelein, J.M. (1998). Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J. 17 3326–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto, A., Piffanelli, P., Nilsson, I., Wallin, E., Panstruga, R., von Heijne, G., and Schulze-Lefert, P. (1999). Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J. Biol. Chem. 274 34993–35004. [DOI] [PubMed] [Google Scholar]
- Eastmond, P.J., and Graham, I.A. (2003). Trehalose metabolism: A regulatory role for trehalose-6-phosphate? Curr. Opin. Plant Biol. 6 231–235. [DOI] [PubMed] [Google Scholar]
- Ferrando, A., Farras, R., Jasik, J., Schell, J., and Koncz, C. (2000). Intron-tagged epitope: A tool for facile detection and purification of proteins expressed in Agrobacterium-transformed plant cells. Plant J. 22 553–560. [DOI] [PubMed] [Google Scholar]
- Fischetti, V.A. (1989). Streptococcal M protein: Molecular design and biological behavior. Clin. Microbiol. Rev. 2 285–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso, G., Giacomelli, L., Ytterberg, A.J., Peltier, J.B., Rudella, A., Sun, Q., and Wijk, K.J. (2004). In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: New proteins, new functions, and a plastid proteome database. Plant Cell 16 478–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, G.W., Berry, G., Liang, X.H., Levine, B., and Herman, B. (1998). Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys. J. 74 2702–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, J.C., Sullivan, J.A., Hibberd, J.M., and Hansen, M.R. (2001). Stromules: Mobile protrusions and interconnections between plastids. Plant Biol. 3 223–233. [Google Scholar]
- Gunning, B.E.S. (2005). Plastid stromules: Video microscopy of their outgrowth, retraction, tensioning, anchoring, branching, bridging, and tip-shedding. Protoplasma 225 33–42. [DOI] [PubMed] [Google Scholar]
- Hatley, M.E., Lockless, S.W., Gibson, S.K., Gilman, A.G., and Ranganathan, R. (2003). Allosteric determinants in guanine nucleotide-binding proteins. Proc. Natl. Acad. Sci. USA 100 14445–14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herwald, H., Cramer, H., Morgelin, M., Russell, W., Sollenberg, U., Norrby-Teglund, A., Flodgaard, H., Lindbom, L., and Bjorck, L. (2004). M protein, a classical bacterial virulence determinant, forms complexes with fibrinogen that induce vascular leakage. Cell 116 367–379. [DOI] [PubMed] [Google Scholar]
- Inaba, T., Alvarez-Huerta, M., Li, M., Bauer, J., Ewers, C., Kessler, F., and Schnell, D.J. (2005). Arabidopsis Tic110 is essential for the assembly and function of the protein import machinery of plastids. Plant Cell 17 1482–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D.T., Froehlich, J.E., and Keegstra, K. (1998). The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J. Biol. Chem. 273 16583–16588. [DOI] [PubMed] [Google Scholar]
- Jang, J.C., Leon, P., Zhou, L., and Sheen, J. (1997). Hexokinase as a sugar sensor in higher plants. Plant Cell 9 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A.M., and Assmann, S.M. (2004). Plants: The latest model system for G-protein research. EMBO Rep. 5 572–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A.M., Ullah, H., and Chen, J.G. (2004). Dual pathways for auxin regulation of cell division and expansion. In Tobacco BY-2 Cells, D. Inze, ed (Berlin: Springer), pp. 180–191.
- Joo, J.H., Wang, S., Chen, J.G., Jones, A.M., and Fedoroff, N.V. (2005). Different signaling and cell death roles of heterotrimeric G protein alpha and beta subunits in the Arabidopsis oxidative stress response to ozone. Plant Cell 17 957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova, T.S., Baumann, C.T., He, L., Wu, X., Grammer, A., Lipsky, P., Hager, G.L., and McNally, J.G. (2003). Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. J. Microsc. 209 56–70. [DOI] [PubMed] [Google Scholar]
- Kenworthy, A.K. (2001). Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods 24 289–296. [DOI] [PubMed] [Google Scholar]
- Kolbe, A., Tiessen, A., Schluepmann, H., Paul, M., Ulrich, S., and Geigenberger, P. (2005). Trehalose 6-phosphate regulates starch synthesis via posttranslational redox activation of ADP-glucose pyrophosphorylase. Proc. Natl. Acad. Sci. USA 102 11118–11123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad, R.J., Young, R.A., Record, R.D., Smith, R.M., Butkerait, P., Manning, D., Jarett, L., and Wolf, B.A. (1995). The heterotrimeric G-protein Gi is localized to the insulin secretory granules of beta-cells and is involved in insulin exocytosis. J. Biol. Chem. 270 12869–12876. [DOI] [PubMed] [Google Scholar]
- Kwok, E.Y., and Hanson, M.R. (2004. a). Plastids and stromules interact with the nucleus and cell membrane in vascular plants. Plant Cell Rep. 23 188–195. [DOI] [PubMed] [Google Scholar]
- Kwok, E.Y., and Hanson, M.R. (2004. b). Stromules and the dynamic nature of plastid morphology. J. Microsc. 214 124–137. [DOI] [PubMed] [Google Scholar]
- Kwok, E.Y., and Hanson, M.R. (2004. c). In vivo analysis of interactions between GFP-labeled microfilaments and plastid stromules. BMC Plant Biol. 4 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok, E.Y., and Hanson, M.R. (2004. d). GFP-labelled Rubisco and aspartate aminotransferase are present in plastid stromules and traffic between plastids. J. Exp. Bot. 55 595–604. [DOI] [PubMed] [Google Scholar]
- Lang, J. (1999). Molecular mechanisms and regulation of insulin exocytosis as a paradigm of endocrine secretion. Eur. J. Biochem. 259 3–17. [DOI] [PubMed] [Google Scholar]
- Lapik, Y.R., and Kaufman, L.S. (2003). The Arabidopsis cupin domain protein AtPirin1 interacts with the G protein alpha-subunit GPA1 and regulates seed germination and early seedling development. Plant Cell 15 1578–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire, K., Van de Velde, S., Van Dijck, P., and Thevelein, J.M. (2004). Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol. Cell 16 293–299. [DOI] [PubMed] [Google Scholar]
- Lorenz, S., Tintelnot, S., Reski, R., and Decker, E.L. (2003). Cyclin D-knockout uncouples developmental progression from sugar availability. Plant Mol. Biol. 53 227–236. [DOI] [PubMed] [Google Scholar]
- Luo, X., Popov, S., Bera, A.K., Wilkie, T.M., and Muallem, S. (2001). RGS proteins provide biochemical control of agonist-evoked [Ca2+]i oscillations. Mol. Cell 7 651–660. [DOI] [PubMed] [Google Scholar]
- Ma, H., Yanofsky, M.F., and Meyerowitz, E.M. (1990). Molecular cloning and characterization of GPA1, a G protein alpha subunit gene from Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 87 3821–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makela, P., McLaughlin, J.E., and Boyer, J.S. (2005). Imaging and quantifying carbohydrate transport to the developing ovaries of maize. Ann. Bot. (Lond.) 96 939–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maness, N.O., and Mcbee, G.G. (1986). Role of placental sac in endosperm carbohydrate import in sorghum caryopses. Crop Sci. 26 1201–1207. [Google Scholar]
- Mason, M.G., and Botella, J.R. (2000). Completing the heterotrimer: Isolation and characterization of an Arabidopsis thaliana G protein gamma-subunit cDNA. Proc. Natl. Acad. Sci. USA 97 14784–14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, M.G., and Botella, J.R. (2001). Isolation of a novel G-protein gamma-subunit from Arabidopsis thaliana and its interaction with Gbeta. Biochim. Biophys. Acta 1520 147–153. [DOI] [PubMed] [Google Scholar]
- Maxfield, F.R., and McGraw, T.E. (2004). Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5 121–132. [DOI] [PubMed] [Google Scholar]
- McLaughlin, J.E., and Boyer, J.S. (2004). Glucose localization in maize ovaries when kernel number decreases at low water potential and sucrose is fed to the stems. Ann. Bot. (Lond.) 94 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges, M., and Murray, J.A. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30 203–212. [DOI] [PubMed] [Google Scholar]
- Moore, B., Zhou, L., Rolland, F., Hall, Q., Cheng, W.H., Liu, Y.X., Hwang, I., Jones, T., and Sheen, J. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300 332–336. [DOI] [PubMed] [Google Scholar]
- Natesan, S.K., Sullivan, J.A., and Gray, J.C. (2005). Stromules: A characteristic cell-specific feature of plastid morphology. J. Exp. Bot. 56 787–797. [DOI] [PubMed] [Google Scholar]
- Pandey, S., and Assmann, S.M. (2004). The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein alpha subunit GPA1 and regulates abscisic acid signaling. Plant Cell 16 1616–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfus-Barbeoch, L., Jones, A.M., and Assmann, S.M. (2004). Plant heterotrimeric G protein function: Insights from Arabidopsis and rice mutants. Curr. Opin. Plant Biol. 7 719–731. [DOI] [PubMed] [Google Scholar]
- Pyke, K.A., and Howells, C.A. (2002). Plastid and stromule morphogenesis in tomato. Ann. Bot. (Lond.) 90 559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensink, W.A., Pilon, M., and Weisbeek, P. (1998). Domains of a transit sequence required for in vivo import in Arabidopsis chloroplasts. Plant Physiol. 118 691–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou-Khamlichi, C., Menges, M., Healy, J.M., and Murray, J.A. (2000). Sugar control of the plant cell cycle: Differential regulation of Arabidopsis D-type cyclin gene expression. Mol. Cell. Biol. 20 4513–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland, F., Winderickx, J., and Thevelein, J.M. (2001). Glucose-sensing mechanisms in eukaryotic cells. Trends Biochem. Sci. 26 310–317. [DOI] [PubMed] [Google Scholar]
- Sheen, J., Zhou, L., and Jang, J.C. (1999). Sugars as signaling molecules. Curr. Opin. Plant Biol. 2 410–418. [DOI] [PubMed] [Google Scholar]
- Siderovski, D.P., and Willard, F.S. (2005). The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int. J. Biol. Sci. 1 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund, G., Basmaciogullari, A., Rouot, B., Marie, J.C., and Rosselin, G. (1999). Cell-specific localization of G protein alpha-subunits in the islets of Langerhans. J. Endocrinol. 162 31–37. [DOI] [PubMed] [Google Scholar]
- Smith, M.D., Fitzpatrick, L.M., Keegstra, K., and Schnell, D.J. (2002). In vitro analysis of chloroplast protein import. In Current Protocols in Cell Biology, K.M. Yamada, ed (New York: John Wiley & Sons), pp. 11.16.11–11.16.21. [DOI] [PubMed]
- Soellick, T.-R., and Uhrig, J. (2001). Development of an optimized interaction-mating protocol for large-scale yeast two-hybrid analyses. Genome Biol. 2 research0052.0051–research0052.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand, A., Asami, T., Alonso, J., Ecker, J.R., and Chory, J. (2003). Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421 79–83. [DOI] [PubMed] [Google Scholar]
- Tiessen, A., Prescha, K., Branscheid, A., Palacios, N., McKibbin, R., Halford, N.G., and Geigenberger, P. (2003). Evidence that SNF1-related kinase and hexokinase are involved in separate sugar-signalling pathways modulating post-translational redox activation of ADP-glucose pyrophosphorylase in potato tubers. Plant J. 35 490–500. [DOI] [PubMed] [Google Scholar]
- Tu, C.J., Schuenemann, D., and Hoffman, N.E. (1999). Chloroplast FtsY, chloroplast signal recognition particle, and GTP are required to reconstitute the soluble phase of light-harvesting chlorophyll protein transport into thylakoid membranes. J. Biol. Chem. 274 27219–27224. [DOI] [PubMed] [Google Scholar]
- Ullah, H., Chen, J.G., Temple, B., Boyes, D.C., Alonso, J.M., Davis, K.R., Ecker, J.R., and Jones, A.M. (2003). The beta-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell 15 393–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah, H., Chen, J.G., Wang, S., and Jones, A.M. (2002). Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol. 129 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah, H., Chen, J.G., Young, J.C., Im, K.H., Sussman, M.R., and Jones, A.M. (2001). Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 292 2066–2069. [DOI] [PubMed] [Google Scholar]
- Vanderbeld, B., and Kelly, G.M. (2000). New thoughts on the role of the beta-gamma subunit in G-protein signal transduction. Biochem. Cell Biol. 78 537–550. [DOI] [PubMed] [Google Scholar]
- Vermeer, J.E., Van Munster, E.B., Vischer, N.O., and Gadella, T.W., Jr. (2004). Probing plasma membrane microdomains in cowpea protoplasts using lipidated GFP-fusion proteins and multimode FRET microscopy. J. Microsc. 214 190–200. [DOI] [PubMed] [Google Scholar]
- Versele, M., de Winde, J.H., and Thevelein, J.M. (1999). A novel regulator of G protein signalling in yeast, Rgs2, downregulates glucose-activation of the cAMP pathway through direct inhibition of Gpa2. EMBO J. 18 5577–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra, R.D. (2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8 135–142. [DOI] [PubMed] [Google Scholar]
- von Heijne, G. (1992). Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225 487–494. [DOI] [PubMed] [Google Scholar]
- Wang, Q., Sullivan, R.W., Kight, A., Henry, R.L., Huang, J., Jones, A.M., and Korth, K.L. (2004). Deletion of the chloroplast-localized Thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol. 136 3594–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.Q., Ullah, H., Jones, A.M., and Assmann, S.M. (2001). G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292 2070–2072. [DOI] [PubMed] [Google Scholar]
- Weiss, C.A., Garnaat, C.W., Mukai, K., Hu, Y., and Ma, H. (1994). Isolation of cDNAs encoding guanine nucleotide-binding protein beta-subunit homologues from maize (ZGB1) and Arabidopsis (AGB1). Proc. Natl. Acad. Sci. USA 91 9554–9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, W., Sheen, J., and Jang, J.C. (2000). The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol. Biol. 44 451–461. [DOI] [PubMed] [Google Scholar]
- Yang, P., Fu, H., Walker, J., Papa, C.M., Smalle, J., Ju, Y.M., and Vierstra, R.D. (2004). Purification of the Arabidopsis 26 S proteasome: Biochemical and molecular analyses revealed the presence of multiple isoforms. J. Biol. Chem. 279 6401–6413. [DOI] [PubMed] [Google Scholar]
- Zhao, J., and Wang, X. (2004). Arabidopsis phospholipase Dalpha1 interacts with the heterotrimeric G-protein alpha-subunit through a motif analogous to the DRY motif in G-protein-coupled receptors. J. Biol. Chem. 279 1794–1800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.