Abstract
Most of the human genome encodes neither protein nor known functional RNA, yet available approaches to seek meaningful information in the “noncoding” sequence are limited. The unique biology of the X chromosome, one of which is silenced in mammalian females, can yield clues into sequence motifs involved in chromosome packaging and function. Although autosomal chromatin has some capacity for inactivation, evidence indicates that sequences enriched on the X chromosome render it fully competent for silencing, except in specific regions that escape inactivation. Here we have used a linguistic approach by analyzing the frequency and distribution of nine base-pair genomic “words” throughout the human genome. Results identify previously unknown sequence differences on the human X chromosome. Notably, the dinucleotide repeats [AT]n, [AC]n, and [AG]n are significantly enriched across the X chromosome compared with autosomes. Moreover, a striking enrichment (>10-fold) of [GATA]n is revealed throughout the 10-Mb segment at Xp22 that escapes inactivation, and is confirmed by fluorescence in situ hybridization. A similar enrichment is found in other eutherian genomes. Our findings clearly demonstrate sequence differences relevant to the novel biology and evolution of the X chromosome. Furthermore, they implicate simple sequence repeats, linked to gene regulation and unusual DNA structures, in the regulation and formation of facultative heterochromatin. Results suggest a new paradigm whereby a regional escape from X inactivation is due to the presence of elements that prevent heterochromatinization, rather than the lack of other elements that promote it.
The inactive X chromosome in mammalian females provides a singular opportunity to study putative sequences involved in the structural and functional transformation of essentially a whole chromosome. The XIST gene on one X chromosome produces a stable nuclear RNA that coats the chromosome, thereby initiating a cascade of chromatin remodeling that permanently silences the chromosome. (for review, see Chadwick and Willard 2003; Hall and Lawrence 2003; Heard 2004). While insertion of Xist transgenes can induce transcriptional inactivation of an autosome (Herzing et al. 1997; Lee and Jaenisch 1997; Marahens et al. 1997; Wutz and Jaenisch 2000), studies of X;autosome translocations show that the capacity for complete, stable autosomal inactivation is compromised (White et al. 1998; Wolff et al. 1998; Sharp et al. 2002; Lyon 2003), as is the capacity to stably bind XIST RNA (Hall et al. 2002b). Furthermore, specific regions of the X chromosome consistently escape silencing (Carrel and Willard 2005). These observations strongly indicate that the X chromosome has features that enhance its ability to be inactivated and that there is sequence specificity to this process.
Sequence motifs involved in chromosome structure or regulation would likely be highly represented throughout the genome, and may comprise motifs that are difficult to discriminate from mere “junk.” It was long ago suggested that repetitive sequences may be involved in promoting chromosome inactivation (Gartler and Riggs 1983), and particular attention has been given to L1 LINE elements based on X;autosome translocation studies, their general enrichment on the X chromosome (for review, see Lyon 2003), and bioinformatic evidence consistent with this hypothesis (Bailey et al. 2000). However, other studies of canonical repeats have concluded that the L1 elements are not likely involved (Chureau et al. 2002; Ke and Collins 2003) or may not be solely responsible (Ross et al. 2005).
In this study, we have taken a different strategy; we have searched for any motifs that are abundant, widely distributed, and specifically enriched on the X chromosome. This will identify specific sequences relevant to X chromosome biology, which may be implicated in such basic processes as chromatin folding, regulation of heterochromatic or euchromatic domains, and recombination. The analyses were performed on genomic sequence masked for known interspersed repeat families (e.g., LINEs, SINEs, and LTRs). Although the copious interspersed repeats may well contribute to genome function, their presence in this analysis would obscure other repeated motifs.
To accomplish this we used a linguistic approach, counting the occurrences and distribution of nine base-pair words in the genomic sequence of all individual human chromosomes, with focus on the X chromosome. We divided the X chromosome into two regions: XE, a 7.5-Mb region at Xp22 that includes the pseudoautosomal region and escapes X inactivation (Carrel and Willard 2005), and XS, the remainder of the chromosome that is largely silenced on the inactive X. Although there are other genes that at least partially escape inactivation scattered throughout XS, the XE region is distinct, in that escape from inactivation is the rule rather than the exception, and XE genes are expressed on the inactive X at levels closer to those on the active X chromosome, in contrast to the lower levels of escape genes on XS (Carrel and Willard 2005). In fact, the almost complete resistance of the XE region to inactivation suggests that it may be distinct not only from XS, but from autosomal chromatin as well, since the latter is at least partially subject to inactivation.
Results
Overview analysis of small word frequenciesin the whole genome
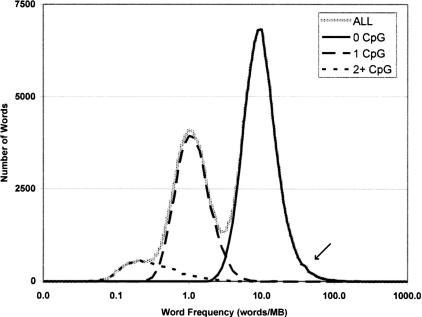
There are 131,072 possible nine base complementary word pairs derived from four letters (ACGT). In the masked human genome (∼1500 Mb), each word would be present roughly 11,000 times in the genome, or ∼7.6 times per Mb, if the frequency of words were random. Figure 1 illustrates the distribution of word frequencies observed. Each possible word is present in the human genome, but there was an extremely wide range of frequencies. While the ubiquitous [A]9 occurs 472,658 times, words containing multiple copies of the underrepresented dinucleotide CpG tend to be quite rare; CGTACGTCG occurs only 47 times in the entire genome. The unique nature of CpG containing words is apparent as they form distinguishable peaks on a frequency distribution histogram (Fig. 1). Of the 10,134 word pairs that occur at low frequencies (between 1 and 1.5 copies per Mb), 96% contain exactly one CpG. In contrast, of 20,765 words that occur at 10-fold higher frequencies, only 0.06% contain any CpG.
Figure 1.
Distribution of word frequencies in the genome. The x-axis represents the frequency of word pairs in the genome, and the y-axis is the number of word pairs that occur at that frequency. The highest peak is largely populated by complex words that contain no CpGs. Words containing two and one CpGs, respectively, populate the first two peaks. The rarest words in the left tail have three or four CpGs, while the shoulder on the right tail is composed of simple sequence, largely mono- and dinucleotide repeats (see arrow).
The other major word class present at frequencies notably deviant from normal frequency distribution consists of words derived from simple sequence repeats (SSRs) or microsatellites (repeats of 1–6-bp units). For example, ATATATATA occurs 604 times per MB, in contrast to the median 9mer word frequency of 5.76 words/MB. The abundance and wide distribution of SSRs, which comprise 3% of the genome (Lander et al. 2001) is not well understood, and while often presumed to be a neutral byproduct of mutation, these sequences have also been speculated to have some regulatory or structural role within the chromosome (e.g., Subramanian et al. 2003b; Ellegren 2004). These observations confirm that our approach accurately identifies words that are statistically and functionally distinctive in the human genome.
Is bulk X chromosome sequence more different from autosomes than individual autosomes are from each other?
Since the X chromosome is subject to different evolutionary forces, its overall sequence content could be distinct from that of autosomes. To assess this, we quantified the extent to which each individual chromosome contained words at frequencies different from the genomic average. This involved summing the differences between the individual chromosomal densities and the mean autosomal density (words/Mb), for each possible word, treating enriched and depleted words (relative to the autosomal average) separately. The bulk deviation in word frequencies for each autosome correlates well with its deviance from average G/C content (Supplemental Fig. 1). The XS segment does not have a great bulk of word frequencies that set it apart from autosomes of similar G/C content. Thus, if certain word types are found enriched in the masked XS sequence, this would not be due to a gross overall difference in sequence content, but would suggest a more subtle, specific enrichment. As will be discussed below, the XE segment and the Y chromosome, however, do appear to have differences from the genomic average beyond what would be expected from their G/C content deviance.
Enrichment of specific SSRs on XS vs. autosomes
Of the 131,072 possible word pairs, 7644 (∼6%) occurred on XS at frequencies significantly different from those in an average autosomal sequence (χ2 analysis, P′ < 0.01, see Methods), but the vast majority of these reflect relatively minor differences that correlate with G/C content. Of the 20 words that are at least twofold enriched on X over the autosomes, many overlap into larger sequences. Physical distribution analysis shows that most of these are derived from a tandem repeat (minisatellite) with 189 well-conserved copies of a 37-bp unit that is both A/T and CpG rich, and which has not been previously described (see Supplemental material for sequence). This sequence is at Xq21.2–21.33 and has a Y homolog at Yp11, which are the boundaries of an Xq/Yp homologous region that is a landmark for a recent evolutionary sex chromosome rearrangement. (Lahn and Page 1999; Tilford et al. 2001). Other words enriched at least twofold were found to be other minisatellite sequences; thus, no well-dispersed words were found to be enriched more than twofold on X.
We adapted our search method to not only identify words with unusual frequencies on X, but to favor more common and widely distributed motifs, screening out those repeated at one or a few sites and rare words, since the lower the copy number of a sequence, the less meaningful enrichment on any individual chromosome would be. Therefore, we ranked words by the difference in word density (words/Mb) on XS and XE vs. autosomes, which takes both abundance and enrichment into account. We also imposed a coefficient of variance cut-off to verify broad physical distribution (see Methods).
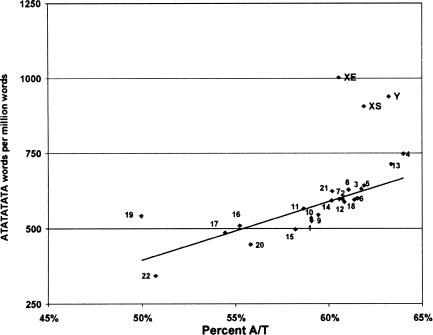
Under these constraints, the most enriched words on XS relative to autosomes are three dinucleotide repeats, [AT]n, [AC]n, and [AG]n. These are enriched between 1.2 and 1.5 times on the X chromosome. This is more striking when one considers that the copy number is very high already on the autosomes. For example, there are 590 words per MB for [AT]n on autosomes compared with about 900 per MB on XS, with quite uniform distribution. Since the X chromosome is rather A/T rich and [AT]n showed more variation among individual autosomes than did AG or AC, we compared the [AT]n density relative with A/T content for all chromosomes. XS and XE (as well as Y) are clearly outliers in terms of [AT]n enrichment, even when compared with similarly A/T-rich, gene-poor chromosomes (Fig. 2); thus, A/T content does not account for this enrichment.
Figure 2.
The density of ATATATATA on chromosomes relative to A/T content. While there is a correlation between [AT]n word density and A/T content (trendline is a linear regression of autosomal values), the enriched [AT]n density on both XE and XS make chromosome X a clear outlier, and chromosome Y as well. The same holds true for the relationship between [AT]n density and gene density.
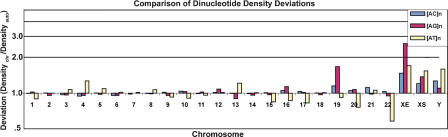
We also performed comparisons of the deviance of the dinucleotide motif word densities from the autosomal mean among all individual chromosomes, (Fig. 3). The X chromosome, both XS and XE, is strikingly more enriched than any other individual autosome for these three dinucleotides. Unlike [AT]n, the [AC]n and [AG]n dinucleotides do not vary with gene density or A/T content, and they are present at similar levels on each of the autosomes, with the exception of chromosome 19, which is anomalous in other ways; it is the most gene-rich chromosome (Grimwood et al. 2004) and has increased density of the various classes of SSRs (Subramanian et al. 2003b). As will be considered in the discussion, each of these dinucleotide sequences has been linked to regulation of individual genes and can form usual DNA structures.
Figure 3.
Individual chromosomal deviation from the autosomal mean frequency of dinucleotide repeat derived 9mer words, expressed as a ratio and plotted on a log scale.
Comparison of XE vs. XS
In considering the biological significance of any enrichment on XS compared with autosomes, it is important to also consider its distribution on XS relative to XE. Although XE and XS occupy the same physical chromosome, their transcriptional behavior under silencing conditions is quite different. The XE region as defined here is one continuous 7.5-Mb block, in which all of the genes escape inactivation, whereas throughout the ∼145 Mb of XS, most, but not all genes are silenced. The XE region also includes the important pseudoautosomal region (PAR), which is homologous to the PAR on the Y chromosome with which it engages in meiotic recombination in males. Given that this region undergoes recombination in both sexes, similar to an autosome, it provides clues as to whether any differences in the repeat content of the X chromosome might be explained by the more limited recombination of XS.
In this respect, it is important to note that the enrichment of dinucleotide repeats is not restricted to the XS region, but is also seen in XE (Fig. 3). This does not preclude the possibility that this motif could have some role related to X inactivation, particularly since we did not find a spatial difference between escape and silenced genes relative to the XIST RNA territory (C.M. Clemson, L.L. Hall, and J.B. Lawrence, in prep.). However, it does indicate that escape of the XE region from inactivation is not due to depletion of dinucleotide repeats. The fact that both XE and XS are enriched for dinucleotide repeats makes the important point that the enrichment is not easily explained by differences in the rate of recombination of XS and autosomes.
Only eight words that can be described with three motifs are significantly (P′ < 0.01) enriched on XS as compared with XE: [AAGGC]n, CCCACCCC, and [CAG]n. However, [AAGGC]n and CCCACCCC are excluded based on their coefficient of variance (V > 100) due to the fact that they are localized large-tandem repeats on XS. The CAG repeat has approximately a twofold enrichment and more even distribution, [CAG]n, but it encodes polyglutamine common in proteins and is highly correlated with gene density, so the enrichment on XS can be attributed to greater gene density than XE (data not shown). Therefore, it is notable that our search found no 9mer or larger words that met the criteria for abundance and distribution that were significantly enriched on XS over XE. This finding is further noteworthy because it contrasts with the distribution of LINE L1 elements in unmasked genomic sequence, which we confirm are enriched (in unmasked sequence) on XS vs. XE (Bailey et al. 2000). We further this result by showing the enrichment of L1 elements on XS vs. all individual autosomes (Supplemental Fig. 2). Interestingly, our analysis shows that the Y chromosome is also enriched in L1 elements.
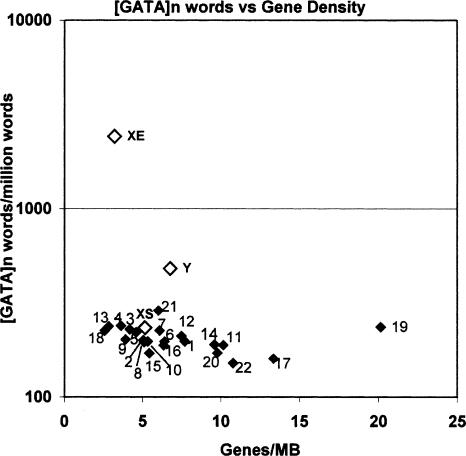
The most striking word frequency difference turned out to be a marked enrichment of a specific motif on XE as compared not only with XS, but also to autosomes. The 9mer word frequency analysis revealed that words representing the tetramer repeat [GATA]n are overwhelmingly enriched in XE. While there is some enrichment also for the [ATCC]n repeat, this was less pronounced and also found in specific segments of other chromosomes (J.A. McNeil and J.B. Lawrence, in prep.), whereas the [GATA]n was enriched on XE over all other autosomes and XS. The [GATA]n repeats constitute a remarkable feature of the XE region, being almost 12 times more frequent on XE than the autosomal mean. Although the X chromosome in its entirety initially appears somewhat enriched with this sequence, about 1.3 times the autosomal average, when one considers our finding that [GATA]n frequency shows an inverse correlation with gene density, the XS is not enriched for [GATA]n over autosomes with similarly low-gene density, in contrast to the dramatic enrichment on XE (Fig. 4).
Figure 4.
Scatter plot of [GATA]n-derived word density vs. gene density for each chromosome.
We scanned the RepBase libraries (which is used by RepeatMasker) for human LINES and SINES and found that these interspersed repeats generally do not have GATA, although some do contain dinucleotide repeats. To ensure that the distribution differences seen for these SSR’s was not related to differences in distribution of interspersed repeats, a limited analysis on unmasked sequence involving XE, XS, and chromosome 7 was performed. The same patterns were seen and the enrichment on XS and XE were still significant.
Detailed analysis of [GATA]n distribution
We examined the genomic distribution of this motif using an alternative method, i.e., fluorescence in situ hybridization. Hybridization of a biotin-labeled 21-bp oligonucleotide probe ([GATA]5G) on a female metaphase spread is shown in Figure 5A. This approach confirms the singular enrichment of this sequence specifically in the XE region, which stands out not only above XS, but over any region of any autosome. The large signal on a segment of Xp is consistent with localization across a several Mb region; in contrast, a smaller spot signal (Fig. 5B) is generated by a hybridization to the ∼7-kb X-Y minisatellite sequence described above. A much lower level of [GATA]n signal is seen on all chromosome arms (excluding large blocks of satellite).
Figure 5.
In situ hybridization of [GATA]4G oligomer (red signals) to human metaphase chromsomes. (A) Female metaphase with DAPI counterstain displayed in reverse contrast. (B) Oligomer hybridization to an ∼7-kb minisatellite; note the small size of the signal relative to the multimegabase regions defined by GATA. (C) Male sex chromosomes.
In hybridizations to male metaphase spreads, a less-intense [GATA]n signal is also seen on the Y chromosome in the pseudoautosomal region at Yp11.3 (Fig. 5C), and to a lesser degree at Yq11, which is populated by many genes and pseudogenes with homologous partners located in XE. The strata included in XE originally were part of a larger PAR, which was shared by X and Y. These strata became independent of the PAR (and no longer able to recombine in male meiosis) due to rearrangements on the Y that translocated much of the sequence from Yp to Yq (Lahn and Page 1999; Ross et al. 2005). The current distribution of [GATA]n on X and Y suggests that the enrichment in XE predates the rearrangement of the strata 4/5 genes on Y, and was therefore a feature of a larger ancestral PAR. The repeats are most prominent in the current PAR, but are still enriched in the remainder of XE and on the portions of Y that are derived from stratas 4 and 5. The same reasoning applies to the enrichment of [AT]n and [AC]n on Y, which show a similar split distribution (data not shown).
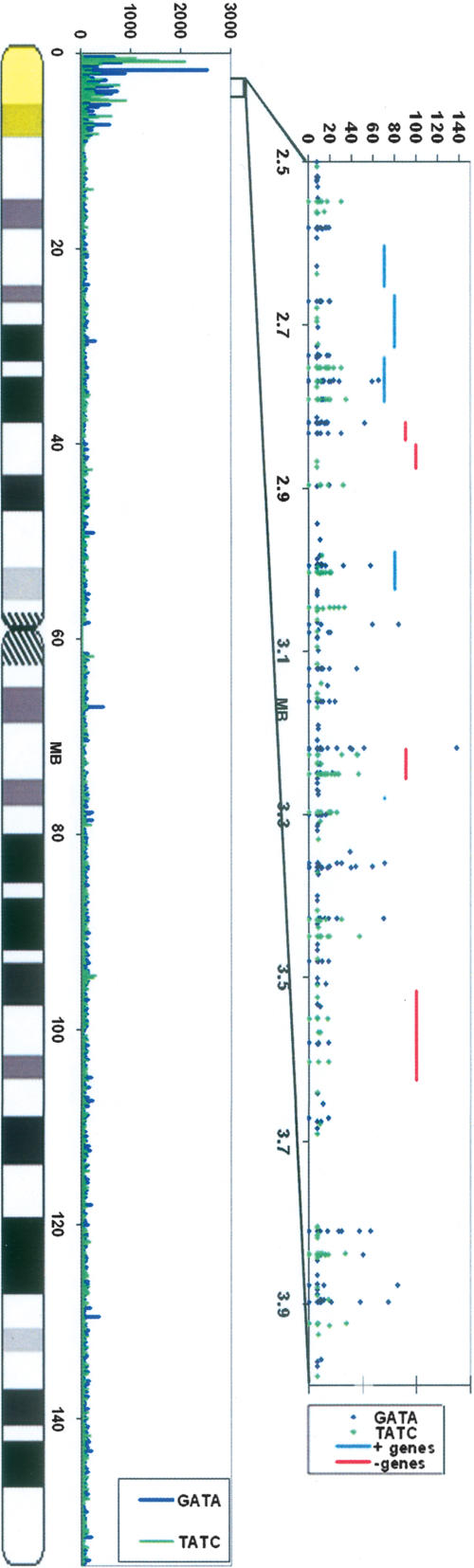
We also examined the distribution of [GATA]n at high resolution along the X chromosome by searching for perfect incidences of GATA repeats of any length, rather than nine base words, and creating a physical map (Fig. 6). This analysis shows that [GATA]n is present at a large number of sites in microsatellites scattered throughout the XE segment. [GATA]n is present both near genes and in intergenic regions, with no apparent relationship between the orientation of genes and [GATA]n/[TATC]n. The enrichment of [GATA]n covers ∼10 MB starting at the p telomere, and encompasses PAR1 and stratas 5 and 4. Although we defined XE conservatively as a 7.5-Mb region based on earlier studies (Carrel et al. 1999), a very recent study (Carrel and Willard 2005) reported a larger 10-Mb region populated entirely by genes escaping inactivation, which corresponds well with the region we find to be enriched with [GATA]n. The [GATA]n distribution on the rest of the X chromosome shows little variance, and therefore does not suggest a clustering of [GATA]n sites in particular regions where isolated genes or small clusters of genes escape inactivation. That such genes might escape by a distinct mechanism is suggested by recent findings (Filippova et al. 2005) showing that individual escape genes on XS are separated from the adjacent silenced genes by CTCF boundary sites, unlike contiguous escape genes in the XE region.
Figure 6.
Physical distribution of [GATA]n on the X chromosome based on NCBI release B35.1 unmasked sequence. The histogram on the left shows the distribution of [GATA]n/[TATC]n-derived word frequencies in 100-kb bins along the chromosome. On the right is a detailed map of the location and length of [GATA]n/[TATC]n tandem repeats in the distal p arm of the X chromosome. Single contiguous repeats are defined as being at least 8 bp long. Interruptions of no more than 4 bp (one repeat unit) of incorrect sequence were allowed if followed by at least two more correct units. Location of genes on the + and − strands are also indicated. The region highlighted in yellow indicates XE in both graphs.
Enrichment of GATA in chimp and dog pseudoautosomal regions
Finally, we examined whether the GATA enrichment seen in the escape region of the human X chromosome is also present in the analogous region of other mammalian species. In this case, the mouse model is less informative because mice (Mus musculus) have a much smaller PAR of different evolutionary origin (Perry et al. 2001) and most of the genes that escape inactivation in the human are silenced in the mouse (Brown and Greally 2003). Many genes found in the human PAR are autosomal in mouse, and there are only two escaping genes in the mouse PAR (plus just five others, including Xist, across the whole chromosome). We found no significant enrichment of GATA in the mouse PAR, although the entire mouse genome is more enriched for GATA.
More informative is the analysis of dogs and chimps, because they have pseudoautosomal regions similar to the human. While not fully characterized, some evidence has shown that genes in this region (in both dog and chimp) escape inactivation (Jegalian and Page 1998), as would be expected to provide equal dosage between males and females. We found that the distribution of GATA and dinucleotides on X in chimps (Pan troglodytes) is similar to humans (data not shown). Perhaps most striking is that a marked enrichment in GATA (approximately sevenfold, Supplemental Fig. 3) is present in dogs (Canis familiaris), which have an X chromosome structure and PAR gene content similar to humans (Kirkness et al. 2003). Dinucleotides in dogs are also enriched on X over autosomes (data not shown).
Discussion
Despite the enormous success in identifying conventional genes within the human genome, knowledge of how to relate “noncoding” genomic sequence to the structure and function of a chromosome is at a primitive stage. Using an open-ended word frequency approach, we identified distinctive sequence features on the X chromosome; these provide new clues to the unique biology of this chromosome, and to the potential role of certain “junk” DNA. Recently there has been increased interest in the abundant microsatellites throughout the human and other genomes (Subramanian et al. 2003b; Ellegren 2004). The idea that SSRs might have functional significance has been discussed (Epplen et al. 1996) and their capacity to adopt nonstandard DNA forms enhances their attraction as candidates in chromosome structure and regulation. Findings here lend credibility to this notion by revealing differences in specific SSR content that correspond to functional differences on the X chromosome. The enrichment of particular SSRs, and not others, suggests specificity that is not easily reconciled with neutral mutational mechanisms involving errors in replication or recombination of highly repetitive DNA.
In our view, the complex biology of chromosome inactivation (and escape from it) is unlikely to be controlled by or dependent upon any one sequence element. For example, there may be motifs that support the propagation of XIST RNA along the chromosome, others that help retain XIST RNA, and still others required for chromatin modifications, DNA methylation, or architectural changes. Substantial attention has been paid to the proposed involvement of LINE L1 elements in these activities (Gartler and Riggs 1983; Lyon 1998; Bailey et al. 2000). We do not interpret our results to rule out a role for LINE elements, but to suggest that motifs other than these interspersed repeats may be involved in X inactivation. In fact, we did not find any widely dispersed words enriched on XS over XE or autosomes at levels that have been seen for L1 (Bailey et al. 2000; Ross et al. 2005), and we have further confirmed the L1 enrichment on XS and showed that it is unique by comparing the L1 densities of all of the individual chromosomes, which had not been done previously. We have shown that transcription of widely dispersed repetitive elements appears to be silenced on Xi, as detected by hybridization to Cot-1 RNA (Hall et al. 2002a; C.M. Clemson, L.L. Hall, and J.B. Lawrence, in prep.); this silencing of repetitive elements themselves, not just protein coding genes, may be intrinsic to the mechanism of chromosome inactivation.
A critical consideration, however, is the pattern of enrichment on XS relative to XE and autosomes that should be expected for candidate sequences involved in X inactivation. While it has generally been presumed that the region at Xp22.2–22.3 would be similar to an autosome and have lower levels of putative “X inactivation motifs,” autosomes show substantially more competence for inactivation when in cis with the XIST gene than the XE region, which is markedly resistant. The most dramatic chromosomal sequence difference identified by our comprehensive search was the ∼11-fold enrichment of GATA repeats scattered widely through the 10-Mb XE chromosome segment. This unique enrichment on XE fits with the singular nature of this region and suggests a new paradigm whereby escape from inactivation may be due to the presence of elements that overcome heterochromatinization, rather than lack of those that promote it.
It is also important to consider the impact of recombination differences, since most of the X chromosome does not recombine in male meiosis, which may impact the evolution of sequence content. The enrichment of L1 LINES on XS over XE, and their enrichment on Y, could be consistent with the possibility that they accumulate due to lower recombination on XS/Y (Smit 1999). Since the PAR region of XE undergoes homologous recombination with Y much like an autosome, then neither the accumulation of GATA on XE nor the accumulation of dinucleotides across the X chromosome (including XE) are likely a consequence of the lower recombination on XS. In male meiosis there is an obligatory recombination event between the PARs of X and Y (Burgoyne 1982); thus it is possible that the GATA accumulation here could facilitate or be related to a high rate of recombination (Lien et al. 2000). Although GATA repeats have not been linked to recombination hotspots, however, the [AG]n has (Myers et al. 2005) and we find [AG]n more modestly, but significantly elevated on the XE.
Although there has been substantial interest in identifying sequences involved in mammalian chromosome inactivation, the dinucleotide repeat enrichment of the human X chromosome was not previously recognized. In fact, microsatellites were suggested to be underrepresented (Jarne et al. 1998). Interestingly, an early study pointed to enrichment of dinucleotide repeats related to sex-chromosome dosage in Drosophila (Lowenhaupt et al. 1989), providing evidence that this may occur in other species. GATA tandem repeats were previously reported as the Bkm satellites associated with sex chromosomes of some reptiles (Singh and Jones 1982). Here we have included an analysis of the GATA repeat, which shows that this striking enrichment is indeed present in other eutherians with an analogous PAR, such as dogs and primates.
Subramanian et al. (Subramanian et al. 2003a) reported that [GATA]n is enriched on the human sex chromosomes, focusing on a localized [GATA]n enrichment on the Y chromosome as potentially involved in the regulation of a domain of Y-linked genes expressed coordinately during gametogenesis. These authors did not, however, point out the specific enrichment in XE or the potential link to the X escape region, but rather discussed more even distribution across the entire X chromosome. We find that XE has ∼11-fold more GATA than XS, and that the enrichment on XE is substantially greater than that on Y (Fig. 5). However, it remains possible that this sequence feature serves a purpose on the Y chromosome, as genes in both the XE region and the Y chromosome must be expressed from a largely heterochromatic environment.
A remarkable feature of the sequence elements identified here is that they all have the capacity to form nonstandard DNA forms, and there is substantial literature linking them to gene regulation. The [GATA]n repeat satisfies the consensus-binding-site motif (WGATAR) for the GATA transcription factor family involved in gene regulation (Patient and McGhee 2002) and recently linked to formation of a higher-order loop in the globin gene (Vakoc et al. 2005). However, the distribution of GATA across the intergenic region of a large chromosome region may reflect a wholly different mechanism than gene-specific regulation through canonical transcription factors. [GATA]n satisfies the motif for a SATB1-binding site, an AT-rich region (with G and C on opposite strands) (Dickinson et al. 1992) that has high base-pair unwinding (Bode et al. 1992) and positions at the bases of chromatin loops (de Belle et al. 1998). More recently, SATB1 sites, which can function in either gene repression or activation, have been directly implicated as “landing platforms” or “entry sites” for chromosome remodeling complexes across broad (∼50 kb) regions (Yasui et al. 2002; Wen et al. 2005).
The dinucleotide motifs also have unusual and labile physical properties, with dual effects of repression or activation. [AG]n has been strongly linked to chromatin regulation and is a polypurine/polypyrimidine motif (PP) capable of forming triplex DNA (Maueler et al. 1998; Ohno et al. 2002). Triplex DNA can link distant sequences and be either a positive or negative regulator of gene expression (Kohwi and Kohwi-Shigematsu 1991). GAGA DNA elements can impact nucleosome packaging (Lehmann 2004), and the Trl (trithorax-like) gene widely involved in developmental regulation encodes the GAGA binding factor. Other evidence shows GAGA is required for silencing by Polycomb group proteins (Strutt et al. 1997), which themselves have recently been linked to mammalian X chromosome inactivation (Silva et al. 2003). X chromosome up-regulation in the male Drosophila involves a smaller number of “chromatin entry sites” believed important in chromatin remodeling (Kelley et al. 1999). Interestingly, [AG]n is present at the two known such sites at the Rox1 and Rox2 RNA genes, and a recent study directly implicates GAGA factor and/or [AG]n in Drosophila X chromosome dosage compensation (Greenberg et al. 2004).
[AT]n and [AC]n have also been linked to specific gene regulation (for example, Rothenburg et al. 2001); these motifs are APPs, which are able to form left-handed or Z-DNA involved in gene expression (Rich and Zhang 2003). A/T rich sequences also show high base-pair unwinding (Bode et al. 1992; Yasui et al. 2002), and ATATAT was initially identified as a core SATB1 contact site within a larger ATC consensus sequence. Thus, the prevalent dinucleotide repeats and their derivatives, particularly for [AT]n, may relate to binding of proteins involved in chromatin organization, such as SATB1 (Dickinson et al. 1992) and SAF-B (Nayler et al. 1998). Recently, a polymorphism in [AC]n and [AG]n repeats has been identified as a “regulatory microsatellite” in voles (Hammock and Young 2005) and, interestingly, the XIST itself is subject to a 450-bp APP sequence 25 kb upstream of the promoter that suppresses promoter activity (Hendrich et al. 1997).
While we focus here on the X chromosome, it is likely that the sequence elements involved in X inactivation are present at significant levels on other chromosomes where they may be involved in the widespread formation of facultative heterochromatin in different regions of the human genome that occurs throughout development. The ubiquitous nature of SSRs throughout the human and other genomes is often taken as indicative of mere “junk.” Yet, in human language, common words such as “to” or “the” are critical for syntax, modifying the meaning of more specific, but less common words (e.g., “to puzzle” vs. “the puzzle”). We suggest that common motifs such as [GATA]n and other SSRs are candidates for common words in the human genome that modify the structure and function of chromosomal domains. Thus, the concept of a “regulatory microsatellite” may apply not only to specific instances of individual genes, but more broadly to the regulation of heterochromatin and euchromatin throughout the genome.
Methods
Genomic sequences used for word-frequency analysis were derived from NCBI release B33. Two regions of the X chromosome, designated XE and XS, were treated as separate chromosomes. The 7.5-MB XE region includes PAR-1 and most of the evolutionary stratum 4 (defined by Lahn and Page 1999) and stratum 5 (defined by Ross et al. 2005), and XS contains the remainder of the chromosome largely subject to inactivation. Interspersed repeats (e.g., SINEs and LINEs) were removed from all sequences using RepeatMasker (A.F.A. Smit and P. Green, http://ftp.genome.washington.edu/RM/RepeatMasker.html, v.2002/05/15) at normal sensitivity, excluding low-complexity sequence (RepeatMasker -nolow -no_is -pa 2). A sliding window was used to tally word frequencies, excluding overlapping identical words and words containing “wildcard” codes (anything other than ACGT). The statistical significance of the differences among proportions for each word was evaluated based on a test of the equivalence of Poisson parameters, number of occurrences per 1,000,000 words, using the χ2 distribution, and P′ < 0.01 significance cutoff. The P-values were corrected to compensate for the large number of tests being performed (P′ = 1-(1-P)k). Distribution analysis on XS was performed by determining the individual word frequencies in 1-MB bins along the chromosome. The coefficient of variance ((stdev*100)/mean) of bin word frequencies was calculated and a cutoff value (V < 100) was used to eliminate words in highly localized satellites. Words were ranked by the difference between regional (XS, XE) word densities and autosomal word density (words/million words). These analyses were performed on a dual processor pentium III computer running GNU/Linux using custom scripts written in GAWK (release 3.1.1).
For fluorescence in situ hybridization, biotinylated oligonucleotide probes were hybridized (5 pM/uL probe in 5% formamide, 2XSSC, 37°C, overnight) to metaphase preparations of normal human peripheral blood lymphocytes that were denatured in 70% formamide, 2XSSC for 2 min. Hybridization was performed as previously described (Clemson et al. 1996; Hall et al. 2002b; Tam et al. 2002) and detected with Texas Red streptavidin and counterstained with DAPI.
Acknowledgments
We recognize the contributions of Meg Byron, Kevin Byron, and John Butler who performed the in situ hybridizations; and David LaPointe and Stephen Baker for assistance with the statistical analysis. We thank Joel Graber for helpful discussions in the initial phases of this project. This work was funded by NIH GM68138 and NIH GM053234
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4627606
References
- Bailey J.A., Carrel L., Chakravarti A., Eichler E.E., Carrel L., Chakravarti A., Eichler E.E., Chakravarti A., Eichler E.E., Eichler E.E. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: The Lyon repeat hypothesis. Proc. Natl. Acad. Sci. 2000;97:6634–6639. doi: 10.1073/pnas.97.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode J., Kohwi Y., Dickinson L., Joh T., Klehr D., Mielke C., Kohwi-Shigematsu T., Kohwi Y., Dickinson L., Joh T., Klehr D., Mielke C., Kohwi-Shigematsu T., Dickinson L., Joh T., Klehr D., Mielke C., Kohwi-Shigematsu T., Joh T., Klehr D., Mielke C., Kohwi-Shigematsu T., Klehr D., Mielke C., Kohwi-Shigematsu T., Mielke C., Kohwi-Shigematsu T., Kohwi-Shigematsu T. Biological significance of unwinding capability of nuclear matrix-associated DNAs. Science. 1992;255:195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- Brown C.J., Greally J.M., Greally J.M. A stain upon the silence: Genes escaping X inactivation. Trends Genet. 2003;19:432–438. doi: 10.1016/S0168-9525(03)00177-X. [DOI] [PubMed] [Google Scholar]
- Burgoyne P.S. Genetic homology and crossing over in the X and Y chromosomes of mammals. Hum. Genet. 1982;61:85–90. doi: 10.1007/BF00274192. [DOI] [PubMed] [Google Scholar]
- Carrel L., Willard H.F., Willard H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Carrel L., Cottle A.A., Goglin K.C., Willard H.F., Cottle A.A., Goglin K.C., Willard H.F., Goglin K.C., Willard H.F., Willard H.F. A first-generation X-inactivation profile of the human X chromosome. Proc. Natl. Acad. Sci. 1999;96:14440–14444. doi: 10.1073/pnas.96.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick B.P., Willard H.F., Willard H.F. Barring gene expression after XIST: Maintaining facultative heterochromatin on the inactive X. Semin. Cell Dev. Biol. 2003;14:359–367. doi: 10.1016/j.semcdb.2003.09.016. [DOI] [PubMed] [Google Scholar]
- Chureau C., Prissette M., Bourdet A., Barbe V., Cattolico L., Jones L., Eggen A., Avner P., Duret L., Prissette M., Bourdet A., Barbe V., Cattolico L., Jones L., Eggen A., Avner P., Duret L., Bourdet A., Barbe V., Cattolico L., Jones L., Eggen A., Avner P., Duret L., Barbe V., Cattolico L., Jones L., Eggen A., Avner P., Duret L., Cattolico L., Jones L., Eggen A., Avner P., Duret L., Jones L., Eggen A., Avner P., Duret L., Eggen A., Avner P., Duret L., Avner P., Duret L., Duret L. Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res. 2002;12:894–908. doi: 10.1101/gr.152902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson C.M., McNeil J.A., Willard H.F., Lawrence J.B., McNeil J.A., Willard H.F., Lawrence J.B., Willard H.F., Lawrence J.B., Lawrence J.B. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Belle I., Cai S., Kohwi-Shigematsu T., Cai S., Kohwi-Shigematsu T., Kohwi-Shigematsu T. The genomic sequences bound to special AT-rich sequence-binding protein 1 (SATB1) in vivo in Jurkat T cells are tightly associated with the nuclear matrix at the bases of the chromatin loops. J. Cell Biol. 1998;141:335–348. doi: 10.1083/jcb.141.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson L.A., Joh T., Kohwi Y., Kohwi-Shigematsu T., Joh T., Kohwi Y., Kohwi-Shigematsu T., Kohwi Y., Kohwi-Shigematsu T., Kohwi-Shigematsu T. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 1992;70:631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- Epplen J.T., Kyas A., Maueler W., Kyas A., Maueler W., Maueler W. Genomic simple repetitive DNAs are targets for differential binding of nuclear proteins. FEBS Lett. 1996;389:92–95. doi: 10.1016/0014-5793(96)00526-1. [DOI] [PubMed] [Google Scholar]
- Filippova G.N., Cheng M.K., Moore J.M., Truong J.P., Hu Y.J., Nguyen D.K., Tsuchiya K.D., Disteche C.M., Cheng M.K., Moore J.M., Truong J.P., Hu Y.J., Nguyen D.K., Tsuchiya K.D., Disteche C.M., Moore J.M., Truong J.P., Hu Y.J., Nguyen D.K., Tsuchiya K.D., Disteche C.M., Truong J.P., Hu Y.J., Nguyen D.K., Tsuchiya K.D., Disteche C.M., Hu Y.J., Nguyen D.K., Tsuchiya K.D., Disteche C.M., Nguyen D.K., Tsuchiya K.D., Disteche C.M., Tsuchiya K.D., Disteche C.M., Disteche C.M. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev. Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Gartler S.M., Riggs A.D., Riggs A.D. Mammalian X-chromosome inactivation. Annu. Rev. Genet. 1983;17:155–190. doi: 10.1146/annurev.ge.17.120183.001103. [DOI] [PubMed] [Google Scholar]
- Greenberg A.J., Yanowitz J.L., Schedl P., Yanowitz J.L., Schedl P., Schedl P. The Drosophila GAGA factor is required for dosage compensation in males and for the formation of the male-specific-lethal complex chromatin entry site at 12DE. Genetics. 2004;166:279–289. doi: 10.1534/genetics.166.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimwood J., Gordon L.A., Olsen A., Terry A., Schmutz J., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Gordon L.A., Olsen A., Terry A., Schmutz J., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Olsen A., Terry A., Schmutz J., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Terry A., Schmutz J., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Schmutz J., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Goodstein D., Couronne O., Tran-Gyamfi M., Couronne O., Tran-Gyamfi M., Tran-Gyamfi M., et al. The DNA sequence and biology of human chromosome 19. Nature. 2004;428:529–535. doi: 10.1038/nature02399. [DOI] [PubMed] [Google Scholar]
- Hall L.L., Lawrence J.B., Lawrence J.B. The cell biology of a novel chromosomal RNA: Chromosome painting by XIST/Xist RNA initiates a remodeling cascade. Semin. Cell Dev. Biol. 2003;14:369–378. doi: 10.1016/j.semcdb.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Hall L.L., Byron M., Sakai K., Carrel L., Willard H.F., Lawrence J.B., Byron M., Sakai K., Carrel L., Willard H.F., Lawrence J.B., Sakai K., Carrel L., Willard H.F., Lawrence J.B., Carrel L., Willard H.F., Lawrence J.B., Willard H.F., Lawrence J.B., Lawrence J.B. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc. Natl. Acad. Sci. 2002a;99:8677–8682. doi: 10.1073/pnas.132468999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L.L., Clemson C.M., Byron M., Wydner K., Lawrence J.B., Clemson C.M., Byron M., Wydner K., Lawrence J.B., Byron M., Wydner K., Lawrence J.B., Wydner K., Lawrence J.B., Lawrence J.B. Unbalanced X;autosome translocations provide evidence for sequence specificity in the association of XIST RNA with chromatin. Hum. Mol. Genet. 2002b;11:3157–3165. doi: 10.1093/hmg/11.25.3157. [DOI] [PubMed] [Google Scholar]
- Hammock E.A., Young L.J., Young L.J. Microsatellite instability generates diversity in brain and sociobehavioral traits. Science. 2005;308:1630–1634. doi: 10.1126/science.1111427. [DOI] [PubMed] [Google Scholar]
- Heard E. Recent advances in X-chromosome inactivation. Curr. Opin. Cell Biol. 2004;16:247–255. doi: 10.1016/j.ceb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Hendrich B.D., Plenge R.M., Willard H.F., Plenge R.M., Willard H.F., Willard H.F. Identification and characterization of the human XIST gene promoter: Implications for models of X chromosome inactivation. Nucleic Acids Res. 1997;25:2661–2671. doi: 10.1093/nar/25.13.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzing L.B.K., Romer J.T., Horn J.M., Ashworth A., Romer J.T., Horn J.M., Ashworth A., Horn J.M., Ashworth A., Ashworth A. Xist has properties of the X-chromosome inactivation centre. Nature. 1997;386:272–275. doi: 10.1038/386272a0. [DOI] [PubMed] [Google Scholar]
- Jarne P., David P., Viard F., David P., Viard F., Viard F. Microsatellites, transposable elements and the X chromosome. Mol. Biol. Evol. 1998;15:28–34. doi: 10.1093/oxfordjournals.molbev.a025844. [DOI] [PubMed] [Google Scholar]
- Jegalian K., Page D.C., Page D.C. A proposed path by which genes common to mammalian X and Y chromosomes evolve to become X inactivated. Nature. 1998;394:776–780. doi: 10.1038/29522. [DOI] [PubMed] [Google Scholar]
- Ke X., Collins A., Collins A. CpG islands in human X-inactivation. Ann. Hum. Genet. 2003;67:242–249. doi: 10.1046/j.1469-1809.2003.00038.x. [DOI] [PubMed] [Google Scholar]
- Kelley R.L., Meller V.H., Gordadze P.R., Roman G., Davis R.L., Kuroda M.I., Meller V.H., Gordadze P.R., Roman G., Davis R.L., Kuroda M.I., Gordadze P.R., Roman G., Davis R.L., Kuroda M.I., Roman G., Davis R.L., Kuroda M.I., Davis R.L., Kuroda M.I., Kuroda M.I. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- Kirkness E.F., Bafna V., Halpern A.L., Levy S., Remington K., Rusch D.B., Delcher A.L., Pop M., Wang W., Fraser C.M., Bafna V., Halpern A.L., Levy S., Remington K., Rusch D.B., Delcher A.L., Pop M., Wang W., Fraser C.M., Halpern A.L., Levy S., Remington K., Rusch D.B., Delcher A.L., Pop M., Wang W., Fraser C.M., Levy S., Remington K., Rusch D.B., Delcher A.L., Pop M., Wang W., Fraser C.M., Remington K., Rusch D.B., Delcher A.L., Pop M., Wang W., Fraser C.M., Rusch D.B., Delcher A.L., Pop M., Wang W., Fraser C.M., Delcher A.L., Pop M., Wang W., Fraser C.M., Pop M., Wang W., Fraser C.M., Wang W., Fraser C.M., Fraser C.M., et al. The dog genome: Survey sequencing and comparative analysis. Science. 2003;301:1898–1903. doi: 10.1126/science.1086432. [DOI] [PubMed] [Google Scholar]
- Kohwi Y., Kohwi-Shigematsu T., Kohwi-Shigematsu T. Altered gene expression correlates with DNA structure. Genes & Dev. 1991;5:2547–2554. doi: 10.1101/gad.5.12b.2547. [DOI] [PubMed] [Google Scholar]
- Lahn B.T., Page D.C., Page D.C. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Devon K., Dewar K., Doyle M., FitzHugh W., Dewar K., Doyle M., FitzHugh W., Doyle M., FitzHugh W., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lee J.T., Jaenisch R., Jaenisch R. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature. 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- Lehmann M. Anything else but GAGA: A nonhistone protein complex reshapes chromatin structure. Trends Genet. 2004;20:15–22. doi: 10.1016/j.tig.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Lien S., Szyda J., Schechinger B., Rappold G., Arnheim N., Szyda J., Schechinger B., Rappold G., Arnheim N., Schechinger B., Rappold G., Arnheim N., Rappold G., Arnheim N., Arnheim N. Evidence for heterogeneity in recombination in the human pseudoautosomal region: High resolution analysis by sperm typing and radiation-hybrid mapping. Am. J. Hum. Genet. 2000;66:557–566. doi: 10.1086/302754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenhaupt K., Rich A., Pardue M.L., Rich A., Pardue M.L., Pardue M.L. Nonrandom distribution of long mono- and dinucleotide repeats in Drosophila chromosomes: Correlations with dosage compensation, heterochromatin, and recombination. Mol. Cell. Biol. 1989;9:1173–1182. doi: 10.1128/mcb.9.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M.F. X-chromosome inactivation: A repeat hypothesis. Cytogenet. Cell Genet. 1998;80:133–137. doi: 10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- Lyon M.F. The Lyon and the LINE hypothesis. Semin. Cell Dev. Biol. 2003;14:313–318. doi: 10.1016/j.semcdb.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Marahens Y., Panning B., Dausman J., Strauss W., Jaenisch R., Panning B., Dausman J., Strauss W., Jaenisch R., Dausman J., Strauss W., Jaenisch R., Strauss W., Jaenisch R., Jaenisch R. Xist deficient mice are defective in dosage compensation but not spermatogenesis. Genes & Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- Maueler W., Kyas A., Keyl H.G., Epplen J.T., Kyas A., Keyl H.G., Epplen J.T., Keyl H.G., Epplen J.T., Epplen J.T. A genome-derived (gaa.ttc)24 trinucleotide block binds nuclear protein(s) specifically and forms triple helices. Gene. 1998;215:389–403. doi: 10.1016/s0378-1119(98)00266-2. [DOI] [PubMed] [Google Scholar]
- Myers S., Bottolo L., Freeman C., McVean G., Donnelly P., Bottolo L., Freeman C., McVean G., Donnelly P., Freeman C., McVean G., Donnelly P., McVean G., Donnelly P., Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Nayler O., Stratling W., Bourquin J.P., Stagljar I., Lindemann L., Jasper H., Hartmann A.M., Fackelmayer F.O., Ullrich A., Stamm S., Stratling W., Bourquin J.P., Stagljar I., Lindemann L., Jasper H., Hartmann A.M., Fackelmayer F.O., Ullrich A., Stamm S., Bourquin J.P., Stagljar I., Lindemann L., Jasper H., Hartmann A.M., Fackelmayer F.O., Ullrich A., Stamm S., Stagljar I., Lindemann L., Jasper H., Hartmann A.M., Fackelmayer F.O., Ullrich A., Stamm S., Lindemann L., Jasper H., Hartmann A.M., Fackelmayer F.O., Ullrich A., Stamm S., Jasper H., Hartmann A.M., Fackelmayer F.O., Ullrich A., Stamm S., Hartmann A.M., Fackelmayer F.O., Ullrich A., Stamm S., Fackelmayer F.O., Ullrich A., Stamm S., Ullrich A., Stamm S., Stamm S. SAF-B protein couples transcription and pre-mRNA splicing to SAR/MAR elements. Nucleic Acids Res. 1998;26:3542–3549. doi: 10.1093/nar/26.15.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M., Fukagawa T., Lee J.S., Ikemura T., Fukagawa T., Lee J.S., Ikemura T., Lee J.S., Ikemura T., Ikemura T. Triplex-forming DNAs in the human interphase nucleus visualized in situ by polypurine/polypyrimidine DNA probes and antitriplex antibodies. Chromosoma. 2002;111:201–213. doi: 10.1007/s00412-002-0198-0. [DOI] [PubMed] [Google Scholar]
- Patient R.K., McGhee J.D., McGhee J.D. The GATA family (vertebrates and invertebrates). Curr. Opin. Genet. Dev. 2002;12:416–422. doi: 10.1016/s0959-437x(02)00319-2. [DOI] [PubMed] [Google Scholar]
- Perry J., Palmer S., Gabriel A., Ashworth A., Palmer S., Gabriel A., Ashworth A., Gabriel A., Ashworth A., Ashworth A. A short pseudoautosomal region in laboratory mice. Genome Res. 2001;11:1826–1832. doi: 10.1101/gr.203001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A., Zhang S., Zhang S. Timeline: Z-DNA: The long road to biological function. Nat. Rev. Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- Ross M.T., Grafham D.V., Coffey A.J., Scherer S., McLay K., Muzny D., Platzer M., Howell G.R., Burrows C., Bird C.P., Grafham D.V., Coffey A.J., Scherer S., McLay K., Muzny D., Platzer M., Howell G.R., Burrows C., Bird C.P., Coffey A.J., Scherer S., McLay K., Muzny D., Platzer M., Howell G.R., Burrows C., Bird C.P., Scherer S., McLay K., Muzny D., Platzer M., Howell G.R., Burrows C., Bird C.P., McLay K., Muzny D., Platzer M., Howell G.R., Burrows C., Bird C.P., Muzny D., Platzer M., Howell G.R., Burrows C., Bird C.P., Platzer M., Howell G.R., Burrows C., Bird C.P., Howell G.R., Burrows C., Bird C.P., Burrows C., Bird C.P., Bird C.P., et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenburg S., Koch-Nolte F., Rich A., Haag F., Koch-Nolte F., Rich A., Haag F., Rich A., Haag F., Haag F. A polymorphic dinucleotide repeat in the rat nucleolin gene forms Z-DNA and inhibits promoter activity. Proc. Natl. Acad. Sci. 2001;98:8985–8990. doi: 10.1073/pnas.121176998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp A.J., Spotswood H.T., Robinson D.O., Turner B.M., Jacobs P.A., Spotswood H.T., Robinson D.O., Turner B.M., Jacobs P.A., Robinson D.O., Turner B.M., Jacobs P.A., Turner B.M., Jacobs P.A., Jacobs P.A. Molecular and cytogenetic analysis of the spreading of X inactivation in X;autosome translocations. Hum. Mol. Genet. 2002;11:3145–3156. doi: 10.1093/hmg/11.25.3145. [DOI] [PubMed] [Google Scholar]
- Silva J., Mak W., Zvetkova I., Appanah R., Nesterova T.B., Webster Z., Peters A.H., Jenuwein T., Otte A.P., Brockdorff N., Mak W., Zvetkova I., Appanah R., Nesterova T.B., Webster Z., Peters A.H., Jenuwein T., Otte A.P., Brockdorff N., Zvetkova I., Appanah R., Nesterova T.B., Webster Z., Peters A.H., Jenuwein T., Otte A.P., Brockdorff N., Appanah R., Nesterova T.B., Webster Z., Peters A.H., Jenuwein T., Otte A.P., Brockdorff N., Nesterova T.B., Webster Z., Peters A.H., Jenuwein T., Otte A.P., Brockdorff N., Webster Z., Peters A.H., Jenuwein T., Otte A.P., Brockdorff N., Peters A.H., Jenuwein T., Otte A.P., Brockdorff N., Jenuwein T., Otte A.P., Brockdorff N., Otte A.P., Brockdorff N., Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev. Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Singh L., Jones K.W., Jones K.W. Sex reversal in the mouse (Mus musculus) is caused by a recurrent nonreciprocal crossover involving the X and an aberrant Y chromosome. Cell. 1982;28:205–216. doi: 10.1016/0092-8674(82)90338-5. [DOI] [PubMed] [Google Scholar]
- Smit A.F. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Strutt H., Cavalli G., Paro R., Cavalli G., Paro R., Paro R. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 1997;16:3621–3632. doi: 10.1093/emboj/16.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S., Mishra R.K., Singh L., Mishra R.K., Singh L., Singh L. Genome-wide analysis of Bkm sequences (GATA repeats): Predominant association with sex chromosomes and potential role in higher order chromatin organization and function. Bioinformatics. 2003a;19:681–685. doi: 10.1093/bioinformatics/btg067. [DOI] [PubMed] [Google Scholar]
- Subramanian S., Mishra R.K., Singh L., Mishra R.K., Singh L., Singh L. Genome-wide analysis of microsatellite repeats in humans: Their abundance and density in specific genomic regions. Genome Biol. 2003b;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam R., Shopland L.S., Johnson C.V., McNeil J.A., Lawrence J.B., Shopland L.S., Johnson C.V., McNeil J.A., Lawrence J.B., Johnson C.V., McNeil J.A., Lawrence J.B., McNeil J.A., Lawrence J.B., Lawrence J.B. Applications of RNA FISH for visualizing gene expression and nuclear archetecture. Oxford University Press; New York.: 2002. [Google Scholar]
- Tilford C.A., Kuroda-Kawaguchi T., Skaletsky H., Rozen S., Brown L.G., Rosenberg M., McPherson J.D., Wylie K., Sekhon M., Kucaba T.A., Kuroda-Kawaguchi T., Skaletsky H., Rozen S., Brown L.G., Rosenberg M., McPherson J.D., Wylie K., Sekhon M., Kucaba T.A., Skaletsky H., Rozen S., Brown L.G., Rosenberg M., McPherson J.D., Wylie K., Sekhon M., Kucaba T.A., Rozen S., Brown L.G., Rosenberg M., McPherson J.D., Wylie K., Sekhon M., Kucaba T.A., Brown L.G., Rosenberg M., McPherson J.D., Wylie K., Sekhon M., Kucaba T.A., Rosenberg M., McPherson J.D., Wylie K., Sekhon M., Kucaba T.A., McPherson J.D., Wylie K., Sekhon M., Kucaba T.A., Wylie K., Sekhon M., Kucaba T.A., Sekhon M., Kucaba T.A., Kucaba T.A., et al. A physical map of the human Y chromosome. Nature. 2001;409:943–945. doi: 10.1038/35057170. [DOI] [PubMed] [Google Scholar]
- Vakoc C.R., Letting D.L., Gheldof N., Sawado T., Bender M.A., Groudine M., Weiss M.J., Dekker J., Blobel G.A., Letting D.L., Gheldof N., Sawado T., Bender M.A., Groudine M., Weiss M.J., Dekker J., Blobel G.A., Gheldof N., Sawado T., Bender M.A., Groudine M., Weiss M.J., Dekker J., Blobel G.A., Sawado T., Bender M.A., Groudine M., Weiss M.J., Dekker J., Blobel G.A., Bender M.A., Groudine M., Weiss M.J., Dekker J., Blobel G.A., Groudine M., Weiss M.J., Dekker J., Blobel G.A., Weiss M.J., Dekker J., Blobel G.A., Dekker J., Blobel G.A., Blobel G.A. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol. Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Wen J., Huang S., Rogers H., Dickinson L.A., Kohwi-Shigematsu T., Noguchi C.T., Huang S., Rogers H., Dickinson L.A., Kohwi-Shigematsu T., Noguchi C.T., Rogers H., Dickinson L.A., Kohwi-Shigematsu T., Noguchi C.T., Dickinson L.A., Kohwi-Shigematsu T., Noguchi C.T., Kohwi-Shigematsu T., Noguchi C.T., Noguchi C.T. SATB1 family protein expressed during early erythroid differentiation modifies globin gene expression. Blood. 2005;105:3330–3339. doi: 10.1182/blood-2004-08-2988. [DOI] [PubMed] [Google Scholar]
- White W.M., Willard H.F., Van Dyke D.L., Wolff D.J., Willard H.F., Van Dyke D.L., Wolff D.J., Van Dyke D.L., Wolff D.J., Wolff D.J. The spreading of X inactivation into autosomal material of an X;autosome translocation: Evidence for a difference between autosomal and X-chromosomal DNA. Am. J. Hum. Genet. 1998;63:20–28. doi: 10.1086/301922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff D.J., Schwartz S., Montgomery T., Zackowski J.L., Schwartz S., Montgomery T., Zackowski J.L., Montgomery T., Zackowski J.L., Zackowski J.L. Random X inactivation in a girl with a balanced t(X;9) and an abnormal phenotype. Am. J. Med. Genet. 1998;77:401–404. doi: 10.1002/(sici)1096-8628(19980605)77:5<401::aid-ajmg9>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Wutz A., Jaenisch R., Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- Yasui D., Miyano M., Cai S., Varga-Weisz P., Kohwi-Shigematsu T., Miyano M., Cai S., Varga-Weisz P., Kohwi-Shigematsu T., Cai S., Varga-Weisz P., Kohwi-Shigematsu T., Varga-Weisz P., Kohwi-Shigematsu T., Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]