Abstract
Many circular genomes have replication termination systems, yet disruption of these systems does not cause an obvious defect in growth or viability. We have found that the replication termination system of Bacillus subtilis contributes to accurate chromosome partitioning. Partitioning of the terminus region requires that chromosome dimers, that have formed as a result of RecA-mediated homologous recombination, be resolved to monomers by the site-specific recombinase encoded by ripX. In addition, the chromosome must be cleared from the region of formation of the division septum. This process is facilitated by the spoIIIE gene product which is required for movement of a chromosome out of the way of the division septum during sporulation. We found that deletion of rtp, which encodes the replication termination protein, in combination with mutations in ripX or spoIIIE, led to an increase in production of anucleate cells. This increase in production of anucleate cells depended on recA, indicating that there is probably an increase in chromosome dimer formation in the absence of the replication termination system. Our results also indicate that SpoIIIE probably enhances the function of the RipX recombinase system. We also determined the subcellular location of the replication termination protein and found that it is a good marker for the position of the chromosome terminus.
DNA replication termination systems have been identified on several circular genomes, including Bacillus subtilis, Escherichia coli, and several plasmids (1, 2). The presence of replication termination systems in diverse organisms suggests an evolutionary advantage for accurate termination of DNA replication. Yet deletion of the chromosomal termination protein genes in a wild-type background has no deleterious effects on growth or development under standard laboratory conditions (1, 3). Here we report that accurate termination of DNA replication contributes to accurate chromosome segregation.
The replication terminator protein (RTP) of B. subtilis is a 29-kDa homodimer (4, 5). Two RTP dimers bind to each Ter site, which consists of a 29-bp imperfect inverted repeat with two overlapping half sites; one of these sites is a strong binding core (B) site, and the other is a weak binding auxiliary (A) site (4). Binding of an RTP dimer to the core site permits cooperative binding of a second dimer to the auxiliary site (6, 7). In B. subtilis, as well as E. coli, the termination complex blocks DNA replication in a polar manner. Replication forks that encounter the B. subtilis RTP-Ter complex from the core side are blocked, although those that encounter RTP-Ter from the auxiliary side are not (8). The termination complexes of B. subtilis and E. coli inhibit helicase-mediated DNA unwinding (9, 10), although the mechanism of helicase inhibition is still debated (11).
Nine Ter sites have been identified in B. subtilis (12). TerI and TerII are located 59 bp apart at ≈172° on the circular 360° chromosome (13, 14). As a result of its slightly asymmetric position, TerI is the termination site that is most often used. TerI encounters the replication fork duplicating from 0° to 172° before TerII can encounter the replication fork duplicating from 360° to 172° (14–16). The replication fork duplicating from 360° to 172° presumably terminates on meeting the fork already halted at TerI. The seven other Ter sites flank TerI and TerII oriented such that they create a replication fork trap in the terminal 10% of the chromosome (12, 14). This trap seems to ensure that a fork that passes through TerI is impeded by the subsequent Ter sites (likewise if a fork passes through TerII).
In bacteria with a single circular chromosome, duplication of a region of the chromosome is closely followed by segregation of the sister regions to opposite halves of the cell. Thus, early replicated regions are partitioned before the bulk of the chromosome is duplicated. Once the two new chromosomes have been completed and partitioned to opposite sides, the cell divides medially. Our current understanding of chromosome partitioning can be simplified into several steps: (i) origin region separation and repositioning; (ii) overall chromosome organization and compaction; and (iii) terminus region separation. This final step includes chromosome decatenation, chromosome dimer to monomer resolution, when necessary, and movement of the termini to either side of midcell before completion of medial septation.
Several genes involved in partitioning have been identified, including spo0J, smc, ripX, spoIIIE, parC, and parE. parC and parE encode the subunits of topoisomerase IV which are required for decatenation (17) and will not be discussed further. Combining some partitioning mutations can cause synthetic effects (18, 19). Therefore, we combined an rtp null mutation with mutations in some of the known partitioning genes. We observed synthetic effects of rtp in combination with spoIIIE and ripX mutations. Both SpoIIIE and RipX have functions that are specifically involved in separating the termini of newly duplicated chromosomes.
SpoIIIE plays a role in postseptational chromosome partitioning. During exponential growth, SpoIIIE enhances accurate chromosome separation when the normal coupling of chromosome partitioning and cell division is disrupted (19, 20). Combining ΔspoIIIE with another partitioning mutant, Δsmc, causes a synthetic lethal phenotype (19). During sporulation, SpoIIIE is required for postseptational partitioning of one chromosome into the smaller forespore compartment (21, 22). SpoIIIE protein localizes to the division septum and has homology to DNA translocation proteins from conjugative plasmids (23). The prevailing model is that SpoIIIE pumps chromosomes out of the way of the division septum.
RipX, the B. subtilis homologue of E. coli XerD, probably acts in the terminus region to resolve chromosome dimers to monomers (24). With circular genomes, dimers can form as a result of homologous recombination between newly duplicated chromosomal regions. Most circular genomes seem to have a site-specific recombinase to resolve such dimers. In the well characterized E. coli system, the dif site is located near the site of replication termination and is acted on by two site-specific recombinases, XerC and XerD (25). The B. subtilis dif site has recently been identified in the terminus region (S. Sciochetti & P. Piggot, personal communication). In E. coli, efficient Xer-mediated dif recombination requires FtsK (26–29), a protein with a carboxyl-terminal domain homologous to SpoIIIE (30). Recent analysis of genome sequences indicates that organisms with Xer homologues all have an FtsK homologue, and those that do not have an Xer homologue do not have an FtsK homologue (31). These findings are consistent with the notion that there is a conserved interaction between Xer and FtsK (31).
In this study we examined both the localization of RTP and the effect of combining Δrtp with some of the known partitioning mutants. Our results indicate that RTP is an excellent marker for chromosome terminus region position in live cells and that RTP-mediated termination of DNA replication can facilitate accurate separation of sister terminus regions.
Materials and Methods
Growth Media and Antibiotics.
Rich medium was LB; defined minimal medium was S7 medium with 1% glucose, 0.1% glutamate, and required amino acids as described (32, 33). Where needed, antibiotics were used at the indicated concentrations: chloramphenicol (to select for the cat gene) 5 μg/ml; erythromycin 0.5 μg/ml and lincomycin 12.5 μg/ml together (to select for the mls gene); spectinomycin (to select for the spc gene) 100 μg/ml; and kanamycin (to select for the kan gene) 5 μg/ml.
Strains.
All strains are derivatives of JH642 which is trpC2 pheA1 (34). The following previously described mutant alleles were used: ΔspoIIIE∷spc (35), spoIIIE36 (21), ΔterC∷cat (3), Δspo0J∷spc (36), Δsmc∷cat (18), ΔripX∷cat (24), and recA260 (cat mls; ref. 37). All mutants were constructed by standard methods (38). The Δrtp∷kan mutation was constructed as follows. A 5′ fragment of rtp (overlapping the ribosome binding site and extending into codon 68) was PCR-amplified with primers oKPL161 and oKPL162 and inserted upstream of kan in pGK67. pGK67 was constructed by inserting kan (PCR-amplified with primers KAN1 and KAN2) from pDG792 (39) into the XbaI site of pGEM-cat (40). A 3′ fragment (extending from within codon 104 to 229 nucleotides past the stop codon) was then PCR-amplified with primers oKPL163 and oKPL164 (primer sequences available on request) and inserted downstream of kan. The resulting plasmid pEX5 (E. Küster-Schöck and A.D.G.) was integrated by double crossover onto the B. subtilis chromosome.
The recA260 mutation is a single crossover insertion-disruption of recA (37). The presence of recA260 was confirmed by sensitivity to mitomycin C (30 ng/ml) on LB agar plates. recA mutants were grown in flasks wrapped in foil to protect them from light. The presence of the ΔripX∷cat in the recA260 (cat mls) mutant was confirmed by recovering each mutation separately. Chromosomal DNA from both the ΔripX∷cat recA260 (cat mls) double and Δrtp∷kan ΔripX∷cat recA260 (cat mls) triple mutant were used to transform wild-type cells selecting for resistance to chloramphenicol (CmR). Transformants were then screened to identify MLS-resistant and MLS-sensitive isolates, and were then tested for ripX (anucleate cell production) and recA (sensitivity to mitomycin-C) phenotypes. Transformants that were CmR- and MLS-sensitive (ripX) were resistant to mitomycin-C and had an anucleate phenotype indicative of the ripX mutation. Transformants that were CmR- and MLS-resistant (recA) were sensitive to mitomycin-C and had an anucleate phenotype indicative of a recA mutation.
yfpmut2 was constructed by site-directed mutagenesis of gfpmut2 (S65A, V68L, S72A; ref. 41), changing amino acid 203 from threonine to tyrosine (T203Y; ref. 42). yfpmut2 was then subcloned in-frame behind a 3′ fragment of rtp (the rtp stop codon was replaced by a XhoI site) in the B. subtilis integration vector pUS19 (43). There is a 5-amino acid linker (LEGSG) between the last amino acid of rtp and the first of yfpmut2. This construct, pGK97, was integrated by single crossover onto the B. subtilis chromosome such that the fusion is the only functional copy of the gene and is under control of the endogenous promoter.
The tandem array of lac operators (lacO) inserted by single crossover at cgeD (44) is marked with cat and was transformed into our strain background by using chromosomal DNA from AT54 (45).
We have found that a carboxyl-terminal fusion of LacI (minus the last 11 codons to reduce tetramerization) to green fluorescent protein (GFP), or its variants, produces a brighter signal than the previously described amino-terminal fusion protein (GFP-LacI; refs. 44 and 46). To construct lacI-gfp and lacI-cfp fusions, the 3′ end of lacI, absent the last 11 codons, was PCR-amplified from pDH88 (47) by using primers oKPL 134 and 135 that added an EcoRI site at the 5′ end and an XhoI site at the 3′ end. The resultant product was then fused in-frame to the 5′ end of gfpmut2 with a five codon linker (LEGSG) to give plasmid pKL153. pKL153 was integrated into strain KPL417 by single crossover into a copy of lacI (driven by the Ppen promoter; ref. 48). The lacI copy had been introduced into the B. subtilis chromosome at the thrC locus with pPL97 (P. Levin, J. Quisel, F. Aragoni, A.D.G., unpublished work), to give strain KPL420. The resulting Ppen-lacI(Δ11)gfpmut2 expression unit was PCR-amplified by using primers oKPL145 and 147 that added EcoRI and BamHI to the 5′ and 3′ ends, respectively, and was then subcloned into pUC9 to give pKL178. A cyan variant of gfp, cfpW7 (49), was swapped for the gfpmut2 of pKL178 by using the XhoI and SphI sites, yielding pKL189. pKL189 was digested with EcoRI and BamHI, and the Ppen-lacI-cfpW7 fragment was inserted into pDG795 (provided by P. Stragier, Institut de Biologie Physico-Chimique, Paris, France; ref. 36) to give pKL190. pKL190 was integrated by double crossover at thrC in the chromosome, resulting in strain KPL682.
Anucleate Cell Determination.
Cells were grown in LB broth at 37°C in flasks in a shaking water bath, except for the Δsmc strains, which were grown in defined minimal medium at 30°C. (The smc phenotype is much more severe in rich medium than in minimal medium; ref. 18.) Samples were taken during midexponential growth, and cells were adhered to poly-L lysine-treated glass slides, fixed with ethanol, and stained with 4′,6-diamidino-2-phenylindole (DAPI, 400–500 ng/ml). Microscopy was performed by using a Nikon E800 microscope equipped with a 100× differential interference contrast (DIC) objective. Images were captured by simultaneous use of DIC optics and fluorescence. The total number of cells and the number of anucleate cells were counted in random fields, and the 95% confidence interval was determined (50).
Live Cell Microscopy.
Cells were grown at 30° in defined minimal medium and prepared for microscopy as described (51). Briefly, an aliquot of cells was removed from an exponentially growing culture and stained with the vital membrane stain FM4–64 (140–280 ng/ml; Molecular Probes) before placement on a pad of 0.5–1% agarose in a solution of 1× T' base (38) with 1 mM MgSO4. The cell outlines (FM4–64) were visualized with a Cy3 filter set, the YFP signal with Chroma filter set # 41029, and the CFP signal with Chroma filter set # 31044. Images were colorized and merged by using Improvision openlab software and cropped in Adobe photoshop 5.5. Images were acquired by using a Nikon E800 microscope equipped with a charge-coupled device camera (Hamamatsu, model C4742–95; Ichinocho, Japan) and Improvision openlab software.
Results
Localization of RTP-YFP in Living Cells.
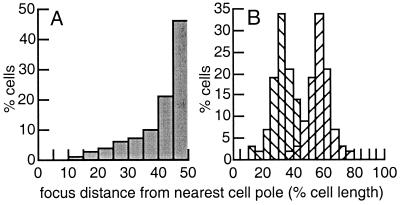
RTP-YFP is a fusion of RTP with a yellow variant of GFP (42) localized as discrete intracellular foci (Fig. 1A). Approximately 80% of cells contained a single RTP-YFP focus located at or near midcell (Fig. 2A). The remaining 20% of cells had two closely juxtaposed foci that were centrally located (Fig. 2B). The RTP-YFP fusion is the only copy of RTP in these cells and does not cause any disturbance in cell growth. Similar results were found with RTP-GFP.
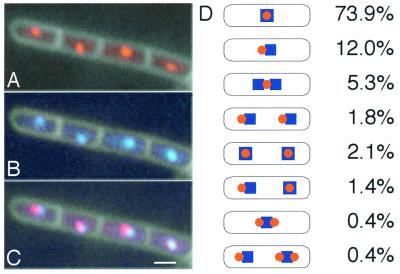
Figure 1.
Chromosome terminus location in live cells. (A) RTP-YFP and cell outline. (B) 181°∷lacO, LacI-CFP, and cell outline. (C) Merged image A and B. (D) Percent of cells belonging to each type of cell pattern in double-labeled cells (283 total cells). The remaining 2.7% of cells counted did not contain signal from both fusions. Orange circles, RTP-YFP; blue squares, LacI-CFP bound to the chromosome terminus at 181°∷lacO. The data are cumulative from two separate experiments.
Figure 2.
RTP-YFP focus position with respect to nearest cell pole. (A) RTP-YFP position in cells with a single focus (230 cells). (B) RTP-YFP position in cells with two foci (58 cells). The distance from mid-focus to nearest cell pole was measured by using Improvision openlab software. The data are cumulative from two separate experiments.
RTP-YFP Colocalizes with the Chromosome Terminus Region.
Previously, the B. subtilis chromosomal terminus region was visualized by inserting a tandem array of lac operators (lacO) at ≈181° in a strain expressing a LacI-GFP fusion (44, 45, 52). We observed RTP-YFP localization in a strain that also carried the 181°∷lacO array and a fusion of LacI to a Cyan variant of GFP (ref. 49; LacI-CFP). We found that in most cells (≈75%) the focus or foci of RTP-YFP was coincident with LacI-CFP bound in the terminus region (Fig. 1 A–D). In almost all of the remaining cells RTP-YFP and LacI-CFP foci were adjacent. We have observed that the lac operator–Lac repressor system, while extremely useful, causes a mild partitioning defect (data not shown; ref. 45). One notable advantage of using RTP-YFP (or RTP-GFP) to localize the terminus region is that strains bearing the fusion appear normal.
Δrtp and Chromosome Partitioning.
The close temporal association between DNA replication and partitioning of the two new copies of a chromosome region led us to speculate that proteins involved in DNA replication might facilitate accurate chromosome partitioning. We found that deletion of rtp did not cause a partitioning defect (Table 1). We postulated that the normal contribution of accurate DNA-replication termination to chromosome partitioning might be masked in a wild-type background. Therefore, we combined Δrtp with several known partitioning mutants.
Table 1.
Anucleate cells
| Relevant genotype | Strain | Anucleates | Total cells | Percentage of anucleates* |
|---|---|---|---|---|
| Wild type | AG174 | 8 | 8,693 | 0.09 (0.06) |
| Δrtp | IRN408 | 6 | 6,687 | 0.09 (0.07) |
| ΔterC† | KPL47 | 35 | 8,364 | 0.42 (0.14) |
| Δspo0J | AG1468 | 29 | 2,048 | 1.4 (0.5) |
| Δrtp Δspo0J | IRN414 | 55 | 3,172 | 1.7 (0.45) |
| Δsmc | RB27 | 94 | 993 | 9.5 (1.8) |
| Δrtp Δsmc | IRN418 | 110 | 1,080 | 10.2 (1.8) |
| ΔspoIIIE | PL422 | 34 | 5,752 | 0.59 (0.2) |
| Δrtp ΔspoIIIE‡ | IRN415 | 158 | 7,606 | 2.08 (0.3) |
| ΔripX | IRN429 | 256 | 2,488 | 10.3 (1.2) |
| Δrtp ΔripX | IRN432 | 369 | 1,782 | 20.7 (1.9) |
| ΔripX ΔspoIIIE§ | IRN436 | 93 | 1,596 | 5.83 (1.15) |
| Δrtp ΔripX ΔspoIIIE | IRN435 | 283 | 1,614 | 17.5 (1.86) |
All data are derived from at least two experiments, except for the Δsmc strains, which are from a single experiment. The 95% confidence interval is shown in parentheses.
We found that the ΔterC mutant SU153 had a mild partitioning defect. ΔterC was constructed by replacement of 11.2 kb of chromosome, spanning 9.7 kb on one side and 1.5 kb on the other side of TerI (removing rtp) with a drug-resistance marker (3). Because this defect is not due to loss of rtp, there may be other genes or DNA sites in this region that have some role in chromosome partitioning.
We observed a similar enhancement of a spoIIIE36 partitioning defect in a Δrtp spoIIIE36 double mutant.
ΔripX spoIIIE36 had a similar percentage of anucleate cells.
Δrtp Had No Effect When Combined with Δspo0J or Δsmc.
Spo0J binds to parS sites located in the origin proximal 20% of the chromosome and is thought to function in origin-region organization and separation (53). A Δrtp Δspo0J double mutant produced essentially the same number of anucleate cells as a Δspo0J single mutant (Table 1).
B. subtilis SMC is involved in partitioning, probably through a role in chromosome organization and compaction (18, 19, 54). Δsmc is temperature-sensitive lethal in rich medium (18). There was no effect of combining Δrtp and Δsmc when cells were grown under conditions permissive for Δsmc. Both the Δsmc and Δrtp Δsmc mutants had ≈10% anucleates (Table 1).
Δrtp Exacerbated the Partitioning Defect of a ΔspoIIIE Mutant.
SpoIIIE is important for moving chromosomes out of the way of the division septum. During exponential growth, this function of SpoIIIE is most readily observed in conditions that perturb chromosome organization (19, 20). We found that Δrtp enhanced anucleate cell production in a ΔspoIIIE mutant. The Δrtp ΔspoIIIE double mutant had an ≈3-fold increase in anucleate cells compared with ΔspoIIIE alone (Table 1). Alone, ΔspoIIIE had an ≈6-fold increase in anucleates compared with wild type.
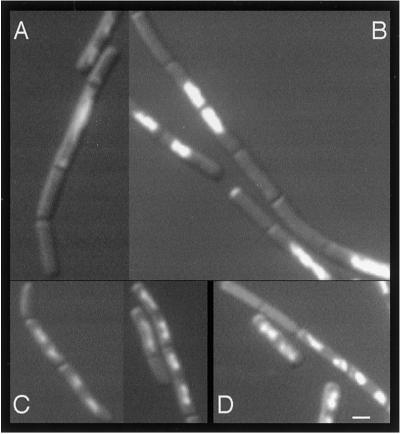
In addition to the quantifiable increase in anucleates in the ΔspoIIIE and Δrtp ΔspoIIIE mutants, there was a characteristic appearance to many of the anucleate cells. We often observed an elongated cell with an unusually large mass of centrally located DNA flanked on one or both sides by a small- to normal-sized anucleate cell (Fig. 3A). We speculate that this defective nucleoid structure results from an initial failure of chromosome separation accompanied by continued DNA replication. Though the resulting large nucleoid mass cannot be partitioned, the division cycle continues unhindered, resulting in an adjacent flanking anucleate cell. This phenotype is very similar to that described for ΔripX strains (see below, ref. 24), and we believe it is a common result of partitioning failure. We have also observed this pattern in the Δspo0J mutant. In addition, we occasionally observed cells where the nucleoid mass was guillotined by a division septum, the so-called “cut” phenotype (19).
Figure 3.
Nucleoid appearance and anucleate cells in various mutants. Samples were taken during exponential growth and stained with DAPI (400–500 ng/ml). (A) Δrtp ΔspoIIIE; (B) Δrtp ΔripX; (C) Δrtp ΔripX recA260; (D) recA260.
Δrtp Enhanced the Partitioning Defect of a ΔripX Mutant.
RipX, the B. subtilis homologue of the E. coli XerD site-specific recombinase (24), acts at a dif site in the Ter region to resolve chromosome dimers to monomers. Like SpoIIIE, RipX is important for proper separation of the chromosome terminus region. We combined Δrtp with both ΔripX and ΔripX ΔspoIIIE.
Both the Δrtp ΔripX double mutant and the Δrtp ΔripX ΔspoIIIE triple mutant had an increased partitioning defect, with ≈2-fold more anucleate cells than the ΔripX single mutant (Table 1). Compared with ΔripX, the ΔripX ΔspoIIIE double mutant had an ≈40% decrease in anucleate cells (Table 1). Previously, the production of anucleate cells by the ΔripX ΔspoIIIE double mutant was reported to be similar to that of the ΔripX single mutant (24). The modest decrease in production of anucleate cells that we observed seems consistent with the previous findings. In addition to the production of anucleate cells and the defective nucleoid structure (Fig. 3B) described above, many of the ΔripX (recA+) mutant cells had abnormally dense or diffuse nucleoids as observed previously (24).
The Enhancing Effect of Δrtp on ΔspoIIIE and ΔripX Anucleate Cell Production Is recA-Dependent.
RecA is required for the primary mode of homologous recombination and, hence, chromosome dimer formation (55). In E. coli, chromosome dimer formation is largely prevented in the absence of recA, and a site-specific recombination system is not needed (56, 57). We found that a B. subtilis recA null (recA260; ref. 37) produced ≈1% anucleates (Table 2, Figs. 3D and 4), but this finding is probably an underestimate of the chromosome defect. As with all our quantitation, we used a very strict definition of “anucleate,” whereby any detectable DAPI-stained material in a cell was interpreted as a nucleated cell. Many recA mutant cells had a decrease in DAPI-stained material compared with wild type. The anucleates in E. coli recA mutants probably result from DNA degradation after failure to repair collapsed DNA replication forks (58, 59). Production of anucleates by DNA degradation is consistent with the absence of the characteristic “cut” phenotype (a division septum cutting the nucleoid) in the B. subtilis recA strains (Fig. 3 and data not shown).
Table 2.
The rtp enhancement of anucleate cell production is recA dependent
| Relevant genotype | Strain* | Anucleates | Total cells | Percentage of anucleates† |
|---|---|---|---|---|
| Wild type | AG174 | 8 | 8,693 | 0.09 (0.06) |
| recA260 | IRN444 | 66 | 6,218 | 1.06 (0.25) |
| Δrtp recA260 | IRN440 | 46 | 6,910 | 0.67 (0.19) |
| ΔspoIIIE recA260 | IRN441 | 39 | 3,622 | 1.08 (0.34) |
| Δrtp ΔspoIIIE recA260 | IRN439 | 59 | 6,040 | 0.98 (0.25) |
| ΔripX recA260 | IRN442 | 63 | 6,654 | 0.95 (0.23) |
| Δrtp ΔripX recA260 | IRN443 | 44 | 4,126 | 1.07 (0.31) |
Strains were handled as described in Materials and Methods, except that the ethanol fixation time was decreased to ≈10 s to preserve cellular structure in strains carrying the recA260 mutation. Identical results were obtained with two different isolates of the ripX recA strains (IRN442 and IRN443), and results shown are cumulative.
All data are derived from at least two experiments. The 95% confidence interval is indicated in parentheses.
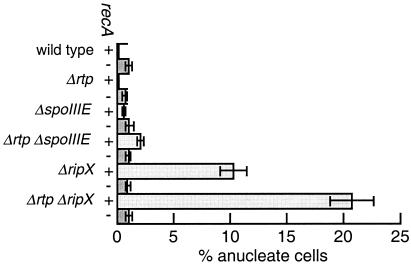
Figure 4.
Effects of recA on production of anucleate cells. Light bars, recA+; dark bars, recA mutant.
The Δrtp ΔspoIIIE recA triple mutant had a partitioning defect similar to that of the recA single mutant, but it was less severe than that of the Δrtp ΔspoIIIE double mutant (Table 2, Fig. 4). In addition to the loss of the rtp-exacerbated anucleate defect, the appearance of the nucleoid in the triple mutant was similar to that in the recA single mutant.
The Δrtp ΔripX recA triple mutant had a partitioning defect similar to that of the recA single mutant (Table 2, Figs. 3 C and D and 4). In addition, the frequency of anucleates and the appearance of the nucleoid in the ΔripX recA double mutant was indistinguishable from that of the recA single mutant (Table 2, Fig. 4). This finding differs from previous work reporting that a recA ΔripX double mutant produces the same number or slightly more anucleates than a ΔripX single mutant (24). We confirmed the presence of both the recA260 and ΔripX mutations in each of our strains (see Materials and Methods). We do not understand the reason for the discrepancy, but suspect that there might be differences because of strain background. The dependence of the ripX defect on recA is in agreement with the phenotype of an E. coli recA xerC double mutant (56), and indicates that, as in E. coli, recA is a primary route for chromosome dimer formation in B. subtilis.
Discussion
By using a fusion of RTP with a variant of GFP (RTP-YFP), we have shown that RTP localizes in discrete foci that correspond to the position of the chromosome terminus region in B. subtilis. Although the newly duplicated origin regions reside on opposite sides of the cell for most of the cell cycle, the terminus region appears to reside at or near midcell (44, 45, 52). Fusion of RTP to GFP variants should prove to be a useful means for visualizing the location of the chromosome terminus region in live cells.
The loss of rtp does not cause a partitioning defect in an otherwise wild-type background. However, our results demonstrate that Δrtp causes an increase in anucleate cell production when combined with mutations that cause defects in partitioning the chromosome terminus region. The rtp-dependent increase in anucleate cell production in both a ΔspoIIIE and a ΔripX background was recA-dependent, indicating that failure to accurately terminate DNA replication perturbs terminus region separation, possibly via an increase in chromosome dimers.
The Absence of rtp Probably Results in an Increase in Chromosome Dimers.
RipX is a site-specific recombinase required for the primary dimer-to-monomer resolution system in B. subtilis (24). The 2-fold increase in anucleates in the Δrtp ΔripX double mutant suggests that deletion of rtp might increase the number of dimers in the population. The recA-dependence of rtp-enhanced anucleate cell production in both spoIIIE and ripX mutants provides stronger evidence that there is an increase in chromosome dimers in the absence of rtp.
We discuss two models to explain how failure to terminate replication accurately in the terminus region might lead to an increase in chromosome dimers. Both models involve the generation of a double-strand end that is highly recombinogenic.
The first model is based on the possibility of overreplication in the absence of RTP. If the 3′ end of the leading strand is not properly terminated, then it could continue on to displace the 5′ end of the lagging strand, resulting in a shift from theta-type replication to rolling circle replication. In fact, inactivation of the replication terminus of plasmid R1 results in such overreplication and destabilization of the plasmid (60). Similar overreplication of the chromosome might result in a DNA structure with a double-strand end. This structure would be a substrate for recA-dependent recombination, leading to the increased probability of chromosome dimer formation.
The second model is based on the proposed generation of a double-strand end after proper RTP-mediated termination, and a role for this end in stimulating dimer-to-monomer resolution. In E. coli, the Ter region has recombinational hotspots independent of site-specific dif recombination (61–63). These hotspots are correlated with the primary Ter sites. It has been proposed that termination of a replication fork results in a “Y” structure that, if cleaved on one of the template strands, gives a recombinogenic double-strand end (63). If the increased frequency of recombination in the Ter region because of proper DNA replication termination normally contributes to chromosome dimer-to-monomer resolution, then the loss of RTP could result in an increase in chromosome dimers.
Functions of SpoIIIE in Chromosome Partitioning.
During sporulation, SpoIIIE is required for postseptational movement of a chromosome into the small forespore compartment (21, 22). During exponential growth, SpoIIIE seems to be an important backup mechanism for partitioning in cells where the two chromosomes fail to separate before medial septum formation (19, 20). SpoIIIE is similar to conjugative plasmid translocation proteins and localizes to the division septum (22, 23). SpoIIIE probably pumps trapped chromosomes out of an essentially complete division septum (22). In addition, the C-terminal region of SpoIIIE is similar to the region of E. coli FtsK that is needed for XerCD function.
There is an apparent paradox in the phenotypes resulting from combining the spoIIIE null mutation with other partitioning mutants. On one hand, ΔspoIIIE causes a decrease in anucleate cells in the ripX mutant. On the other hand, ΔspoIIIE enhances the production of anucleates in the Δrtp mutant. We suspect that SpoIIIE has two functions, one of which is to move trapped chromosomes out of the way of the division septum, and the other is to facilitate RipX-mediated resolution of chromosome dimers to monomers.
We speculate that the unresolved dimers that are formed in the absence of RipX are often moved out of the way of the septum by SpoIIIE. Presumably, pumping the two termini into one cell results in a partitioning disaster with one chromosome caught in the septum. In the absence of SpoIIIE, there is probably more time to resolve chromosome dimers to monomers, presumably via RecA-dependent homologous recombination. In this way, the absence of SpoIIIE can partly suppress the effects of the ripX mutation.
In contrast, there is enhanced production of anucleates in the Δrtp ΔspoIIIE double mutant. Though spoIIIE is not required for RipX-dependent dimer resolution, we propose that SpoIIIE facilitates RipX-mediated resolution of chromosome dimers. In the Δrtp ΔspoIIIE mutant, RipX would, therefore, be less effective at dimer resolution. The carboxyl-terminal domain of E. coli FtsK is homologous to SpoIIIE (30) and is required for efficient chromosome dimer resolution via dif recombination (26–29). Like SpoIIIE, FtsK localizes to the division septum (64). The amino terminal portion of FtsK is required for cell division (26, 64). B. subtilis has a second gene, ytpT, homologous to spoIIIE, but ytpT null mutants and ΔspoIIIE ΔytpT double mutants are not appreciably different from ΔspoIIIE mutants (J. C. Lindow, R. A. Britton, and A.D.G., unpublished data). Apparently, B. subtilis has not developed the tight coupling between dimer resolution, postseptational partitioning, and cell division revealed by various E. coli ftsK mutants. We suspect that B. subtilis may tolerate delays in chromosome partitioning better than E. coli, perhaps because B. subtilis often grows in chains, and daughter cells remain adjacent to each other for longer periods of time.
Replication Proteins and Chromosome Partitioning.
Accurate termination of a round of DNA replication at chromosomal Ter sites seems to facilitate accurate separation of the terminus regions of the chromosome in the final phase of partitioning. Bacteria with a single circular chromosome partition newly duplicated chromosome regions to opposite sides of the cell before the entire chromosome has been replicated. This tight temporal association between DNA replication and chromosome partitioning indicates that proteins involved in DNA replication might facilitate chromosome partitioning. B. subtilis has a centrally located, stationary replisome (51). We have suggested that bacteria may have taken advantage of the motor properties of DNA polymerase and used a stationary replisome to facilitate movement of newly duplicated chromosome regions in opposite directions away from midcell (51). Several genes have been identified that play a role in accurate chromosome partitioning, and combining partitioning mutants often produces synthetic effects. We expect other partitioning genes remain to be discovered and that some, like rtp, may have known roles in DNA replication.
Acknowledgments
We thank Stephen Sciochetti and Patrick Piggot for the ΔripX mutant, Gerry Wake for the ΔterC deletion strain SU153, R. Yasbin for the recA260 mutant, Aurelio Teleman, Chris Webb, and Richard Losick for the 181°(cgeD)∷lacO strain, Roger Tsien for the W7 variant of CFP, Petra Levin for the spoIIIE mutants, and Elke Küster-Schöck for pEX5. We thank Gerry Wake, Stephen Sciochetti, Tania Baker, Rob Britton, and Kate Bacon Schneider for comments on the manuscript and all of the members of our lab, particularly Rob Britton, for helpful discussions. This work was supported by Public Health Services Grant GM41934 and the Merck/MIT Collaborative Program.
Abbreviations
- RTP
replication terminator protein
- lacO
lac operators
- RTP-YFP
a fusion of RTP with a yellow variant of green fluorescent protein
- GFP
green fluorescent protein
- DAPI
4′,6-diamidino-2-phenylindole
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011506098.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011506098
References
- 1.Baker T A. Cell. 1995;80:521–524. doi: 10.1016/0092-8674(95)90504-9. [DOI] [PubMed] [Google Scholar]
- 2.Bussiere D E, Bastia D. Mol Microbiol. 1999;31:1611–1618. doi: 10.1046/j.1365-2958.1999.01287.x. [DOI] [PubMed] [Google Scholar]
- 3.Iismaa T P, Wake R G. J Mol Biol. 1987;195:299–310. doi: 10.1016/0022-2836(87)90651-6. [DOI] [PubMed] [Google Scholar]
- 4.Lewis P J, Ralston G B, Christopherson R I, Wake R G. J Mol Biol. 1990;214:73–84. doi: 10.1016/0022-2836(90)90147-E. [DOI] [PubMed] [Google Scholar]
- 5.Bussiere D E, Bastia D, White S W. Cell. 1995;80:651–660. doi: 10.1016/0092-8674(95)90519-7. [DOI] [PubMed] [Google Scholar]
- 6.Langley D B, Smith M T, Lewis P J, Wake R G. Mol Microbiol. 1993;10:771–779. doi: 10.1111/j.1365-2958.1993.tb00947.x. [DOI] [PubMed] [Google Scholar]
- 7.Carrigan C M, Pack R A, Smith M T, Wake R G. J Mol Biol. 1991;222:197–207. doi: 10.1016/0022-2836(91)90206-l. [DOI] [PubMed] [Google Scholar]
- 8.Smith M T, Wake R G. J Mol Biol. 1992;227:648–657. doi: 10.1016/0022-2836(92)90214-5. [DOI] [PubMed] [Google Scholar]
- 9.Khatri G S, MacAllister T, Sista P R, Bastia D. Cell. 1989;59:667–674. doi: 10.1016/0092-8674(89)90012-3. [DOI] [PubMed] [Google Scholar]
- 10.Kaul S, Mohanty B K, Sahoo T, Patel I, Khan S A, Bastia D. Proc Natl Acad Sci USA. 1994;91:11143–11147. doi: 10.1073/pnas.91.23.11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duggin I G, Andersen P A, Smith M T, Wilce J A, King G F, Wake R G. J Mol Biol. 1999;286:1325–1335. doi: 10.1006/jmbi.1999.2553. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths A A, Andersen P A, Wake R G. J Bacteriol. 1998;180:3360–3367. doi: 10.1128/jb.180.13.3360-3367.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrigan C M, Haarsma J A, Smith M T, Wake R G. Nucleic Acids Res. 1987;15:8501–8509. doi: 10.1093/nar/15.20.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths A A, Wake R G. J Bacteriol. 2000;182:1448–1451. doi: 10.1128/jb.182.5.1448-1451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss A S, Wake R G. J Mol Biol. 1983;171:119–137. doi: 10.1016/s0022-2836(83)80349-0. [DOI] [PubMed] [Google Scholar]
- 16.Weiss A S, Wake R G. Cell. 1984;39:683–689. doi: 10.1016/0092-8674(84)90475-6. [DOI] [PubMed] [Google Scholar]
- 17.Adams D E, Shekhtman E M, Zechiedrich E L, Schmid M B, Cozzarelli N R. Cell. 1992;71:277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- 18.Britton R A, Lin D C, Grossman A D. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Britton R A, Grossman A D. J Bacteriol. 1999;181:5860–5864. doi: 10.1128/jb.181.18.5860-5864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharpe M E, Errington J. Proc Natl Acad Sci USA. 1995;92:8630–8634. doi: 10.1073/pnas.92.19.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu L J, Errington J. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- 22.Wu L J, Lewis P J, Allmansberger R, Hauser P M, Errington J. Genes Dev. 1995;9:1316–1326. doi: 10.1101/gad.9.11.1316. [DOI] [PubMed] [Google Scholar]
- 23.Wu L J, Errington J. EMBO J. 1997;16:2161–2169. doi: 10.1093/emboj/16.8.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sciochetti S A, Piggot P J, Sherratt D J, Blakely G. J Bacteriol. 1999;181:6053–6062. doi: 10.1128/jb.181.19.6053-6062.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill T M. In: Features of the Chromosomal Terminus Region. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Vol. 2. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1602–1614. [Google Scholar]
- 26.Liu G, Draper G C, Donachie W D. Mol Microbiol. 1998;29:893–903. doi: 10.1046/j.1365-2958.1998.00986.x. [DOI] [PubMed] [Google Scholar]
- 27.Steiner W, Liu G, Donachie W D, Kuempel P. Mol Microbiol. 1999;31:579–583. doi: 10.1046/j.1365-2958.1999.01198.x. [DOI] [PubMed] [Google Scholar]
- 28.Yu X C, Weihe E K, Margolin W. J Bacteriol. 1998;180:6424–6428. doi: 10.1128/jb.180.23.6424-6428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Recchia G D, Aroyo M, Wolf D, Blakely G, Sherratt D J. EMBO J. 1999;18:5724–5734. doi: 10.1093/emboj/18.20.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Begg K J, Dewar S J, Donachie W D. J Bacteriol. 1995;177:6211–6222. doi: 10.1128/jb.177.21.6211-6222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Recchia G D, Sherratt D J. Mol Microbiol. 1999;34:1146–1148. doi: 10.1046/j.1365-2958.1999.01668.x. [DOI] [PubMed] [Google Scholar]
- 32.Vasantha N, Freese E. J Bacteriol. 1980;144:1119–1125. doi: 10.1128/jb.144.3.1119-1125.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaacks K J, Healy J, Losick R, Grossman A D. J Bacteriol. 1989;171:4121–4129. doi: 10.1128/jb.171.8.4121-4129.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perego M, Spiegelman G B, Hoch J A. Mol Microbiol. 1988;2:689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- 35.Pogliano K, Hofmeister A E, Losick R. J Bacteriol. 1997;179:3331–3341. doi: 10.1128/jb.179.10.3331-3341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ireton K, Gunther N W, IV, Grossman A D. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheo D, Bayles K W, Yasbin R. Biochimie (Paris) 1992;74:755–762. doi: 10.1016/0300-9084(92)90148-8. [DOI] [PubMed] [Google Scholar]
- 38.Harwood C R, Cutting S M. Molecular Biological Methods for Bacillus. Chichester, U.K.: Wiley; 1990. [Google Scholar]
- 39.Guerout-Fleury A M, Shazand K, Frandsen N, Stragier P. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 40.Youngman P, Poth H, Green B, York K, Olmedo G, Smith K. In: Methods for Genetic Manipulation, Cloning, and Functional Analysis of Sporulation Genes in Bacillus Subtilis. Smith I, Slepecky R, Setlow P, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 65–87. [Google Scholar]
- 41.Cormack B P, Valdivia R H, Falkow S. Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 42.Ormo M, Cubitt A B, Kallio K, Gross L A, Tsien R Y, Remington S J. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 43.Benson A K, Haldenwang W G. J Bacteriol. 1993;175:2347–2356. doi: 10.1128/jb.175.8.2347-2356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Webb C D, Teleman A, Gordon S, Straight A, Belmont A, Lin D C, Grossman A D, Wright A, Losick R. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- 45.Teleman A A, Graumann P L, Lin D C H, Grossman A D, Losick R. Curr Biol. 1998;8:1102–1109. doi: 10.1016/s0960-9822(98)70464-6. [DOI] [PubMed] [Google Scholar]
- 46.Robinett C C, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont A S. J Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henner D J. Methods Enzymol. 1990;185:223–228. doi: 10.1016/0076-6879(90)85022-g. [DOI] [PubMed] [Google Scholar]
- 48.Yansura D G, Henner D J. Proc Natl Acad Sci USA. 1984;81:439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heim R, Tsien R Y. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 50.Hogg R V, Tanis E A. Probability and Statistical Inference. New York: Macmillan; 1983. [Google Scholar]
- 51.Lemon K P, Grossman A D. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- 52.Webb C D, Graumann P L, Kahana J A, Teleman A A, Silver P A, Losick R. Mol Microbiol. 1998;28:883–892. doi: 10.1046/j.1365-2958.1998.00808.x. [DOI] [PubMed] [Google Scholar]
- 53.Lin D C, Grossman A D. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 54.Moriya S, Tsujikawa E, Hassan A K, Asai K, Kodama T, Ogasawara N. Mol Microbiol. 1998;29:179–187. doi: 10.1046/j.1365-2958.1998.00919.x. [DOI] [PubMed] [Google Scholar]
- 55.Dubnau D. In: Genetic Exchange and Homologous Recombination. Sonenshein A L, Hoch J A, Losick R, editors. Washington, DC: Am. Soc. Microbiol.; 1993. pp. 555–584. [Google Scholar]
- 56.Blakely G, Colloms S, May G, Burke M, Sherratt D. New Biol. 1991;3:789–798. [PubMed] [Google Scholar]
- 57.Kuempel P L, Henson J M, Dircks L, Tecklenburg M, Lim D F. New Biol. 1991;3:799–811. [PubMed] [Google Scholar]
- 58.Skarstad K, Boye E. J Bacteriol. 1993;175:5505–5509. doi: 10.1128/jb.175.17.5505-5509.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cox M M. Genes Cells. 1998;3:65–78. doi: 10.1046/j.1365-2443.1998.00175.x. [DOI] [PubMed] [Google Scholar]
- 60.Krabbe M, Zabielski J, Bernander R, Nordstrom K. Mol Microbiol. 1997;24:723–735. doi: 10.1046/j.1365-2958.1997.3791747.x. [DOI] [PubMed] [Google Scholar]
- 61.Louarn J M, Louarn J, Francois V, Patte J. J Bacteriol. 1991;173:5097–5104. doi: 10.1128/jb.173.16.5097-5104.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Louarn J, Cornet F, Francois V, Patte J, Louarn J M. J Bacteriol. 1994;176:7524–7531. doi: 10.1128/jb.176.24.7524-7531.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horiuchi T, Fujimura Y, Nishitani H, Kobayashi T, Hidaka M. J Bacteriol. 1994;176:4656–4663. doi: 10.1128/jb.176.15.4656-4663.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang L, Lutkenhaus J. Mol Microbiol. 1998;29:731–740. doi: 10.1046/j.1365-2958.1998.00958.x. [DOI] [PubMed] [Google Scholar]