Abstract
A crucial step in the RNA interference (RNAi) pathway involves the assembly of RISC, the RNA-induced silencing complex. RISC initially recognizes a double-stranded short interfering RNA (siRNA), but only one strand is finally retained in the functional ribonucleoprotein complex. The non-incorporated strand, or ‘passenger' strand, is removed during the assembly process and most probably degraded thereafter. In this report, we show that the passenger strand is cleaved during the course of RISC assembly following the same rules established for the siRNA-guided cleavage of a target RNA. Chemical modifications impairing the cleavage of the passenger strand also impair the cleavage of a target RNA in vitro as well as the silencing of a reporter gene in vivo, suggesting that passenger strand removal is facilitated by its cleavage during RISC assembly. Interestingly, target RNA cleavage can be rescued if an otherwise non-cleavable passenger strand shows a nick at the scissile phosphodiester bond, which further indicates that the cleavage event per se is not essential.
Keywords: passenger strand, RNAi, Argonaute, RISC
Introduction
RNA interference (RNAi) is a post-transcriptional gene-silencing phenomenon that occurs in many eukaryotic organisms following stimulation by double-stranded RNA (dsRNA; Fire et al, 1998). In general, the RNAi response (reviewed by Filipowicz, 2005) can be divided into two distinct steps. The first step, called the assembly phase, comprises the recognition of a dsRNA molecule and its processing into ∼21-nucleotide (nt) RNA molecules, termed short interfering RNAs (siRNAs), by the RNase III-like enzyme Dicer (Hammond et al, 2000; Zamore et al, 2000; Bernstein et al, 2001). siRNAs are then shuttled into the RNA-induced silencing complex (RISC), a ribonucleoprotein complex composed of an Argonaute protein (Elbashir et al, 2001a; Lee et al, 2004; Pham et al, 2004; Tomari et al, 2004a, 2004b) and a single-stranded guide RNA. The selection of the RNA strand to be incorporated is governed by the thermodynamic profile of the siRNA duplex termini (Khvorova et al, 2003; Schwarz et al, 2003). In the second phase, also known as the effector phase, RISC uses this single-stranded RNA molecule as a guide to endonucleolytically cleave complementary RNAs (Hammond et al, 2000; Nykänen et al, 2001; Elbashir et al, 2001a; Martinez et al, 2002). Although RISC-mediated target RNA cleavage is very well studied (Haley & Zamore, 2004; Liu et al, 2004; Martinez & Tuschl, 2004; Meister et al, 2004; Schwarz et al, 2004; Song et al, 2004), the final steps of the assembly process of RISC are still a matter of debate, especially how the ‘guide' strand is separated from the passenger strand.
Whereas in mammalian cells only few studies exist on the assembly of RISC (Pham & Sontheimer, 2005), this process has been extensively studied in Drosophila melanogaster (Okamura et al, 2004; Tomari et al, 2004a, 2004b), in which the unwinding of the siRNA has been shown to depend on the presence of Ago2 (Okamura et al, 2004; Tomari et al, 2004b). Such studies have so far focused on the siRNA strand that is incorporated into RISC, referred to as the ‘guide strand'. In contrast, the fate of the non-incorporated strand, or ‘passenger strand', has mostly been neglected. In this report, we investigate the role and fate of the passenger strand during RISC assembly in HeLa cells both in vitro and in vivo, and focus on its implications on silencing. We show that for its efficient removal, the passenger strand has to be cleavable at its ‘natural' site, that is, at 10 nt from the 5′-phosphate of the guide strand. If this cleavage step is blocked, the siRNA duplex still becomes loaded into RISC, but target RNA cleavage is severely impaired owing to the non-efficient removal of the passenger strand.
Results and Discussion
Modified passenger strands impair target RNA cleavage
It was previously shown that affinity-purified human RISC is able to cleave synthetic, short, non-capped RNAs (Martinez & Tuschl, 2004). Interestingly, a short target RNA containing a 2′-O-methyl ribose at guanosine 9 (G9, the nucleotide immediately upstream of the cleavage site) was poorly cleaved, probably owing to steric hindrance caused by the bulky methyl group. A deoxyribose at G9 and 2′-O-methyl ribose groups 1 and 2 nt downstream of the cleavage site were, however, well tolerated.
We reasoned that RISC, during its assembly on the guide strand, might regard a passenger strand as its first RNA target, in a similar manner as affinity-purified RISC recognizes and cleaves a short target RNA. In this model, a chemically modified short RNA that cannot be cleaved by affinity-purified RISC should also not be cleaved when present as a passenger strand in an siRNA. We used HeLa cytoplasmic extracts and monitored the effect of these and other modifications on target RNA cleavage when present on the passenger strand of an siRNA duplex.
We observed a greater than sixfold reduction in the cleavage of a complementary target RNA when the passenger strand contained a phosphorothioate bond between G9 and A10, a modification that was shown to impair cleavage of a substrate RNA (Schwarz et al, 2004), or featured a 2′-O-methyl ribose at G9 (Fig 1A, and Fig 1B, lanes 2,3). However, 2′-O-methyl ribose groups positioned 1 or 2 nt downstream of the cleavage site (A10 and A11) did not impair target RNA cleavage, as was the case for an unmodified siRNA (Fig 1A, and Fig 1B, lanes 1,4,5). A similar, ∼6-fold reduction in target RNA cleavage was obtained with a passenger strand, in which the adenine residue downstream of the predicted cleavage site (A10) was substituted by a 4-thio-uridine (4S-U; Fig 1C, and Fig 1D, lane 2). This modification, when present in a short target RNA, abolished cleavage by affinity-purified RISC, most probably owing to a geometrical distortion in base pairing (supplementary Fig S1 online). 4S-U residues at non-central positions did not severely interfere with the cleavage of the target RNA (Fig 1C, and Fig 1D, lanes 1,3,4). Finally, we tested the effect of passenger strands featuring mismatches at different positions on the cleavage of a guide-complementary target RNA. A central 4-nt mismatch reduced the cleavage efficiency by a factor of 3 when compared with a full complementary siRNA (Fig 1E, and Fig 1F, lane 2), whereas non-central mismatches did not significantly affect the efficiency of cleavage (Fig 1E, and Fig 1F, lanes 1,3,4). These results indicate that modifications at the putative cleavage site on the passenger strand severely impair the assembly of functional RISC. Interestingly, 2′-O-methyl modifications at central positions on the guide strand had no effect on target RNA cleavage (data not shown), underlining that the impairment of target RNA cleavage is due to the passenger strand being rendered non-cleavable.
Figure 1.
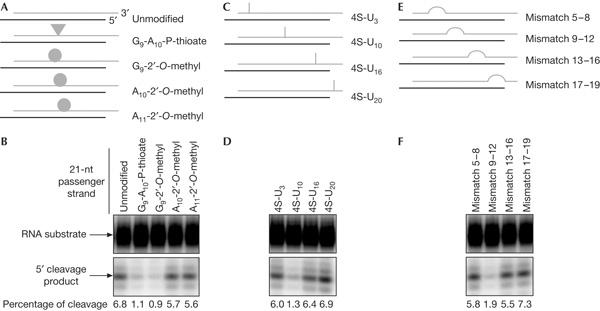
Chemical modifications on the passenger strand impair cleavage of a target RNA. (A) Graphical representation of short interfering RNAs (siRNAs) containing unmodified or chemically modified passenger strands. Passenger strands are shown in grey and guide strands in black in this report. 2′-O-methyl modifications are depicted as circles and phosphorothioates as triangles. (B) Phosphorimaging of cleavage reactions using siRNAs depicted in (A) resolved on a 6% denaturing polyacrylamide gel. Arrows point to the RNA substrate and the labelled 5′-cleavage product. The percentage of cleaved target RNA is indicated at the bottom of the gel. (C) Graphical representation of siRNAs containing 4S-U substitutions (indicated as bars) at different positions on the passenger strand. (D) Phosphorimaging of cleavage reactions using siRNAs depicted in (C). (E) Graphical representation of siRNAs containing 4-nt mismatches at different positions on the passenger strand. (F) Phosphorimaging of cleavage reactions using siRNAs depicted in (E).
The passenger strand is cleaved during RISC assembly
As (i) RISC properly recognizes siRNAs featuring non-cleavable passenger strands (supplementary Fig S2A,B online), and (ii) Ago2 interacts with both strands of the duplex (supplementary Fig S2C,D online), we reasoned that the inhibition of target RNA cleavage might be explained by RISC being unable to effectively remove a passenger strand that cannot be cleaved, rather than failing to assemble on a modified siRNA. In principle, it could also be the case that a non-cleavable passenger strand acted as a suicide target on re-binding functional RISC. We can, however, rule out this possibility, as a non-cleavable passenger strand is not released intact from the original siRNA (supplementary Fig S3 online). To provide conclusive evidence for the cleavage of the passenger strand during RISC assembly, we set out to detect the predicted 9-nt cleavage product. We performed a time-course analysis using HeLa cytoplasmic extracts and 5′-phospho-radiolabelled siRNAs, the passenger strands of which were left unmodified, or were modified either with a 2′-O-methyl ribose at G9 or with a phosphorothioate bond between G9 and A10 (Fig 2A). The unmodified passenger strand yielded the expected 9-nt cleavage product after only 5 min of incubation, reached a maximum at 15 min and decreased thereafter (Fig 2D, left panel). In sharp contrast, the modified passenger strands were not cleaved (Fig 2D, left panel). Similarly, a passenger strand featuring a central 4-nt mismatch (Fig 2B) failed to yield a 9-nt cleavage product (Fig 2D, central panel). However, passenger strand cleavage could be rescued in the latter case by restoring the complementarity on the guide strand (Fig 2D, central panel). It is worth noting that the profiles of the 9-nt cleavage product differ from the concomitant 3′–5′ exonucleolytic degradation of the passenger strand, which leads to the steady accumulation of <21 nt species during the course of the reaction (supplementary Fig S4 online). Moreover, this unspecific degradation is present in all cases, irrespective of whether the passenger strand is being cleaved or not.
Figure 2.
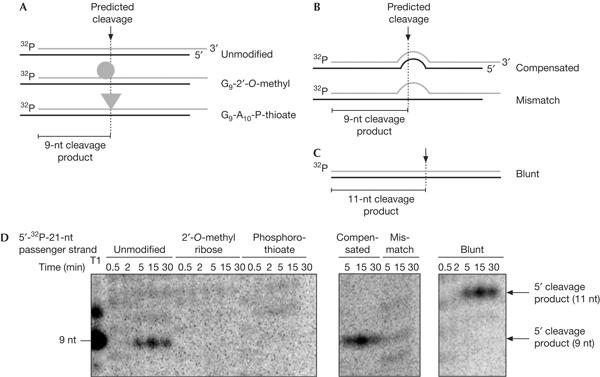
The passenger strand is cleaved during RNA-induced silencing complex assembly. (A) Graphical representation of short interfering RNAs (siRNAs) composed of unmodified guide strands and unmodified or chemically modified passenger strands. In all cases, the passenger strands were 5′ phospho-radiolabelled (indicated by ‘32P'). The dotted line depicts the position where cleavage is predicted to take place on the passenger strand, that is, between guanosine-9 (G9) and adenosine-10 (A10). The expected 9-nt cleavage product is indicated. (B) Graphical representation of an siRNA formed by a 5′-phospho-radiolabelled, unmodified passenger strand (for the sequence, see ‘mismatch 9–12' in the legend of Fig 1E) and guide strands, the sequence of which either compensates for the 4-nt mutation in the passenger strand or leads to a central 4-nt mismatch. The dotted line depicts the predicted cleavage position. (C) Graphical representation of a blunt siRNA formed by an unmodified guide strand and a 5′-phospho-radiolabelled, unmodified passenger strand, in which the sequence has been shifted 2 nt towards the 5′ end. The dotted line depicts the predicted cleavage position, which has also shifted by 2 nt. The expected 11-nt cleavage product is indicated. (D) Phosphorimaging analysis of a time-course cleavage reaction resolved in a 15% denaturing gel electrophoresis. The region of the gel corresponding to sizes between 8 and 12 nt has been enhanced for an optimal visualization of the cleavage products. The picture of the whole gel is depicted in supplementary Fig S4 online.
Interestingly, the cleavage of the passenger strand correlates temporally with the interaction of Ago2 and the siRNA (compare Fig 2D with supplementary Fig S2D online). As soon as Ago2 contacts the passenger strand of the siRNA, the 9-nt cleavage product emerges, supporting our hypothesis that the passenger strand is cleaved in the course of RISC assembly.
It is well established that the cleavage position on a target RNA is located 10 nt from the 5′ end of the guide strand (Elbashir et al, 2001b). To test whether this rule also applies to the cleavage of the passenger strand, we extended the 5′ end of the guide strand by 2 nt, resulting in a blunt-ended siRNA (Fig 2C). This duplex cleaved a complementary target RNA with a 2-nt shift (data not shown). The cleavage site on the passenger strand was also shifted by 2 nt, now generating an 11-nt cleavage product (Fig 2D, right panel). This argues that RISC, during assembly, cleaves the passenger strand by a mechanism similar to that by which functional RISC cleaves a complementary target RNA.
Bypassing the passenger strand cleavage event
Why does the cleavage of the passenger strand facilitate the generation of functional RISC? A nick in the passenger strand is expected to result in a drastic change in the thermodynamic profile of the siRNA duplex, which could sustain the removal of the passenger strand. We therefore generated an siRNA by annealing two passenger strands of 9 and 12 nt length to a 21-nt guide strand, a so-called ‘9+12', nicked siRNA (Fig 3A). This siRNA faithfully interacted with Ago2 (data not shown) and guided target RNA cleavage as efficiently as an siRNA containing a conventional 21-nt passenger strand did (Fig 3B, lanes 2,10). This result indicates that the Ago2-mediated cleavage event per se appears to be not essential for the generation of functional RISC, as it can be bypassed by a pre-existing nick. It thus argues against a model in which the cleavage event imposes a conformational change that would facilitate the removal of the passenger strand.
Figure 3.
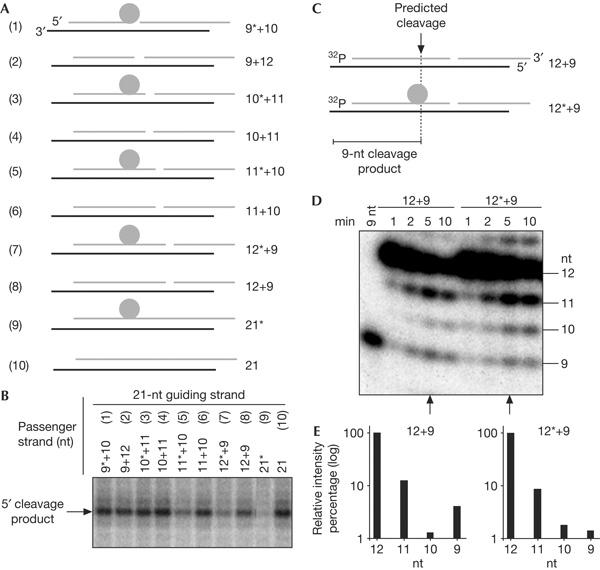
Functional RNA-induced silencing complex assembles on a short interfering RNA in which an otherwise non-cleavable passenger strand is nicked at the putative cleavage site. (A) Graphical representation of short interfering RNAs (siRNAs) formed by one (21 nt) or two passenger strands (x+y) of different lengths leading to a nicked passenger strand of 21 nt. Numbers on the right refer to the length of the strands in nucleotides. The asterisk indicates a 2′-O-methyl ribose at G9. (B) Phosphorimaging analysis of a cleavage reaction resolved in a 6% denaturing gel electrophoresis showing the efficiency of target RNA cleavage as a function of the position of a nick on the passenger strand and the presence or absence of a 2′-O-methyl ribose at the cleavage position. (C) Graphical representation of siRNAs formed by two passenger strands (12 nt+9 nt), in which the 12-nt fragment has been 5′ phospho-radiolabelled (indicated by ‘32P'), and either left unmodified (duplex 12+9) or modified with a 2′-O-methyl ribose at position G9 (duplex 12*+9). Numbers on the right refer to the length of the strands in nucleotides. The dotted line depicts the position where cleavage is predicted to take place on the passenger strand, that is, between G9 and A10. The expected 9-nt cleavage product is indicated. (D) Phosphorimaging analysis of a time-course cleavage reaction resolved in a 15% denaturing gel electrophoresis. A phosphorylated, 9-nt RNA oligonucleotide was used as a marker. Note the different pattern between the cleavable and non-cleavable passenger strand. Whereas unspecific 3′-to-5′ exonucleolytic degradation of the non-cleavable passenger strand leads to the steady accumulation of <12 nt species during the course of the reaction, the cleavable passenger strand, which is subjected to a similar degradation, shows a clear enrichment of the 9-nt species. Arrows indicate the time point at which the quantification in (E) was performed. (E) Quantification of the phosphorimaging analysis in (D). Bars represent the relative intensity of the bands, the sizes of which are indicated along the x axis. The 9-nt cleavage product accumulates only when using the unmodified, cleavable 12-nt fragment of the passenger strand.
We also generated a duplex ‘9*+12', in which a 2′-O-methyl group was placed at the G9 position (the cleavage site) of the 9-nt passenger strand (Fig 3A). This duplex efficiently cleaved the target RNA, demonstrating that a 2′-O-methyl group at the cleavage site has no effect on the removal of the cleaved passenger strand (Fig 3B, lane 1). More importantly, the complete rescue of target RNA cleavage by providing a nick on the cleavage site of an otherwise non-cleavable passenger strand highlights the importance of passenger strand cleavage during RISC assembly.
Interestingly, we found that displacing the nick stepwise further downstream along the passenger strand and, additionally, maintaining its non-cleavable character (Fig 3A, 10*+11, 11*+10, 12*+9) led to a reduction in target RNA cleavage, as the distance between the nick and the 2′-O-methyl group increased (Fig 3B, lanes 3,5,7). In contrast, duplexes with unmodified passenger strands (Fig 3A, 10+11, 11+10, 12+9) guided efficient cleavage of the target RNA (Fig 3B, lanes 4,6,8), underlining the relevance of passenger strand cleavage at the ‘natural' position, that is, the phosphodiester bond between nt 9 and 10 counting from its 5′ end. An intact, 21-nt non-cleavable passenger strand showed the most pronounced impairment of target RNA cleavage compared with a 21-nt cleavable passenger strand (Fig 3B, lanes 9, 10, respectively). We proposed that the inhibition of target RNA cleavage observed for the 12*+9 duplex should correlate with the inability to cleave the 12-nt, modified passenger strand into a 9-nt cleavage product. Indeed, specific accumulation of the 9-nt cleavage product was observed only when using the unmodified 12+9 duplex and not the modified 12*+9 duplex (Fig 3C–E).
These findings raise a couple of interesting questions. As a central 4-nt mismatch on the siRNA also impaired the removal of the passenger strand, an explanation has to be found for how microRNAs (miRNAs) with several internal bulges, even at the putative cleavage site, are loaded into microRISC (miRISC). Furthermore, Ago1, Ago3 and Ago4 have been shown to be associated with mature miRNAs (Liu et al, 2004; Meister et al, 2004). As in mammals only Ago2 is known to harbour endonucleolytic activity, how do the catalytically inactive Argonaute proteins remove the passenger strand? Clearly, a bypass pathway should exist, which allows the assembly of miRISC in the absence of slicer activity. During miRNA maturation, an auxiliary factor might assist in the loading of miRISC independently of the presence of a catalytically active Ago protein. A different possibility might rely on the reduced thermodynamic stability of miRNA duplexes, as, in contrast to siRNAs, miRNA duplexes feature bulges of non-paired bases at different positions. Interestingly, cleavage assays carried out at 37°C rescued target RNA cleavage guided by an siRNA containing a central, 4-nt mismatch on the passenger strand (Fig 4A,B). In contrast, siRNAs featuring fully complementary passenger strands and showing 2′-O-methyl or phosphorothioate modifications at the cleavage site showed a minor rescue (Fig 4A,B).
Figure 4.
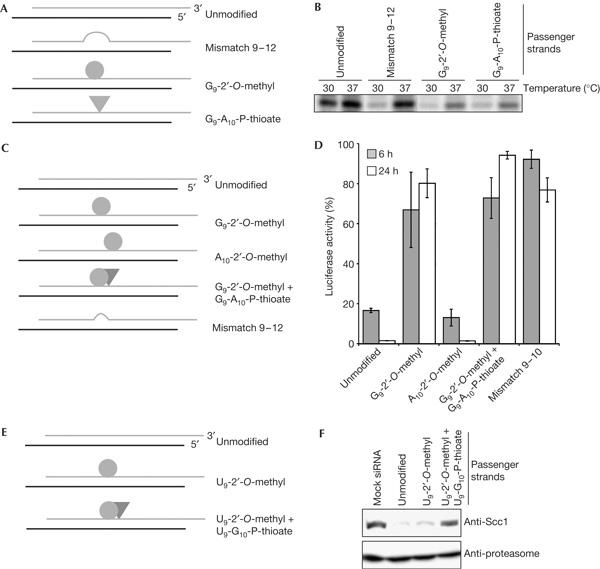
In vitro and in vivo analysis of a putative bypass mechanism for RNA-induced silencing complex assembly. (A) Graphical representation of short interfering RNA (siRNAs) containing unmodified or chemically modified passenger strands. (B) Phosphorimaging analysis of a cleavage reaction resolved in a 6% denaturing gel electrophoresis. (C) Same as (A). (D) Luciferase activities measured after 6 or 24 h after transfecting HeLa cells with siRNAs depicted in (C). (E) Same as (A). (F) Western blot analysis of a knockdown experiment in HeLa cells 24 h after transfecting HeLa cells with siRNAs depicted in (E).
Finally, we tested the effect of siRNAs containing modified passenger strands in vivo. We co-transfected HeLa cells with a plasmid containing the firefly luciferase gene together with siRNAs harbouring differently modified passenger strands (Fig 4C) and analysed luciferase activities normalized to cells transfected with a mock siRNA. An unmodified siRNA effectively silenced the reporter gene, whereas we observed a major impairment in silencing with a passenger strand containing a 2′-O-methyl ribose at position 9 (Fig 4D), certifying that cleavage of the passenger strand also has a role in vivo. The same modification at position 10, however, did not affect the knockdown efficiency. The most severe impairment in silencing was caused by a passenger strand containing both a 2′-O-methyl ribose at position 9 and a phosphorothioate bond between nt 9 and 10. A passenger strand with a central mismatch at positions 9 and 10 was also very inefficient in silencing after 6 h. However, in contrast to all other modifications that impaired the knockdown, silencing improved slightly after 24 h, probably owing to a putative bypass mechanism.
We also tested silencing of an endogenous gene, human cohesin (Scc1), using both unmodified and modified duplexes (Fig 4E). A passenger strand featuring a 2′-O-methyl ribose at position 9 led to a less efficient silencing than an unmodified passenger strand (Fig 4F). However, after enhancing the non-cleavable character by adding a phosphorothioate bond between nt 9 and 10 of the passenger strand, a severe impairment was detected (Fig 4F).
In conclusion, we show that the cleavage of the passenger strand is indeed a prerequisite for effective assembly of functional human RISC. Our results may also provide valuable clues for the findings in D. melanogaster, in which RISC assembly has been shown to depend on the presence of Ago2 (Okamura et al, 2004; Tomari et al, 2004a, 2004b). The results from HeLa cytoplasmic extracts favour a model in which efficient and fast removal of the passenger strand is dependent on the catalytic activity of Ago2. We also propose that removal of the cleaved passenger strand during RISC assembly follows similar rules as the removal of the cognate target RNA after cleavage: RISC, during its assembly, has to be liberated from the passenger strand that would otherwise block the recognition of a target RNA. In turn, functional RISC, as every multiple turnover enzyme (Hutvágner & Zamore, 2002; Haley & Zamore, 2004; Martinez & Tuschl, 2004), has to guarantee product release only after catalysis.
Note: During the submission process, Matranga et al, Rand et al (Cell, Immediate Early Publication, 3 November 2005) and Miyoshi et al (Genes and Development, December 2005) reported a similar function of the passenger strand in Drosophila. Kraynack & Baker (RNA, Epub ahead of print, 21 November 2005) report that for some siRNA sequences, passenger strands fully modified with 2′-O-methyl residues do not impair silencing.
Methods
Short interfering RNA duplexes. The sequence of the passenger strand was 5′-CGUACGCGGAAUACUUCGAAA-3′. The sequence of the guide strand was 5′-UCGAAGUAUUCCGCGUACGUG-3′. All duplexes contained symmetric 2-nt overhangs at the 3′ end. Duplexes were annealed to a final concentration of 10 μM in 100 mM KOAc, 2 mM Mg(OAc)2 and 30 mM Hepes (pH 7.4), and dilutions were made thereof. Oligoribonucleotides were obtained from Dharmacon Research Inc. (Lafayette, CO, USA) and PROLIGO Primers and Probes (Paris, France) and were generously provided by Professor Tom Tuschl (The Rockefeller University).
In the case of 4S-U-modified passenger strands in Fig 1C, the base at the positions indicated was exchanged by a 4S-U residue.
The sequences of the passenger strands in Fig 1E were 5′-CGUAGCGCGAAUACUUCGAAA-3′ for ‘mismatch 5–8', 5′-CGUACGCGAGUAACUUCGAAA-3′ for ‘mismatch 9–12', 5′-CGUACGCGGAAUAUUCAGAAA-3′ for ‘mismatch 13–16' and 5′-CGUACGCGGAAUACUUAGCAA-3′ for ‘mismatch 17–19'.
The sequence of the passenger strand in Fig 2C was 5′-CGUACGCGAGUAACUUCGAAA-3′. To restore complementarity, the sequence of the guide strand was changed accordingly.
To generate the blunt siRNA duplex in Fig 2C, the sequence of the guide strand was changed to 5′-UUUCGAAGUAUUCCGCGUACG-3′.
The sequences of the duplexes used in the knockdown experiment in Fig 4E,F were taken from Hirota et al (2004).
The sequence of the passenger strands used in the luciferase assay (Fig 4C,D) was 5′-CUUACGCUGAGUACUUCGAAA-3′. For the duplex ‘mismatch 9–10', the sequence was 5′-CUUACGCUACGUACUUCGAAA-3′. The sequence of the guide strand was 5′-UCGAAGUACUCAGCGUAAGUG-3′.
When indicated in the figure legends, 5′ phospho-radiolabelling of the respective strands was performed as described previously (Martinez & Tuschl, 2004).
Cleavage reactions. Cleavage reactions were performed as described previously (Martinez et al, 2002), with the exceptions that MgCl2 was used at 5 mM and that the final concentration of siRNAs was adjusted to 10 nM. HeLa cytoplasmic extracts were provided by Professor Reinhard Luhrmann (Department of Cellular Biochemistry, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany) and by Paragon Bioservices Inc. (Baltimore, MD, USA). The substrate RNA is 32P-cap-labelled (Elbashir et al, 2001b). Radiolabelled markers were generated by partial RNase T1 digestion of the 32P-cap-labelled RNA substrate.
Passenger strand cleavage experiments were performed as cleavage reactions, the difference being the absence of 32P-cap-labelled substrate RNA.
Cell culture and transfections. Cell culture and transfections were performed as described previously (Hirota et al, 2004). HeLa cells were seeded at a density of 4 × 104 cells per 12-well plate, 1 day before transfection. siRNA duplexes were used at a final concentration of 200 nM. At 24 h after transfection, the cells were washed with cold PBS and lysed directly in 1 × SDS–polyacrylamide gel electrophoresis sample buffer. For luciferase assays, HeLa cells were cultured in 24-well plates and transfected with 100 nM siRNAs, 0.2 μg of pGL3 promoter (Pp-luc) and 0.01 μg of phRL-TK control vector (Rr-luc; Promega, Madison, WI, USA) for internal standardization. We used sextuplicates for each condition. Cells were collected and extracts were assayed 6 and 24 h after transfection.
Western blotting. Samples were sonicated and boiled before being loaded on an 8% SDS–polyacrylamide gel electrophoresis. Gels were blotted onto polyvinylidene difluoride membranes using the semi-dry method. Membranes were blocked in 3% non-fat dry milk in Tris-buffered saline+0.05% Tween. Antibodies against human cohesin (Scc1) and the proteasome were used at a concentration of 2 μg/ml (Sumara et al, 2002). Secondary antibodies were from JacksonImmunoResearch (Sigma Aldrich, West Grove, PA, USA).
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/7400637-s1.pdf).
Supplementary Material
Supplementary Information
Acknowledgments
We thank A. Kuras, G. Obernosterer, D. Pezic and S. Weitzer, members of the laboratory, for encouragement and suggestions during the completion of this work, Professor R. Schroeder and C. Ribeiro for critically reading the manuscript, H. Manninga for the synthesis of 4-thio-uridine-modified siRNAs and for comments on the manuscript and Y. Dorsett for discussions. J.M. is a Junior Group Leader at IMBA. P.J.F.L. is funded by the Boehringer Ingelheim Fonds PhD Scholarship and S.L.A. is funded by the Vienna Biocenter PhD program and partly by Fonds zur Förderung der Wissenschaftlichen Forschung through WK001. IMBA is the Institute of Molecular Biotechnology supported by the Austrian Academy of Sciences.
References
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T (2001a) RNA interference is mediated by 21 and 22 nt RNAs. Genes Dev 15: 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T (2001b) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J 20: 6877–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W (2005) RNAi: the nuts and bolts of the RISC machine. Cell 122: 17–20 [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Haley B, Zamore PD (2004) Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol 11: 599–606 [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296 [DOI] [PubMed] [Google Scholar]
- Hirota T, Gerlich D, Koch B, Ellenberg J, Peters JM (2004) Distinct functions of condensin I and II in mitotic chromosome assembly. J Cell Sci 117: 6435–6445 [DOI] [PubMed] [Google Scholar]
- Hutvágner G, Zamore PD (2002) A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060 [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW (2004) Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81 [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Martinez J, Tuschl T (2004) RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev 18: 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Lührmann R, Tuschl T (2002) Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T (2004) Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Nykänen A, Haley B, Zamore PD (2001) ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107: 309–321 [DOI] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC (2004) Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev 18: 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham JW, Sontheimer EJ (2005) Molecular requirements for RNA-induced silencing complex assembly in the Drosophila RNA interference pathway. J Biol Chem 280: 39278–39283 [DOI] [PubMed] [Google Scholar]
- Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ (2004) A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 117: 83–94 [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Tomari Y, Zamore PD (2004) The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr Biol 14: 787–791 [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L (2004) Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305: 1434–1437 [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redemann N, Nigg EA, Peters JM (2002) The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol Cell 9: 515–525 [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD (2004a) RISC assembly defects in the Drosophila RNAi mutant armitage. Cell 116: 831–841 [DOI] [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD (2004b) A protein sensor for siRNA asymmetry. Science 306: 1377–1380 [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


