Abstract
The post-translational modification of histones regulates many cellular processes, including transcription, replication and DNA repair. A large number of combinations of post-translational modifications are possible. This cipher is referred to as the histone code. Many of the enzymes that lay down this code have been identified. However, so far, few code-reading proteins have been identified. Here, we describe a protein-array approach for identifying methyl-specific interacting proteins. We found that not only chromo domains but also tudor and MBT domains bind to methylated peptides from the amino-terminal tails of histones H3 and H4. Binding specificity observed on the protein-domain microarray was corroborated using peptide pull-downs, surface plasma resonance and far western blotting. Thus, our studies expose tudor and MBT domains as new classes of methyl-lysine-binding protein modules, and also demonstrates that protein-domain microarrays are powerful tools for the identification of new domain types that recognize histone modifications.
Keywords: lysine methylation, protein-domain arrays, tudor domain
Introduction
Covalent modification of histones is important for the regulation of transcription and chromatin dynamics (Ehrenhofer-Murray, 2004). These covalent modifications are deposited in a combinatorial manner, predominantly on the amino-terminal tails of the core histones. This combinatorial assortment of phosphorylation, acetylation and methylation on these tails has been termed the ‘histone code' (Jenuwein & Allis, 2001). The complexity of this code is further enhanced by the fact that there are three forms of lysine methylation (mono-, di- and tri-) and two forms of arginine methylation (mono- and di-; Aletta et al, 1998). It has been demonstrated that different codes are associated with active and suppressed transcriptional states, which inturn results in the recruitment of distinct protein complexes that affect chromatin structure. A subset of proteins within these complexes harbour conserved protein domains, which are believed to be responsible for mediating these interactions. These include chromo domains that bind to methylated lysine residues and bromo domains that bind to acetylated lysine residues (Jenuwein & Allis, 2001).
Histone acetylation is associated with an active transcriptional state, whereas lysine methylation can be either repressive or activating, depending on the site of the post-translational modification. In support of the idea that distinct methyl-lysine marks recruit different proteins that in turn specify different transcriptional responses, it has been shown that lysine 9 methylation on histone H3 (H3K9me) results in the chromo domain-dependent recruitment of HP1 (Bannister et al, 2001; Lachner et al, 2001). Likewise, specific chromo domain-mediated interactions occur between H3K27me and Polycomb (Fischle et al, 2003; Min et al, 2003), and between H3K4me and Chd1 (Pray-Grant et al, 2005). Recently, the WD40 repeats of WDR5 were shown to directly associate with H3K4me (Wysocka et al, 2005). Thus, methyl marks on histone tails are not read by chromo domain-containing proteins alone.
Chromo domains and bromo domains are usually found in proteins that are associated with chromatin. There are several other domain types that are also found predominantly in chromatin-associated proteins, including tudors, PhDs, SANTs, SWIRMs, MBTs and PWWPs. Indeed, some of these domain types (tudors, MBTs and PWWPs) are structurally related to chromo domains and have been collectively called the ‘royal family' (Maurer-Stroh et al, 2003). It has been suggested that the ‘royal family' of protein domains may have functional similarities. In keeping with this idea, the tudor domain of the protein mutated in spinal muscular atrophy, the SMN protein, binds to a symmetrically dimethylated arginine motif (Friesen et al, 2001), and recent studies have shown that the tudor domain of the double-stranded break-sensing protein, 53BP1, can bind to H3K79me2 (Huyen et al, 2004).
To screen for protein domains that possess the ability to ‘read' the modification status of the histone tails, we have taken a protein-domain microarray approach. Similar microarray approaches have been used to identify protein–protein interactions, including interactions that are sensitive to arginine methylation and are phospho-serine dependent (Espejo et al, 2002; Liu et al, 2002). The arrays used in the above-mentioned studies focused on signal-transduction issues and harboured WW, SH3, SH2, PDZ and 14.3.3 domains. To address the subject of modified histone tails binding to protein domains, we generated a microarray that focused on domains found in chromatin-associated proteins. This chromatin-associated domain array (CADOR) chip contains bromo, chromo, tudor, PhD, SANT, SWIRM, MBT, CW and PWWP domains fused to glutathione S-transferase (GST).
To identify novel methyl-lysine-dependent protein–protein interactions, we have probed the CADOR chip with fluorophore-tagged N-terminal peptides from histones H3 and H4 that vary in their degree and position of methylation. The well-documented interaction between the chromo domains of HP1 (α, β and γ) and H3K9me peptides was detected. In addition, novel methyl-dependent interactions are seen with a chromo domain (CDY1), tudor domains (53BP1, C20orf104 and JMJD2A) and MBT domains (CGI-72 and L(3)MBTL), thus demonstrating the feasibility of this approach for identifying proteins that read the histone code.
Results
Detection of known methyl-dependent interactions
To identify potential proteins that can bind to histone tails in a modification-dependent manner, we generated protein microarrays using domains found predominantly in chromatin-associated proteins. To generate these CADOR chips, 109 different protein domains were cloned as GST fusions (Fig 1A,B) and spotted onto nitrocellulose-coated glass slides (Fig 1C). To establish that the binding integrity of the domains has been maintained, the array was first probed with a symmetrically arginine methylated peptide from the splicing factor SmD3 (SmD3-Rme2s), which has previously been demonstrated to bind to the tudor domain of SMN (Friesen et al, 2001). As expected, we see specific binding to SMN, as well as novel interactions with the tudor domains of TDRD3 and a Schizosaccharomyces pombe protein (Fig 1C). The unmethylated SmD3 peptide does not bind to any fusion proteins (data not shown). This raises the possibility that asymmetrically arginine methylated peptide may also bind to tudor domains. It has also been reported that the tudor domains of the double-stranded break-sensing protein, 53BP1, can bind to H3K79me2 (Huyen et al, 2004). The CADOR chip was thus also probed with a peptide from histone H3 that harbours the dimethylated K79 residue. In this case, methyl-dependent binding to the tudor domain C20orf104 (red oval) was observed, but not that of 53BP1 (blue oval). In addition, the unmethylated peptide bound several domains.
Figure 1.
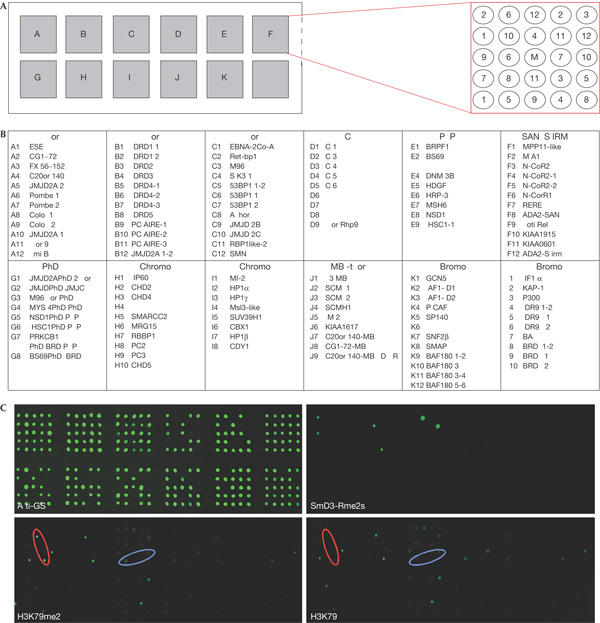
Methylated peptides bind to tudor domains on the CADOR chip. (A) A collection of 109 glutathione S-transferase (GST) fusion proteins were arrayed in duplicate on a nitrocellulose slide. The layout of the array is shown. The middle position (M) contains GST alone as a background indicator. (B) The arrayed GST fusion proteins are listed. The accession numbers and regions cloned can be found in the supplementary information online. (C) The array was first probed with an anti-GST antibody and detected with a fluorescein isothiocyanate-conjugated secondary antibody to establish roughly equal loading. The array was subsequently probed with Cy3-labelled peptides, including a peptide from SmD3 that harbours symmetrically dimethylated arginine residues and peptides harbouring K79 from histone H3 in dimethylated and unmethylated forms. The H3K79me2 peptide shows a high degree of methyl-independent binding. A methyl-dependent interaction is seen with the tudor domain of C20orf104 (red oval). The tudor domains of 53BP1 are marked (blue oval).
Detection of novel methyl-dependent interactions
Next, to identify novel protein domains that could ‘read' the various post-translational modifications on histone tails, we probed the CADOR chip with peptides that were mono-, di- or tri-methylated at lysines 4 and 9 of histone H3 and lysine 20 of histone H4 (Fig 2). The binding of the HP1 chromo domain to the H3K9 methyl mark is well accepted, and this interaction is clearly seen with all three HP1 variants. In addition, a novel interaction between the CDY1 chromo domain and the H3K9me2 and H3K9me3 peptides is observed. Only the H3K9 methyl mark possesses the ability to bind to the arrayed chromo domains, and no chromo domain interactions are seen with methylated peptides harbouring H3K4 or H4K20.
Figure 2.
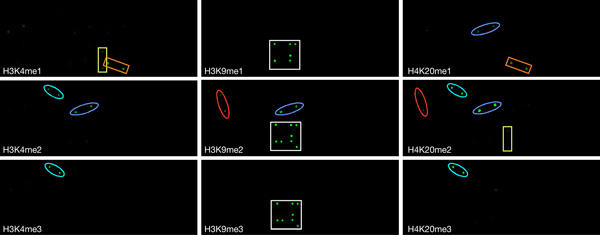
Probing the CADOR chip with a series of methylated peptides from the tails of histones H3 and H4. The protein-domain microarray was probed with Cy3-labelled peptides. The unmethylated H3(1–18) and H4(11–28) peptides showed no binding (data not shown). Chromo domain interactions are blocked with a white square. Tudor domain interactions are highlighted with ovals: C20orf104 (red), 53BP1 (blue) and JMJD2A (turquoise). MBT domain interactions are marked with rectangles: CGI-72 (orange) and L(3)MBTL (yellow).
Tudor domain binding is seen at all three methylation sites tested (H3K4, H3K9 and H4K20; Fig 2), and not only the site of methylation but also the degree of methylation (mono-, di- or tri-) is important for binding. The tudor domains of 53BP1 preferentially bind to the di-methylated state of all three sites. The tudor domain of C20orf104 shows a similar binding profile to 53BP1. The tudor domains of JMJD2A bind to di- and tri-methylated H3K4 and H4K20.
MBT domains bind most strongly to a mono-methylated lysine mark. The MBT domains of both L(3)MBTL and CGI-72 show this ability at H3K4me1. The MBT domain of CGI-72 also binds to H4K20me1, and although L(3)MBTL does not bind to the mono-methylated mark here, it binds to H4K20me2.
To independently validate the domain–peptide interactions that we detected with the CADOR chip, we used the more traditional approaches of peptide pull-down (Fig 3) and surface plasmon resonance (supplementary Fig S1 online). These reciprocal approaches confirmed the methyl-dependant domain interactions first detected on the CADOR chip (Fig 2). The binding data are summarized in Table 1.
Figure 3.
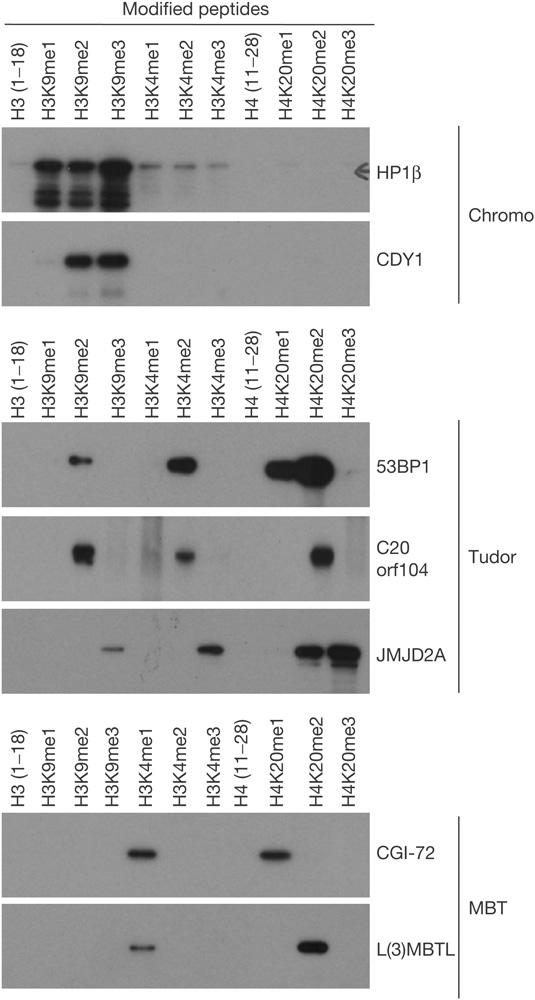
A pull-down-based approach detects methyl-dependent interactions between domains (chromo, tudor and MBT) and methylated histone tail peptides. Biotinylated peptides were immobilized on streptavidin beads and used to pull down the indicated glutathione S-transferase fusion proteins.
Table 1.
Summary of interaction detected in the pull-down assay
| H3K9me1 | H3K9me2 | H3K9me3 | H3K4me1 | H3K4me2 | H3K4me3 | H4K20me1 | H4K20me2 | H4K20me3 | H3K79me2* | SmD3-Rme2s | |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Chromo domains | |||||||||||
| HP1α |
√ |
√ |
√ |
— |
— |
— |
— |
— |
— |
— |
— |
| HP1β |
√ |
√ |
√ |
— |
— |
— |
— |
— |
— |
— |
— |
| HP1γ |
√ |
√ |
√ |
— |
— |
— |
— |
— |
— |
— |
— |
| CDY1 |
— |
√ |
√ |
— |
— |
— |
— |
— |
— |
— |
— |
|
Tudor domains | |||||||||||
| SMN |
— |
— |
— |
— |
— |
— |
— |
— |
— |
— |
√ |
| TDRD3 |
— |
— |
— |
— |
— |
— |
— |
— |
— |
— |
√ |
| Pombe-1 |
— |
— |
— |
— |
— |
— |
— |
— |
— |
— |
√ |
| 53BP1(1,2)‡ |
— |
√ |
— |
— |
√ |
— |
√ |
√ |
— |
— |
— |
| C20orf104 |
— |
√ |
— |
— |
√ |
— |
— |
√ |
— |
√ |
— |
| JMJD2A(1,2)‡ |
— |
— |
√ |
— |
— |
√ |
— |
√ |
√ |
— |
— |
|
MBT domains | |||||||||||
| CGI-72 |
— |
— |
— |
√ |
— |
— |
√ |
— |
— |
— |
— |
| L(3)MBTL(2,3)‡ |
— |
— |
— |
√ |
— |
— |
— |
√ |
— |
— |
— |
| *This peptide shows a degree of methyl-independent binding. | |||||||||||
| ‡If multiple domains are present in a protein, the numbers in parentheses indicate the domains in this particular clone. | |||||||||||
Tudor and MBT domains bind core histones
We have found that the peptides from the N-terminal tails of histones H3 and H4 have the ability to bind to a subset of tudor domains. To establish that these domains not only bind to peptides but also can interact with histones as a whole, we performed far western assays with GST–tudor fusion proteins on core histone. Core histones were isolated from G9a wild-type and null embryonic stem cells and from Suv39h double-null mouse embryonic fibroblasts (MEFs; Lachner et al, 2001; Tachibana et al, 2002). G9a-null cells lack the di-methyl mark on H3K9, and Suv39h double-null cells are not tri-methylated at H3K9 (Fig 4A—western). As a control in this assay, we see that HP1β binds to histone H3, and that the binding is reduced to histone H3 isolated from both the G9a-null and Suv39h double-null cells. The HP1β chromo domain binds to mono-, di- and tri-methylated H3K9 (Figs 2 and 3), and the loss of any one methylated form reduces this binding to the histone, but does not eliminate it (Fig 4A—far western). The chromo domains of HP1β and CDY1 show similar binding profiles on both the CADOR array and by pull-down (Figs 2 and 3). This similarity is again seen in the far western experiment, in which the CDY1 chromo domain binds only to histone H3, and this binding is sensitive to lysine 9 methylation by Suv39h and G9a. Furthermore, full-length versions of these two chromo domain-containing proteins colocalize in Suv39h wild-type but not double-knockout MEFs, when co-transfected as DsRed–HP1β and green fluorescent protein (GFP)–CDY1 fusions (Fig 4B). Probings with the tudor domains of JMJD2A, C20orf104 and 53BP1 show that they all bind to histones H3 and H4, but not to H2A or H2B (Fig 4A—far western), as would be predicted from the domain and peptide pull-down experiments (Figs 2 and 3). Interestingly, binding of the JMJD2A tudor domains to histones H3 and H4 requires the presence of the Suv39h enzymes. This could be explained by the recent findings that tri-methylation of H3K9 is required for the subsequent tri-methylation of H4K20 (Schotta et al, 2004), and suggests that the JMJD2A protein has repressor activity.
Figure 4.
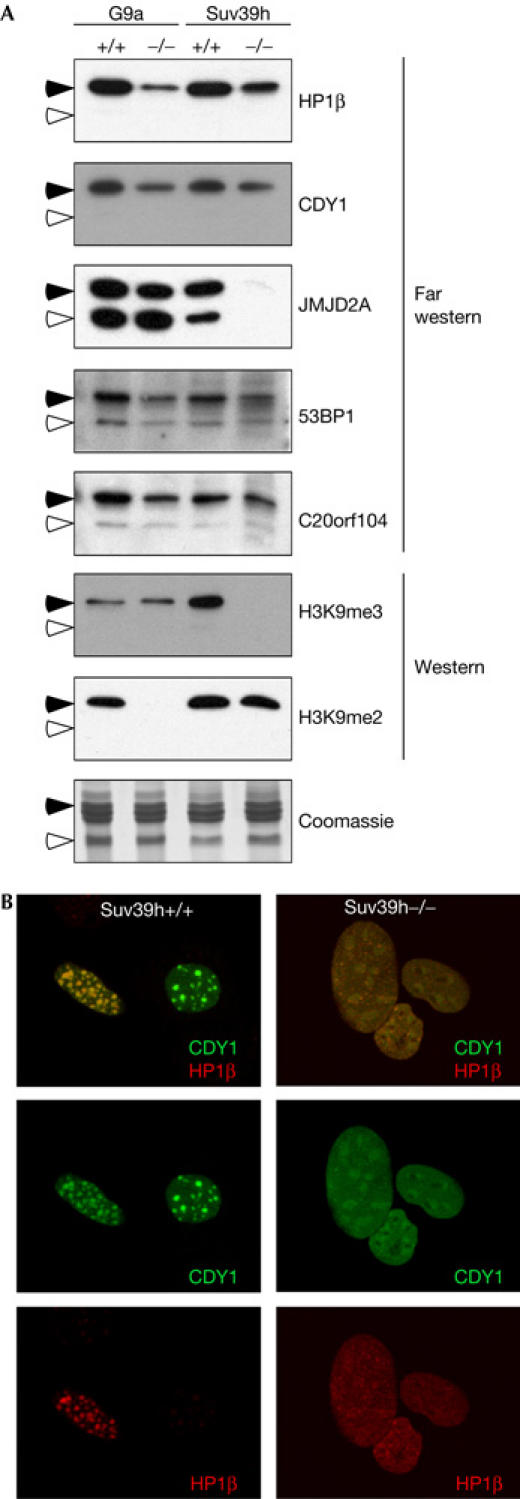
Tudor and chromo domains bind to histones. (A) Tudor domains bind to histones H3 and H4, and CDY1 binds to histone H3. Core histones were acid purified from G9a wild-type and knockout cells and Suv39h wild-type and double-knockout cells. Core histones were subjected to far western analysis by incubation with the indicated glutathione S-transferase (GST) fusion proteins. Western analysis was performed with methyl-specific antibodies to confirm the null status of the cell lines. The arrowheads denote the possition of histones H3 (closed) and H4 (open). Further description of western analysis is given in the supplementary information online. (B) Green fluorescent protein (GFP)–CDY1 and DsRed–HP1β were co-transfected into Suv39h wild-type and double-knockout mouse embryonic fibroblasts (MEFs). GFP–CDY1 and DsRed–HP1β colocalize to heterochromatin. This colocalization is lost in Suv39h−/− MEFs.
Discussion
Here we describe the use of a protein-domain microarray approach to screen for chromatin-associated domains that specifically recognize histone H3 and H4 tail peptides methylated to varying degrees on specific lysine residues. It is important to note that interactions may be missed when using this approach. This could occur if the GST fusion protein has not retained its structure under these relatively harsh conditions, or if two domains within the same protein (or in the same complex) are binding different marks on the histone tails, thus stabilizing an interaction due to an avidity effect. However, using this approach, we identified six novel methyl-dependent interactions between domains and histone tails. The chromo domain of CDY1 can interact with di- and tri-methylated H3K9. CDY1 is found on the Y chromosome and has been implicated in the process of spermatogenesis, and there is a strong association between the loss of CDY1 function and male infertility (Machev et al, 2004).
From this study, tudor domains have emerged as a new domain type that can bind to histone tails. The tudor domains of JMJD2A bind most strongly to di- and tri- methylated H4K20, and also to H3K4me3 and H3K9me3 (Figs 2 and 3). JMJD2A is a member of the JMJD2 gene family. Three of the six JMJD2 family members harbour two tudor domains (Katoh, 2004). Notably, JMJD2A (KIAA0677) was recently identified as a component of the N-CoR corepressor complex (Yoon et al, 2003), and the tudor domains of JMJD2A are required for its repressor activity (Zhang et al, 2005). In addition, JMJD2A binds the retinoblastoma protein (Rb) and has been implicated in the repression of E2F-regulated promoters (Gray et al, 2005). This is consistent with our finding that the tudor domains of JMJD2A lose their ability to bind to histones H3 and H4 in the absence of Suv39h activity (Fig 4)—a signal that is implicated in gene silencing (Lachner et al, 2001). 53BP1 is involved in sensing DNA double-stranded breaks (Charier et al, 2004; Stucki & Jackson, 2004). The tudor domains of 53BP1 were recently shown to bind to di-methylated lysine 79 on histone H3 (Huyen et al, 2004). Using a domain array approach, we do not see methyl-dependent binding of this peptide to the tudor domains of 53BP1 (Fig 1C). However, we do see binding of these tudor domains to other di-methylated peptides, including H3K4me2, H3K9me2 and H4K20me2 (Fig 2). The strongest relative binding of the 53BP1 tudor domains is to H4-2mK20 (supplementary Fig S1 online). This is in keeping with the recent finding that in S. pombe, there is a genetic link between H4K20 methylation and Crb2, the homologue of 53BP1 (Sanders et al, 2004). A similar profile of binding is seen for the single tudor domain of C20orf104. The C20orf104 has been identified as a tumour antigen (Behrends et al, 2003).
The MBT domain-containing proteins CGI-72 and L(3)MBTL bind to the mono-methylated mark on H3K4. CGI-72 also binds to the mono-methylated form of H4K20, and L(3)MBTL binds to H4K20me2 (Figs 2 and 3). L(3)MBTL is a member of the Polycomb group proteins, which associates with condensed chromosomes during mitosis (Koga et al, 1999) and possesses transcriptional repressor activity (Boccuni et al, 2003). In addition, deletions of the L(3)MBTL locus are associated with myeloid malignancies (Li et al, 2004). CGI-72 has not been studied, but it is structurally similar to C20orf104.
This study uses a novel approach to identify protein domains that can ‘read' post-translational modifications laid down on histone tails, suggesting that the ‘royal family' of protein domains are an important class of methyl-dependent protein interaction domains. Using this approach, it is likely that other domain–peptide interactions will be detected, as more of the histone code is used to probe this array. Furthermore, this approach will allow us to investigate the importance of not only the degree of methylation but also the combinations of different post-translational modifications that promote or inhibit specific interactions, thus getting at the crux of the histone code.
Methods
Cloning and purification of GST fusion proteins. The complementary DNAs encoding the domains listed in Fig 1B (see supplementary information online) were cloned into the pGEX-6P1 vectors by PCR using a human cDNA library (Origene, Rockville, MD, USA), and verified by DNA sequencing. GST fusion proteins were purified as described previously (Espejo et al, 2002). The full open reading frame of CDY1 was cloned into pEGFP (Clontech, Palo Alto, CA, USA) and HP1β was cloned into DsRed2 (Clontech).
Generation of protein microarray, peptide synthesis and labeling. The generation of protein microarrays has been described (Espejo et al, 2002). Peptides were synthesized by the W.M. Keck Center (New Haven, CT, USA). Methylated and unmethylated forms of the following peptides were synthesized: histone H3 (1–18)—acetyl-ARTKQTARKSTGGKAPRK-biotin, histone H4 (11–28)—acetyl-GKGGAKRHRKVLRDNIQGK-biotin, histone H3 (70–88)—acetyl-LVREIAQDFKTDLRFQSSK-biotin and SmD3 biotin-KGRGRGRRGGRGQNSASRGGSQR-cooh (all arginines are symmetrically dimethylated). Biotinylated peptides were labelled as described previously (Espejo et al, 2002).
Peptide pull-downs. Biotinylated peptides (20 μg) were immobilized on 10 μl of streptavidin beads (Sigma, St Louis, MO, USA) in 200 μl of binding buffer (50 mM Tris–HCl pH 7.5, 15 mM NaCl, 1 mM EDTA, 2 mM dithiothreitol and 0.5% NP-40) at 4°C. The next day, the beads were washed three times with binding buffer and then incubated with 25 μg of GST fusion protein for 2.5 h with rotation at 4°C. After five washes with binding buffer, the beads were boiled in protein loading buffer, fractionated by 10% SDS–polyacrylamide gel electrophoresis and subjected to western blot analysis using an anti-GST antibody.
Far western blotting. Core histones were acid purified (Butler et al, 1986) from G9a and Suv39h knockout and wild-type cell lines, and 4 μg of each sample was run on SDS–polyacrylamide gel electrophoresis, and then transferred onto a PVDF membrane. Blots were blocked in PBS–Tween 20 containing 5% non-fat dry milk, and then incubated with 5 μg/ml of the indicated GST fusion protein in the blocking buffer overnight at 4°C. The blots were then probed with an anti-GST antibody followed by anti-rabbit–horseradish peroxidase and then subjected to enhanced chemiluminescence (Amersham, Uppsala, Sweden) detection. Western analysis was performed with antibodies specific to H3K9me2 (Upstate, Charlottesville, VA, USA; Cat# 07-212) and H3K9me3 (Upstate; Cat# 07-442). Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/7400625-s1.pdf).
Supplementary Material
Supplementary Information
Acknowledgments
We thank T. Jenuwein for the Suv39h double-null cells and Y. Shinkai for the G9a-null cells. M.T.B. is supported by National Institutes of Health (NIH) grant DK62248, and in part by an institutional centre grant ES07784 and pilot funding from ES011047. Y.Z. is supported by an NIH grant GM68804.
References
- Aletta JM, Cimato TR, Ettinger MJ (1998) Protein methylation: a signal event in post-translational modification. Trends Biochem Sci 23: 89–91 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 [DOI] [PubMed] [Google Scholar]
- Behrends U et al. (2003) Novel tumor antigens identified by autologous antibody screening of childhood medulloblastoma cDNA libraries. Int J Cancer 106: 244–251 [DOI] [PubMed] [Google Scholar]
- Boccuni P, MacGrogan D, Scandura JM, Nimer SD (2003) The human L(3)MBT polycomb group protein is a transcriptional repressor and interacts physically and functionally with TEL (ETV6). J Biol Chem 278: 15412–15420 [DOI] [PubMed] [Google Scholar]
- Butler AP, Byus CV, Slaga TJ (1986) Phosphorylation of histones is stimulated by phorbol esters in quiescent Reuber H35 hepatoma cells. J Biol Chem 261: 9421–9425 [PubMed] [Google Scholar]
- Charier G, Couprie J, Alpha-Bazin B, Meyer V, Quemeneur E, Guerois R, Callebaut I, Gilquin B, Zinn-Justin S (2004) The Tudor tandem of 53BP1: a new structural motif involved in DNA and RG-rich peptide binding. Structure (Camb) 12: 1551–1562 [DOI] [PubMed] [Google Scholar]
- Ehrenhofer-Murray AE (2004) Chromatin dynamics at DNA replication, transcription and repair. Eur J Biochem 271: 2335–2349 [DOI] [PubMed] [Google Scholar]
- Espejo A, Cote J, Bednarek A, Richard S, Bedford MT (2002) A protein-domain microarray identifies novel protein–protein interactions. Biochem J 367: 697–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S (2003) Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev 17: 1870–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G (2001) SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell 7: 1111–1117 [DOI] [PubMed] [Google Scholar]
- Gray SG et al. (2005) Functional characterization of JMJD2A, a histone deacetylase- and retinoblastoma-binding protein. J Biol Chem 280: 28507–28518 [DOI] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD (2004) Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432: 406–411 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Katoh M (2004) Identification and characterization of JMJD2 family genes in silico. Int J Oncol 24: 1623–1628 [PubMed] [Google Scholar]
- Koga H, Matsui S, Hirota T, Takebayashi S, Okumura K, Saya H (1999) A human homolog of Drosophila lethal(3)malignant brain tumor (l(3)mbt) protein associates with condensed mitotic chromosomes. Oncogene 18: 3799–3809 [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120 [DOI] [PubMed] [Google Scholar]
- Li J, Bench AJ, Vassiliou GS, Fourouclas N, Ferguson-Smith AC, Green AR (2004) Imprinting of the human L3MBTL gene, a polycomb family member located in a region of chromosome 20 deleted in human myeloid malignancies. Proc Natl Acad Sci USA 101: 7341–7346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MY, Cai S, Espejo A, Bedford MT, Walker CL (2002) 14-3-3 interacts with the tumor suppressor tuberin at Akt phosphorylation site(s). Cancer Res 62: 6475–6480 [PubMed] [Google Scholar]
- Machev N et al. (2004) Sequence family variant loss from the AZFc interval of the human Y chromosome, but not gene copy loss, is strongly associated with male infertility. J Med Genet 41: 814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP (2003) The Tudor domain ‘Royal Family': Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci 28: 69–74 [DOI] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM (2003) Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev 17: 1823–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR III, Grant PA (2005) Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433: 434–438 [DOI] [PubMed] [Google Scholar]
- Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T (2004) Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119: 603–614 [DOI] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T (2004) A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 18: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M, Jackson SP (2004) Tudor domains track down DNA breaks. Nat Cell Biol 6: 1150–1152 [DOI] [PubMed] [Google Scholar]
- Tachibana M et al. (2002) G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16: 1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD (2005) WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121: 859–872 [DOI] [PubMed] [Google Scholar]
- Yoon HG, Chan DW, Reynolds AB, Qin J, Wong J (2003) N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol Cell 12: 723–734 [DOI] [PubMed] [Google Scholar]
- Zhang D, Yoon HG, Wong J (2005) JMJD2A is a novel N-CoR-interacting protein and is involved in repression of the human transcription factor achaete scute-like homologue 2 (ASCL2/Hash2). Mol Cell Biol 25: 6404–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


