Abstract
We studied the structural dynamics of chicken myosin V by combining the localization power of fluorescent imaging with one nanometer accuracy (FIONA) with the ability to detect angular changes of a fluorescent probe. The myosin V was labeled with bifunctional rhodamine on one of its calmodulin light chains. For every 74 nm translocation, the probe exhibited two reorientational motions, associated with alternating smaller and larger translational steps. Molecules previously identified as stepping alternatively 74-0 nm were found to actually step 64-10 nm. Additional tilting often occurred without full steps, possibly indicating flexibility of the attached myosin heads or probing of their vicinity. Processive myosin V molecules sometimes shifted from the top to the side of actin, possibly to avoid an obstacle. The data indicate marked adaptability of this molecular motor to a nonuniform local environment and provide strong support for a straight-neck model of myosin V in which the lever arm of the leading head is tilted backwards at the prepowerstoke angle.
Keywords: fluorescence, head-to-head coordination, myosin V, polarization, single molecule
Introduction
Myosin V is a two-headed motor protein responsible for actin-based intracellular transport and positioning of certain membrane bound vesicles, endoplasmic reticulum and mRNA-containing particles (Reck-Peterson et al, 2000; Vale, 2003). Recent technical advances in single-molecule biophysics have revealed various details about the mechanism of myosin V's activity. Total internal reflection fluorescence (TIRF) microscopy showed that myosin V is processive (exhibits dozens of mechanical steps before detaching from actin, Sakamoto et al, 2000) and optical trap experiments indicated that the molecule takes ∼37 nm strides toward the barbed end of actin (Mehta et al, 1999; Rief et al, 2000). More recent optical trap studies have shown that the 37-nm displacement is composed of an active stroke that is only ∼20–25 nm (Moore et al, 2001; Veigel et al, 2002); the remainder of the stride is presumably due to a thermally driven search for an appropriately oriented actin-binding site. Polarized TIRF microscopy of myosin V molecules labeled on their calmodulin (CaM) light chains detected tilting of the molecule's neck region on each step (Forkey et al, 2003), as expected if the light chain domain serves as a mechanical lever arm. Fluorescence imaging at one nanometer accuracy (FIONA) showed that the two heads step ‘hand-over-hand' to maintain association with actin during processive motility (Yildiz et al, 2003; Snyder et al, 2004). The trailing head is thought to translate 74 nm (twice the stride length) to become the leading head at the completion of each step (Yildiz et al, 2003; Churchman et al, 2005; Warshaw et al, 2005).
In the polarized TIRF and FIONA experiments, several populations of myosin V molecules were detected, presumably differing on which of the six light chain subunits on one of the heads was labeled with the fluorophore. In the FIONA study, alternating step distances of 52-23-52…, 42-33-42…, or 74-0-74… nm characterized each individual molecule. In some cases, the string of alternating distances was broken by a step with an unexpected value and then the alternating pattern would resume. In the polarized TIRF microscopy study, almost half of the molecules that were translocating did not seem to tilt whereas the other half tilted through a sufficient angular range (taking angular ambiguities in the experimental measurement into account) to explain the 37 nm stride length.
These observations led us to consider the two models for the structural configuration of the heads drawn in Figure 1. In the ‘straight-neck' model, the pivot for tilting of the CaM neck domain is between the CaM molecules and actin, that is within the motor domain, so that no major structural rearrangement takes place within the neck itself when the corresponding head makes a step. In this model, alternating large and small steps are expected and no 74-0-74-type molecules should arise, regardless of the particular CaM labeling site. Every CaM subunit should tilt on each step, again regardless of labeling position. In the ‘bent-neck' model (Figure 1B), the CaM nearest the motor domain is tilted toward the barbed end in both leading and trailing positions, and the structural changes (e.g., a strong kink) are farther from actin than the first light chain. This model predicts that 74-0-74-type molecules will not exhibit fluorophore tilting if the label is on a CaM situated below the kink (towards actin). The nontilting molecules in the polarized TIRF experiments were therefore grouped under the ‘bent-neck' model. Recent experiments with myosin V with eGFP on the head (Snyder et al, 2004), combined with the observations of Yildiz et al (2003) also led to speculation of a bend between the first and second light chains on the lever arm. Electron micrographs showed some myosin V molecules bound to actin with leading heads bent forward (Walker et al, 2000). Later detailed studies, however, showed that configurations with sharply bent CaM domains are rare, and that the majority of leading heads have their CaMs and the C-terminal region of their motor domains (the converter domains) tilted backward at the prepower-stroke position (Burgess et al, 2002), as in the ‘straight-neck' model.
Figure 1.
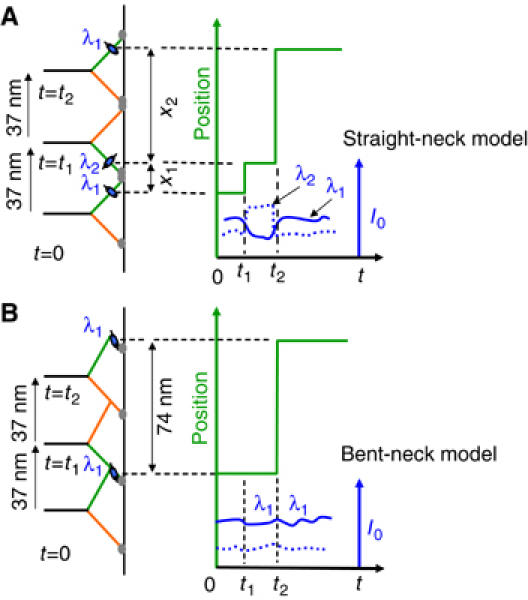
Cartoons illustrating (A) the straight-neck and (B) the bent-neck or telemark-skier models. According to this simple picture, the straight-neck model requires the fluorescent probe (blue oval) on calmodulin (i) to move a distance >0 and <74 nm every time the stalk moves 37 nm and (ii) to alter its orientation between some angle λ1 and another λ2 at each step. Therefore, every step must be accompanied by change in intensities of polarized fluorescence (I0, right side). In contrast, the bent-neck model predicts that when the labeled CaM is below the bend (i) the stepping pattern is 74-0 and (ii) no large variations occur in I0 since the probe orientation does not change upon a step.
Myosin V has been identified as a transport motor and/or tether for pigmented granules in mammalian melanocytes (Reck-Peterson et al, 2000) and amphibian melanophores (Rogers and Gelfand, 1998). It positions smooth endoplasmic reticulum in squid axoplasm (Tabb et al, 1998) and in dendridic spines of mouse brain Purkinje cells (Dekker-Ohno et al, 1996). In yeast, one myosin V, Myo2p, is involved in moving vacuolar and secretory vesicles while another, Myo4p, transports mRNA from mother to bud during cell division (Bertrand et al, 1998; Reck-Peterson et al, 2000). There is evidence that the yeast myosin V isoforms are nonprocessive (Reck-Peterson et al, 2001). Each of these roles requires specificity of cargo interaction and defined localization in a crowded in vivo environment. Although vesicles are transported over long distances along both microtubule and actin cytoskeletal filaments (Al-Haddad et al, 2001; Gross et al, 2002; Rodionov et al, 2003), the number of myosin V molecules actively transporting an individual vesicle within the cell may be as few as one or two in some instances (Snider et al, 2004). To transport a cargo to a specified region in the cell with various obstacles densely placed in its path (Heuser and Kirschner, 1980; Medalia et al, 2002), myosin V needs to display strongly processive motility but still maintain flexibility. It presumably has to alter its course along a filament, to switch to other filaments, and to function in cooperation with other cytoplasmic and vesicular transport proteins (Brown, 1999; Mallik and Gross, 2004). The complex milieu and behavior suggest that myosin V should possess multiple mechanisms that allow coordination not only between its two actin-binding heads but also with its surroundings.
Several biochemical, mechanical and structural experiments have elucidated details about head-to-head coordination. These studies have suggested that in double-headed attachment complexes with actin, intramolecular strain modulates the kinetics to maintain the mechano-chemical cycles of the heads out of synchrony so that both do not detach at the same time (De La Cruz et al, 1999; Baker et al, 2004; Coureux et al, 2004; Rosenfeld and Sweeney, 2004; Uemura et al, 2004; Veigel et al, 2005; Vilfan, 2005). Less is known about myosin V's interaction with components in its environment and its ability to adjust its path and negotiate through the complex and crowded cellular territory.
Each myosin V molecule is a dimer of two N-terminal globular ‘heads', two light chain domains, an α-helical coiled-coil, and a C-terminal domain that interacts with the cargo. The head binds actin, hydrolyzes ATP, and generates motion. The light chain domain or so-called ‘lever arm' has six consensus ‘IQ motifs' that, in chicken brain myosin V, probably bind an essential light chain (ELC) in position 1 (numbering from the motor domain) and one additional ELC and four CaM subunits in positions 2–5 (De La Cruz et al, 2000; Espindola et al, 2000; Terrak et al, 2005).
To establish a direct relationship between tilting and stepping of the CaMs and thus resolve whether the neck regions of actively translocating myosin V molecules bend, and to determine whether they play any role in adjusting the molecule's path along actin, it is necessary to measure nm-resolved translation and tilting simultaneously. Using the combined polarized TIRF and FIONA technique described here, we have simultaneously monitored the orientational and translational motions of individual fluorescent probes and studied the motions of bifunctional rhodamine (BR) rigidly attached to a CaM in the neck region of myosin V. The results strongly favor the ‘straight-neck' model (Figure 1A) and reveal various unexpected modes of motion that depart from the strict hand-over-hand alternating steps. These variations reveal new mechanisms that may enable myosin V to negotiate flexibly within its inhomogeneous environment and perform as an efficient intracellular transporter.
Results and discussion
Alternating steps and tilts
BR probes were attached at low stoichiometry to a CaM subunit on a whole myosin V, isolated from chicken brains. The two reactive groups of the dye rigidly attach it to the CaM, limiting its rotation relative to the protein. The orientation of the dye therefore reflects the orientation of the CaM. The labeled myosin V processively translocated along actin attached to the microscope slide surface at 150 or 500 nM ATP. Recordings of active myosin V were obtained in a prism-type TIRF microscope (see Materials and methods). The sample was illuminated in successive frames by an evanescent wave switched every 150 or 500 ms (depending on the [ATP]) between beams propagating in the laboratory x–z and y–z planes (z is the microscope optical axis) and with polarizations along the y- and x-axis, respectively (see Materials and methods for details). A CCD camera triggered the beam switching shutters and captured images of the rhodamine fluorescence. Following previous FIONA experiments (Yildiz et al, 2003; Snyder et al, 2004), images of individual fluorophores acquired in each CCD frame were fit with a two-dimensional Gaussian function to determine the position and emission intensity of the probe (see Supplementary Figure S2 and the accompanying text). The center of the Gaussian function fit to each image was marked as the probe position and the height of the fit above the background signal as its peak intensity, I0.
Figure 2A shows the position and intensity data of a BR-labeled myosin V translocating at 9.6 nm/s at [ATP]=150 nM. Data taken with excitations along x- and y-axis are shown in green and red symbols, respectively. Since the probe is excited with polarized beams, its fluorescence intensity depends on its orientation relative to the incident polarization. In Figure 2A, every step taken by the myosin V is associated with a large change of the normalized peak intensity, I0/I0Max, in one or both channels. The intensity variations are not due to blinking of the fluorophore since they are strongly correlated with the stepping of myosin V (dashed lines), and they often entail complementary changes in both intensity channels. Instead, the variations indicate tilting of the BR probe when the motor protein walks along actin. The anticorrelated two- to four-fold modulation in I0 suggests that the probe reorientations are mainly in the x–y plane with the motor walking along the side of the filament, which was aligned at 20° to the x-axis. Since the BR is tightly bound to the lever arm, the intensity changes indicate orientational changes of the labeled lever arm.
Figure 2.
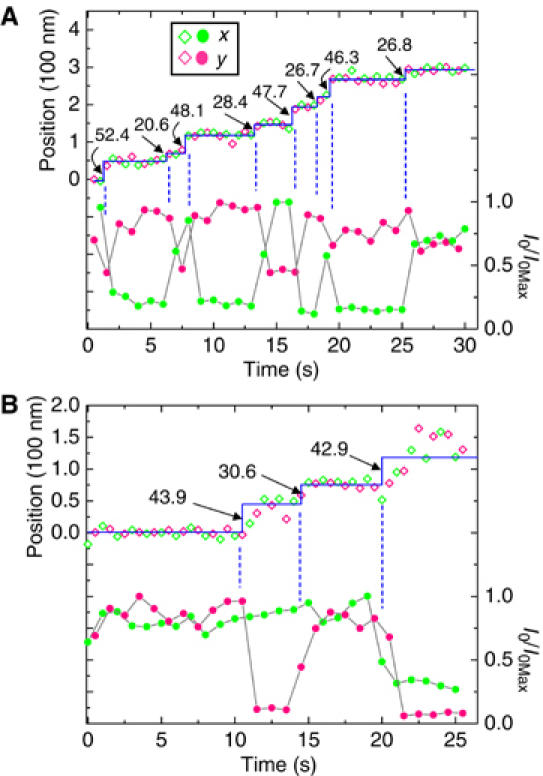
(A) Position (◊) and intensity (•) data of a myosin V tagged with a BR dye on one lever arm and measured with two polarized beams directed along the x- (green symbols) and y-axis (red symbols). I0Max is the maximum fluorescence intensity excited by the individual beams. The solid line is an intensity-weighted fit through the position data. Dashed vertical lines highlight the strong correlation between translational steps taken by the labeled lever arm and its orientational changes. In this case, the angular change occurs mostly in the x–y plane. (B) Another myosin molecule with BR bound to a different site on the neck. Positional and angular changes are again highly correlated. The probe tilts primarily in the x–z plane with the molecule walking along the x direction. The symbol key in (A) applies to both data sets.
Due to the tilting motions and the polarized illumination, two consecutive I0 values can be quite different (e.g., between 8 and 14 s). As a result, the precision in determining the probe's position, which depends on the number of photons collected (Yildiz et al, 2003), can vary between consecutive CCD frames (also see Supplementary Figure S2). In order to correctly account for the different precisions, the mean position within a period between transitions is determined by a weighted-average,
![]()
where ri denotes the position and I0i the corresponding intensities. The errors on the mean dwell positions in this trace, taking the weighting into account (Bevington, 1969), are <±2.5 nm. The solid staircases plotted in the figures are the intensity-weighted fits to the position data from both polarizations, yielding the indicated steps alternately averaging 49±3 and 25±3 nm in Figure 2A. These values are consistent with CaM labeling of the myosin on the fourth IQ position from the motor domain. The motor protein executes eight steps ‘hand-over-hand' in ∼30 s, covering a distance of 293 nm. No significant bias in the apparent position is associated with alternations of the excitation polarization in this example.
Data from another molecule, shown in Figure 2B, exhibit 43-30-type stepping and correlated intensity variations mainly for the signal from y-directed excitation. This behavior is most compatible with BR-CaM labeling at the fifth or sixth light chain position, and probe tilting mainly in the x–z plane. The actin in this example lays parallel to the x-axis.
We now return to the issue of ‘straight-neck' versus ‘bent-neck' models. Figure 3 shows four separate myosin V molecules with larger and smaller steps at [ATP]=150 nM (Figure 3A and B) and 500 nM (Figure 3C and D). Using the large intensity variations (dashed lines) as timing marks to identify steps shows that each rotation of the probe is associated with a step, alternating between 63±2 and 10±1 nm in panels A and B, and 65±3 and 12±2 nm in panels C and D. In the analysis used for our previous studies (Yildiz et al, 2003), the stepping pattern of these motors would be categorized as the 74-0 type. However, when the present simultaneously recorded polarized intensity data are used to mark transitions, the intensity-weighted average positions before and after each mark show that they are associated with steps. The accompanying probe reorientations implicate lever arm tilting and enhance the possibility of identifying the small steps.
Figure 3.
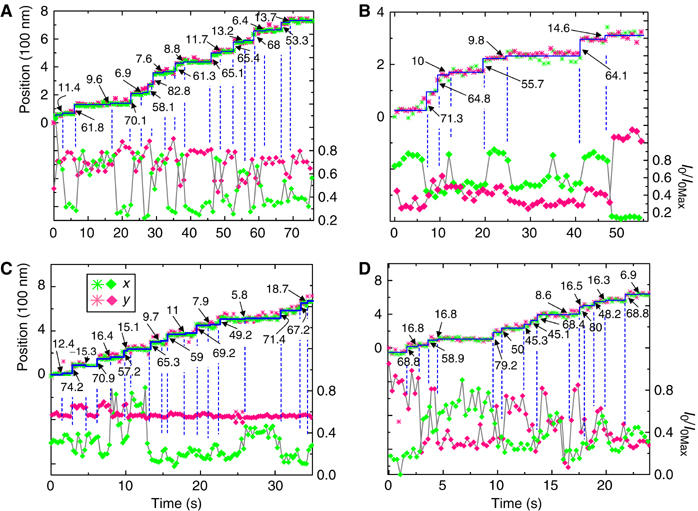
Position (*) and intensity (⧫) data at 150 nM ATP (A, B) and 500 nM ATP (C, D). Data in (A) and (B) are acquired every 0.5 s while that in (C) and (D) are taken every 0.15 s. The traces demonstrate that ‘74' nm steps observed previously with myosin V are actually composed of 64 and 10 nm steps. The large and small steps are marked next to the position traces. Although at low [ATP], the small steps can be usually identified from the position records alone, reliance on the polarization data becomes more critical at the higher [ATP] in (C) and (D). The records in (C) and (D) have been truncated for clarity. Dashed vertical lines mark the intensity changes that coincide with steps. The processivity directions for panels (A–D) are approximately 135°, 290°, 90°, and 0°, respectively, measured clockwise with respect to the microscope x-axis.
We have examined 68 such large and small steps that yielded overall mean values of 64.3±0.7 and 11.7±1 nm (±s.e.m.), respectively. In >90% of cases, both the ∼64 nm steps and the ∼10 nm steps in this category were statistically significant (P<0.01). As shown in Figure 1A, the larger of the two alternating step distances corresponds to a movement of the trailing head to the leading position. Thus, we identify the previously undetected small steps as displacements of the leading (nonstepping) lever arm when the (unlabeled) trailing head swings forward. The present data therefore show that the previously categorized 74-0 molecules actually follow a 64-10 stepping pattern. Moreover, application of a hidden Markov model to myosin V FIONA data acquired with unpolarized excitation also confirms that the alternating steps are indeed 64 and 10 nm (Dr Fred Sigworth, private communication). These molecules are likely to have the label in one of the two CaM positions closest to the motor domain. It should be pointed out that the sketch in Figure 1 is a simplified picture of processivity, showing the motor's configuration only in its state dwelling with both heads bound to actin. The time resolution of our experiments blurs out events faster than ∼200 ms, such as the multiple directed and diffusional motions that presumably take place during a step (Veigel et al, 2002; Spudich and Rock, 2002). Therefore, conformational changes of the lever arm that persist for <200 ms are not reported by our measurements.
For ∼200 individual myosin V molecules studied here, three major populations of stepping pattern: 44-30, 50-25, and 64-10 were evident, presumably corresponding to different labeled CaM positions. Assuming a straight lever arm 25 nm long, with six light chains (3.6 nm each) plus a 3 nm rotating converter domain, the expected stepping for a molecule tagged on the sixth, fourth and second CaM are 40-34, 51-23, and 61-13, very close to the three observed patterns. Each of the observed populations may also be a mixture of two subpopulations of myosin V labeled on adjacent sites.
Since the exact probe position is not known for each molecule, the bent-neck model with a kink between the first and second light chains is formally possible, if IQ position one is never exchanged. In that case, however, to span 36 nm, the part of the lever arm beyond the kink (⩽5 × 3.6 nm=18 nm) would lie almost parallel to the actin filament—an unlikely position. Thus, the lack of molecules with truly 0 nm steps and the observation that the lever arm reorients at virtually every step strongly support the ‘straight-neck' model for chicken myosin V, as shown in Figure 1A. This conclusion agrees with that from single particle electron micrographic image processing studies on mouse myosin V (Burgess et al, 2002).
Departures from strict hand-over-hand alternation
Azimuthal shift around actin. The recordings presented so far were clearly categorized into regularly alternating classes, such as 50-24, and represented probe reorientations mostly in one of three planes: x–y, x–z or y–z. Some motility records, however, show departures from both these traits. They display two or more sequential ⩾20 nm steps whose sizes lie outside the alternating pattern and whose polarizations imply a shift between two different planes.
The molecular motor that produced Figure 4A was labeled on the first or second CaM, judging by the stepping patterns at the beginning and end of the trace. Polarized excitation beams sometimes produce low fluorescence in one channel giving rise to increased noise and, occasionally, an apparent shift in the probe position. However, the intensity-weighted averaging procedure mentioned above guides the fit (solid line) through the data points with highest accuracy (largest numbers of photons) and allows determination of the position, 〈r〉w, to within ±4 nm. The motility starts with lever arm orientations largely in the x–z plane, as indicated by the direct correlation of the two intensity traces and the larger deflection of the beam propagating along the y-axis (red). At ∼10 s, the intensity traces suddenly become anticorrelated and the deflections in the other trace (green) increase, indicating that the orientation switched toward the x–y plane.
Figure 4.

Records at 500 nM ATP showing how simultaneous position and polarization information reveals myosin V moving from top to side of a actin filaments. Both molecules are 64-10 nm steppers but while the molecule in (A) swings right at ∼18 s, that in (B) turns left around actin at ∼52 s. Some steps are left unmarked for clarity. A sideways transition of the myosin leads to a jumbled stepping pattern and altered relative changes in fluorescent intensity (see text). After the transition is complete, the apparent step size returns but the position data has moved from one set of grid lines (solid) to another (dashed) that are shifted by 20 nm. Occasionally, probe localization experienced systematic error due to low fluorescence intensity with alternating input polarizations, displacing the position data (A). (C) Differences in (i) the layout of the actin binding sites and (ii) patterns of probe reorientations, for a myosin walking on top versus on the side of actin. The binding sites for the motor domains are 36 nm apart both on top (blue monomers) and on the side (red monomers) of actin. But owing to the helical structure of actin, the blue and red lattices are shifted relatively by four actin monomers, corresponding to 22 nm. A shift of three monomers of the binding sites, or 16.5 nm, is consistent with our data as well. After the myosin makes a right azimuthal turn around actin, the position data jumps from one lattice to the other undergoing a shift of ∼20 nm. The actin in (A) is aligned at 330° and that in (B) at 280°, with the x-axis. If the actin is laid along the x-axis (A), then the probe tilts largely in the x–z plane while the motor walks on top of actin but switches to an x–y tilting when the molecule makes the transition to the side.
The 56-40-50 nm steps between 12 and 22 s do not fit into the usual alternating large and small pattern (64-10-64…). As shown in the previous figures, alternating large and small steps are expected for a myosin walking in a fixed plane. But if the motor were to rotate 90° to the right around actin in this interval (viewed along actin toward the barbed end), then an extra 18 nm (half of the crossover length of actin) of stepping distance would be expected. In the cartoon in Figure 4C, left panel, it would mean a transition from the array of blue actin binding sites (marked by solid lines) to the array of red sites (marked by dashed lines), displaced by four actin monomers or ∼22 nm, due to the helical structure of actin. Indeed, an overlay of two 74 nm grids in Figure 4A shifted by 20 nm (solid and dashed lines) shows that the molecule overstepped by approximately this amount near 18 s. The normal step sizes are eventually re-established after the changeover, suggesting that the molecule walks relatively straight after it changes planes. The dye's polarization pattern, however, remains anticorrelated (in x–y plane) as expected from Figure 4C (right panel) for a motor that continues to walk on the side of actin. The motor in Figure 3D briefly shows similar deviation from its usual stepping behavior between 12 and 17 s with corresponding intensities changing from anticorrelated modulation (t<10 s) to correlated variations (15 s<t<22 s). The switch of the intensity pattern from anti- to directly correlated modulations signals the myosin's transition from the side to the top of the actin.
Figure 4B shows another example of a myosin V making a transition from the top of actin to one side. However, in this case, the transition is to the left, as evidenced by a shift of the motor's position data from the solid grid to the dashed grid set back by 20 nm. The sudden switch of BR probe's tilting at ∼52 s from the y–z plane to the x–y plane further corroborates rotation of the myosin around its substrate.
In contrast to our results, experiments on the motion of unconstrained myosin V have shown that the motor cargo typically traces a left-handed spiral (Ali et al, 2002) and that actin propelled in a gliding assay by myosin V also twirls leftward with a long pitch (Beausang et al, 2006). Our data, showing examples of left- and right-handed azimuthal swings, demonstrate adaptability of the protein during translocation. The abruptness of this motion, rather than along a smooth helical route, suggests that it may occur on an encounter with an obstacle in its preferred path.
‘Extra' reorientations. Some recordings of myosin V orientation showed clear rotational motions that were not associated with normal translational steps. We call these reorientation events ‘extra' since they were detected in addition to the expected lever arm motions that produced alternating 44-30, 50-25, or 64-10 nm types of stepping. For example, in Figure 5A, translational steps of 24, 48, and 26 nm suggest this molecule to be of the 50-25 type. All three steps are signaled by changes in the intensity signals (vertical lines). In addition, the probe unexpectedly reorients around 6 and 16 s (vertical arrows). Figure 5 is a gallery of traces containing such unexpected orientation changes. In each case, typical alternating steps with correlated tilting are marked by dashed vertical lines. Extra realignments of the lever arm are marked by vertical arrows. The extra reorientations are unambiguous as they produce complementary changes in I0 excited by the x- and y-polarized illumination. Corresponding displacements marked in the figure, though only ∼5 nm, are all statistically significant (P<0.05).
Figure 5.
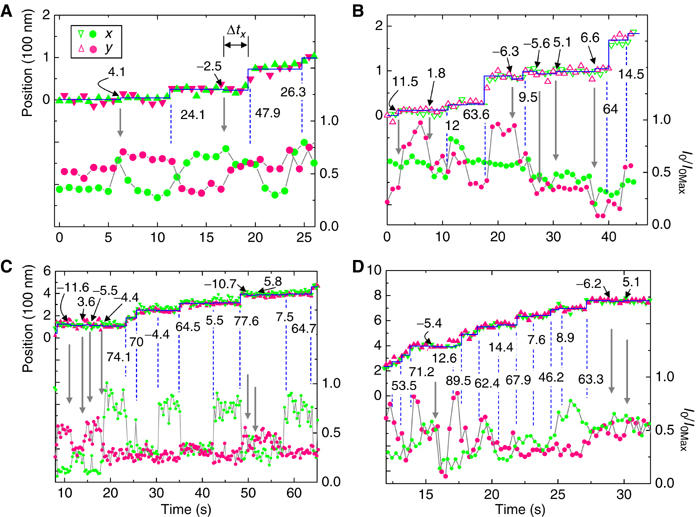
Traces from several recordings demonstrating angular motions of the lever arm beyond those directly linked to myosin stepping. Every full step taken by the proteins in (A–D) is accompanied by changes in polarized fluorescence intensity (dashed lines), signaling a change in the lever arm orientation. However, sometimes extra angular changes occur (vertical arrows) resulting in small (∼±5–10 nm) movements of the lever arm lasting for a short time, Δtx (shown in A). These forward and backward displacements are indicated above each position trace. The symbol legend in (A) applies to all four panels. (A , B) recorded at 150 nM ATP; (C, D) at 500 nM ATP.
The rotational motions associated with <10 nm steps have intensity changes as pronounced as those that occur with normal steps and were detected in ∼70% of the molecules studied. Two conventions were followed in identifying these events to insure conservative counting: (i) tilts were labeled as ‘extras' only after two regular tilts had been tallied for every 74 nm distance moved (i.e., after the entire recording had been classified under one of the three types of stepping), and (ii) only those events that cause the probe reorientation to persist for two or more video frames were accepted. It is unlikely that these extra realignments are caused by mobility of incompletely bound bifunctional probes because, according to mass spectrometric analysis (Forkey et al, 2003), >95% of the labeled CaMs had their two inserted Cys residues crosslinked by the bifunctional probe. Photophysical blinking of BR probes (Moerner, 1997) can also be ruled out since intensity changes from x- and y-polarized excitation are sometimes complementary. Electron microscopy (see Supplementary Figure S3) indicates that exchange of the native CaMs for labeled ones does not cause any major structural disruption to the myosin V molecule.
That the ∼±5 nm displacements are real shifts of the fluorescent probe's center-of-mass and not relics of random dye rotations is supported by the fact that regardless of the dye's plane of rotation (x–y, x–z, or y–z), the average movement is always the characteristic ±5 nm. One might expect that an artifactual effect of tilting on apparent position would cause the size of the displacements to strongly depend on the plane of probe reorientations. The dependence of the tilt kinetics on [ATP] and 2,3-butanedione monoxime (BDM) (only ‘extra' tilts see below) also makes these events very unlikely to be artifacts. The same arguments apply to the 10 nm motions in 64-10 molecules.
In order to relate the extra reorientations to the biochemical cycle of myosin V, we studied their dependence on ATP concentration and addition of BDM, a myosin inhibitor. Reducing [ATP] from 500 to 150 nM decreased the velocity of motility from 17.5 to 6 nm/s by increasing the waiting time until the next ATP bound. The effect of BDM on myosin is somewhat complex. McKillop et al (1994) found that BDM weakens muscle myosin's binding to actin. Ostap (2002) reported that 10 mM BDM does not inhibit myosin V ATPase, but Uemura et al (2004) found that 100 mM BDM inhibits the actomyosin V ATPase by slowing the release of ADP. We found that BDM reduces the average motility rate of myosin V in the TIRF assay by ∼20% at 150 nM ATP and ∼30% at 500 nM ATP, consistent with slowing of ADP release rate from ∼12.6 to ∼3.4 s−1.
The presence of BDM increased the fraction of extra tilt events per 37 nm translocation from ∼10 to ∼25% (not shown) at both 150 and 500 nM ATP. Histograms of the displacements that result from the extra reorientations at the two ATP concentrations and in the presence and absence of 100 mM BDM (Figure 6A and B here and Supplementary Figure S4A and B) show that, irrespective of [ATP] or [BDM], the displacements associated with the reorientations average ∼±5 nm. The data include all three populations of myosin V (54-20, 44-30, and 64-10 nm type molecules). It is interesting to note here that neighboring binding sites on actin are ∼5.5 nm apart. Identifying the trailing head as the one making the larger displacements, we find no correlation between the position of the labeled head (leading or trailing) and the direction of the 5 nm forward or backward extra steps. However, at both 150 and 500 nM ATP, the presence of BDM appears to raise the fraction of backwards displacements, perhaps by suppressing ADP release or by reducing myosin's affinity for actin (Figure 6 and Supplementary Figure S4).
Figure 6.
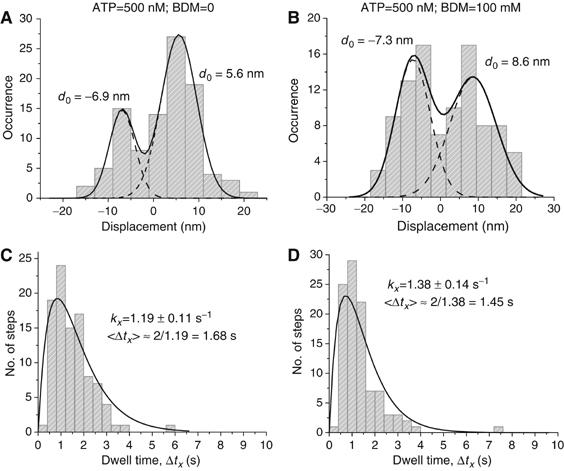
(A, B) Histograms of displacements detected from extra reorientation events at [ATP]=500 nM and [BDM]=0 or 100 mM. The data could be fitted by a sum of two Gaussian functions, yielding the indicated average displacement values, d0. On average, the lever arm movements due to the extra reorientations are ∼±5 nm with standard errors of the means ∼±0.8 nm. BDM appears to enhance the proportion of backward displacements. (C, D) Corresponding dwell times, Δtx, of the extra realignments shown in (A) and (B). Both sets can be well-fit with an expression, AkxΔtx exp(−kxΔtx), describing a convolution of two processes in series with equal rates. Here A is a normalization constant and kx is the rate constant used as a fitting parameter. The rate constant associated with the extra reorientations increases with [ATP] (compare with Supplementary Figure S4), correspondingly decreasing the average dwell time 〈Δtx〉. kx is higher than the ATPase rate at the two ATP concentrations considered.
After an extra tilt, the next event is either another extra tilt or a full step of the trailing head. The dwell time, Δtx, is defined as the time from the extra tilt until its termination (see Figure 5A). Histograms of Δtx (Figure 6C and D and Supplementary Figure S4C and D) are reasonably well fit by the expression, P(Δtx)∝Δtx exp (−kxΔtx), corresponding to a model in which two stochastic processes, with the same rate constant, are completed in series before the termination of the dwell time. A model involving two separate exponential components, with individually adjustable rate constants, did not fit the data better.
The rate constant, kx, determined from the fits, increases 3.3-fold with increasing ATP in the absence of BDM, and 2.8-fold at 100 mM BDM. At 150 nM ATP (Supplementary Figure S4), addition of BDM decreased kx by ∼35%, whereas at 500 nM ATP BDM increased kx by ∼15% (Figure 6). It is illuminating to compare these rate constants to the corresponding ATP binding rates, ∼0.14 s−1 at 150 nM ATP and ∼0.45 s−1 at 500 nM ATP (De la Cruz et al, 1999). The rate constants for the extra reorientations at the two ATP concentrations are 0.43 and 1.2 s−1 (Figure 6 and Supplementary Figure S4). Thus, the extra events are about three times shorter than the dwell time between steps that translocate myosin V upon nucleotide binding.
The dependence of the rate constant kx on [ATP] suggests that the duration of the extra tilts is linked to ATP binding, but they do not last as long as expected. We propose that the observed extra reorientations produce tiny shifts of the lever arm signaling myosin V testing its vicinity before taking the next full step. An average movement of ∼5 nm hints at hopping between neighboring actin sites. Such a mechanism might be used to detect obstacles in the protein's path, such as imperfections on actin, nonprocessive actin binding proteins, or other motors. In double-headed attachments, sustained tilting of the leading or trailing head is energetically unfavorable (Vilfan, 2005), but reversible rotational motions in the CaM light chain region might test the structural stability of the actomyosin complex, and thus could contribute to the coordination between the two heads to insure high processivity. A mechanism for testing the vicinity of the molecule and adjusting its path, such as the ∼90° azimuthal shifts described above, would aid the motor in carrying large cargoes, such as a membrane bound vesicle, through the viscous and inhomogeneous cytoplasm of a cell. As mentioned in the Introduction, roles ascribed to myosin V (Reck-Peterson et al, 2000), and the apparently very small numbers of molecules operating on an individual cargo particle (Snider et al, 2004) suggest the requirement for such agile processivity.
In summary, we have developed a technique for simultaneously measuring the position and orientation of a fluorescent probe. We investigated the structural dynamics of individual fluorescently labeled chicken myosin V molecules. The simultaneous measurements allowed detection of and differentiation between two major types of lever arm reorientations. The first kind of swing is coincident with steps that translocate the motor protein along actin. This class of orientational changes shows that the alternating 74-0 nm stepping pattern previously assigned to some myosin V molecules is actually a 64-10 nm pattern. The results provide strong support for a model of processive motility in which lever arm of the leading attached head is relatively straight and oriented at the prepower stroke angle. The correlated step and reorientation data also led to identification of transitions of the myosin from the top of the actin to the side, and vice versa. Our studies also revealed a second class of lever arm reorientations. Although often as large as the first kind, these orientational changes occur without producing myosin V stepping. Rather, these intriguing movements of the lever arm are associated with very small position changes. These events are likely linked to the motor protein probing its immediate surroundings prior to executing a full step. Previous studies have shown how coordination between the heads of myosin V helps the protein maintain a high duty ratio. Our experiments identify a new mechanism, in which the lever arms examine their local environment and probably contribute towards making myosin V a highly adaptable motor protein.
Materials and methods
Sample preparation
Construction of sample chamber is described in detail in Snyder et al (2004). Quartz microscope slides were used instead of glass in order to reduce background fluorescence from impurities. Whole myosin V was extracted from chick brain. CaM on the light chain domain of the proteins were labeled with single BR probes (average stoichiometry ∼0.4 Br-CaM per double-headed myosin V) as described by Forkey et al (2003). Rabbit muscle actin was purified according to Pardee and Spudich (1982). F-actin filaments were immobilized on the nitrocellulose-coated bottom surface (see Supplementary Figure S1B) of the quartz slide following Forkey et al (2003), and BR-labeled myosin V solution containing ATP and deoxygenation agents was flowed into the chamber. The protocol for preparing the final solution follows that of Yildiz et al (2003). Experiments with BDM and ATP were conducted on the same sample within 10 min after measurements with only ATP. BDM (Sigma, Lot 092K1722) was prepared as a 200 mM stock in KMg50-MOPS buffer (10 mM MOPS, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 1 mM DTT, final pH 7.0) and used within an hour (Ostap, 2002).
Polarized TIRF
Linearly polarized 532 nm laser light (CrystaLaser, Model GCL-075-L) was passed through a quarter-wave plate and split by a polarizing beam-splitter (see Supplementary Figure S1A). Each beam was then passed through a mechanical shutter (Uniblitz, Model LS672), a spatial filter, and a 5 × beam-expander. The expanded beams were then sent through a sheet polarizer in each optical pathway, producing two beams, each of power ≈9 mW, propagating in the x–z and y–z planes (z is the microscope optical axis), and with horizontal polarization. Each beam was focused through an octagonal prism onto the sample by two independently positioned lenses with focal lengths of 7 cm. Total internal reflection (TIR) of the polarized light at the quartz-sample interface was utilized to excite BR molecules attached to myosin V. The two beams entering the prism from the x and y directions produced polarizations of the evanescent waves along the y- and x-axis, respectively. Fluorescence was collected with a × 60 objective (Olympus, NA=1.2, water) on an inverted Olympus IX-70 microscope fitted with an additional × 1.5 magnifier. Images were captured with a frame transfer CCD camera (Micromax 512BFT, Roper Scientific, 512 × 512 × 13 μm pixels; or iXon DV887-BV, Andor Technology, 512 × 512 × 16 μm pixels). The excitation polarization was synchronized with the camera so that alternating frames captured images with one of the two polarized incident beams. Integration time for a frame was fixed at 0.5 s for 150 nM ATP and 0.15 s for 500 nM ATP in order to capture sufficient images during each dwell interval to determine the position and orientation. Details about the data acquisition and method of analyses are described in Yildiz et al (2003) and Snyder et al (2004). Briefly, the distribution of fluorescence emission of an individual molecule in each frame was fitted with a 2D Gaussian function and the molecule's position identified from the center of the function. The peak intensity of the fitted function was taken as the fluorescence intensity. Fitting was carried out in successive frames and the motility and orientation of the myosin V determined from the collective data.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Legends to Supplementary Figures
Acknowledgments
SS thanks Evan Graves and Ahmet Yildiz for many helpful discussions. We are grateful to Mr Huy Pham for preparation and exchange of the proteins. The work was supported by NIH Grants AR44420 and GM068625 to PRS and AR26846 to YEG. Support from the DOE, Division of Materials Sciences (under Award No. DEFG02-91ER45439), through the Frederick Seitz Materials Research Laboratory at the University of Illinois at Urbana-Champaign to PRS is acknowledged.
References
- Al-Haddad A, Shonn MA, Redlich B, Blocker A, Burkhardt JK, Yu H, Hammer JA III, Weiss DG, Steffen W, Griffiths G, Kuznetsov SA (2001) Myosin Va bound to phagosomes binds to F-actin and delays microtubule-dependent motility. Mol Biol Cell 12: 2742–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali MY, Uemura S, Adachi K, Itoh H, Kinosita K Jr, Ishiwata S (2002) Myosin V is a left-handed spiral motor on the right-handed actin helix. Nat Struct Biol 9: 464–467 [DOI] [PubMed] [Google Scholar]
- Baker JE, Krementsova EB, Kennedy GG, Armstrong A, Trybus KM, Warshaw DM (2004) Myosin V processivity: multiple kinetic pathways for head-to-head coordination. Proc Natl Acad Sci USA 101: 5542–5546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausang JF, Schroeder HW III, Gilmour JA, Goldman YE (2006) Twirling of actin by myosins II and V. Biophys J 90: 587a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM (1998) Localization of ASH1 mRNA particles in living yeast. Mol Cell 2: 437–445 [DOI] [PubMed] [Google Scholar]
- Bevington PR (1969) Data Reduction and Error Analysis for the Physical Sciences. New York, USA: McGraw-Hill Book Company [Google Scholar]
- Brown SS (1999) Cooperation between microtubule- and actin-based motor proteins. Annu Rev Cell Dev Biol 15: 63–80 [DOI] [PubMed] [Google Scholar]
- Burgess S, Walker M, Wang F, Sellers JR, White HD, Knight PJ, Trinick J (2002) The prepower stroke conformation of myosin V. J Cell Biol 159: 983–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Ökten Z, Rock RS, Dawson JF, Spudich JA (2005) Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proc Natl Acad Sci USA 102: 1419–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coureux PD, Sweeney HL, Houdusse A (2004) Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J 23: 4527–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz EM, Wells AL, Rosenfeld SS, Ostap EM, Sweeney HL (1999) The kinetic mechanism of myosin V. Proc Natl Acad Sci USA 96: 13726–13731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz EM, Wells AL, Sweeney HL, Ostap EM (2000) Actin and light chain isoform dependence of myosin V kinetics. Biochemistry 39: 14196–14202 [DOI] [PubMed] [Google Scholar]
- Dekker-Ohno K, Hayasaka S, Takagishi Y, Oda S, Wakasugi N, Mikoshiba K, Inouye M, Yamamura H (1996) Endoplasmic reticulum is missing in dendritic spines of Purkinje cells of the ataxic mutant rat. Brain Res 714: 226–230 [DOI] [PubMed] [Google Scholar]
- Espindola FS, Suter DM, Partata LB, Cao T, Wolenski JS, Cheney RE, King SM, Mooseker MS (2000) The light chain composition of chicken brain myosin-Va: calmodulin, myosin-II essential light chains, and 8-kDa dynein light chain/PIN. Cell Motil Cytoskeleton 47: 269–281 [DOI] [PubMed] [Google Scholar]
- Forkey JN, Quinlan ME, Shaw M, Corrie JET, Goldman YE (2003) Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization. Nature 422: 399–404 [DOI] [PubMed] [Google Scholar]
- Gross SP, Tuma MC, Deacon SW, Serpinskaya AS, Reilein AR, Gelfand VI (2002) Interactions and regulation of molecular motors in Xenopus melanophores. J Cell Biol 156: 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser JE, Kirschner MW (1980) Filament organization revealed in platinum replicas of freeze-dried cytoskeletons. J Cell Biol 86: 212–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallik R, Gross SP (2004) Molecular motors: strategies to get along. Curr Biol 14: R971–R982 [DOI] [PubMed] [Google Scholar]
- McKillop DFA, Fortune NS, Ranatunga KW, Geeves MA (1994) The influence of 2,3-butanedione 2-monoxime (BDM) on the interaction between actin and myosin in solution and in skinned muscle fibres. J Muscle Res Cell Motil 15: 309–318 [DOI] [PubMed] [Google Scholar]
- Medalia O, Weber I, Frangakis AS, Nicastro D, Gerisch G, Baumeister W (2002) Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science 298: 1209–1213 [DOI] [PubMed] [Google Scholar]
- Mehta AD, Rock RS, Rief M, Spudich JA, Mooseker MS, Cheney RE (1999) Myosin-V is a processive actin-based motor. Nature 400: 590–593 [DOI] [PubMed] [Google Scholar]
- Moerner WE (1997) Polymer luminescence: those blinking single molecules. Science 277: 1059–1060 [Google Scholar]
- Moore JR, Krementsova EB, Trybus KM, Warshaw DM (2001) Myosin V exhibits a high duty cycle and large unitary displacement. J Cell Biol 155: 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostap EM (2002) 2,3-Butanedione monoxime (BDM) as a myosin inhibitor. J Muscle Res Cell Motil 23: 305–308 [DOI] [PubMed] [Google Scholar]
- Pardee JD, Spudich JA (1982) Purification of muscle actin. Methods Cell Biol 24: 271–289 [DOI] [PubMed] [Google Scholar]
- Reck-Peterson SL, Provance DW Jr, Mooseker MS, Mercer JA (2000) Class V myosins. Biochim Biophys Acta 1496: 36–51 [DOI] [PubMed] [Google Scholar]
- Reck-Peterson SL, Tyska MJ, Novick PJ, Mooseker MS (2001) The yeast class V myosins, Myo2p and Myo4p, are nonprocessive actin-based motors. J Cell Biol 153: 1121–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rief M, Rock RS, Mehta AD, Mooseker MS, Cheney RE, Spudich JA (2000) Myosin-V stepping kinetics: a molecular model for processivity. Proc Natl Acad Sci USA 97: 9482–9486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov V, Yi J, Kashina A, Oladipo A, Gross SP (2003) Switching between microtubule- and actin-based transport systems in melanophores is controlled by cAMP levels. Curr Biol 13: 1837–1847 [DOI] [PubMed] [Google Scholar]
- Rogers SL, Gelfand VI (1998) Myosin cooperates with microtubule motors during organelle transport in melanophores. Curr Biol 8: 161–164 [DOI] [PubMed] [Google Scholar]
- Rosenfeld SS, Sweeney HL (2004) A model of myosin V processivity. J Biol Chem 279: 40100–40111 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Amitani I, Yokota E, Ando T (2000) Direct observation of processive movement by individual myosin V molecules. Biochem Biophys Res Commun 272: 586–590 [DOI] [PubMed] [Google Scholar]
- Snider J, Lin F, Zahedi N, Rodionov V, Yu CC, Gross SP (2004) Intracellular actin-based transport: how far you go depends on how often you switch. Proc Natl Acad Sci USA 101: 13204–13209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder GE, Sakamoto T, Hammer JA III, Sellers JR, Selvin PR (2004) Nanometer localization of single green fluorescent proteins: evidence that myosin V walks hand over-hand via telemark configuration. Biophys J 87: 1776–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA, Rock RS (2002) A crossbridge too far. Nat Cell Biol 4: E8–E10 [DOI] [PubMed] [Google Scholar]
- Tabb JS, Molyneaux BJ, Cohen DL, Kuznetsov SA, Langford GM (1998) Transport of ER vesicles on actin filaments in neurons by myosin V. J Cell Sci 111: 3221–3234 [DOI] [PubMed] [Google Scholar]
- Terrak M, Rebowski G, Lu RC, Grabarek Z, Dominguez R (2005) Structure of the light chain-binding domain of myosin V. Proc Natl Acad Sci USA 102: 12718–12723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura S, Higuchi H, Olivares AO, De La Cruz EM, Ishiwata S (2004) Mechanochemical coupling of two substeps in a single myosin V motor. Nat Struct Mol Biol 11: 877–883 [DOI] [PubMed] [Google Scholar]
- Vale RD (2003) The molecular motor toolbox for intracellular transport. Cell 112: 467–480 [DOI] [PubMed] [Google Scholar]
- Veigel C, Schmitz S, Wang F, Sellers JR (2005) Load-dependent kinetics of myosin-V can explain its high processivity. Nat Cell Biol 7: 861–869 [DOI] [PubMed] [Google Scholar]
- Veigel C, Wang F, Bartoo ML, Sellers JR, Molloy JE (2002) The gated gait of the processive molecular motor, myosin V. Nat Cell Biol 4: 59–65 [DOI] [PubMed] [Google Scholar]
- Vilfan A (2005) Elastic lever-arm model for myosin V. Biophys J 88: 3792–3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker ML, Burgess SA, Sellers JR, Wang F, Hammer JA III, Trinick J, Knight PJ (2000) Two-headed binding of a processive myosin to F-actin. Nature 405: 804–807 [DOI] [PubMed] [Google Scholar]
- Warshaw DM, Kennedy GG, Work SS, Krementsova EB, Beck S, Trybus KM (2005) Differential labeling of myosin V heads with quantum dots allows direct visualization of hand-over-hand processivity. Biophys J 88: L30–L32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR (2003) Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science 300: 2061–2065 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Legends to Supplementary Figures


