Abstract
Class I histone deacetylases (HDACs) are ubiquitous enzymes that repress gene expression by deacetylating histone tails and promoting chromatin compaction. Pro-inflammatory agents activate programmes of gene expression through transcription factors such as nuclear factor-κB (NF-κB), even in the context of ubiquitous HDAC activity. How this is accomplished remains unknown. We found that cells treated with the pro-inflammatory cytokine tumour necrosis factor-α rapidly and substantially reduced HDAC1 protein levels without affecting other class I HDACs. In addition, HDAC1 depletion occurred through protein degradation, required IKK2 activity and resulted in increased transcription from both NF-κB-associated and unassociated gene promoters. Our study suggests that the activation of programmes of gene expression by pro-inflammatory agents requires global changes in specific critical epigenetic regulators such as HDAC1.
Keywords: chromatin, histone deacetylases, IKK2, NF-κB, TNF-α
Introduction
Histone deacetylases (HDACs) are enzymes that catalyse the removal of acetyl groups from acetyllysine residues in both histones and other cellular proteins (de Ruijter et al, 2003). A total of 18 human HDACs, grouped into four classes on the basis of their sequence homology, have been identified. Of these, the class I members HDAC1, HDAC2, HDAC3 and HDAC8 are ubiquitous, predominantly nuclear proteins that usually exist as members of multiprotein complexes involved in transcriptional repression. Little is known with regard to the regulation of class I HDACs, although examples of their regulation through transcriptional, post-translational, complex composition and subcellular transport mechanisms exist (Sengupta & Seto, 2004).
One of the primary functions of HDAC proteins is to maintain transcriptionally silent chromatin (Cheung et al, 2000). HDACs achieve this through the removal of acetyl groups from the amino-terminal tails of histone proteins, especially histones H3 and H4. Although general class I/II/IV HDAC inhibition by chemical inhibitors (HDACi) appreciably alters the expression of only a few genes (Glaser et al, 2003), there is evidence that mammalian class I HDACs, and especially HDAC1, are critical repressors of genes involved in cell proliferation, including the cyclin-dependent kinase inhibitors p21WAF1/CIP1 and p27KIP1 (Lagger et al, 2002).
Pro-inflammatory agents, including the cytokines tumour necrosis factor-α (TNF-α) and interleukin-1β (IL-1β) and the endotoxin lipopolysaccharide (LPS), activate a host of proliferative, anti-apoptotic, immunoregulatory and inflammatory genes through the activation of the ubiquitous transcription factor nuclear factor-κB (NF-κB; Ghosh et al, 1998). Canonical NF-κB (p65RelA/p50) normally resides in a latent form in the cell cytoplasm as a complex with the inhibitor protein IκBα. Following binding of pro-inflammatory agents to their respective extracellular receptors, various signal transduction pathways lead to the phosphorylation of IκBα by the IκB kinase (IKK) signalosome (IKK1/IKK2/NEMO), dissociation of the NF-κB/IκBα complex, degradation of ubiquitinated IκBα by the 26S proteasome and translocation of NF-κB to the nucleus, where it binds to its target genes and activates their transcription. Notably, one of its target genes encodes IκBα, thereby allowing for a time-dependent, negative feedback regulation of NF-κB activity (Ghosh et al, 1998). In addition, the IKKs have only recently been identified as having a role in other signalling cascades (Finnberg & El-Deiry, 2004), thus adding to the complexity of their responses to pro-inflammatory signals.
The activation of programmes of gene expression is a complicated process, especially when regulated by an erstwhile latent transcription factor such as NF-κB. How this protein is able to access regions of condensed chromatin and deliver transcriptional activation in the face of ubiquitous HDAC transcriptional repression is essentially unknown. To address this question, we investigated the regulation of class I HDACs following cell treatment with the pro-inflammatory cytokine TNF-α.
Results and Discussion
TNF-α depletes HDAC1 but not other class I HDACs
To determine how pro-inflammatory cytokines regulate HDACs, we first investigated the effects of TNF-α on class I HDAC protein levels in both breast carcinoma (ZR-75-1) and immortalized normal breast (76N-E6) cells. Surprisingly, results of western blotting showed a rapid, substantial depletion of HDAC1 protein in ZR-75-1 nuclear extracts and a substantial but transient HDAC1 depletion in 76N-E6 nuclear extracts (Fig 1). Significant changes in protein levels were not observed for the other class I HDACs in either ZR-75-1 or 76N-E6 nuclear extracts, although an atypical presence of HDAC2 in cytoplasmic extracts and post-translational modifications of HDAC8 in both 76N-E6 extracts were observed. Western blots with the cytoplasmic protein paxillin and the nuclear protein poly(ADP-ribose) polymerase (PARP) demonstrated the purity of these extracts. Similar HDAC1 depletion phenomena were found in other cell types, including breast carcinoma (MCF-7 and SK-BR-3), melanoma (MeWo), non-small-cell lung carcinoma (NCI-H522 and NCI-H1792) and ovarian epithelial carcinoma (SK-OV-3.ip1) cell lines, although there were notable differences in their kinetics and persistence (see supplementary Fig S1 online). Taken together, class I HDAC depletion by TNF-α seems to be a general phenomenon among different cell types, but is specific for HDAC1.
Figure 1.
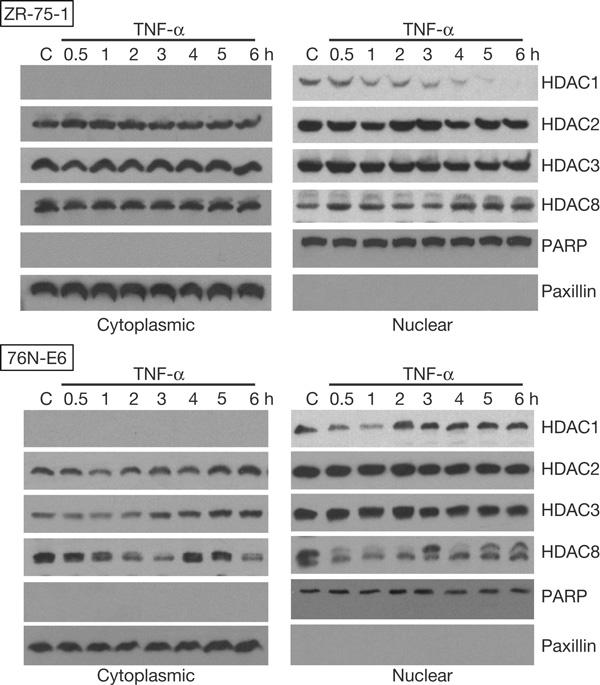
Histone deacetylase (HDAC)1 protein depletion occurs after treatment of cells with tumour necrosis factor-α (TNF-α). Breast carcinoma (ZR-75-1) and immortalized normal breast (76N-E6) were treated with 20 ng/ml recombinant human TNF-α for the indicated durations and then collected. Western blots of SDS–polyacrylamide gel electrophoresis-resolved cytoplasmic and nuclear extract proteins from these cells probed with different class I HDAC and control protein antibodies and visualized by chemiluminescence are shown. PARP, poly(ADP-ribose) polymerase.
HDAC1 depletion occurs through protein degradation
Focusing on TNF-α-mediated HDAC1 depletion, we sought to determine how this phenomenon occurs. Some class I HDACs, particularly HDAC3, are known to shuttle between the cytoplasm and nucleus (Baek et al, 2002). However, this was not the cause for the observed HDAC1 depletion, as no cytoplasmic HDAC1 accumulation was observed following TNF-α treatment in any cell type (Fig 1). Likewise, transcriptional or RNA stability changes did not seem to be responsible for the apparent HDAC1 protein depletion, as HDAC1 messenger RNA levels in ZR-75-1 cells were relatively unchanged following TNF-α treatment, as determined by real-time PCR (Fig 2A). Although no changes in HDAC1 post-translational modifications were discernable by SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and western blotting (Fig 1), we nonetheless investigated potential post-translational mechanisms for HDAC1 depletion through the use of various small molecule inhibitors. Treatment of ZR-75-1 cells with the proteasomal inhibitor MG-132 appreciably increased the intrinsic nuclear level of HDAC1 protein and inhibited HDAC1 depletion by TNF-α (Fig 2B). This was probably a direct effect, as both TNF-α and MG-132 treatments increased the levels of ubiquitinated HDAC1 (Fig 2C). Conversely, treatment with the general caspase inhibitor Z-VAD-FMK had no effect on the intrinsic HDAC1 level or the process of HDAC1 depletion by TNF-α (Fig 2B). Treatment with the protein synthesis inhibitor cycloheximide significantly reduced the intrinsic HDAC1 level, but did not show continued decreases with increasing time. In addition, HDAC1 depletion by TNF-α was not appreciably inhibited in cycloheximide-treated cells, suggesting that protein synthesis is not absolutely required for this phenomenon. The moderate half-life of HDAC1 was confirmed by a pulse–chase experiment, which also showed that HDAC1 depletion by TNF-α was primarily the result of accelerated protein degradation and not reduced protein synthesis (Fig 2D). Finally, the specific IKK2 kinase inhibitor BMS 345541 did not change the intrinsic HDAC1 level, but completely inhibited TNF-α-mediated HDAC1 depletion (Fig 2B). Note that in the case of all the small molecule inhibitors, their effects were not the result of a change in the kinetics of HDAC1 depletion by TNF-α. Taken together, our data suggest that intrinsic HDAC1 levels are regulated by ubiquitination and proteasomal activity, whereas TNF-α-dependent HDAC1 depletion requires IKK2 activity and is mediated by the proteasome.
Figure 2.
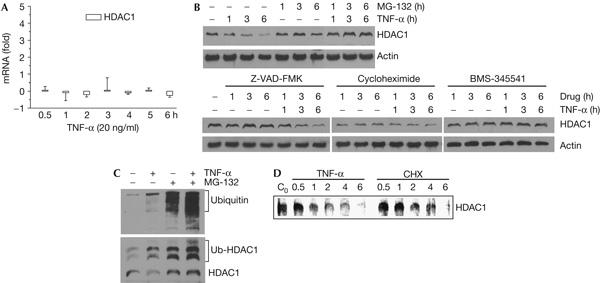
Histone deacetylase 1 protein depletion occurs through protein degradation. (A) ZR-75-1 cells were treated with tumour necrosis factor-α (TNF-α) for the indicated durations and RNA was purified and reverse transcribed into complementary DNA. The cDNA samples were used as templates for real-time quantitative PCR amplification of histone deacetylase (HDAC)1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). HDAC1 amplifications were normalized against those of GAPDH, and the fold changes in the number of transcripts were determined and plotted as bar graphs. (B) ZR-75-1 cells were treated with 20 ng/ml TNF-α, 10 μM MG-132, 10 μM Z-VAD-FMK, 5 μg/ml cycloheximide and 25 μM BMS-345541 for the indicated durations and then collected. Western blots of whole-cell extract proteins probed with HDAC1 and β-actin antibodies are shown. (C) HDAC1 protein was immunoprecipitated from ZR-75-1 cells treated with 20 ng/ml TNF-α and/or 10 μM MG-132 for 4 h, as indicated, western blotted, probed with ubiquitin antibodies (top panel) and later stripped and re-probed with HDAC1 antibodies (bottom panel). (D) ZR-75-1 cells were pulsed with [35S]methionine for 1 h and then chased in rich medium containing either 20 ng/ml TNF-α or 5 μg/ml cycloheximide (CHX) for the indicated durations before collection. HDAC1 protein was immunoprecipitated from whole-cell extracts, resolved by SDS–polyacrylamide gel electrophoresis and radiolabelled protein visualized by a phosphoimager.
TNF-α-mediated HDAC1 depletion is dependent on IKK2
Inhibition of TNF-α-mediated HDAC1 depletion by the specific IKK2 kinase inhibitor BMS 345541 suggested a role for the NF-κB signalling pathway in this phenomenon. Likewise, the data with the proteasomal inhibitor MG-132 support an NF-κB involvement, as proteasome inhibition can elevate cellular IκBα levels and suppress free NF-κB (Ghosh et al, 1998). Investigating the effects of the NF-κB-activating pro-inflammatory agents on HDAC depletion, we found that the cytokine IL-1β and the bacterial endotoxin LPS caused specific HDAC1 depletion in ZR-75-1 cells (Fig 3A). However, the duration and intensity of these effects differed from those seen with TNF-α and could reflect differences in the signal transduction pathways activated by these pro-inflammatory agents. Treatment with the TNF-related apoptosis-inducing ligand TRAIL also caused a depletion of HDAC1. This result was however complicated by the fact that the TRAIL treatment induced the cleavage of PARP, consistent with an induction of apoptosis. Thus, HDAC1 depletion in TRAIL-treated cells could be due to the loss of membrane integrity and generalized proteolysis, characteristic of apoptotic cells. PARP cleavage was not observed in TNF-α-, IL-1β- or LPS-treated ZR-75-1 cells (Figs 1A and 3A), suggesting that HDAC1 depletion caused by these treatments is mediated by other mechanisms (for example, proteasomal degradation).
Figure 3.
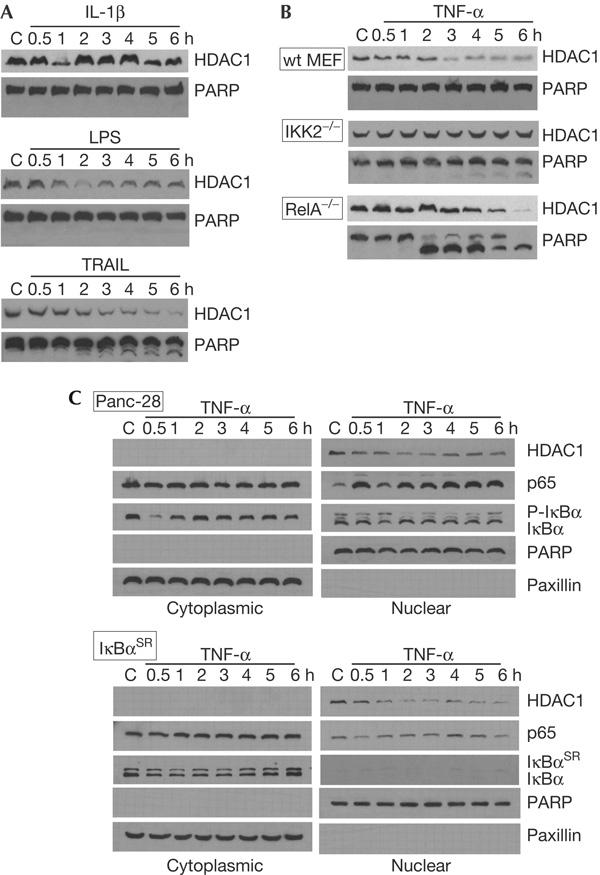
Histone deacetylase 1 protein depletion requires IKK2. (A) ZR-75-1 cells were treated with 5 ng/ml interleukin-1β (IL-1β), 10 μg/ml lipopolysaccharide (LPS) or 50 ng/ml TNF-related apoptosis-inducing ligand (TRAIL) for the indicated durations and then collected. Western blots of nuclear extract proteins probed with histone deacetylase (HDAC)1 and poly(ADP-ribose) polymerase (PARP) antibodies are shown. (B) Wild-type (wt) mouse embryo fibroblasts (MEFs) and their complementary cells containing homozygous deletions of IKK2−/− or RelA−/− were treated with 20 ng/ml tumour necrosis factor-α (TNF-α) for the indicated durations. Western blots of nuclear extract proteins probed with HDAC1 and PARP antibodies are shown. (C) Normal Panc-28 pancreatic cancer cells or those constitutively expressing the IκBα super-repressor were treated with 20 ng/ml TNF-α for the durations indicated, then collected. Western blots of cytoplasmic and nuclear extract proteins probed for HDAC1, p65, IκBα, PARP and paxillin are shown.
To further verify the involvement of the NF-κB activation pathway in HDAC1 depletion, experiments were carried out with mouse embryo fibroblast (MEF) cell lines containing homozygous deletions in IKK2 and RelA. We observed a rapid and marked HDAC1 depletion in nuclear extracts from TNF-α-treated wild-type MEFs but not in IKK2−/− MEFs (Fig 3B). RelA−/− MEFs are inherently sensitive to TNF-α, which resulted in significant PARP cleavage and apoptosis and an associated depletion of HDAC1. HDAC1 levels did not seem to decrease until at least the start of PARP cleavage; however, this result should be interpreted with caution. IKK2−/− MEFs, which are significantly less sensitive to TNF-α than RelA−/− MEFs, showed less PARP cleavage and no change in HDAC1 levels. These experiments strongly support an involvement of IKK2 in the TNF-α-mediated depletion of HDAC1, but are less convincing with regard to a role for NF-κB in this process. To clarify the latter, experiments were carried out with cells that constitutively express the super-repressor mutant form of IκBα. These cells inhibited the activation of NF-κB by TNF-α but did not appreciably inhibit HDAC1 depletion (Fig 3C). Thus, NF-κB may not be directly involved in HDAC1 depletion.
Global and local consequences of HDAC1 depletion
As expected from the feedback regulation of NF-κB and IκBα by TNF-α (Ghosh et al, 1998), we observed an oscillatory increase of cytoplasmic protein levels of p65 and IκBα, which peaked at 2 and 3 h in ZR-75-1 cells (Fig 4A). We observed that the levels of nuclear p53 and cytoplasmic p21 proteins also peaked at the same time points. Real-time PCR measurements showed an increase in the p53 and p21WAF1/CIP1 mRNA levels that preceded the changes in their respective protein levels, consistent with transcriptional activation of both genes (Fig 4B). The peak activation of these genes at similar time points suggests a global change in the gene expression profile in ZR-75-1 cells following TNF-α treatment, which may involve changes in specific epigenetic regulators such as HDAC1. However, a causal relationship between these phenomena and HDAC1 is less clear, as upregulation of these genes often preceded complete HDAC1 depletion. More probably, a functional loss of HDAC1 activity (for example, following dissociation from multiprotein complexes) may precede HDAC1 degradation and evident depletion.
Figure 4.
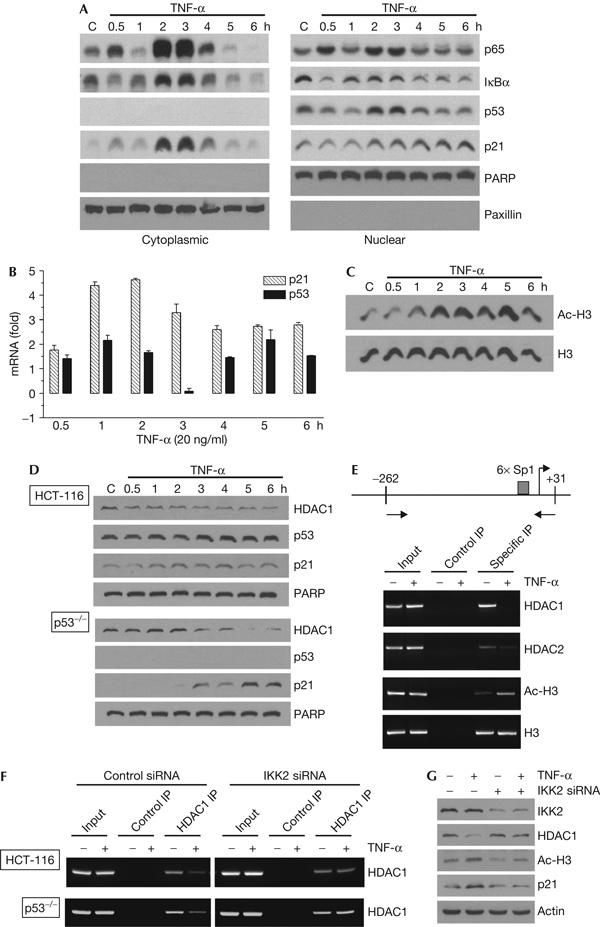
Histone deacetylase 1 protein depletion affects general histone acetylation and specific gene expression. (A) ZR-75-1 cells were treated with 20 ng/ml tumour necrosis factor-α (TNF-α) for the indicated durations, after which nuclear and cytoplasmic extracts were obtained. Extract proteins were resolved on SDS–polyacrylamide gel electrophoresis gels, blotted and probed for p65, IκBα, p53, p21, poly(ADP-ribose) polymerase (PARP) and paxillin. (B) RNA was purified from these cells and reverse transcribed into complementary DNA. The cDNA samples were used as templates for real-time quantitative PCR amplification of p21, p53 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Both p21 and p53 amplifications were normalized against those of GAPDH, and the fold changes in the number of transcripts were determined and plotted as bar graphs. (C) Western blots of nuclear extract proteins from the experiment described in (A) were also probed for acetyl histone H3 (Ac-H3) and histone H3 (H3). (D) Normal HCT-116 colon cancer cells or those containing a homozygous deletion of the p53 gene (p53−/−) were treated with 20 ng/ml of TNF-α for the indicated durations, collected and nuclear extracts were made. Western blots of these proteins probed with histone deacetylase (HDAC)1, p53, p21 and PARP antibodies are shown. (E) ZR-75-1 cells were either treated (+) or not treated (−) with 20 ng/ml of TNF-α for 3 h and processed for chromatin immunoprecipitation (ChIP) assays. The antibodies used were HDAC1, HDAC2, acetyl histone H3, histone H3 and a non-immunogenic control. Top panel: schematic representation of the WAF1/CIP1 promoter, including the locations of the primers used for ChIP. Bottom panel: immunoprecipitation (IP) inputs and PCR-amplified products resolved on 2% agarose gels and visualized by ethidium bromide staining and UV fluorescence. (F) Normal and p53−/− HCT-116 cells were treated with 20 nM control (left panels) or IKK2 short interfering RNA (siRNA; right panels) for 48 h, then with 20 ng/ml TNF-α for 3 h, as indicated, and processed for ChIP assays. The antibodies used were HDAC1 and a non-immunogenic control. (G) Whole-cell extracts from above p53−/− HCT-116 cells were western blotted and probed with antibodies against IKK2, HDAC1, acetyl histone H3, p21 and β-actin.
One obvious consequence of cellular HDAC1 depletion would be an increase in cellular acetylated histone levels (for example, acetylated histone H3), as is the case when cells are treated with HDACi (Richon et al, 2004). We observed that TNF-α treatment significantly increased the acetylated histone H3 levels in 2 h after exposure, whereas total histone H3 levels were unaffected in ZR-75-1 cells (Fig 4C). Note that these effects need not be exclusively the result of HDAC1 depletion, given that transcriptionally active NF-κB is known to deliver histone acetyltransferases such as p300/CBP to the chromatin, thereby increasing acetylated histone H3 levels on many genes (Li & Verma, 2002).
WAF1/CIP1 is one of the few genes the expression of which is significantly enhanced when cells are treated with a variety of HDACi (Glaser et al, 2003). This may also be the case with TNF-α-mediated HDAC1 depletion. However, ZR-75-1 cells contain wild-type p53, which is a strong transcriptional activator of WAF1/CIP1 (El-Deiry et al, 1993). To test this, experiments were carried out in colon cancer cells lacking p53. In these cells, TNF-α treatment led to a robust accumulation of nuclear p21 protein, coincident with HDAC1 depletion (Fig 4D). Thus, HDAC1 depletion does not require p53; neither does the absence of p53 diminish WAF1/CIP1 activation.
Upregulation of p21WAF1/CIP1 mRNA levels after TNF-α treatment may in part be the result of reduced HDAC activity on the WAF1/CIP1 promoter resulting from HDAC1 depletion. To test this hypothesis, we carried out chromatin immunoprecipitation (ChIP) assays to determine the levels of acetylated histone H3 and HDAC1 and HDAC2 proteins on the WAF1/CIP1 promoter after TNF-α treatment. Levels of total histone H3 protein served as a control. TNF-α caused a complete loss of detectable HDAC1 protein associated with the WAF1/CIP1 promoter and, to a lesser extent, a reduction of HDAC2 protein levels (Fig 4E). This loss of HDAC2 protein is not surprising, because a significant fraction of cellular HDAC2 protein is normally associated with HDAC1 in multiprotein transcriptional repressor complexes such as Sin3, NuRD and CoREST (Sengupta & Seto, 2004). Finally, we found an increase in acetylated histone H3 levels associated with the WAF1/CIP1 promoter, consistent with a local reduction in HDAC activity. Similar ChIP experiments with cells lacking p53 or with reduced IKK2 protein levels confirmed that p53 is not fully responsible for the reduction of HDAC1 associated with the WAF1/CIP1 promoter, but that IKK2 is absolutely required for the TNF-α-dependent phenomenon (Fig 4F). Likewise, IKK2 was required for p21 upregulation and increased histone H3 acetylation in TNF-α-treated p53−/− cells (Fig 4G). Taken together, our data suggest that the cellular depletion of HDAC1 results in significant local consequences for specific gene promoters and their transcriptional strength and in global changes in acetylated histone levels, thus speaking of the importance of HDAC1 in regulating the expression of many genes responsive to pro-inflammatory agents. However, HDACs are known to have both positive and negative effects on cellular responses to these agents (Blanchard & Chipoy, 2005). Thus, HDAC1 depletion may be only a part of this complex process.
Speculation
HDAC1 depletion seems to be a general phenomenon, occurring in normal and transformed cells derived from several tissue types. Although our studies showed that HDAC1 depletion is mediated by IKK2 as part of its response to pro-inflammatory signals, global changes in HDAC1 levels could also be important for the activation of other gene expression programmes, especially those that require a rapid induction of large numbers of genes and/or involve latent transcription factors. In support of this hypothesis, we have obtained preliminary evidence that the pleiotropic cytokines interferon-α and interferon-γ, which primarily act through Janus kinase activation of latent STAT (signal transducer and activator of transcription) transcription factors and do not involve IKK2 or NF-κB, also cause HDAC1 elimination in different cell types (D.H. Yan & Y.N.V.G., unpublished observations). Thus, HDAC depletion may be a general consequence of the activation of gene expression programmes by different signalling pathways and their associated transcriptional activators.
Methods
Cell culture. The breast cancer cell lines MCF-7, SK-BR-3 and ZR-75-1 were obtained from ATCC (Manassas, VA, USA). Other cell lines were obtained from colleagues at M.D. Anderson and Johns Hopkins. ZR-75-1 cells were propagated in RPMI-1640 medium supplemented with 10% FBS, 76N-E6 cells in DFCI-1 medium and the other cell lines in DMEM with 10% FBS. Cells (106/10 cm dish) were incubated for 36–48 h to allow doubling before subjecting to further cytokine or drug treatments.
Antibodies, cytokines and drugs. HDAC1, HDAC3, p53 and PARP antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA), β-actin antibody was from Sigma (St Louis, MO, USA), HDAC2, HDAC8, acetyl histone H3 and histone H3 antibodies were from Upstate Cell Signaling Solutions (Charlottesville, VA, USA), p21 and ubiquitin antibodies were from Cell Signaling Technology (Beverly, MA, USA) and paxillin antibody was from NeoMarkers (Fremont, CA, USA). TNF-α was obtained from Biosource International (Camarillo, CA, USA) and LPS was from Sigma. IL-1β was a gift from Dr E.A. Grimm (M.D. Anderson). MG-132, Z-VAD-FMK and cycloheximide were obtained from Sigma and BMS-345541 was from Calbiochem (SanDiego, CA, USA).
Western blotting. Nuclear and cytoplasmic extracts were prepared according to the procedure outlined by Schreiber et al (1989), whereas whole-cell extracts were NP-40 lysates. Typically, 50 μg extracts were resolved on 10% SDS–PAGE gels, transferred to nitrocellulose membranes, blotted with the appropriate antibody and detected using a chemiluminescence detection kit (SuperSignal; Pierce, Rockford, IL, USA).
Real-time reverse transcription–PCR. Total RNA was extracted by using an RNAqueous kit (Ambion, Austin, TX, USA) and complementary DNA was synthesized by using the RETROscript kit (Ambion) according to the manufacturer's instructions. Real-time reverse transcription–PCR experiments were carried out in an iCycler (Bio-Rad, Hercules, CA, USA) using the cDNA templates and TaqMan probe-based gene expression assay primer–probe mixes appropriate for HDAC1, p21, p53 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Applied Biosystems, Foster City, CA, USA). PCR conditions for each of the amplicons were optimized using serial dilutions of the control template, and efficiencies of the reactions were established. The fold changes in the treated samples compared with controls were quantitatively determined by using the following formula: fold change = efficiencyδCt, where δCt is the threshold cycle difference between control and treated samples normalized against GAPDH.
Pulse–chase experiments. ZR-75-1 cells were maintained for 1 h in cysteine- and methionine-deficient medium, pulsed for 1 h with 30 μCi/ml [35S]methionine and then chased with medium containing 2 mM of cysteine and methionine each.
Short interfering RNA transfections. Transfections of cells with 20 nM IKK2 or scrambled control short interfering RNAs (Dharmacon, Lafayette, CO, USA) were carried out according to the manufacturer's instructions. Transfected cells were incubated for 48 h before subsequent cytokine or drug treatment.
Chromatin immunoprecipitation. ChIP assays were carried out with the Upstate ChIP kit according to the manufacturer's instructions. Specific chromatin complexes were immunoprecipitated with HDAC1, HDAC2, acetyl histone H3 and histone H3 antibodies. A nonspecific antibody served as a non-immunogenic control. PCR amplification of the WAF1/CIP1 promoter was carried out with the primers sp1A-p21 (5′-CAGCGCACCAACGCAGGCG-3′) and sp1B-p21 (5′-CAGCTCCGGCTCCACAAGGA-3′) for 20 cycles. The amplified products were run on a 2% agarose gel and the data were quantified using a gel analysis system (Alpha Innotech, San Leandro, CA, USA). Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/7400613-s1.pdf).
Supplementary Material
Supplementary Information
Acknowledgments
We thank P. Chiao, E. Grimm, M.-C. Hung, K. Keyomarsi, R. Lotan and B. Vogelstein for cell lines and reagents. This work was supported by Grant W81XWH-04-1-0610 from the US Army Breast Cancer Research Program and by funds from the Commonwealth Cancer Foundation for Research.
References
- Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG (2002) Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-κB and β-amyloid precursor protein. Cell 110: 55–67 [DOI] [PubMed] [Google Scholar]
- Blanchard F, Chipoy C (2005) Histone deacetylase inhibitors: new drugs for the treatment of inflammatory diseases? Drug Discov Today 10: 197–204 [DOI] [PubMed] [Google Scholar]
- Cheung WL, Briggs SD, Allis CD (2000) Acetylation and chromosomal functions. Curr Opin Cell Biol 12: 326–333 [DOI] [PubMed] [Google Scholar]
- de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB (2003) Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J 370: 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B (1993) WAF1, a potential mediator of p53 tumor suppression. Cell 75: 817–825 [DOI] [PubMed] [Google Scholar]
- Finnberg N, El-Deiry WS (2004) Activating FOXO3a, NF-κB and p53 by targeting IKKs: an effective multi-faceted targeting of the tumor-cell phenotype? Cancer Biol Ther 3: 614–616 [DOI] [PubMed] [Google Scholar]
- Ghosh S, May MJ, Kopp EB (1998) NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol 16: 225–260 [DOI] [PubMed] [Google Scholar]
- Glaser KB, Staver MJ, Waring JF, Stender J, Ulrich RG, Davidsen SK (2003) Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther 2: 151–163 [PubMed] [Google Scholar]
- Lagger G et al. (2002) Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J 21: 2672–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Verma IM (2002) NF-κB regulation in the immune system. Nat Rev Immunol 2: 725–734 [DOI] [PubMed] [Google Scholar]
- Richon VM, Zhou X, Secrist JP, Cordon-Cardo C, Kelly WK, Drobnjak M, Marks PA (2004) Histone deacetylase inhibitors: assays to assess effectiveness in vitro and in vivo. Methods Enzymol 376: 199–205 [DOI] [PubMed] [Google Scholar]
- Schreiber E, Matthias P, Muller MM, Schaffner W (1989) Rapid detection of octamer binding proteins with ‘mini-extracts', prepared from a small number of cells. Nucleic Acids Res 17: 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta N, Seto E (2004) Regulation of histone deacetylase activities. J Cell Biochem 93: 57–67 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


