Abstract
Background
The proportion of muscle fibre types and their size affect muscularity as well as functional properties of the musculature and meat quality. We aimed to identify QTL for microstructural muscle properties including muscle fibre size, their numbers and fibre type proportions as well as biophysical parameters of meat quality and traits related to body composition, i.e. pH, conductivity, area of M. longissimus dorsi and lean meat content. A QTL scan was conducted in a porcine experimental population that is based on Duroc and Berlin Miniature Pig.
Results
Least square regression interval mapping revealed five significant and 42 suggestive QTL for traits related to muscle fibre composition under the line-cross model as well as eight significant and 40 suggestive QTL under the half-sib model. For traits related to body composition and biophysical parameters of meat quality five and twelve significant plus nine and 22 suggestive QTL were found under the line-cross and half-sib model, respectively. Regions with either significant QTL for muscle fibre traits or significant QTL for meat quality and muscularity or both were detected on SSC1, 2, 3, 4, 5, 13, 14, 15, and 16. QTL for microstructural properties explained a larger proportion of variance than did QTL for meat quality and body composition.
Conclusion
Microstructural properties of pig muscle and meat quality are governed by genetic variation at many loci distributed throughout the genome. QTL analysis under both, the line-cross and half-sib model, allows detecting QTL in case of fixation or segregation of the QTL alleles among the founder populations and thus provide comprehensive insight into the genetic variation of the traits under investigation. Genomic regions affecting complex traits of muscularity and meat quality as well as microstructural properties might point to QTL that in first instance affect muscle fibre traits and by this in second instance meat quality. Disentangling complex traits in their constituent phenotypes might facilitate the identification of QTL and the elucidation of the pleiotropic nature of QTL effects.
Background
The muscular system is the most prominent active part of the locomotor apparatus and accounts for most of the body mass. Muscle tissue largely contributes to the metabolism as well: muscle represents a major site of daily energy consumption, thermoregulation and storage of energy and amino acids. As an important source of proteins for human nutrition, skeletal muscle of farm animals is of interest. Each muscle consists of three main fibre types, slow-twitch oxidative, fast-twitch oxido-glycolytic and fast-twitch glycolytic fibres [1], which are characterised by different microstructural, biochemical and metabolic properties. The number and size of the muscle fibres are major factors determining growth and weight of each muscle. In man, muscle fibre type composition varies considerably due to inherited factors and environmental effects that contribute to phenotypic variation at 45 and 40%, respectively, while the remaining 15% is explained by the error component related to muscle sampling and technical variance [2]. The proportion of type I and II fibres (slow and fast twitch fibres) has been related to insulin resistance and diabetes II predisposition, obesity and body mass index, as well as fat catabolism and capacity to gain or loss weight, i.e. the constitution [3-5]. Muscle fibre composition is to some extent predictive of obesity [6].
Correspondingly, the proportion of muscle fibre types is correlated to growth performance, meatiness and fatness and affects post mortem development of muscle to meat and thus meat quality traits in pigs [7-10]. The rate and extent of post-mortem pH decline is higher in fast-twitch glycolytic muscles with higher glycogen content. Cross sectional area of muscle fibres is negatively correlated to meat tenderness [7-10]. In pig, cattle, mouse, sheep, and horse coefficients of heritability for muscle structure traits were estimated to lie between h2 = 0.20 and 0.60 [10-14].
QTL for muscle fibre traits in pigs were detected on chromosome 8 and chromosomes 1, 2, 6, 14 and X [15,16]. Here we report on the identification of QTL for micro-structural muscle properties, i.e. fibre number, fibre size and fibre type proportions including proportion of defect fibres in a segregating population of pigs derived from Duroc and Berlin Miniature Pig, i.e. breeds being divergent with regard to traits related to growth, muscularity and fatness. Moreover, we identified QTL for traits related to body composition as well as biophysical muscle properties that are technological parameters of meat quality in pig breeding.
Results
Traits measured and markers genotyped are detailed in Tables 1 and 2. Results of the QTL analyses are compiled in Tables 3 to 6, in which all QTL exceeding the 5% chromosome-wide significance threshold for suggestive linkage are included.
Table 1.
Means and standard deviations of traits related to muscle fibre size and distribution, muscularity and meat quality measured in M. longissimus dorsi at 13th/14th rib on F2 animals of the DUMI resource population
| Trait | Definition | Mean ± SD | |
| 1 | Fib/mm2 | mean number of fibres per mm2 | 366 ± 99 |
| ToF# | total number of fibres at section of M. l. d. [×1000] | 803.5 ± 203.5 | |
| Diamean | mean diameter of all fibres [μm] | 58.52 ± 7.93 | |
| DiaAnF | mean diameter of angular fibres [μm] | 23.27 ± 7.09 | |
| DiaGiF | mean diameter of giant fibres [μm] | 14.05 ± 34.47 | |
| ProAnF | relative proportion of angular fibres [%] | 0.90 ± 0.94 | |
| ProGiF | relative proportion of giant fibres [%] | 0.05 ± 0.19 | |
| DiaSTO/red | mean diameter of STO/red fibres [μm] | 50.98 ± 7.13 | |
| DiaFTO/im | mean diameter of FTO/intermediate fibres [μm] | 50.19 ± 8.24 | |
| DiaFTG/w | mean diameter of FTG/white fibres [μm] | 62.32 ± 9.32 | |
| ProSTO/red | relative proportion of STO/red fibres [%] | 18.68 ± 6.19 | |
| ProFTO/im | relative proportion of FTO/intermediate fibres [%] | 11.34 ± 4.23 | |
| ProFTG/w | relative proportion of FTG/white fibres [%] | 69.02 ± 6.49 | |
| 2 | DiaSTO | mean diameter of STO fibres [μm] | 51.60 ± 7.01 |
| DiaFTO | mean diameter of FTO fibres [μm] | 49.31 ± 7.44 | |
| DiaFTG | mean diameter of FTG fibres [μm] | 61.91 ± 9.19 | |
| ProSTO | relative proportion of red fibres [%] | 15.93 ± 5.40 | |
| ProFTO | relative proportion of intermediate fibres [%] | 12.35 ± 4.34 | |
| ProFTG | relative proportion of white fibres [%] | 71.34 ± 6.90 | |
| CapSTO | mean number of capillaries at STO fibre | 1.48 ± 0.46 | |
| CapFTO | mean number of capillaries at FTO fibre | 0.96 ± 0.34 | |
| CapFTG | mean number of capillaries at FTG fibre | 0.55 ± 0.17 | |
| Capmean | mean number of capillaries at fibres | 0.75 ± 0.23 | |
| Cap/mm2 | mean number of capillaries per mm2 | 82.19 ± 21.86 | |
| 3 | Diared | mean diameter of red fibres [μm] | 50.47 ± 7.21 |
| Diaim | mean diameter of intermediate fibres [μm] | 50.91 ± 8.81 | |
| Diaw | mean diameter of white fibres [μm] | 62.66 ± 9.44 | |
| Prored | relative proportion of red fibres [%] | 20.95 ± 5.90 | |
| Proim | relative proportion of intermediate fibres [%] | 10.50 ± 3.98 | |
| Prow | relative proportion of white fibres [%] | 67.11 ± 5.45 | |
| 4 | FOM | Fat-O-Meater: lean meat content according to regression | 33.74 ± 8.62 |
| FOM_M | FOM_meat: depth of M. glutaeus medius | 40.01 ± 8.71 | |
| MAML | loin eye area: area of M. l. d. at 13th/14th rib [cm2] | 23.30 ± 4.38 | |
| pH1ML | pH1-loin: pH-value 45 min post mortem | 6.46 ± 0.25 | |
| pH24ML | pH24-loin: pH-value 24 hours post mortem | 5.56 ± 0.13 | |
| C1ML | conductivity1-loin: conductivity 45 min post mortem [mS/cm] | 3.44 ± 0.53 | |
| C24ML | conductivity24-loin: conductivity 24 hours post mortem [mS/cm] | 3.49 ± 1.22 | |
| MCOpto | meat colour, Opto Star: meat colour 24 hours post [%] | 69.40 ± 6.29 |
1fibre number and size, fibre type proportions determined by NADH-TR alone and combined NADH-TR/ATPase reaction (n = 308)
2fibre size, fibre type proportions determined by combined NADH-TR/ATPase reaction (n = 140)
3fibre size, fibre type proportions determined by NADH-TR reaction (n = 168)
4carcass and meat quality traits of standard performance test (n = 863) [ZDS]
Table 2.
Markers used in the QTL analysis and genetic map as established for the DUMI resource population (sex average, Kosambi cM)
| Chromosome | Coverage1 [cM] | Markers and genetic distances [cM] | ||||||||
| SSC1 | 16.4 – 140.5 (144.0) | SW1515 | 33.0 | SW1851 | 33.4 | S0155 | 8.3 | RLN* | 61.6 | SW1301 |
| SSC2 | 0.0 – 74.82 (132.1) | SW2443 | 42.7 | FTH1* | 20.2 | SW240 | 23.0 | STS2* | 13.8 | C3* |
| 18.3 | SW1564 | 14.2 | BHMT* | 13.5 | S0226 | |||||
| SSC3 | 17.8 – 102.2 (129.3) | SW72 | 50.8 | S0164 | 25.7 | SW2570 | 33.2 | S0002 | ||
| SSC4 | 4.1 – 120.0 (130.1) | S0227 | 47.2 | S0001 | 14.3 | STS3* | 4.6 | CRH* | 1.3 | STS1* |
| 1.5 | STS4* | 28.3 | S0214 | 36.6 | S0097 | |||||
| SSC5 | 8.4 – 102.9 (114.4) | SW1482 | 59.3 | SW1134 | 40.0 | IGF1 | 34.6 | SW378 | ||
| SSC6 | 7.3 – 102.0 (165.7)3 | S0035 | 24.1 | HP* | 39.4 | S0087 | 10.4 | SW1067 | 9.7 | SW193 |
| 5.8 | S0300 | 5.9 | TGFB1* | 26.9 | S0220 | 35.0 | LEP* | |||
| 8.5 | S0059 | 16.7 | S0003 | |||||||
| SSC7 | 3.7 – 134.9 (156.6) | S0025 | 24.4 | S0064 | 23.5 | DQB* | 23.5 | BF* | 15.5 | S0102 |
| 15.8 | SW175 | 36.2 | S0115 | 30.2 | S0101 | |||||
| SSC8 | 0.0 – 112.3 (127.7) | SW2410 | 79.5 | S0086 | 24.7 | S0144 | 23.8 | SW61 | ||
| SSC9 | 11.1 – 96.5 (138.5) | SW21 | 26.3 | SW911 | 33.1 | SW54 | 17.1 | S0109 | 32.9 | S0295 |
| SSC10 | 0. 0 – 124.1 (124.1) | SW830 | 77.7 | S0070 | 49,3 | ITIH2* | 36.1 | SW2067 | ||
| SSC11 | 14.1 – 76.2 (84.9) | SW2008 | 32.0 | S0071 | 32.0 | S0386 | 34.0 | SW703 | ||
| SSC12 | 6.6 – 108.3 (113.1) | S0143 | 49.6 | SW874 | 42.6 | SW605 | ||||
| SSC13 | 1.6 – 79.3 (126.2) | S0219 | 40.0 | SW344 | 36.6 | SW398 | ||||
| SSC14 | 7.4 – 111.5 (111.5) | SW857 | 53.4 | S0007 | 30.4 | VIN* | 37.8 | SWC27 | ||
| SSC15 | 1.3 – 107.4 (111.8) | S0355 | 35.3 | SW1111 | 48.5 | SW936 | 33.5 | SW1119 | ||
| SSC16 | 0.0 – 92.6 (93.2) | S0111 | 51.2 | S0026 | 42.4 | S0061 | ||||
| SSC17 | 0.0 – 94.0 (97.0) | SW335 | 34.6 | SW840 | 35.1 | SW2431 | ||||
| SSC18 | 5.0 – 57.6 (57.6) | SW1023 | 23.9 | SW787 | 43.0 | SWR414 | ||||
1relative position of flanking markers of the set used in the present study on public linkage map (USDA-MARC v2); 2S0226 not covered by USDA-MARC v2, but SW14, which is closely linked to S0226 (PigMaP v1.5); 3S0035 at 0.0 cM and S0003 at 144.5 cM in the International Workshop 1 SSC6 integrated map with a total length of 166.0 cM; * biallelic markers
Table 3.
Evidence for QTL significant at the 5% chromosome-wide level for traits related to muscle fibre distribution by chromosome obtained by F2 analysis. Estimated significance levels (F-value), position, % of F2 variance explained by each QTL, and gene effects.
| Trait | SSC | Position [cM] | F- Value | % Variance1 | Additive | Dominance | ||
| Effect2 | S.E. | Effect2 | S.E. | |||||
| DiaAnF | 1 | 3 | 8.4*** | 11.8 | 2.64 | 1.39 | -7.04 | 1.80 |
| ProSTO | 1 | 93 | 5.5* | 10.4 | -0.99 | 1.45 | 12.97 | 3.99 |
| DiaFTG | 1 | 114 | 5.0* | 9.5 | -5.86 | 2.53 | 11.77 | 5.47 |
| DiaFTG | 2 | 63 | 9.4*** | 17.8 | -7.40 | 1.90 | -10.07 | 5.91 |
| DiaFTO | 2 | 63 | 4.7* | 8.8 | -4.86 | 1.68 | -4.86 | 5.06 |
| Diamean | 2 | 66 | 9.5*** | 17.9 | -7.04 | 1.78 | -8.85 | 5.58 |
| ToF# | 2 | 141 | 5.9* | 4.9 | -78.2 | 23.7 | 37.5 | 37.7 |
| Fib/mm2 | 2 | 145 | 6.3* | 4.4 | -34.0 | 9.7 | 67.8 | 14.3 |
| ProGiF | 3 | 0 | 6.0* | 11.2 | 0.03 | 0.02 | -0.09 | 0.03 |
| Fib/mm2 | 3 | 37 | 6.9** | 3.7 | -51.5 | 14.1 | -36.9 | 27.2 |
| CapSTO | 3 | 57 | 5.7* | 10.7 | 0.07 | 0.07 | 0.34 | 0.11 |
| ProFTG | 4 | 18 | 6.4* | 10.7 | -2.47 | 1.35 | -5.76 | 2.58 |
| ProFTG/w | 4 | 26 | 7.8** | 5.7 | -2.98 | 0.77 | -1.00 | 1.57 |
| Prow | 4 | 33 | 5.9* | 9.3 | -2.66 | 0.97 | 2.93 | 1.73 |
| DiaFTG/w | 4 | 79 | 4.9* | 3.2 | 2.38 | 1.56 | 5.54 | 3.04 |
| Fib/mm2 | 4 | 96 | 5.4* | 10.2 | 4.11 | 15.11 | -53.30 | 17.83 |
| Diamean | 4 | 96 | 7.6** | 14.2 | 0.08 | 1.89 | 7.52 | 2.23 |
| DiaFTG | 4 | 96 | 8.6*** | 16.3 | 0.57 | 2.14 | 8.82 | 2.53 |
| Diared | 5 | 0 | 6.4* | 10.2 | 1.52 | 2.08 | -8.85 | 3.45 |
| Diared | 7 | 168 | 5.6* | 8.9 | 2.67 | 0.93 | 2.89 | 1.43 |
| DiaSTO | 9 | 108 | 4.3* | 8.0 | 2.84 | 0.99 | 0.37 | 1.36 |
| Capmean | 10 | 54 | 5.2* | 9.8 | -0.07 | 0.05 | -0.32 | -0.19 |
| CapSTO | 10 | 57 | 6.4** | 12.1 | -0.21 | 0.08 | -0.49 | 0.18 |
| Proim | 11 | 21 | 4.2* | 6.7 | 1.79 | 0.67 | 0.84 | 1.26 |
| ProAnF | 11 | 69 | 4.9* | 9.2 | 0.01 | 0.14 | -0.90 | 0.29 |
| Fib/mm2 | 12 | 90 | 4.3* | 6.8 | -40.1 | 16.9 | -40.5 | 24.6 |
| Diared | 12 | 90 | 5.6* | 8.9 | 2.89 | 1.07 | 3.03 | 1.57 |
| ToF# | 12 | 92 | 6.0** | 9.7 | -92.3 | 32.8 | -92.1 | 45.1 |
| DiaSTO/red | 12 | 92 | 4.6* | 3.0 | 1.85 | 0.72 | 1.71 | 0.98 |
| Prored | 14 | 51 | 7.3** | 11.6 | -3.87 | 1.23 | 3.11 | 1.99 |
| Prow | 14 | 54 | 4.3* | 6.6 | 1.78 | 1.14 | -3.72 | 1.74 |
| ProSTO/red | 14 | 54 | 6.1* | 4.2 | -1.99 | 0.71 | 1.61 | 1.20 |
| Fib/mm2 | 14 | 93 | 4.9* | 7.9 | -24.8 | 28.6 | 20.0 | 69.4 |
| ToF# | 14 | 98 | 7.6** | 12.4 | -19.9 | 54.2 | 461.3 | 120.6 |
| Diaim | 14 | 102 | 4.2* | 6.8 | -0.57 | 1.95 | -11.89 | 4.09 |
| Diaw | 14 | 102 | 7.9*** | 12.7 | 1.60 | 1.96 | -15.65 | 4.09 |
| ToF# | 15 | 56 | 5.7* | 10.8 | -98.3 | 29.0 | -7.1 | 66.9 |
| ProAnF | 15 | 57 | 5.7* | 9.2 | 0.01 | 0.22 | -1.66 | 0.51 |
| DiaGiF | 15 | 69 | 5.7** | 9.1 | 21.66 | 7.11 | 21.37 | 14.11 |
| DiaSTO | 15 | 81 | 5.4* | 10.2 | 3.15 | 1.06 | -1.98 | 1.64 |
| DiaFTO | 15 | 84 | 5.8** | 11.0 | 2.70 | 1.08 | -3.48 | 1.58 |
| ProGiF | 16 | 93 | 4.5* | 8.5 | -0.05 | 0.02 | -0.04 | 0.03 |
| Diamean | 18 | 0 | 4.5* | 8.5 | -2.94 | 1.02 | -1.72 | 1.56 |
| DiaSTO | 18 | 0 | 5.6* | 10.6 | -2.87 | 0.88 | -1.33 | 1.34 |
| DiaFTO | 18 | 0 | 4.7* | 8.9 | -2.47 | 0.96 | -2.74 | 1.47 |
| DiaSTO/red | 18 | 0 | 4.8* | 3.2 | -2.00 | 0.68 | -1.24 | 0.99 |
| ProGiF | 18 | 48 | 5.5* | 10.4 | -0.06 | 0.03 | -0.13 | 0.05 |
*: significant at the 5% chromosome-wide level, **: significant at the 1% chromosome-wide level, ***: significant at the 5% genome-wide level
1the fraction of phenotypic variance in the F2 explained by a QTL; calculated as the proportion of residual variance of the statistical models with and without the QTL effect
2positive values of additive genetic and dominance effects imply higher trait values forced by the Duroc allele;
Table 6.
Evidence for QTL significant at the 5% chromosome-wide level for traits related to muscularity and meat quality by chromosome obtained by half-sib analysis. Estimated significance levels (F-value), position, % of F2 variance explained by each QTL, and gene effects.
| Trait | SSC | Position [cM] | F-Value | % Variance |
| pH1ML | 1 | 99 | 3.5* | 2.1 |
| FOM | 2 | 20 | 6.6**** | 3.8 |
| MAML | 2 | 20 | 10.0**** | 5.8 |
| FOM_M | 2 | 20 | 2.7* | 1.6 |
| C1ML | 2 | 24 | 3.8** | 1.8 |
| MCOpto | 3 | 0 | 7.6**** | 5.6 |
| pH1ML | 3 | 24 | 3.0* | 1.7 |
| pH1ML | 4 | 12 | 3.3* | 1.9 |
| MAML | 4 | 71 | 3.2* | 1.9 |
| FOM | 4 | 73 | 5.7**** | 3.3 |
| MCOpto | 5 | 9 | 6.6*** | 4.9 |
| FOM | 6 | 0 | 4.7*** | 2.7 |
| C24ML | 6 | 24 | 4.6* | 2.7 |
| MAML | 6 | 32 | 5.4**** | 3.1 |
| MCOpto | 6 | 170 | 4.5* | 3.3 |
| pH1ML | 8 | 128 | 3.3* | 2.0 |
| MCOpto | 10 | 128 | 2.9* | 1.7 |
| C24ML | 11 | 36 | 3.6* | 2.1 |
| pH1ML | 11 | 72 | 3.0* | 1.7 |
| pH24ML | 13 | 0 | 3.4* | 2.0 |
| MCOpto | 13 | 43 | 8.0**** | 5.9 |
| FOM_M | 14 | 56 | 2.7* | 1.6 |
| C24ML | 14 | 120 | 3.9* | 2.2 |
| pH24ML | 15 | 48 | 5.4*** | 3.1 |
| C1ML | 15 | 88 | 3.0* | 1.4 |
| FOM | 16 | 0 | 5.2**** | 3.0 |
| FOM_M | 16 | 8 | 3.8** | 2.2 |
| C1ML | 16 | 20 | 4.5*** | 2.3 |
| MCOpto | 16 | 42 | 4.5* | 3.3 |
| MCOpto | 17 | 0 | 3.8* | 2.8 |
| C24ML | 17 | 0 | 5.2*** | 3.0 |
| FOM | 17 | 12 | 2.7* | 1.6 |
| pH1ML | 17 | 68 | 3.1* | 1.8 |
| FOM | 18 | 0 | 3.1* | 1.8 |
*: significant at the 5% chromosome-wide level, **: significant at the 1% chromosome-wide level, ***: significant at the 5% genome-wide level, ****: significant at the 1% genome-wide level
1the fraction of phenotypic variance in the F2 explained by a QTL; calculated as the proportion of residual variance of the statistical models with and without the QTL effect
Analysis under the line-cross model assumes that the founder lines were fixed for different QTL alleles. For 30 traits related to muscle fibre composition 47 estimated QTL positions were detected with at least suggestive significance (p < 0.05 for each chromosome, i.e. roughly one false positive per genome). Out of these five reached the genome-wide p <0.05 significance thresholds. For eight traits related to body composition and meat quality 14 computed QTL positions were found with five exceeding the 5% or 1% genome-wide significance threshold. Analysis under the half-sib model does not make the assumption of fixation of QTL alleles in the founder lines. It showed 48 QTL for microstructural muscle properties and 34 for traits of muscularity and biophysical parameters, with 20 reaching genome-wide significance. Estimates of the degree of phenotypic variation explained by the QTL range between some 2% for suggestive QTL for meat quality and carcass traits and 24% for a QTL for proportion of giant fibres on SSC12.
The telomeric region of SSC1 contained a QTL for the diameter of angular fibres (DiaAnF) reaching 5% genome-wide significance under the line-cross model. Estimated additive and dominance effects indicated an overdominant QTL (dominance effect > additive genetic effect) with the Miniature Pig allele causing higher trait values (Table 3). The same region showed a suggestive QTL for proportion of giant fibres (ProGiF) under the half-sib model (Table 4). The distal region of SSC1 exhibited a number of suggestive QTL related to the proportion and diameter of slow twitch oxidative (STO) and fast twitch glycolytic (FTG) fibres and fibre number (Tables 3 and 4).
Table 4.
Evidence for QTL significant at the 5% chromosome-wide level for traits related to muscle fibre distribution by chromosome obtained by half-sib analysis. Estimated significance levels (F-value), position, % of F2 variance explained by each QTL, and gene effects.
| Trait | SSC | Position [cM] | F- Value | % Variance1 |
| ProGiF | 1 | 0 | 8.7* | 24.3 |
| ProFTG/w | 1 | 108 | 4.3** | 6.2 |
| Diared | 1 | 117 | 3.9** | 11.0 |
| Fib/mm2 | 1 | 120 | 3.4* | 9.5 |
| Diaw | 1 | 120 | 4.1** | 11.5 |
| ProSTO/red | 1 | 124 | 4.2** | 6.0 |
| Prored | 1 | 136 | 4.5** | 12.6 |
| ToF# | 2 | 40 | 4.8** | 15.0 |
| ProFTO | 2 | 100 | 5.8*** | 18.2 |
| CapSTO | 2 | 123 | 5.6*** | 17.5 |
| Proim | 3 | 10 | 3.3* | 9.1 |
| ProFTO | 3 | 16 | 3.8* | 11.9 |
| DiaAnf | 4 | 13 | 4.6** | 12.9 |
| Prow | 4 | 19 | 3.2* | 9.0 |
| ProFTO/im | 4 | 64 | 3.4* | 4.8 |
| ProFTG/w | 4 | 80 | 4.5* | 6.4 |
| DiaFTO/im | 4 | 80 | 4.3** | 6.2 |
| DiaGiF | 4 | 129 | 4.7* | 13.1 |
| ProGiF | 4 | 132 | 6.2*** | 8.8 |
| ToF# | 5 | 75 | 4.3* | 12.2 |
| Prow | 7 | 139 | 3.4* | 9.5 |
| ProFTO | 8 | 55 | 6.0*** | 18.9 |
| ProAnF | 8 | 97 | 5.1* | 15.9 |
| ProFTG | 8 | 127 | 5.4** | 17.0 |
| Fib/mm2 | 9 | 50 | 3.9** | 4.1 |
| DiaFTG/w | 9 | 72 | 3.3* | 3.2 |
| ProAnF | 10 | 11 | 4.3* | 13.5 |
| ToF# | 11 | 80 | 6.0** | 17.3 |
| Diaim | 11 | 83 | 4.0** | 11.1 |
| Diared | 11 | 85 | 3.9** | 11.0 |
| Diaw | 11 | 86 | 4.7** | 13.1 |
| Fib/mm2 | 11 | 91 | 5.2*** | 5.9 |
| Prored | 11 | 98 | 3.5* | 9.9 |
| Prow | 11 | 98 | 3.4* | 9.5 |
| ProGiF | 12 | 38 | 10.3*** | 24.4 |
| DiaGiF | 12 | 56 | 7.4*** | 17.5 |
| Capmean | 13 | 54 | 3.2* | 10.0 |
| DiaFTG/w | 13 | 60 | 3.9* | 5.6 |
| Fib/mm2 | 13 | 62 | 4.1** | 5.9 |
| CapSTO | 13 | 66 | 4.0* | 12.7 |
| ToF# | 13 | 76 | 3.2* | 9.3 |
| ToF# | 14 | 24 | 3.9* | 12.3 |
| ProGiF | 15 | 32 | 12.7**** | 15.5 |
| DiaGiF | 15 | 34 | 6.4** | 15.1 |
| Prored | 16 | 12 | 3.1* | 8.6 |
| ProGiF | 16 | 74 | 4.3* | 13.6 |
| Fib/mm2 | 17 | 22 | 3.1* | 9.7 |
| ProAnF | 17 | 58 | 4.1** | 5.9 |
*: significant at the 5% chromosome-wide level, **: significant at the 1% chromosome-wide level, ***: significant at the 5% genome-wide level, ****: significant at the 1% genome-wide level
1the fraction of phenotypic variance in the F2 explained by a QTL; calculated as the proportion of residual variance of the statistical models with and without the QTL effect
On SSC2 highly significant QTL for meatiness (FOM, MAML) were detected in the proximal region in marker interval SW2443-FTH1 by half-sib analysis (Table 6); according to line-cross model a QTL for mean fibre diameter (Diamean) mapped to the intermediate region of SSC2 (SW240-STS2) that explained a considerable high proportion of phenotypic variation (Table 3); distal of these are QTL with genome-wide significance and strong effects on proportion of slow twitch fibres and their capillarisation (ProSTO and CapSTO) (C3-SW1564, half-sib model, Table 4).
The telomeric region of SSC3 contained a highly significant QTL for meat colour (MCOpto) as detected by half-sib and line-cross analysis (Tables 5, 6). Suggestive QTL for microstructural muscle traits map slightly more distal (Tables 3, 4).
Table 5.
Evidence for QTL significant at the 5% chromosome-wide level for traits related to muscularity and meat quality by chromosome obtained by F2 analysis. Estimated significance levels (F-value), position, % of F2 variance explained by each QTL, and gene effects.
| Trait | SSC | Position [cM] | F-Value | % Variance1 | Additive | Dominance | ||
| Effect2 | S.E. | Effect2 | S.E. | |||||
| pH24ML | 1 | 21 | 6.9* | 1.8 | -0.027 | 0.007 | -0.003 | 0.013 |
| C1ML | 2 | 56 | 5.88* | 1.4 | 0.146 | 0.045 | 0.098 | 0.118 |
| FOM | 3 | 58 | 7.9** | 2.1 | 1.66 | 0.49 | 1.69 | 0.78 |
| MCOpto | 3 | 0 | 13.4**** | 3.6 | -1.74 | 0.35 | 0.57 | 0.51 |
| FOM | 4 | 80 | 19.0**** | 5.1 | 3.21 | 0.53 | 0.06 | 0.89 |
| MAML | 4 | 78 | 13.8**** | 3.7 | 1.27 | 0.25 | 0.47 | 0.42 |
| pH24ML | 4 | 90 | 6.4* | 1.7 | -0.022 | 0.007 | -0.021 | 0.012 |
| C1ML | 4 | 26 | 5.4* | 1.5 | 0.021 | 0.039 | -0.248 | 0.076 |
| MAML | 6 | 175 | 11.4*** | 3.0 | -0.99 | 0.23 | 0.52 | 0.35 |
| C24ML | 6 | 138 | 6.1* | 1.5 | -0.273 | 0.083 | -0.285 | 0.184 |
| pH1ML | 13 | 76 | 5.9* | 1.6 | 0.038 | 0.012 | 0.021 | 0.016 |
| pH24ML | 15 | 63 | 5.7* | 1.5 | -0.031 | 0.009 | 0.026 | 0.024 |
| MCOpto | 15 | 117 | 8.0*** | 2.2 | -1.27 | 0.37 | -0.89 | 0.50 |
| pH24ML | 18 | 66 | 4.8* | 1.3 | -0.019 | 0.007 | -0.250 | 0.012 |
*: significant at the 5% chromosome-wide level, **: significant at the 1% chromosome-wide level, ***: significant at the 5% genome-wide level, ****: significant at the 1% genome-wide level
1the fraction of phenotypic variance in the F2 explained by a QTL; calculated as the proportion of residual variance of the statistical models with and without the QTL effect
2positive values of additive genetic and dominance effects imply higher trait values forced by the Duroc allele;
According to the line-cross analysis the intermediate region of SSC4 (STS3-S0214) carried a highly significant QTL for lean meat content, FOM, and eye muscle area, MAML (Table 5) as well as some suggestive QTL for fibre type composition and meat quality (Tables 3, 4, 5). The more proximal area of SSC4 (S0227-S0001) carried suggestive QTL for the proportion of fast twitch fibres (Prow, ProFTG, ProFTG/w), as revealed by the F2 and half-sib model (Tables 3, 4). According to the half-sib model the region between S0214 and S0097 of SSC4 bore QTL for the number and size of giant fibres (Table 4).
Chromosomes 5, 6 and 7 did not show any or just suggestive QTL for microstructural muscle properties. Loci controlling lean meat content segregated on SSC6 in the marker interval S0035-S0087 in the half-sib model and between S0059 and S0003 in the line-cross model (Tables 5, 6). For SSC8 using the half-sib model a significant QTL for proportion of fast twitch oxidative fibres (ProFTO) was detected in the marker interval SW2410-S0086 (Table 4).
While on SSC9 and 10 there were no significant QTL for muscle fibre traits SSC11 had a significant QTL for fibre number per mm2 (Fib/mm2) as detected by half-sib analysis between markers S0386 and SW703 (Table 4). According to the half-sib analysis loci affecting proportion and size of giant fibres (ProGiF, DiaGiF) segregated on SSC12 close to marker SW874 (Table 4). No QTL reaching genome-wide significance mapped to SSC13.
The distal part of SSC14 (Vin-SWC27) exhibited a significant QTL for fibre diameter (Diaw). Dominance effects are larger than additive genetic effects with Duroc alleles decreasing the trait value (Table 3). The same interval harboured a suggestive QTL for meat conductivity (C24ML) while the region of SSC14 close to marker S0007 showed suggestive QTL for proportion of different fibre types and FOM shown by line-cross and half sib analysis, respectively (Tables 3, 6).
SSC15 showed significant QTL for proportion and diameter of giant fibres (ProGiF, DiaGiF) as revealed by half-sib analysis between markers S0355 and SW1111 as well as suggestive QTL for size and proportion of other fibre types shown by line-cross analysis in interval SW1111-SW936 (Tables 3, 4). Also close to marker SW1111 there was a significant QTL for ph24ML detected by the half-sib analysis (Table 6). Even more distal (SW936-SW1119) a significant QTL for meat colour was found in the line cross model analysis (Table 5).
The telomeric regions of SSC16 and SSC17 contained significant QTL for meat quality (C1ML, C24ML) and carcass traits (FOM) under the half-sib model (Table 6). SSC 18 carried no QTL reaching the genome-wide significance for the traits analysed.
Discussion
A genome scan was performed in a F2 experimental crossbred population using a marker set covering more than 80% of the porcine linkage map (USDA-MARC v2, ArkDB [17]) for traits related to muscle fibre type composition and meat quality applying line-cross and half-sib analysis. The study showed that microstructural properties of pig muscle as well as meat quality traits are governed by genetic variation at many loci distributed throughout the genome. This study revealed more QTL exceeding the 5% chromosome-wide significant threshold than expected by chance when analysing 38 traits and 18 autosomes. The proportion of phenotypic variation explained was higher for QTL for microstructural properties, ranging between 3.0 and 24.4%, than for meat quality and carcass traits (1.3–5.9%). This indicates higher power of the analyses for the fibre type traits. Muscle fibre type composition is known to affect meat quality with larger fibre diameters and higher proportion of white fibres resulting in poor meat quality [7-10]. Muscle fibre traits have moderate to high heritabilities (h2 = 0.20 – 0.59) compared to meat quality traits (h2 = 0.15 – 0.32) [10]. Thus large effects of QTL for fibre traits might reflect the larger impact of genetic variation on these traits than on meat quality. It should be noticed that the number of animals with muscle fibre phenotypes available for the present study was lower than the number of animals with technological meat quality phenotypes. Therefore the power of the analysis might have not been sufficient to detect even more QTL for fibre traits with lower effects that probably exist. In a genome scan conducted in a resource population based on Japanese Wild Boar and Large White Nii and coworkers (2005) [16] identified QTL for proportion of muscle fibre types and their proportion of relative area of skeletal muscle that explained 5 to 7% of phenotypic variation – similar to QTL for meat colour and pH. The large effects of QTL for fibre traits obtained in the present study may also be due to higher variation in fibre type traits than meat quality traits in the DUMI population. Coefficients of variation for fibre size were close to 15%, for proportion of STO and FTO fibres exceeded 30%, while it was up to 3% for meat pH, 10 and 15% for meat colour and conductivity 45 min post mortem; just coefficient of variation of conductivity 24 hours post mortem exceeded 30%. Coefficient of variation even exceeded 100% for traits related to giant and angular fibres. It should be noticed that angular and giant fibres occurred at a low frequency and thus the traits have distorted distribution, making the estimation of QTL less reliable. The coefficients of variation for fibre type proportion obtained by Nii et al. (2005) [16] were lower (ca. 22%) than in the DUMI population but still higher than coefficients of variation of meat quality traits like pH.
Thus, by looking at the microstructural properties of muscle rather than measuring complex meat quality traits one gets closer to the genes' effects. Moreover, higher phenotypic variation of these traits facilitates the identification of QTL. In consequence, the QTL analysis for both (1) complex traits related to biophysical muscle/meat parameters and (2) microstructural muscle properties – know to be responsible for a large proportion of variation in the aforementioned traits – at the same time, represents a model case that allows dissecting genomic regions, which control these traits, and in which variation in the meat phenotype is probably associated to genes that in first line affect muscle fibre traits.
We found more QTL using the half-sib model than the line-cross model indicating that the founders of the DUMI population are not fixed for different alleles at many of the QTL. In F2 resource populations of Chinese Meishan and commercial pig lines de Koning et al. (2001) [18] as well as Bidanel et al. (2001) [19] also applied line-cross and half-sib models and were able to identify additional QTL due to the use of both. The authors discuss that the use of both models allows investigating different assumptions about QTL genotypes in founder populations that are obviously both true for some QTL in resource populations typically used in farm animals. The line-cross model assuming that different QTL alleles are fixed in founder populations is very powerful when this assumption corresponds to the true state of nature of the QTL and it is quite robust to limited deviations from this ideal situation, even though it tends to underestimate QTL effects in such situations [20]. The half-sib model is more general with no assumption about the number and frequency of QTL alleles in founder populations and probably more realistic for many QTL. This is in line with QTL studies made in commercial pig populations and their crosses showing that even in these selected populations there is still a considerable amount of genetic variation at loci affecting traits of interest [21-23].
In pig breeding biophysical muscle properties like pH and conductivity are measured in routine to monitor meat quality mainly in terms of water binding capacity. At the cellular level, a high proportion of glycolytic muscle fibres, large fibre diameters, low vascularisation and reduced mitochondrial activity are associated with low water binding capacity [24,25]. The present study focuses on these fibre type traits representing cellular factors on meat quality. At the molecular level, pathways involved in energy and calcium homeostasis are of importance. For example, genetic variation of ryanodine receptors, calcium-release channels, has major impact on pH and conductivity in pork and muscle activities such as protein metabolism, differentiation, and growth as well as pathophysiological conditions seen in dystrophinopathies, Brody's disease, and malignant hyperthermia in human [26]. Due to their great economical importance body composition traits, especially lean meat content, are also recorded in routine in pig breeding. Correspondingly, there are many efforts to identify QTL responsible for the variation in technological parameters of meat quality and in body composition traits. Genome scans were conducted in different experimental and commercial populations and revealed QTL effects on all 18 autosomes and chromosome X as currently summarized on the Pig Quantitative Trait Loci (QTL) database (PigQTLdb) [27,28]. Here, we report QTL for meat quality traits with genome-wide significance on SSC3, 5, 13, 15, 16 and 17 and QTL for traits related to lean meat content on SSC2, 4, 6, and 16.
On SSC3 QTL with effect on pH value, meat colour, and juiciness have been found in several experimental populations [18,29-32]. The QTL effects on pH value were validated and confirmed in commercial lines [21,33]. These results together with our data indicate that loci affecting meat colour and meat quality traits related to water binding capacity, like pH value and conductivity, segregate in many populations including commercial breeds and are located on the proximal region of the p-arm of SSC3. Skeletal muscle myosin regulatory light chain 2 (HUMMLC2B) represents a positional functional candidate gene [34]. Malek et al. (2001) [15] detected QTL for meat colour on SSC5, however these map to the distal end close to marker SW378, while the QTL reported in the present study is at the opposite telomer close to marker SW1482. De Koning and co-worker (2001) [18] reported on a QTL for meat colour on SSC13 that corresponds to our finding. On SSC15 effects on pH value and colour matching our QTL have been reported by others [15,18]. Also QTL for glycogen content and/or glycolytic potential were shown [15]. The QTL region comprises PRKAG3, (protein kinase, AMP-activated, y-(3)-subunit), that has been shown to be the causal gene for variation in glycogen content in the muscle and the resulting meat quality [35,36]. To some extent these traits reflect the fibre type distribution for which we found a number of QTL on SSC15. Geldermann et al. (2003) [37] and Paszek et al. (2001) [29] mapped QTL for pH and conductivity on SSC16 in vicinity to those estimated by us. On SSC17 QTL for meat colour, juiciness, lactate concentration, and glycolytic potential were shown by Malek et al. (2001) [15].
The finding of a QTL for muscularity, i.e. FOM and eye muscle area, on SSC2 corresponds to reports of Jeon et al. (1999) [38] and Nezer et al. (1999) [39] on paternally expressed QTL affecting muscle growth. The authors highlighted IGF2 (insulin like growth factor 2) as the positional candidate gene. Subsequent analyses led to the identification of the causal mutation in intron 3 of IGF2 that modulates its expression [40]. Effects of the IGF2 polymorphisms have been confirmed in other populations [41,42] and IGF2 is also a likely candidate for the QTL shown in the present study. The QTL for body composition on SSC4 represent the first ever published in pigs [43]. The QTL have been confirmed in several populations [44]. In a discordant sibpair analysis using AFLP markers we also identified QTL for body composition traits on SSC4 and proposed CRH (corticotropin releasing hormone) gene as a functional positional candidate gene [45]. QTL for muscularity have been previously shown in the central and distal region of SSC6 [30,46], however the QTL for muscularity proximal on SSC6 shown in the present study are new, as well as the QTL for lean meat content on SSC16.
Recently, Nii and coworkers (2005) [16] report on QTL for muscle fibre traits and meat quality in a Japanese Wild Boar × Large White intercross on SSC 1, 2, 6, 14, and X. Discrimination of type I, IIA, or IIB fibres was based on myosine ATP method after alkaline preincubation expected to reveal phenotypes corresponding to ours. Nii and co-worker (2005) [16] found genome-wide significant QTL for type I fibres on SSC1 and 14 in vicinity of the regions where we found chromosome-wide significant QTL for slow twitch and red fibres, respectively. According to Nii et al. [16], positional candidate genes for the QTL on SSC14 are genes of the calcineurin signalling pathway that is involved in muscle fibre type switch, namely PPP3CC (protein phosphatase 3 catalytic subunit γ isoform, calcineurin A γ), PPP3CB (protein phosphatase 3 catalytic subunit β isoform, calcineurin A β) and NFAM1 (NFAT activation module 1). Moreover, QTL for type IIA, IIB and intermediate, white, and fast twitch fibres on the intermediate region of SSC2 and proximal on SSC14 have been found by us and Nii (2005) [16]. QTL for proportion of type I fibres were detected on SSC8.
In mice genome-wide significant QTL for weight of muscles with different muscle fibre type composition were detected on chromosome 3, 8, and 17 that were muscle-specific but not muscle fibre type specific [47]. According to the actual comparative map, these regions correspond to the proximal part of SSC6, q-arm of SSC14 and central region of SSC7. In that region of SSC6 we did not detect QTL for muscle fibre composition but for FOM and MAML.
Thus the results correspond in indicating QTL for size of particular muscle, M. gastrognemius in mice and M. longissimus dorsi in pig, however the first is a mixed fiber muscle while the second consists mainly of fast fibres. The mouse QTL on chromosome 8 affects size of the fast muscle extensor digitorum. QTL in the corresponding porcine genome regions are for fibre number and diameter. We found no significant QTL on SSC7. Studies of the potential role of the myostatin pathway on muscle strength in human revealed linkage of markers close to myostatin (GDF8) on HSA2, to CDKN1A (cyclin-dependent kinase inhibitor 1A) on HSA6, and to myogenic factor 3 (MYOD1) on HSA11, as well as markers on HSA12 and HSA13 [48,49] to potential QTL. In cattle and mouse myostatin has been shown to affect muscle mass [50,51]. Porcine myostatin maps to SSC15q23 [52] and it is a positional functional candidate gene for the QTL for muscle fibre type traits and biophysical muscle properties identified here. The QTL regions on HSA6, 11, 12, and 13 correspond to the proximal half of SSC7, p-arm of SSC2 (MYOD1 at 2p14-17; [53]), SSC 5, and SSC11. We found QTL for fibre diameters and fibres number on SSC2 and 11. Genes of the myostatin pathway are functional candidates for traits related to muscle strength and dynamic properties as well as fibre composition traits in man and pig. Other established functional candidate genes for muscle fibre traits that represent positional candidates for the QTL detected in this study [54,55], i.e. MEF2C (MADS box transcription enhancer factor 2, polypeptide C; SSC2), MEF2D (MADS box transcription enhancer factor 2, polypeptide D; SSC4), PPARGC1A, (peroxisome proliferative activated receptor, gamma, coactivator 1, alpha; SSC8) and PPARG (peroxisome proliferator-activated receptor gamma 2; SSC13).
In particular, those genomic regions are of interest that exhibit both (1) QTL for body composition and biophysical muscle properties, which are used as technological parameters of meat quality to select in breeding routine, and (2) QTL for muscle fibre number and distribution traits, which are more strictly genetically controlled but affect growth, body composition and meat quality to a large extent.
Taking into account QTL with genome-wide significance we found correspondence between the QTL for pH24ML and the QTL for ProGiF on SSC15. It should be noticed that giant fibres occurred at a low frequency and thus the trait has distorted distribution, making the estimation of QTL less reliable. Taking a further look at regions with either genome-wide significant QTL for fibre type traits or genome-wide significant QTL for meat quality reveals further correspondence on SSC1 (pH24ML and DiaAnF), on SSC2 (C1ML and Diamean and DiaFTG; Figure 1), on SSC3 (MCopto and ProGif, Proim, ProFTO), on SSC5 (MCopto and Diared), on SSC13 (MCopto and Fib/mm2), on SSC14 (C24ML and Diaw), and on SSC16 (C1ML and Prored). Looking at all QTL detected even more regions appear with QTL for fibre type distribution traits and meat quality traits on SSC4, 8, 11, and 17. With regard to the relationship between meat quality, lean meat content and fibre type distribution traits p-arm of SSC2 and proximal region of q-arm of SSC4 are of interest. On SSC2 and SSC4 we found genome-wide significant QTL for FOM and area of M longissimus on the one hand and fibre diameter (DiaFTG, Diamean) on the other hand that might again indicate a common genetic background. Also on SSC14, 16, 17, and 18 we found QTL for body composition and muscle fibre traits. On SSC7 and 9 we only found QTL for muscle fibre type distribution traits. SSC6 showed genome-wide significant QTL for FOM and MAML and for meat quality traits, but no QTL for muscle fibre traits. Nii et al. (2005) [16] found QTL for meat colour and hematin content on SSC6 close to QTL for type II fibres.
Figure 1.
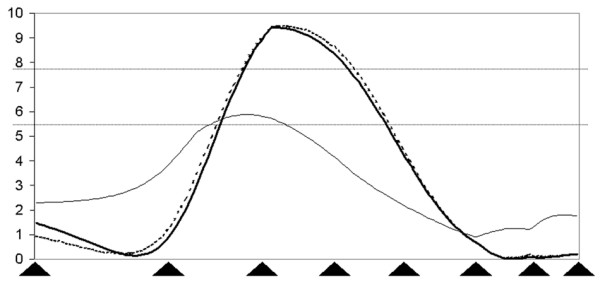
Plot of the F-ratio from least square interval mapping for evidence of QTL for DIAFTG ( ) and DIAmean (
) and DIAmean ( ) as well as C1ML (
) as well as C1ML ( ) on SSC2. The x-axis indicates the relative position on the linkage map. The y-axis represents the F-value. Arrows on the x-axis indicate the position of markers (SW2443, FTH1, SW240, STS2, C3, SW1564, BHMT, S0226). Lines indicate the 5% genome-wide and 5% chromosome-wide significance thresholds.
) on SSC2. The x-axis indicates the relative position on the linkage map. The y-axis represents the F-value. Arrows on the x-axis indicate the position of markers (SW2443, FTH1, SW240, STS2, C3, SW1564, BHMT, S0226). Lines indicate the 5% genome-wide and 5% chromosome-wide significance thresholds.
Conclusion
Microstructural properties of pig muscle and meat quality are governed by genetic variation at many loci distributed throughout the genome. The number and type of muscle fibres affect body composition and muscle properties and are to a large extent determined prenatal by genetic factors. The application of linkage analyses, which are a priori hypothesis-free, on traits of high heritability increases the power of the approach. Disentangling complex traits in their constituent phenotypes might facilitate the identification of QTL and the elucidation of the pleiotropic nature of QTL effects. QTL analysis under both, the line-cross and half-sib model, allows detecting QTL that are fixed or segregating among the founder populations and thus provide comprehensive insight into the genetic variation of the traits under investigation. The map-based data provided here will facilitate the identification of genes directly affecting muscle fibre traits and indirectly meat quality traits especially when combined together with current attempts to identify genes expressed during myogenesis [56,57]. In those regions harbouring QTL for muscle fibre traits and QTL for meat quality and body composition traits, i.e. on SSC1, SSC15 (with QTL for pH and size and proportion of defect fibres), on SSC2, SSC4 and on SSC14 (with QTL for conductivity, muscularity and fibre size) and on SSC3 (with QTL for meat colour and fibre proportion) effects on meat quality traits and body composition might be the result of genetic variation primarily affecting muscle fibre traits.
Methods
Animals
Analyses were done in an experimental population based on reciprocal crossbreeding of Duroc and Berlin Miniature Pig breeds that is a three-generation porcine F2 population (DUMI population). In detail, a Duroc boar was mated to four Berlin Miniature Pig dams and a Berlin Miniature Pig boar was mated to five Duroc dams to produce 43 F1 dams and 5 F1 boars. F1 boars and dams were kept at two places to finally produce 905 F2-animals in total. Thirty-three full sib-families comprising 469 F2-individuals born from 32 sows and four boars were kept and performance tested on the research farm of the Institute of Animal Science in Berlin Dahlem, Humboldt University of Berlin (up to day 100) and at the performance test station of the federal country Brandenburg (day 100 to day 200). At the research farm Frankenforst of the Institute of Animal Breeding and Genetics, University of Bonn, 436 F2 animals of 21 full sib-families were born from 11 sows and three boars, raised and performance tested. F2 piglets were weaned at about 6 weeks of age and kept in flat decks until day 100 and subsequently in single pens until slaughter at 200 days of age. The F2-animals of the DUMI population have obese carcasses with lean meat content of about 34% (FOM). Mean daily gain (weight gain during performance test; day 110 to 200) was 470 ± 117 g with weight at day 200 ranging between 29 and 104 kg [58].
Phenotypes
Traits related to body composition as well as biophysical muscle properties are defined and listed in Table 1 together with their mean values. Correlations between muscle fibre traits and meat quality ranged between 0.1 and 0.3 [58]. Muscle fibre characteristics of the longissimus muscle were determined by microscopic image analyses after histochemical fibre type differentiation. The samples were taken immediately post mortem at the 13th/14th rib, frozen in liquid nitrogen and stored at -70°C. Serial cross-sections (12 μm) were obtained in a cryostat microtom (-20°C) in order to be processed for the following histochemical reactions:
The definitions of muscle fibre distribution traits as well as their mean values are given in Table 5. In order to differentiate the three main fibre types "red", "intermediate", "white" and "slow twitch oxidative = STO", "fast twitch oxidative = FTO", "fast twitch glycolytic = FTG", respectively, the samples were stained either by the NADH tetrazolium reductase reaction (NADH-TR) alone or by the combined NADH-TR/ATPase reaction described by Horak (1983) [59]. While using just the NADH-TR reaction allows differentiating fibres according to their high, moderate or low oxidative enzyme activity, the combined method of NADH-TR and acid stable myofibrillar ATPase reaction indicates the contractile properties of the fast or slow fibre type, respectively [60]. In addition, the hyper-contracted giant fibres were classified by their typical oval or round shape and/or their large size independent of their specific enzyme activity [61]. Giant fibres display a condition where single fibre loose their connection within the muscle tissue and therefore contract maximal. Occurrence of giant fibres is correlated with inferior meat quality [58,61]. The angular fibres were classified and measured based on their characteristic right-angled shape with concave borders, described by Walasik et al. (2000) [62].
For the identification of the capillaries the alkaline phosphatase reaction was used detecting this marker enzyme of endothelial cells as described by Josza et al. (1993) [63].
The staining results of the fibre type classification by the NADH-TR reaction and by the combined NADH-TR/ATPase reactions and the identification of the capillaries, giant fibres, and angular fibres are shown in Figure 2.
Figure 2.
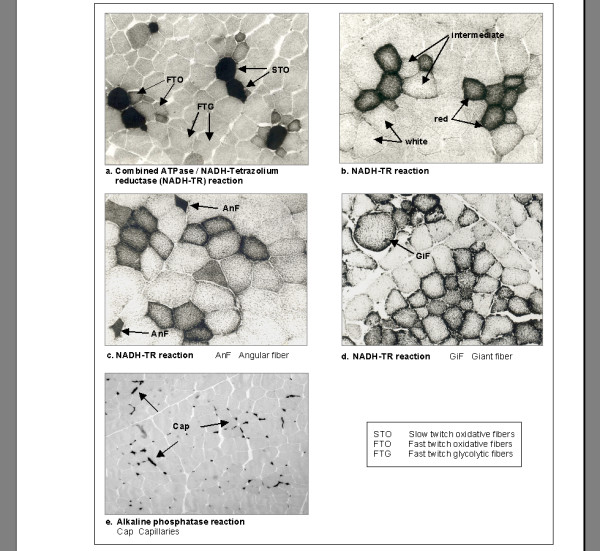
Cross sections of fibres in M. longissimus dorsi. Combined ATPase/NADH-TR reaction was used to identify STO, FTO and FTG fibres (a) while NADH-TR reaction was used to identify red, intermediate and white fibres as well as angular and giant fibres (b, c, d). For the identification of the capillaries the alkaline phosphatase reaction was used (e). Magnification: 200× (a-d); 100× (e).
The quantitative microscopic determination of fibre type proportion, fibre size and capillar density was done on 400 fibre cross sections per animal by using the image analysis systems AMBA (IBSB, Berlin, Germany) [64] and semiautomatic computer-aided image analyser MFA (muscle fibre analyser) [65]. Taking into account of the special structure of the longissimus muscle and the clustered slow twitch fibres, all fibres of the primary bundles, which were completely visible, were studied. These bundles were randomly selected over the slides.
In total 308 F2 animals were examined for microstructural muscle traits. QTL analysis for these traits involving differentiation of fibre types based on staining intensity was performed separately for the two subsets of the material with phenotypic evaluation using either NADH-TR (n = 168) or combined NADH-TR/ATPase reaction (n = 140). In the pig muscle fibre typings based solely on metabolic properties (NADH-TR) or combined determination of metabolic and myofibrillar ATPase stability are highly corresponding [66]. Therefore QTL analysis was also performed for the whole data set with traits named with the suffix "STO/red", "FTO/im" and "FTG/w".
Markers and QTL-analysis
Altogether the animals of the DUMI population were genotyped at 88 loci covering the porcine autosomes with mean interval size of 30.7 cM. The set of markers includes 72 microsatellites and 16 biallelic markers. Linkage analysis was performed using the program CRI MAP, version 2.4 [67]. The order of markers and the genetic distances between them are given in Table 2.
The QTL analysis was done with the program QTL express accessible via internet [68,69] by interval mapping based on least square regression analysis developed for three generation F2 populations and half-sib families [70,71]. Additive genetic effects were estimated at 1 cM intervals as half of the difference of the trait value between homozygous carriers of the Duroc and the Miniature Pig alleles; i.e. positive values for the additive of genetic effects point to a higher trait value for homozygous carriers of the Duroc allele. Dominance effects are estimated as the difference between the trait value of heterozygous individuals and the mean trait value observed for homozygous animals. Subsequently, additive and dominant coefficients at fixed positions in the genome of each F2 animal and their phenotypic values were regressed onto the additive and dominance coefficients in intervals of 1 cM. Least square regression models used for QTL analysis included along with additive and dominance coefficients for the putative QTL the fixed effects of family, parity and sex as well as slaughter weight as a co-variable, which were found to affect almost all traits analyses in previous analyses of variance ignoring any molecular genetic information.
Also a paternal half-sib analysis was accomplished making no assumptions on the relative frequencies of the QTL alleles in the founder populations. Therefore the F2-animals were treated as paternal half-sib families and the probability for the occurrence of a paternal allele was estimated in intervals of 1 cM. The probabilities of inheritance of distinct paternal gametic phases were regressed onto allele substitution effects at the putative QTL. The regression model included sex and litter as fixed effects and slaughter weight as co variable.
Significance thresholds at the 5 and 1% level were determined empirically by permutation for individual chromosomes [72]. Chromosome-wide 1 and 5% significance thresholds became genome-wide significance thresholds after Bonferroni correction for 18 autosomes of the haploid porcine genome.
Authors' contributions
KW significantly contributed to the concept and design of the study and data analysis and wrote the manuscript. IF performed phenotypic analyses and helped in drafting the manuscript. TH was largely involved in design and coordination of the study. EM carried out genotyping and assisted statistical analysis and preparing the manuscript. KS made substantial contribution to the design of the study, coordination of data collection and helped in drafting the manuscript. SP provided input to design of the study, performed molecular genetic and statistical analyses and assisted drafting the manuscript and revising it critically for scientific content.
Acknowledgments
Acknowledgements
This work was supported by funds provided by Federal Ministry of Education and Research (BMBF), Ministry of the Environment, Conservation, Agriculture and Consumer Protection of the State of North Rhine-Westphalia (MUNLV) and the EU Project PorDictor (QLK-2000-01363) (coordinator K. Wimmers).
Contributor Information
Klaus Wimmers, Email: wimmers@fbn-dummerstorf.de.
Ilse Fiedler, Email: fiedler@fbn-dummerstorf.de.
Torsten Hardge, Email: hardge@ing.boehringer-ingelheim.com.
Eduard Murani, Email: murani@fbn-dummerstorf.de.
Karl Schellander, Email: karl.schellander@itz.uni-bonn.de.
Siriluck Ponsuksili, Email: s.wimmers@fbn-dummerstorf.de.
References
- Peter JB, Barnard RJ, Edgerton VR, Gillespie CA, Stempel KE. Metabolic profiles of three fiber types of skeletal muscle in guinea pigs and rabbits. Biochemistry. 1972;11:2627–2633. doi: 10.1021/bi00764a013. [DOI] [PubMed] [Google Scholar]
- Simoneau JA, Bouchard C. Genetic determinism of fiber type proportion in human skeletal muscle. FASEB J. 1995;9:1091–1095. doi: 10.1096/fasebj.9.11.7649409. [DOI] [PubMed] [Google Scholar]
- Simoneau JA, Bouchard C. Skeletal muscle metabolism and body fat content in men and women. Obes Res. 1995;3:23–29. doi: 10.1002/j.1550-8528.1995.tb00117.x. [DOI] [PubMed] [Google Scholar]
- Simoneau JA, Kelley DE. Altered glycolytic and oxidative capacities of skeletal muscle contribute to insulin resistance in NIDDM. J Appl Physiol. 1997;83:166–171. doi: 10.1152/jappl.1997.83.1.166. [DOI] [PubMed] [Google Scholar]
- Simoneau JA, Veerkamp JH, Turcotte LP, Kelley DE. Markers of capacity to utilize fatty acids in human skeletal muscle: relation to insulin resistance and obesity and effects of weight loss. FASEB J. 1999;13:2051–2060. doi: 10.1096/fasebj.13.14.2051. [DOI] [PubMed] [Google Scholar]
- Tanner CJ, Barakat HA, Dohm GL, Pories WJ, MacDonald KG, Cunningham PR, Swanson MS, Houmard JA. Muscle fiber type is associated with obesity and weight loss. Am J Physiol Endocrinol Metab. 2002;282:1191–1196. doi: 10.1152/ajpendo.00416.2001. [DOI] [PubMed] [Google Scholar]
- Lengerken vG, Maak S, Wicke M, Fiedler I, Ender K. Suitability of structural and functional traits of skeletal muscle for the genetic improvement of meat quality in pigs. Arch Tierz. 1994;37:133–143. [Google Scholar]
- Fiedler I, Ender K, Wicke M, Maak S, Lengerken vG, Meyer W. Structural and functional characteristics of muscle fibres in pigs with different malignant hyperthermia susceptibility (MHS) and different meat quality. Meat Sci. 1999;53:9–15. doi: 10.1016/S0309-1740(99)00030-3. [DOI] [PubMed] [Google Scholar]
- Rehfeldt C, Stickland N, Fiedler I, Wegner J. Environment and genetic factors as sources of variation in skeletal muscle fibre number. Basic Appl Myol. 1999;9:235–253. [Google Scholar]
- Larzul C, Lefaucheur L, Ecolan P, Gogue J, Talmant A, Sellier P, Le Roy P, Monin G. Phenotypic and genetic parameters for longissimus muscle fibre characteristics in relation to growth, carcass, and meat quality traits in large white pigs. J Anim Sci. 1997;75:3126–3137. doi: 10.2527/1997.75123126x. [DOI] [PubMed] [Google Scholar]
- Rehfeldt C, Bünger L, Dietl G, Fiedler I, Wegner J. Zur Erblichkeit von Muskelstrukturmerkmalen und ihren genetisch begründeten Beziehungen zu Wachstum und Belastbarkeit bei Labormäusen. Arch Tierz. 1988;31:185–195. [Google Scholar]
- Snyman MA, Cloete SW, Olivier JJ. Genetic and phenotypic correlations of total weight of lamb weaned with body weight, clean fleece weight and mean fibre diameter in three South African Merino flocks. Livest Prod Sci. 1998;55:157–162. doi: 10.1016/S0301-6226(98)00119-5. [DOI] [Google Scholar]
- Rivero JL, Barrey E. Heritabilities and genetic and phenotypic parameters for gluteus medius muscle fibre type composition, fibre size and capillaries in purebred Spanish horses. Livest Prod Sci. 2001;72:233–241. doi: 10.1016/S0301-6226(01)00221-4. [DOI] [Google Scholar]
- Fiedler I, Dietl G, Rehfeldt C, Wegner J, Ender K. Muscle fibre traits as additional selection criteria for muscle growth and meat quality in pigs – results of a simulated selection. J Anim Breed Genet. 2004;121:331–344. doi: 10.1111/j.1439-0388.2004.00466.x. [DOI] [Google Scholar]
- Malek M, Dekkers JC, Lee HK, Baas TJ, Prusa K, Huff-Lonergan E, Rothschild MF. A molecular genome scan analysis to identify chromosomal regions influencing economic traits in the pig. II. Meat and muscle composition. Mamm Genome. 2001;12:637–645. doi: 10.1007/s003350020019. [DOI] [PubMed] [Google Scholar]
- Nii M, Hayashi T, Mikawa S, Tani F, Niki A, Mori N, Uchida Y, Fujishima-Kanaya N, Komatsu M, Awata T. Quantitative trait loci mapping for meat quality and muscle fiber traits in a Japanese wild boar × Large White intercross. J Anim Sci. 2005;83:308–315. doi: 10.2527/2005.832308x. [DOI] [PubMed] [Google Scholar]
- ArkDB http://www.thearkdb.org/
- de Koning DJ, Harlizius B, Rattink AP, Groenen MA, Brascamp EW, van Arendonk JA. Detection and characterization of quantitative trait loci for meat quality traits in pigs. J Anim Sci. 2001;79:2812–2819. doi: 10.2527/2001.79112812x. [DOI] [PubMed] [Google Scholar]
- Bidanel JP, Milan D, Iannuccelli N, Amigues Y, Boscher MY, Bourgeois F, Caritez JC, Gruand J, Le Roy P, Lagant H, Quintanilla R, Renard C, Gellin J, Ollivier L, Chevalet C. Detection of quantitative trait loci for growth and fatness in pigs. Genet Sel Evol. 2001;33:289–309. doi: 10.1051/gse:2001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonso L, Haley CS. Power of different F2 schemes for QTL detection in livestock. Anim Sci. 1998;66:1–8. [Google Scholar]
- Evans GJ, Giuffra E, Sanchez A, Kerje S, Davalos G, Vidal O, Illan S, Noguera JL, Varona L, Velander I, Southwood OI, de Koning DJ, Haley CS, Plastow GS, Andersson L. Identification of quantitative trait loci for production traits in commercial pig populations. Genetics. 2003;164:621–627. doi: 10.1093/genetics/164.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal O, Noguera JL, Amills M, Varona L, Gil M, Jimenez N, Davalos G, Folch JM, Sanchez A. Identification of carcass and meat quality quantitative trait loci in a Landrace pig population selected for growth and leanness. J Anim Sci. 2005;83:293–300. doi: 10.2527/2005.832293x. [DOI] [PubMed] [Google Scholar]
- Nagamine Y, Haley CS, Sewalem A, Visscher PM. Quantitative trait loci variation for growth and obesity between and within lines of pigs (Sus scrofa) Genetics. 2003;164:629–635. doi: 10.1093/genetics/164.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocks L, Klont RE, Buist W, Greef de K, Tieman M, Engel B. The effects of selection of pigs on growth rate vs leanness on histological characteristics of different muscle. J Anim Sci. 2000;78:1247–1254. doi: 10.2527/2000.7851247x. [DOI] [PubMed] [Google Scholar]
- Gellerich FN, Trumbeckaite S, Müller T, Deschauer M, Chen Y, Gizatullina Z, Zierz S. Energetic depression caused by mitochondial dysfunction. Mol Cell Biochem. 2004;256–257:391–405. doi: 10.1023/B:MCBI.0000009885.34498.e6. [DOI] [PubMed] [Google Scholar]
- Berchtold MW, Brinkmeier H, Muntener M. Calcium ion in skeletal muscle: its crucial role for muscle function, plasticity, and disease. Physiol Rev. 2000;80:1215–1265. doi: 10.1152/physrev.2000.80.3.1215. [DOI] [PubMed] [Google Scholar]
- PigQTLdb http://www.animalgenome.org/QTLdb/
- Hu ZL, Dracheva S, Jang W, Maglott D, Bastiaansen J, Rothschild MF, Reecy JM. A QTL resource and comparison tool for pigs: PigQTLDB. Mamm Genome. 2005;16:792–800. doi: 10.1007/s00335-005-0060-9. [DOI] [PubMed] [Google Scholar]
- Paszek AA, Wilkie PJ, Flickinger GH, Miller LM, Louis CF, Rohrer GA, Alexander LJ, Beattie CW, Schook LB. Interval mapping of carcass and meat quality traits in a divergent swine cross. Anim Biotech. 2001;12:155–165. doi: 10.1081/ABIO-100108342. [DOI] [PubMed] [Google Scholar]
- Ovilo C, Clop A, Noguera JL, Oliver MA, Barragan C, Rodriguez C, Silio L, Toro MA, Coll A, Folch JM, Sanchez A, Babot D, Varona L, Perez-Enciso M. Quantitative trait locus mapping for meat quality traits in an Iberian × Landrace F2 pig population. J Anim Sci. 2002;80:2801–2808. doi: 10.2527/2002.80112801x. [DOI] [PubMed] [Google Scholar]
- Sato S, Oyamada Y, Atsuji K, Nade T, Sato S, Kobayashi E, Mitsuhashi T, Nirasawa K, Komatsuda A, Saito Y, Terai S, Hayashi T, Sugimoto Y. Quantitative trait loci analysis for growth and carcass traits in a Meishan × Duroc F2 resource population. J Anim Sci. 2003;81:2938–2949. doi: 10.2527/2003.81122938x. [DOI] [PubMed] [Google Scholar]
- Zuo B, Xiong YZ, Su YH, Deng CY, Zheng R, Jiang S. Mapping quantitative trait loci for meat quality on pig chromosome 3,4 and 7. Asian-Australian J Anim Sci. 2003;16:320–324. [Google Scholar]
- de Koning DJ, Pong-Wong R, Varona L, Evans GJ, Giuffra E, Sanchez A, Plastow G, Noguera JL, Andersson L, Haley CS. Full pedigree quantitative trait locus analysis in commercial pigs using variance components. J Anim Sci. 2003;81:2155–2163. doi: 10.2527/2003.8192155x. [DOI] [PubMed] [Google Scholar]
- Davoli R, Fontanesi L, Cagnazzo M, Scotti E, Buttazzoni L, Yerle M, Russo V. Identification of SNPs, mapping and analysis of allele frequencies in two candidate genes for meat production traits: the porcine myosin heavy chain 2B (MYH4) and the skeletal muscle myosin regulatory light chain 2 (HUMMLC2B) Anim Genet. 2003;34:221–225. doi: 10.1046/j.1365-2052.2003.00992.x. [DOI] [PubMed] [Google Scholar]
- Milan D, Jeon JT, Looft C, Amarger V, Robic A, Thelander M, Rogel-Gaillard C, Paul S, Iannuccelli N, Rask L, Ronne H, Lundstrom K, Reinsch N, Gellin J, Kalm E, Roy PL, Chardon P, Andersson L. A mutation in PRKAG3 associated with excess glycogen content in pig skeletal muscle. Science. 2000;288:1248–1251. doi: 10.1126/science.288.5469.1248. [DOI] [PubMed] [Google Scholar]
- Ciobanu D, Bastiaansen J, Malek M, Helm J, Woollard J, Plastow G, Rothschild M. Evidence for new alleles in the protein kinase adenosine monophosphate-activated gamma(3)-subunit gene associated with low glycogen content in pig skeletal muscle and improved meat quality. Genetics. 2001;159:1151–1162. doi: 10.1093/genetics/159.3.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldermann H, Müller E, Moser G, Reiner G, Bartenschlager H, Cepica S, Stratil A, Kuryl J, Moran C, Davoli R, Brunsch C. Genome-wide linkage and QTL mapping in porcine F2 families generated from Pietrain, Meishan and Wild Boar crosses. J Anim Breed Genet. 2003;120:363–393. doi: 10.1046/j.0931-2668.2003.00408.x. [DOI] [Google Scholar]
- Jeon JT, Carlborg O, Tornsten A, Giuffra E, Amarger V, Chardon P, Andersson-Eklund L, Andersson K, Hansson I, Lundstrom K, Andersson L. A paternally expressed QTL affecting skeletal and cardiac muscle mass in pigs maps to the IGF2 locus. Nat Genet. 1999;21:157–158. doi: 10.1038/5938. [DOI] [PubMed] [Google Scholar]
- Nezer C, Moreau L, Brouwers B, Coppieters W, Detilleux J, Hanset R, Karim L, Kvasz A, Leroy P, Georges M. An imprinted QTL with major effect on muscle mass and fat deposition maps to the IGF2 locus in pigs. Nat Genet. 1999;21:155–156. doi: 10.1038/5935. [DOI] [PubMed] [Google Scholar]
- Van Laere AS, Nguyen M, Braunschweig M, Nezer C, Collette C, Moreau L, Archibald AL, Haley CS, Buys N, Tally M, Andersson G, Georges M, Andersson L. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature. 2003;425:832–836. doi: 10.1038/nature02064. [DOI] [PubMed] [Google Scholar]
- Jungerius BJ, van Laere AS, Te Pas MF, van Oost BA, Andersson L, Groenen MA. The IGF2-intron3-G3072A substitution explains a major imprinted QTL effect on backfat thickness in a Meishan × European white pig intercross. Genet Res. 2004;84:95–101. doi: 10.1017/S0016672304007098. [DOI] [PubMed] [Google Scholar]
- Estelle J, Mercade A, Noguera JL, Perez-Enciso M, Ovilo C, Sanchez A, Folch JM. Effect of the porcine IGF2-intron3-G3072A substitution in an outbred Large White population and in an Iberian × Landrace cross. J Anim Sci. 2005;83:2723–2728. doi: 10.2527/2005.83122723x. [DOI] [PubMed] [Google Scholar]
- Andersson L, Haley CS, Ellegren H, Knott SA, Johansson M, Andersson K, Andersson-Eklund L, Edfors-Lilja I, Fredholm M, Hansson I, Hakansson J, Lundström K. Genetic mapping of quantitative trait loci for growth and fatness in pigs. Science. 1994;263:1771–1774. doi: 10.1126/science.8134840. [DOI] [PubMed] [Google Scholar]
- Walling GA, Visscher PM, Andersson L, Rothschild MF, Wang L, Moser G, Groenen MA, Bidanel JP, Cepica S, Archibald AL, Geldermann H, de Koning DJ, Milan D, Haley CS. Combined analyses of data from quantitative trait loci mapping studies. Chromosome 4 effects on porcine growth and fatness. Genetics. 2000;155:1369–1378. doi: 10.1093/genetics/155.3.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmers K, Murani E, Ponsuksili S, Yerle M, Schellander K. Detection of quantitative trait loci for carcass traits in the pig by using AFLP. Mamm Genome. 2002;13:206–210. doi: 10.1007/s00335-001-3052-4. [DOI] [PubMed] [Google Scholar]
- Grindflek E, Szyda J, Liu Z, Lien S. Detection of quantitative trait loci for meat quality in a commercial slaughter pig cross. Mamm Genome. 2001;12:299–304. doi: 10.1007/s003350010278. [DOI] [PubMed] [Google Scholar]
- Lionikas A, Blizard DA, Vandenbergh DJ, Glover MG, Stout JT, Vogler GP, McClearn GE, Larsson L. Genetic architecture of fast- and slow-twitch skeletal muscle weight in 200-day-old mice of the C57BL/6J and DBA/2J lineage. Physiol Genomics. 2003;16:141–152. doi: 10.1152/physiolgenomics.00103.2003. [DOI] [PubMed] [Google Scholar]
- Huygens W, Thomis MA, Peeters MW, Aerssens J, Janssen R, Vlietinck RF, Beunen G. Linkage of myostatin pathway genes with knee strength in humans. Physiol Genomics. 2004;17:264–270. doi: 10.1152/physiolgenomics.00224.2003. [DOI] [PubMed] [Google Scholar]
- Huygens W, Thomis MA, Peeters MW, Aerssens J, Vlietinck R, Beunen GP. Quantitative Trait Loci for human muscle strength: linkage analysis of myostatin pathway genes. Physiol Genomics. 2005, May 24. [DOI] [PubMed]
- Grobet L, Martin LJ, Poncelet D, Pirottin D, Brouwers B, Riquet J, Schoeberlein A, Dunner S, Menissier F, Massabanda J, Fries R, Hanset R, Georges M. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet. 1997;17:71–74. doi: 10.1038/ng0997-71. [DOI] [PubMed] [Google Scholar]
- Bunger L, Ott G, Varga L, Schlote W, Rehfeldt C, Renne U, Williams JL, Hill WG. Marker-assisted introgression of the Compact mutant myostatin allele MstnCmpt-dl1Abc into a mouse line with extreme growth effects on body composition and muscularity. Genet Res. 2004;84:161–173. doi: 10.1017/S0016672304007165. [DOI] [PubMed] [Google Scholar]
- Sonstegard TS, Rohrer GA, Smith TP. Myostatin maps to porcine chromosome 15 by linkage and physical analyses. Anim Genet. 1998;29:19–22. doi: 10.1046/j.1365-2052.1998.00229.x. [DOI] [PubMed] [Google Scholar]
- Cepica S, Yerle M, Stratil A, Schroffel J, Redl B. Regional localization of porcine MYOD1, MYF5, LEP, UCP3 and LCN1 genes. Anim Genet. 1999;30:476–478. [PubMed] [Google Scholar]
- Wang YX, Zhang CL, Yu RT, Cho HK, Nelson MC, Bayuga-Ocampo CR, Ham J, Kang H, Evans RM. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2004;2:e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Wimmers K, Murani E, Schellander K, Ponsuksili S. Combining QTL- and expression-analysis: identification of functional positional candidate genes for meat quality and carcass traits. Arch Tierz. 2005;48:23–31. [Google Scholar]
- Zhao SH, Nettleton D, Liu W, Fitzsimmons C, Ernst CW, Raney NE, Tuggle CK. Complementary DNA macroarray analyses of differential gene expression in porcine fetal and postnatal muscle. J Anim Sci. 2003;81:2179–2188. doi: 10.2527/2003.8192179x. [DOI] [PubMed] [Google Scholar]
- Fiedler I, Nürnberg K, Hardge T, Nürnberg G, Ender K. Phenotypic variations of muscle fibre and intramuscular fat traits in Longissimus muscle of F2 population Duroc × Berlin Miniature Pig and relationships to meat quality. Meat Sci. 2003;63:131–139. doi: 10.1016/S0309-1740(02)00075-X. [DOI] [PubMed] [Google Scholar]
- Horak V. A successive histochemical staining for succinate dehydrogenase and „reserved" ATPase in a single section for the skeletal muscle fibre typing. Histochemistry. 1983;78:545–553. doi: 10.1007/BF00496207. [DOI] [PubMed] [Google Scholar]
- Wegner J, Fiedler I, Klosowska D, Klosowski B, Ziegan B. Veränderungen der Muskelfasertypenverteilung im M. longissimus dorsi von Ebern während des Wachstums, dargestellt mit verschiedenen histochemischen Methoden. Anat Histol Embryol. 1993;22:355–359. doi: 10.1111/j.1439-0264.1993.tb00231.x. [DOI] [PubMed] [Google Scholar]
- Sosnicki A. Association of micrometric traits on meat quality, fattening and slaughter traits in the pig. J Anim Sci. 1987;64:1412–1418. doi: 10.2527/jas1987.652584x. [DOI] [PubMed] [Google Scholar]
- Walasik K, Klosowska D, Grzeskowiak G. Occurrence of pathological changed in M. longissimus lumborum of some crosses of pigs with share of Hampshire breed. Rocz Nauk, Zootechn. 2000;6:233–237. [Google Scholar]
- Josza L, Lehto MU, Jarvinen M, Kvist M, Reffy A, Kannus P. A comparative-study of methods for demonstration and quantification of capillaries in skeletal-muscle. Acta Histochem. 1993;94:89–96. doi: 10.1016/S0065-1281(11)80344-2. [DOI] [PubMed] [Google Scholar]
- Roth K, Wenzelides K. Das interaktive Bildverarbeitungssystem AMBA/R. Gegenbaurs Morphol Jahrb. 1989;135:25–32. [PubMed] [Google Scholar]
- Beyersdorfer G, Ohlerich M, Wegner J. A semi-automated device for measuring muscle-fibers at the microscopic cross-section slide. Z Mikrosk Anat Forsch. 1985;99:671–675. [PubMed] [Google Scholar]
- Picard B, Lefaucheur L, Berri C, Duclos MJ. Muscle fibre ontogenesis in farm animal species. Reprod Nutr Dev. 2002;42:415–431. doi: 10.1051/rnd:2002035. [DOI] [PubMed] [Google Scholar]
- Green P, Falls K, Crooks S. Documentation for CRI-MAP, Version 2.4. 1990.
- QTLexpress http://qtl.cap.ed.ac.uk/
- Seaton G, Haley CS, Knott SA, Kearsey M, Visscher PM. QTL Express: mapping quantitative trait loci in simple and complex pedigrees. Bioinformatics. 2002;18:339–340. doi: 10.1093/bioinformatics/18.2.339. [DOI] [PubMed] [Google Scholar]
- Haley CS, Knott SA, Elsen JM. Mapping quantitative trait loci in crosses between outbred lines using least squares. Genetics. 1994;136:1195–1207. doi: 10.1093/genetics/136.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott SA, Elsen JM, Haley CS. Methods for multiple marker mapping of quantitative trait loci in half-sib populations. Theor Appl Genet. 1996;93:71–80. doi: 10.1007/s001220050249. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]


