Summary
Workshop on Centrosomes and Spindle Pole Bodies
Keywords: basal body, centrosome, microtubule organizing centres, mitosis, spindle pole body
Introduction
This series of meetings (Palazzo, 2002; Stearns & Winey, 1997) began as a joint ASCB/EMBO venture in 1997 with a meeting that was a landmark event, not only because a minor field had grown to fill an entire meeting, but also because key presentations defined the course of the field for years to come. The first biochemical mass spectrometric analysis of the budding yeast spindle pole body (SPB) by John Kilmartin and Matthias Mann marked the dawn of a new era in centrosome and SPB studies (Wigge et al, 1998). Similarly, Jeff Salisbury's clear demonstration of centrosome amplification in tumour samples, coupled with the work of Bill Brinkley, Stephen Doxsey and others, was part of a resurrection of Theodor Boveri's turn-of-the-century hypothesis that centrosome amplification could lead to genetic instability and cancer (Brinkley, 2001). However, all was not harmonious, as elegant work describing spindle assembly in centrosome-free Xenopus extracts (Heald et al, 1996), coupled with studies showing Drosophila development in the absence of functional centrosomes, sparked a key debate at the 1997 meeting that persists to this day. The root of this controversy lies in the ability of spindles to form and apparently function without centrosomes. Mass spectrometry continued to set the pace at the second meeting in 2002, as Erich Nigg and Mann's groups defined a set of centrosome components that have become the staple diet for the community ever since (Andersen et al, 2003). However, more contention arose at the second meeting as a result of the stunning laser ablation studies by Alexy Khodjakov that challenged another of Boveri's hypotheses—that centrioles can only be made in the presence of an existing centriole template. After a three-year wait, eager ‘pole-heads' returned to Heidelberg in September 2005, to be swept into another deluge of information and insights provided by a third fast-paced and exciting meeting.

The third EMBO workshop on Centrosomes and Spindle Pole Bodies, organized by M. Bornens, S.J. Doxsey, M. Knop and J. Raff, was held at the European Molecular Biology Laboratory in Heidelberg, Germany, between 23 and 27 September 2005.
Controlling centriole duplication
It is well known that centrosomes, basal bodies and SPBs must nucleate and organize microtubule arrays in a desired architecture to fulfil the required function of the cell type in question. Importantly, the replication and separation of centrioles during cell division must be coordinated with the needs of the individual cell. For most cells, this means that a single structure, the centriole pair, is duplicated once per cell cycle to generate two structures that can sit at either end of a bipolar mitotic spindle. As we know remarkably little about centrosome duplication beyond the fact that it requires cyclin-dependent kinase 2 (Cdk2)/cyclin A/E activity, there was great excitement at the presentations on the polo-like kinase 4 (Plk4)/Sak by E. Nigg (Martinsried, Germany) and M. Bettencourt-Dias (D. Glover's laboratory, Cambridge, UK). Their depletion of this key centrosomal kinase from human (Plk4) and Drosophila (Sak) cells reduced the number of centrioles at spindle poles (Fig 1A,B). Interestingly, overproduction of Plk4/Sak pushed centriole numbers over the normal limit of four (Fig 1D,E), solidifying the central role of this kinase in the control of centriole duplication. A second manifestation of altered centriole number from the fly Sak mutants (Fig 1B,F) was a reduction in the number of sperm axonemes, consistent with the hypothesis that an inability to assemble centrioles (basal bodies) should result in a failure to assemble flagella (Fig 1G). This key role for Plk4/Sak generates two obvious questions: how is the kinase instructed when to drive duplication and what molecular mechanism promotes duplication? The realization that polo kinases are sometimes targeted to substrates that have been previously phosphorylated by Cdks (Elia et al, 2003) could address the first question. One attractive idea is that Cdk2/cyclin A/E phosphorylation of key centrosome substrates generates Plk4/Sak landing pads for the transient recruitment of this second kinase to promote duplication. J. Maller (Denver, CO, USA) has identified a Cdk2/cyclin E centrosome targeting motif that could enable this hypothesis to be tested. His work has defined a centrosome localization sequence (CLS) in cyclin E that competes with cyclin A and removes the latter from the centrosome. Interestingly, the cyclin E CLS could still bind to centrosomes in cells derived from Cdk2-knockout mice. K. O'Connell's group (Bethesda, MD, USA) might have insights into the second question of how Plk4/Sak works through their genetic analysis of Zyg1 (perhaps the closest relative to Plk4 in Caenorhabditis elegans). They isolated several mutations that both compensate for deficiencies in Zyg1 and confer a phenotype in wild-type backgrounds. Excitingly, one such Szy (suppressor of Zyg1) mutation leads to the generation of a large number of microtubule cylinders that are resistant to nocodazole throughout the cytoplasm when crossed away from the starting Zyg1 mutation. The uncanny similarities between these aggregates and centrioles led O'Connell to speculate that the product of this Szy5 gene might inhibit centriole formation until the appropriate time when the process of centriole assembly is allowed to proceed—the logic being that mutation compromises Szy5 activity to a sufficient degree that a crippled kinase can overcome the crippled inhibitor.
Figure 1.
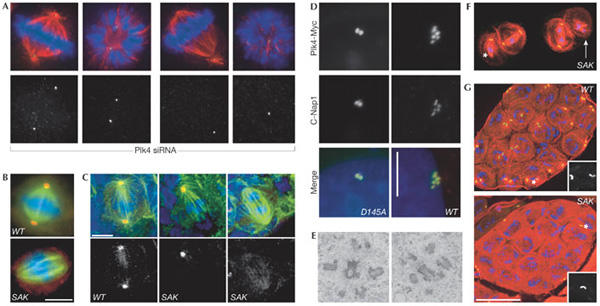
Plk4/Sak regulates centrosome duplication. (A) Small interfering RNA targeting of human Plk4 induces progressive aberrations in spindle morphology and a stepwise reduction in centriole numbers (green, centrin; red, α-tubulin; blue, DNA). (B,C) Drosophila Sak is required for centrosome duplication in S2 cells (B) and larval brain cells from Sak flies (C) (green, α-tubulin; blue, DNA; red, γ-tubulin). Scale bars, 5 μm. (D) Staining for the centriole marker C-Nap1 (red) and the transfected Plk4–myc fusion protein (green) reveals an increase in centriole number following overexpression of wild-type (WT), but not catalytically inactive (D154A) Plk4–Myc in human U2OS cells. Scale bar, 10 μm. (E) A transmission electron micrograph of Plk4–Myc-expressing U2OS cells 48 h after induction of transcription shows the centriole content of the cells. (F) Primary Sak spermatocytes in meiosis I stained to reveal microtubules (red), GFP-PACT (a centriolar marker) and DNA (blue). The asterisk marks an unfocused pole that lacks a centriole whereas a highly disorganized spindle that completely lacks centrioles is indicated by the white arrow. (G) Wild-type and SAK primary spermatocyte cysts stained as in (F). There are 16 cells in each cyst but only the wild-type cyst has a centriole pair in each cell. Scale bar, 50 μm. Panels (B,C,F,G) are reprinted from Bettencourt-Dias et al (2005) with permission from Elsevier. Panels (A,D,E) reprinted from Habedanck et al (2005) with permission from Nature Publishing Group.
Centriole assembly
The dogma in the field dictates that after a cue for duplication has been given, a new centriole appears perpendicular to an existing centriole (Brinkley, 2001). An impressive collaboration between The Max Planck Institute in Dresden (Germany) and Leica Microsystems Inc. (Wetzlar, Germany), has led to the design and assembly of the EMPACT2+RTS machine for high-pressure freezing of C. elegans embryos under microscopic observation. This technology has allowed T. Müller-Reichert, L. Pelletier and A. Hyman (Dresden, Germany) to follow embryos by video microscopy in real time from fertilization to their first division, and to fix the embryos by high-pressure freezing at precisely defined points in the centriole duplication cycle. After freezing, E. O'Toole (Boulder, CO, USA) examined serial plastic sections by electron microscope tomography, which resulted in a series of breathtaking ultrastructural insights into the assembly of the simple C. elegans centriole (Fig 2A). The accuracy of the timing of fixation enabled Müller-Reichert, Pelletier and Hyman to correlate the structural data with the recruitment and requirement of the distinct centrosome components—spindle assembly 4 (SAS4), SAS5 and SAS6, which the P. Gönczy (Lausanne, Switzerland), K. Oegema (La Jolla, CA, USA) and Hyman laboratories have shown to be crucial for centriole duplication. The elegance of the study left no doubt about the power of this approach to correlate ultrastructure with gene function, a correlation that could not have been imagined just a few years ago. Furthermore, Gönczy, Glover and J. Raff (Cambridge, UK) showed that SAS function is conserved in flies and humans, which leads to the question of how much information can be revealed about the duplication of the more complex human centrioles, using the C. elegans model? It will be important to determine whether the ultrastructural details defined during the assembly of the nine-singlet microtubule centriole in C. elegans can also be found in the process of duplication of the nine-triplet microtubule centriole found in animals, ciliates, and some algae (Fig 2). Simply put, the question of whether the barrel structure that precedes centriole microtubule assembly in C. elegans can be found in other organisms is a crucial challenge.
Figure 2.
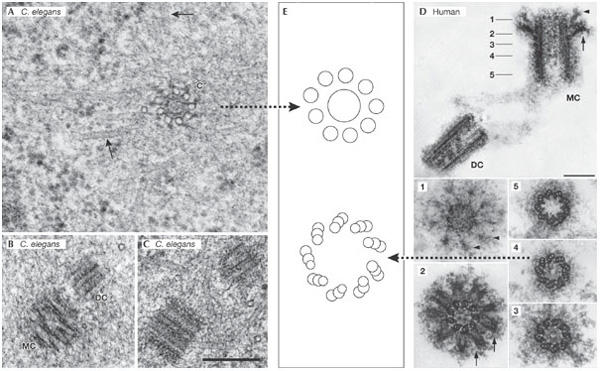
Human and worm centriolar structure: similar design, different complexity. (A–C) Centrioles of the Caenorhabditis elegans embryo. Scale bar, 250 nm. (A) Transmission electron micrograph of a prometaphase centriole (c) with singlet microtubules in cross section. Spindle microtubules are indicated by arrows. (B) Mother centriole (MC) and daughter centriole (DC) in prometaphase. (C) A centriole pair in interphase. (D) A human centrosome isolated from the human KE37 lymphoblastic cell line that was used for the proteome analysis by Andersen et al (2003). The five numbered panels below correspond to sections through the MC at the points indicated in the upper panel. The arrowheads and the arrows identify the distal and sub-distal appendages, respectively. Scale bars, 0.2 mm. (E) Cartoons depicting the microtubule architecture of the indicated centrioles. (A–C) were generously provided by T. Müller-Reichert (Dresden, Germany) and (D) by M. Bornens (Paris, France).
De novo centriole synthesis
Following the inspirational work from Wallace Marshall and Joel Rosenbaum in Chlamydomonas (Marshall et al, 2001), A. Khodjakov (Albany, NY, USA) continued his pursuit of the question he asked at the 2002 meeting: what happens when you use a laser to ablate the centrosome in mammalian cells? The answer that centrioles can form by de novo synthesis without the need for a pre-existing template has now been consolidated by his extensive analyses, and K. Sluder's (Worcester, MA, USA) microsurgery experiments also provided compelling support for de novo synthesis. In the Sluder experiments, half of the cells that had their centrosomes removed during the G2 phase of the cell cycle could be followed through at least two rounds of division and these cells generated centrioles de novo. Khodjakov and Sluder's studies showed that de novo assembly results in unusually high numbers of centrioles, indicating that these have the ability to generate far more centrioles at any given time than the carefully controlled one-to-two replication process. How do cells inhibit the assembly of many centrioles if that potential exists? Khodjakov proposed that the presence of a single centriole inhibits de novo centriole assembly, not unlike the insight that O'Connell proposed for Szy5. This is an important concept, but difficult to understand, as it seems that it is the actual architectural ensemble—the molecular organization—of the centriole that somehow provides this inhibitory assembly signal. Khodjakov speculated that the de novo pathway is activated in every cell cycle, however only those pro-centrioles that dock on mature centrioles are stabilized and ultimately develop into centrioles.
Centriole amplification after S-phase arrest
As much can be learned by studying abnormal versions of a particular event, centrosome proliferation in cells that have been arrested in S-phase is frequently used to gain insight into the events of centriole duplication that accompany normal cell-cycle progression. The original studies of Ron Balczon and Brinkley, showing that Chinese hamster ovary cells arrested in S-phase by treatment with hydroxyurea (Balczon et al, 1995), were extended by R. Kuriyama (Minneapolis, MN, USA), who found a considerable degree of cell-type specificity for the ability of cells to engage in overduplication in response to hydroxyurea. E. Bourke (C. Morrison's laboratory, Galway, Ireland) described the dependency of DNA damage-induced centrosome amplification on the conserved damage checkpoint kinase Chk1. By contrast, depletion of Chk2 led to centrosome amplification even in the absence of DNA damage. The connection between checkpoint control and centrosome amplification was extended by J. Parvin's (Boston, MA, USA) observations that repression of breast cancer 1 (Brca1) function by RNA interference or inhibitory peptides that compromised the ubiquitin ligase function of the Brca1 complex led to centrosome amplification. As this was only seen in breast-tissue-derived cell lines, Parvin suggested that the role of the Brca1 complex might be undertaken by other E3 ubiquitin ligases in other cell/tissue types. The involvement of key oncogenic proteins, such as Brca1, in centrosome amplification received considerable support from several posters at the meeting from more clinically orientated groups that are using centrosome amplification as a prognostic marker to monitor and grade tumorigenesis. Surprisingly, a search for potential targets for Brca1 led to the discovery of two lysine residues in γ-tubulin that seem to have specific effects on the duplication and activity of centrosomes. The relationship of γ-tubulin—once thought to serve only as a template in microtubule nucleation—to other complex processes such as centrosome duplication provides another set of challenges to the field.
Centriole composition
Although the mass spectrometric characterization of centrosome composition by the Nigg–Mann collaboration removed one of the last taboos of mitosis research by revealing some of the elusive components of the centriole, it took us from a state of virtual ignorance to one of information overload (Andersen et al, 2003). Several groups have started to see through this list to the logical pathway that builds a centriole. S. Bahe (Martinsried, Germany) and Nigg presented a thorough characterization of a protein called rootletin, which forms striking fibres emanating from the proximal ends of centrioles. Rootletin is distantly related to C-Nap1, a protein that A. Fry (Leicester, UK) and Nigg have previously shown to be important for centriole cohesion. Like C-Nap1, rootletin associates with centrosomes in interphase, is required for centriole cohesion and binds to the mitotic NIMA-related kinase 2 (Nek2).
In a tour de force to match the seminal study establishing that Spc110 acts as a spacer to link microtubules to the SPB, J. Kilmartin's (Cambridge, UK) keynote address presented an elegant analysis of another conserved centrosome component called Sfi1. Kilmartin originally identified Sfi1 by purifying proteins that associated with one of the long-time favourites of the spindle pole community, the highly conserved calmodulin-related molecule, centrin. Sfi1 has a series of consecutive centrin-binding sites with globular regions at either end of the molecule. Focusing on the yeast Sfi1, Kilmartin solved the crystal structure of a subset of Sfi1 repeats, and by using rotary shadowing electron microscopy to determine the length of the repeats, thereby suggested a model for the arrangement of Sfi1 in the bridge and in SPB duplication. M. Bornens (Paris, France) presented a detailed characterization of a molecule with an Sfi1 centrin-binding motif he called p65 that is required for the full extension of the pro-centrioles to form full-length centrioles. Several other presentations characterized new centriole and SPB components, including a report from C. Gonzalez (Barcelona, Spain) about the cloning of a centriolar protein in Drosophila, which has a main role in pericentriolar material recruitment. Fusion of this protein to the green fluorescent protein (GFP) revealed centrioles and basal bodies in all cell lineages at any stage of the cell cycle throughout development. This and other fusion proteins shown in the mass spectrometric analysis of Drosophila centrosomes by B. Lange (Berlin, Germany), will become important tools in the study of centrosomal function in differentiating cells in this powerful genetic system.
Recruitment of γ-tubulin to form nucleating sites
Because it nucleates the tubulin polymers that will become microtubules, γ-tubulin lies at the core of microtubule organizing centre (MTOC) function. It is known that γ-tubulin is found as one member of a large γ-tubulin ring complex (γ-TuRC), which also contains several γ-ring proteins (GRIPs) that are associated with centrosomes or are found in the cell cytoplasm. Two GRIPs (GCP2 and GCP3) are also part of a smaller γ-tubulin-containing complex that is found in the cytoplasm and known as the γ-TuSC. Budding yeast can survive with this smaller complex without γ-TuRC, but it is unclear whether this reflects the divergence of its γ-tubulin amino-acid sequence from more typical sequences, or if the γ-TuSC alone is sufficient to nucleate a functional network of microtubules. However, data presented in Heidelberg suggest the latter. B. Raynaud-Messina (Toulouse, France) found that depletion of Drosophila γ-TuSC blocked microtubule nucleation, whereas less severe phenotypes developed from the depletion of γ-TuRC. Her data suggest that the γ-TuSC can form a functional nucleating complex and that γ-TuRC complexes nucleate distinct arrays or confer distinct properties on the microtubules they nucleate. This idea was echoed in another system. Although the fission yeast γ-TuSC components are essential, earlier work from Takashi Toda and Kathy Gould's groups and new work presented by K. Sawin (Edinburgh, UK) show that homologues of the γ-TuRC-specific components are not essential. The organization of microtubules in fission yeast differs from the centrosomenucleated array of higher eukaryotes, because additional interphase microtubule bundles, which emanate from sites on the nuclear envelope, complement the microtubules that are nucleated from the SPB. Furthermore, γ-TuSC components are seen along these interphase bundles, suggesting that microtubule nucleation probably occurs in the bundles. However, the γ-TuRC-specific molecules are required alongside conserved interphase microtubule bundle components, described by Sawin, S. Venkatram (Nashville, TN, USA) and P. Tran (Philadelphia, PA, USA), to ensure that these interphase bundles are appropriately robust and have the proper dynamics. Work from these researchers showed that when γ-TuRC-specific components are missing, there are fewer interphase bundles than normal and the dynamics of these bundles is altered without having a significant impact on spindle microtubules. By contrast, loss of γ-TuSC components compromises microtubule nucleation from all MTOCs.
Another genetic system gave further insights into the diversity of γ-tubulin function as C. Wiese (Wisconsin, WI, USA) showed that Drosophila γ-tubulin can also ‘call the tunes'. She used isotype-specific antibodies to distinguish between the γ-tubulins γ-tub23C and γ-tub37CD, and also showed distinct distributions for each molecule and an inability of the 23C variant to associate with γ-TuRCs in embryos when γ-tub37CD was present. The basis for this distinction may lie in the fact that each γ-tubulin binds a distinct Grip84 (GCP2) splice variant.
RNA in centrosomes?
M. Alliegro (New Orleans, LA, USA) presented yet more controversial work with R. Palazzo (Troy, NY, USA) about a question that has haunted the field for some time: does the centrosome contain nucleic acids? Random prime polymerase chain reaction of isolated clam centrosomes revealed the presence of unique RNAs that could not be accounted for when searching through existing databases. Close scrutiny of one of these RNAs revealed that it localizes to the centrosome in clam oocytes, is translated and contains a reverse transcriptase domain. Given the controversial history of this aspect of centrosome research, the fact that no previous studies claiming to demonstrate centrosomes or basal body nucleic-acid content have withstood the test of time, and that studies by Roy Gould and Gary Borisy have shown that mammalian cell centrosomes can be riddled with virus particles (Gould & Borisy, 1977), the presentation met with an appropriate mix of caution, enthusiasm and intense scrutiny. Further work will be required to convince the field of the significance of this initial study.
Beyond centrioles: basal bodies, flagella and cilia
One of the key confusions for the uninitiated in centriole biology is the fact that centrioles can be highly variable in structure (Wheatley, 1982). For example, C. elegans centrioles are barrel-shaped arrays of nine-singlet microtubules, those of syncitial Drosophila have nine-doublet microtubules, whereas centrioles of Chlamydomonas and higher systems, including animals, contain nine-triplet microtubules (Fig 2). Even greater complexity is encountered when looking at basal bodies and further out into the cilium and flagellum (Fig 3A). The only real way to understand how such a variety of forms can be generated, and why the simple ones do not need the baggage of the complex ones, is to take each system apart and compile a list of their components for systematic functional analysis. This is exactly what the groups of M. Winey (Boulder, CO, USA), P. Dupuis-Williams (Orsay, France), K. Gull (Oxford, UK), P. McKean (Lancaster, UK), S. Dutcher (St Louis, MO, USA), W. Marshall (San Francisco, CA, USA), and G. Pazour (Worcester, MA, USA) presented for the ciliates Tetrahymena and Paramecium, the Trypanosome parasite and the alga Chlamydomonas. Their studies revealed considerable differences between these morphologically related structures.
Figure 3.
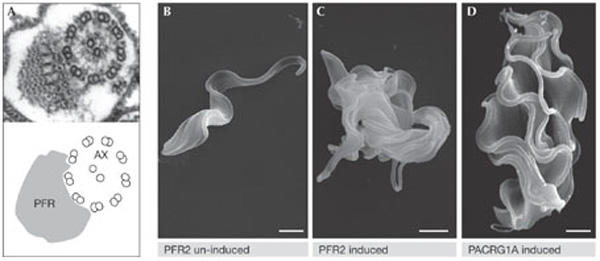
Disruption of flagellar structure blocks cytokinesis of Trypanosomas brucei. (A) A transmission electron micrograph showing the 9 + 2 structure of the axoneme (AX) alongside the paraflagellar rod (PFR). (B–D) Scanning electron micrographs showing bloodstream-form T. brucei before (B) and after ablation of the flagellar proteins PFR2 (C) and PACRG1A (D). Images were kindly provided by K. Gull (Oxford, UK).
The challenge of the task ahead was put in stark relief by the realization from studies of Chlamydomonas (Pazour, Dutcher and Marshall) that the isolated flagellum contains 21 kinases and 11 phosphatases. The studies also showed that there are similarities between the flagellum of this simple model system and the mammalian cilium and primary cilium. As many human diseases, including polycystic kidney disease, develop from problematic ciliogenesis, model systems have again proven to be highly informative in the characterization of human disease. Perhaps as remarkable as this conservation, is the divergence that has accompanied the specialization of particular organisms. For example, 208 of the T. brucei flagella components described by Gull and McKean are restricted to other trypanosomatids, whereas Dupuis-Williams showed how the reliance of Paramecium on basal bodies to build one of the most complex microtubule cytoskeletons has led to the proliferation of an extraordinary number of basal-body-associated tubulin family members. The idea that the reliance of parasites such as Trypanosomes and Leishmania on a highly specialized flagellum is a potential Achilles heel that could be exploited for therapeutic benefit was made apparent when Gull showed that bloodstream T. brucei cells cannot divide if flagellar PFR or axonemal proteins are ablated (Fig 3). Most importantly, from a health and animal welfare point of view, this lethality is specific to the mammalian bloodstream forms that are responsible for 60,000 deaths and 100,000 disabilities per year in sub-Saharan Africa (Wellcome Trust, 2005). Therefore, basic characterization of an intellectually appealing system is moving rapidly into high-throughput screening for small molecules to cure one of the biggest killers in the developing world.
Control
A second emerging medical interest in centrosomes and spindle poles, stems from the realization that many of the pathways that determine cell fate are coordinated at the pole. It is assumed that the resulting cross talk integrates a variety of otherwise unconnected pathways to ensure that the correct cell fate is pursued. This has led to the growing belief that intervention in this coordination of pathways at the pole could have great potential in the treatment of cancer. Significant energy is being invested in research into the organization of particular pathways that regulate entrance and exit from mitosis, because these are particularly tractable in genetic systems. However, S. Doxsey (Worcester, MA, USA) consolidated Sluder's observation that the centrosome is required for transit of the restriction point from G1, by showing that depletion of a large number of centriole components also arrested cells in G1.
The involvement of the SPB in the control of cytokinesis was raised at the 1997 workshop by Viesturs Simanis and Iain Hagan, who used Schizosaccharomyces pombe to show that the septum initiation network is regulated from one SPB that harbours active Spg1 GTPase (Sohrmann et al, 1998). Fission yeast was then swiftly overtaken by budding yeast through elegant work from groups such as Elmar Schiebel, Ray Deshaies and Angelika Amon, who studied the equivalent budding yeast network—the mitotic exit network (MEN). At this meeting, E. Schiebel (Heidelberg, Germany) presented findings from his collaboration with G. Pereira's group (Heidelberg, Germany), showing the role of Kin4 in restraining mitotic exit when spindles are misorientated—with the observation from Steve Elledge's group that the yeast homologue of metazoan Plk1, Cdc5, regulated mitotic exit (Hu et al, 2001). Schiebel also showed that polo kinase responds to misalignment of the spindle and associates with the nucleators/ anchors of these cytoplasmic microtubules, Spc72 and the protein that acts as a scaffold for the MEN, Nud1. The idea that elements of this pathway might be conserved in higher systems, was given greater impetus by the account of human Mob4 by A. Tavares (Lisbon, Portugal). Mob proteins were first identified through their association of the founder member, Mob1, with the budding yeast MEN kinase Dbf2. Tavares showed that human Mob4 not only binds to the human homologues of Dbf2 (nuclear Dbf2-related (NDR) kinases) but also, similar to Dbf2, is located at the centrosome and required for some aspect of cytokinesis. Mob4 seems to mimic a centriole component described by Doxsey, called Centriolin—which is in part homologous to Nud1 and Cdc11 of both yeasts—as it is required for the final stages of abscission. Doxsey furthered his studies by showing that Centriolin is found not only on the centriole appendages but also in a ring structure at the midbody that is inherited by one of the two daughter cells as a scar, in much the same way that bud scars are retained in the older cells of Saccharomyces cerevisiae. Interestingly, these midbody scars/rings of centriolin persist through several replication cycles and so provide a counting mechanism for the number of divisions a particular cell has undergone. Furthermore, in an expansion of earlier work by Matthieu Piel and Michel Bornens, which showed that the mother centriole correlates with abscission (Piel et al, 2001), Doxsey has now found that the docking and fusion of GFP-labelled secretory vesicles at the midbody is required for abscission. Vesicle delivery is asymmetric, coming from one of the two daughter cells, and it will be exciting to see if centrosome age has a role in this process. A final spin on mitotic kinases and the centrosome came from an interesting study of Cdk11 by R. Giet (Rennes, France). The messenger RNA-processing function of the 110-kDa version of this kinase has been studied in some detail, but Giet showed that a second 58-kDa molecule, which is produced from the same transcript by use of an internal ribosome entry site in late G2 and throughout mitosis, associates with the centrosome and is required for spindle formation. This is a particularly intriguing finding because both polo and Cdk/cyclin B kinases can be targeted to substrates provided they have been phosphorylated by Cdks (Elia et al, 2003; Mimura et al, 2004). The assumption for mammalian cells has been that Cdk2/cyclin A or Cdk1/cyclin B1/2 is responsible for promoting the recruitment of polo and perhaps of each other, to specific targets. However, the discovery of Cdk11 as a third class of mitotic Cdk that could work alongside Cdk1 and Cdk2 now expands the repertoire of possibilities.
Asymmetry
A recurrent theme in the structure and function of discrete MTOCs is asymmetry. Only one of the four centrioles in the two centrosomes of a G2 cell has appendages and the others will mature with a discrete hierarchy (Vorobjev & Chentsov, 1982). In fission yeast, the recruitment of the NIMA-related kinase Fin1 shows that fungal SPBs, similar to centrioles, take at least one-and-a-half cell cycles to mature, thus a particular SPB will be in one of four states (Grallert et al, 2004). However, it has not been clear how or why such asymmetry should occur in unicellular fungi. An elegant, and particularly thorough, study from M. Knop (Heidelberg, Germany) defined fundamental differences between each of the four SPBs that are produced during meiosis in budding yeast. In response to carbon limitation, fewer spores are formed in accordance with the degree of starvation (Fig 4). Knop's studies showed that the gradient in the number of spores produced reflects modifications on the SPBs, which enable them to nucleate the prospore membrane that generates the spore wall. Intriguingly, the system favours dyad formation in which heterozygosity is ensured if only two spores are to be formed. Thus, because the mating-type loci are centromere-linked—when the cell can only make two progeny—these two will have opposite mating types and will therefore be able to mate immediately after sporulation. This design means that if a gene is next to a centromere and is essential, inactivation of heterozygosity of a gene can be tolerated. Conversely, if it lies further away, there is a chance that recombination will shuffle the pack so that a dyad meiosis cannot save the day as it produces a homozygous-lethal. Indeed, Knop showed that the viability of heterozygotes outstripped that of homozygotes in co-culture experiments, suggesting that SPB-driven selection is occurring. This may account for their report that essential genes are enriched in centromere-linked regions of the genome. Therefore, organization of the genome can now be added to the microtubule cytoskeleton and cell cycle as being influenced by SPB functions.
Figure 4.
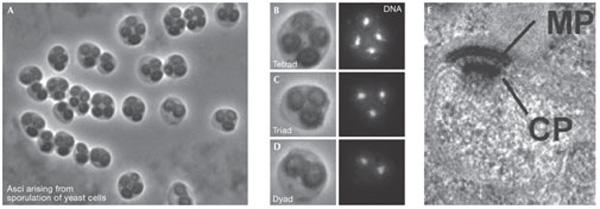
Nutrient levels control the differentiation of the outer face of budding yeast spindle pole bodies to generated the meiotic plaque during sporulation. (A–D) Sporulating yeast cells adjust the number of spores they form according to the level of carbon sources available producing asci (= sporulated yeast cells) with only 1, 2 (B), or 3 (C) spores instead of the normal number of 4 (D). There is a corresponding reduction in the number of nuclei as some of the genomes are aborted after the meiotic divisions have occurred. (E) The formation of a regulated number of spores depends entirely on the assembly of the meiotic plaque (MP), a meiosis-II-specific structure of the spindle pole body (SPB). The MP is assembled while attached to the central plaque (CP) on the cytoplasmic side of the SPB. MP assembly at a subset of SPBs is regulated by a dynamic self-organizing system, which is controlled by the abundance of MP components and age-related differences between the SPBs. CP central plaque. Scale bar, 200 nm. Images were kindly provided by M. Knop (Heidelberg, Germany) and reprinted from Taxis et al (2005) with permission from The Rockefeller University Press.
Centrosomes: who needs them?
The most surprising presentation at this third workshop harked back to the first, as Raff described another mutation in Drosophila that, like the study of centrosomin (Cnn) that Thomas Kaufman presented at the 1997 meeting, affected core centrosome functions, but had a minor impact on fly development. Despite the absence of any apparent centrosomes, flies that lacked Drosophila SAS4 developed normally, although a detailed analysis of whether embryos of these flies are truly developing without centrosomes has not yet been completed. However, as the flies are left as apparently quivering creatures unable to effectively execute sexual reproduction, most attendees concurred with the stance aired by John Kilmartin at the first meeting, that he was happy that his own cells were still using their own centrosomes.
Conclusions
Coming away from this third meeting, the participants were awed by the progress of this field over the past eight years. We have moved on from the wish to identify centrosome components that preceded the first meeting in 1997, to what is now a field with an overflowing treasure chest of centrosome and SPB components. We are also beginning to see detailed accounts of the macromolecular interactions at the atomic level during SPB duplication. Importantly, the work presented at this meeting clearly repositioned the centrosome as a ‘central organizer' for the regulation of many cell processes. The continued demonstration of the role of the centrosome and related structures in disease—not only diseases of fertility, reproduction and cancer as first appreciated, but also unexpected diseases of the kidney, blindness, those caused by parasites, and others—gives greater impetus to the journey in centrosome and SPB biology. What is truly admirable in this field is that, in spite of the intense pace and clear intellectual intensity and competition that the participants have experienced during the past eight years, most remain generous and continue to share openly ideas and reagents. Clearly, this is driving the field forward at an extraordinary pace. This is a field that is experiencing the transition from youth to maturity, and which shows no signs of a reduced trajectory, but is now experiencing the best of times.

Iain M. Hagan

Robert E. Palazzo
Acknowledgments
We thank those we have consulted about the text and apologize to those whose work we have been unable to cite owing to space restrictions.
References
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–574 [DOI] [PubMed] [Google Scholar]
- Balczon R, Bao L, Zimmer WE, Brown K, Zinkowski RP, Brinkley BR (1995) Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J Cell Biol 130: 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley BR (2001) Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol 11: 18–21 [DOI] [PubMed] [Google Scholar]
- Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB (2003) The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell 115: 83–95 [DOI] [PubMed] [Google Scholar]
- Gould RR, Borisy GG (1977) The pericentriolar material in Chinese hamster ovary cells nucleates microtubule formation. J Cell Biol 73: 601–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A, Krapp A, Bagley S, Simanis V, Hagan IM (2004) Recruitment of NIMA kinase shows that maturation of the S. pombe spindle-pole body occurs over consecutive cell cycles and reveals a role for NIMA in modulating SIN activity. Genes Dev 18: 1007–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA (2005) The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol 7: 1140–1146 [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E (1996) Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382: 420–425 [DOI] [PubMed] [Google Scholar]
- Hu F, Wang Y, Liu D, Li Y, Qin J, Elledge SJ (2001) Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell 107: 655–665 [DOI] [PubMed] [Google Scholar]
- Marshall WF, Vucica Y, Rosenbaum JL (2001) Kinetics and regulation of de novo centriole assembly. Implications for the mechanism of centriole duplication. Curr Biol 11: 308–317 [DOI] [PubMed] [Google Scholar]
- Mimura S, Seki T, Tanaka S, Diffley JF (2004) Phosphorylation-dependent binding of mitotic cyclins to Cdc6 contributes to DNA replication control. Nature 431: 1118–1123 [DOI] [PubMed] [Google Scholar]
- Palazzo RE (2002) Centrosome and spindle pole body dynamics: A review of the EMBO/EMBL Conference on Centrosomes and Spindle Pole Bodies, Heidelberg, September 13–17, 2002. Cell Motil Cytoskeleton 54: 148–154 [DOI] [PubMed] [Google Scholar]
- Piel M, Nordberg J, Euteneuer U, Bornens M (2001) Centrosome dependent exit of cytokinesis in animal cells. Science 291: 1550–1553 [DOI] [PubMed] [Google Scholar]
- Sohrmann M, Schmidt S, Hagan I, Simanis V (1998) Asymmetric segregation on spindle poles of the Schizosaccharomyces pombe septum-inducing protein kinase Cdc7p. Genes Dev 12: 84–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Winey M (1997) The cell center at 100. Cell 91: 303–309 [DOI] [PubMed] [Google Scholar]
- Taxis C, Keller P, Kavagiou Z, Jensen LJ, Colombelli J, Bork P, Stelzer EH, Knop M (2005) Spore number control and breeding in Saccharomyces cerevisiae: a key role for a self-organizing system. J Cell Biol 171: 627–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev IA, Chentsov YS (1982) Centrioles In the cell-cycle. 1. Epithelial cells. J Cell Biol 98: 938–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust (2005) Three of a Kind.15 July London, UK: Wellcome Trust http://www.wellcome.ac.uk/doc_WTX026055.html
- Wheatley D (1982) The centriole: a central enigma in cell biology. Amsterdam: Elsevier Biomedical
- Wigge PA, Jensen ON, Holmes S, Soues S, Mann M, Kilmartin JV (1998) Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol 141: 967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]


