Abstract
Recent data from Drosophila suggest that a substantial fraction of amino acid substitutions observed between species are beneficial. If these beneficial mutations are on average partially recessive, then the rate of protein evolution is predicted to be faster for X-linked genes compared to autosomal genes (the “faster-X” hypothesis). We test this prediction by comparing rates of protein substitutions between orthologous genes, taking advantage of variations in chromosome fusions within the genus Drosophila. In members of the Drosophila melanogaster species group, the chromosomal arm 3L segregates as an ordinary autosome (i.e., two homologous copies in both males and females). However, in the Drosophila pseudoobscura species group, this chromosomal arm has become fused to the ancestral X chromosome and is hemizygous in males. The faster-X hypothesis predicts that protein evolution should be faster for genes on this chromosomal arm in the D. pseudoobscura lineage, relative to the D. melanogaster lineage. Here we combine new sequence data for 202 gene fragments in Drosophila miranda (in the pseudoobscura species group) with the completed genomes of D. melanogaster, D. pseudoobscura, and Drosophila yakuba to show that there are no detectable differences in rates of amino acid evolution for orthologous X-linked and autosomal genes. Our results imply that the contribution of the faster-X (if any) to the large-X effect on reproductive isolation in Drosophila is not due to a generally faster rate of protein evolution. The lack of a detectable faster-X effect in these species suggests either that beneficial amino acids are not partially recessive on average, or that adaptive evolution does not often use newly arising amino acid mutations.
While beneficial mutations are the raw material for adaptive evolution by natural selection, little is known about their nature. For example, it is unclear if most adaptation draws primarily on newly arising mutations (e.g., Braverman et al. 1995), on previously neutral polymorphisms (Dykhuizen and Hartl 1980; Innan and Kim 2004), or on a pool of deleterious polymorphisms in mutation–selection balance (Orr and Betancourt 2001). The intensity of selection felt by a new mutation is scaled by its degree of dominance, h. It is also unknown whether beneficial alleles have on average partially recessive effects on fitness (i.e., their dominance coefficient h < 0.5) or are partially dominant (h > 0.5) and thus more readily visible to selection when heterozygous. In heterogametic sexual species, genes on the X chromosome generally lack a homologous copy in males (i.e., they are hemizygous). If the average intensity of selection on beneficial mutations is the same on the X chromosome and autosomes, Charlesworth et al. (1987) showed that newly arising partially recessive beneficial mutations have a higher probability of fixation if arising on the X chromosome. Therefore, if mutation rates to beneficial alleles are equal, the rate of adaptive evolution will be higher under these conditions for genes on the X chromosome than for autosomal loci. This predicted phenomenon has been called the “faster-X” effect.
The faster-X theory has been proposed as a possible explanation for several biological phenomena. Among these is the observation in Drosophila, and other taxa, that the X chromosome harbors a disproportionately large number of loci causing reproductive isolation between closely related species (called the “large X effect”) (for review, see Coyne and Orr 2004). While the weight of current evidence appears to favor the dominance theory (Turelli and Orr 1995) for the large-X effect, faster-X evolution remains a possible, and not mutually exclusive, factor contributing to the large-X effect (Charlesworth et al. 1987; Naveira 2003; Tao and Hartl 2003; Coyne et al. 2004). Recent studies have found that a substantial fraction of amino acid divergence between members of the Drosophila melanogaster species group is driven by positive selection. This adaptive amino acid evolution seems to be a general property of most genes (Fay et al. 2002; Smith and Eyre-Walker 2002; Bierne and Eyre-Walker 2004; Sawyer et al. 2003; Andolfatto 2005), as well as specific genes underlying reproductive isolation (Ting et al. 1998; Presgraves et al. 2003; Barbash et al. 2004). This suggests that a major component of the faster-X effect on reproductive isolation could be a byproduct of an overall faster rate of adaptive protein evolution for genes on the X chromosome. In addition, the X chromosomes of Drosophila simulans (Begun and Whitley 2000) and humans (Payseur et al. 2002), as well as the Z chromosome of chickens (Sundström et al. 2004), harbor reduced levels of nucleotide variability compared to expectations based on autosomal genes. These results have been interpreted as evidence for a faster rate of adaptive evolution on the X (or Z) chromosomes. The rationale behind this is that the more frequent fixation of partially recessive advantageous mutations on X (and the shorter time to fixation) will result in a greater reduction in variability at linked neutral sites (i.e., the hitchhiking effect) relative to autosomes.
Two recent studies in Drosophila tested for faster protein evolution on X, using DNA divergence data (Betancourt et al. 2002; Counterman et al. 2004). The rationale of these studies is that, if a substantial fraction of amino acid substitutions are beneficial (as mentioned above) and are on average partially recessive, then the rate of protein evolution should be faster for X-linked genes compared to autosomal ones. The rate of protein evolution is usually measured by the rate of amino acid substitutions compared to synonymous substitutions (dN/dS), to account for differences in mutation rates among loci. Betancourt et al. (2002) used DNA divergence data between Drosophila melanogaster and Drosophila simulans for a total of 254 published sequences and did not find a significant difference in rates of protein evolution between X-linked and autosomal genes. However, this test may lack power since genes can vary widely in their level of functional constraint (and therefore in dN/dS). Thus, even if a particular gene is more likely to evolve faster when X-linked, the average rate of protein evolution of X-linked genes might not statistically exceed the rate of autosomal ones.
Counterman et al. (2004) proposed a more robust test for faster-X evolution, by comparing rates of protein evolution between orthologous genes that vary in whether they are autosomal or X-linked, taking advantage of chromosome fusions in the genus Drosophila (Muller 1940). In members of the D. melanogaster species group, the chromosomal arm 3L (Muller’s element D) is segregating as an ordinary autosome (i.e., two homologous copies in both males and females). In the Drosophila pseudoobscura species group, however, this chromosomal arm has become fused to the ancestral X chromosome (Muller’s element A) and is hemizygous in males (see Fig. 1). This test therefore allows a comparison of rates of divergence at orthologous loci that are X-linked in one species group and autosomal in another. Counterman et al. (2004) compared a relatively small number of gene fragments (between 25 and 110) in three or four species of these two groups to test for faster-X evolution and found a marginally significant faster-X effect in one comparison but not the other. Here, we follow a similar approach to test whether the X chromosome shows a higher rate of protein evolution by taking advantage of the X/autosome fusion in two members of the D. pseudoobscura species group relative to the D. melanogaster species group. We significantly increase the number of genes analyzed by using the genome sequences of D. melanogaster, Drosophila yakuba, and D. pseudoobscura (comparing a total of 2646 genes) and obtaining new sequence information for >200 gene fragments from Drosophila miranda.
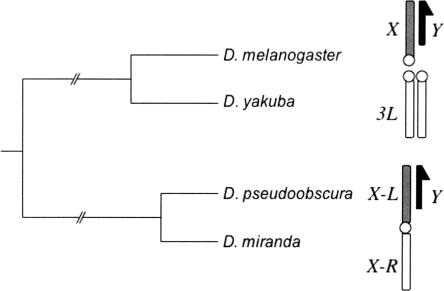
Figure 1.
Karyotypes of the D. melanogaster and obscura species groups. In the obscura group, which includes D. pseudoobscura and D. miranda, a translocation of Muller’s element D (white) to the X chromosome (gray) results in a karyotype where autosomal loci in the melanogaster group are X-linked in the obscura group. This new X chromosome was created ∼13 Mya. In this figure, the neo-sex chromosomes in D. miranda (which result from rearrangements between the Y chromosome and an autosome) are not shown for simplicity.
Results
In Drosophila species, chromosome arms are sometimes referred to as Muller elements. In D. melanogaster, this labeling begins with the X chromosome (Muller element A), and continues through arms 2L, 2R, and so on. In this study, we focus on elements A and D. In the D. melanogaster karyotype, element D is the left arm of the (autosomal) third chromosome, 3L. In D. pseudoobscura and D. miranda, element D has fused to the ancestral X chromosome (element A) and has thus become the right arm of the X chromosome (chromosome XR) (see Fig. 1.) In this paper, we use the notation X/XL to refer to element A, and 3L/XR to refer to element D, making it clear which element has remained X-linked in all species studied (element A = X/XL), and which has been translocated and only is X-linked in the pseudoobscura group (element D = 3L/XR).
Whole chromosome analysis (three-species comparison)
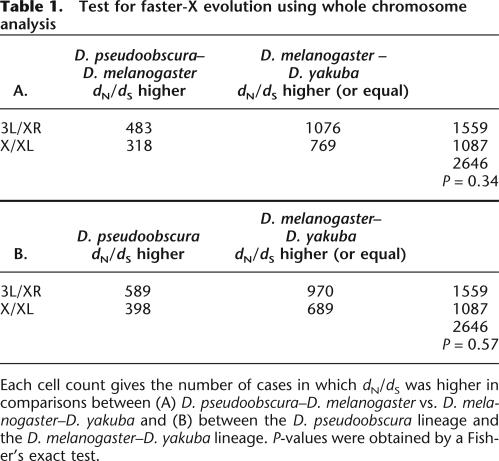
If proteins evolve faster when X-linked, genes located on 3L/XR should show a higher rate of protein evolution (dN/dS) in the D. pseudooboscura lineage, where this element is X-linked compared to the D. melanogaster/D. yakuba lineage, where it is autosomal (see Fig. 1). We used the genome sequences of these three species to test for faster-X evolution using orthologous gene comparisons similar to the three-species test performed by Counterman et al. (2004). To account for lineage-specific effects (e.g., a generally lower rate of amino acid or synonymous evolution in one lineage), it is necessary to contrast dN/dS of loci from 3L/XR with genes from another chromosome. Following Counterman et al. (2004), we chose chromosome X/XL for this purpose, whose genes are X-linked in all possible comparisons. We annotated 1087 genes on X/XL and 1559 genes on 3L/XR in the D. yakuba genome. Table 1A presents a contingency table comparing dN/dS between D. pseudoobscura and D. melanogaster to dN/dS between D. melanogaster and D. yakuba. The rationale of this test is that, if there is a general faster-X effect on amino acid substitutions, then dN/dS will be systematically higher between D. melanogaster/D. pseudoobscura than between D. melanogaster/D. yakuba for loci on 3L/XR compared to loci on X/XL. Table 1A shows that this is not the case (P = 0.44, Fisher’s exact test), suggesting that X-linkage does not generally increase the rate of protein evolution.
Table 1.
Test for faster-X evolution using whole chromosome analysis
Each cell count gives the number of cases in which dN/dS was higher in comparisons between (A) D. pseudoobscura–D. melanogaster vs. D. melanogaster–D. yakuba and (B) between the D. pseudoobscura lineage and the D. melanogaster–D. yakuba lineage. P-values were obtained by a Fisher’s exact test.
However, the above test of the faster-X hypothesis could lack statistical power for several reasons. First, the rate of synonymous substitutions dS is saturated between D. melanogaster and D. pseudoobscura (Bergman et al. 2002; Richards et al. 2005). This means that there is little information about the true value of dN/dS between the two species, and these estimates have wide confidence intervals. Secondly, substitutions on the lineage leading to D. melanogaster are counted twice, once in the comparison with D. pseudoobscura, and again in the comparison to D. yakuba (Fig. 1). The problem of nonindependence can be addressed by directly estimating dN and dS per branch on the three-species phylogeny. In Table 1B, we compare the dN/dS on the branch leading to D. pseudoobscura to the sum of dN/dS on the branches leading to D. melanogaster and D. yakuba for both X/XL and 3L/XR. Again, there is no detectable acceleration of dN/dS on the lineage leading to D. pseudoobscura for genes on 3L/XR compared to genes on X/XL (Table 1B) (P = 0.56, Fisher’s exact test). A third factor contributing to a lack of power is the fact that chromosome 3R has been fused to the X chromosome in D. pseudoobscura for a relatively short time period (∼13 million years [Myr]) compared to the total tree length (50–110 Myr) (Richards et al. 2005). Thus, the “background noise” of amino acid substitutions accumulating on this chromosome before it became an X chromosome might be too large to detect the faster-X effect using this type of comparison.
Paired comparisons between four species
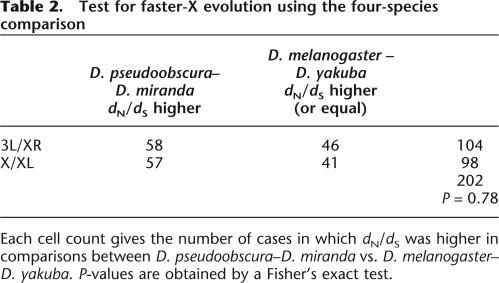
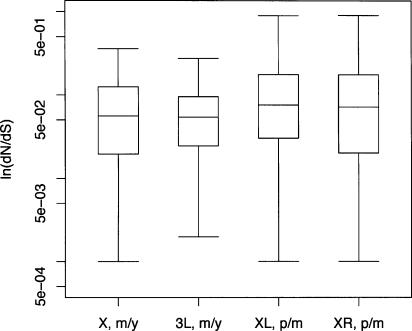
Given the large amount of “background noise” expected in the three-species comparisons, a more robust test of the faster-X hypothesis is to compare rates of substitution among orthologous loci that have historically always been X-linked in one comparison and autosomal in another (see Counterman et al. 2004). In D. pseudoobscura and its close relative D. miranda, the X chromosome is metacentric with two large, euchromatic arms (labeled XL and XR). In D. melanogaster and D. yakuba, the X is acrocentric and homologous to XL, and the autosomal arm 3L is homologous to the right arm of the pseudoobscura/miranda X. We sequenced and annotated 98 loci on X/XL, and 104 genes on 3L/XR in D. miranda, to test whether X-linked genes show a higher rate of protein evolution compared to autosomal genes. Table 2 summarizes the results of the paired comparison of the dN/dS analysis as a contingency table. The fraction of comparisons where the rate of protein evolution dN/dS was greater in D. pseudoobscura–D. miranda than in D. yakuba–D. melanogaster was 0.58 for X/XL and 0.55 for 3L/XR. This difference is not statistically significant, using a Fisher’s exact test (P = 0.78) (see Table 2). The distributions of the dN/dS ratios are shown in Figure 2. Kolmogorov-Smirnov tests reveal no differences in the distributions, either between species pairs or between chromosome arms.
Table 2.
Test for faster-X evolution using the four-species comparison
Each cell count gives the number of cases in which dN/dS was higher in comparisons between D. pseudoobscura–D. miranda vs. D. melanogaster–D. yakuba. P-values are obtained by a Fisher’s exact test.
Figure 2.
Summaries of the distribution of dN/dS ratios for the 202 loci analyzed in the four-species comparison. Rectangles surround the quantiles of the distribution (25-th, 50-th, and 75-th percentiles), with lines extending out to the 2.5-th and 97.5-th percentiles. (m/y) dN/dS between D. melanogaster and D. yakuba; (p/m) dN/dS between D. pseudoobscura and D. miranda.
Integration of divergence and expression data
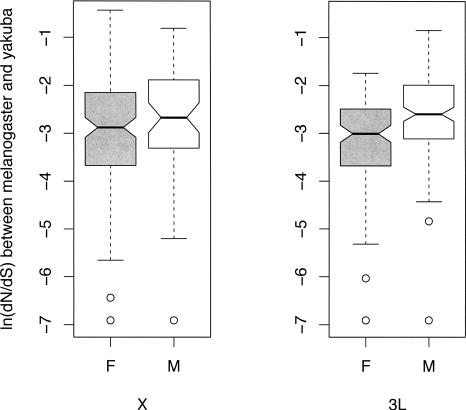
The analyses presented above are only concerned with differences in rates of divergence in X-linked versus autosomal genes, and suggest no difference between the two. However, such large-scale analyses may obscure faster-X evolution that may be restricted to certain classes of genes, such as X-linked loci that are male biased in expression, which are known to evolve at higher rates than female-biased or non-sex-biased genes (Zhang et al. 2004). Since a faster-X would be driven by selection in males (in the XY sex chromosome system), it follows that genes expressed specifically in males may be preferred targets for male-specific adaptive protein evolution on the X. Of the 2646 genes that we annotated on X and 3L in D. yakuba, we identified 436 as significantly sex biased in expression, using gene expression data from Parisi et al. (2003). On the X, there were 52 male-biased and 132 female-biased genes. On 3L, there were 142 male-biased and 110 female-biased genes. Repeating the three-species comparison using only male- or female-biased genes does not change any of our conclusions. That is, we find no evidence for a faster-X effect for either class of genes (data not shown). Interestingly, however, male-biased genes always appear to evolve faster than female-biased genes. Figure 3 shows boxplots of dN/dS between D. melanogaster and D. yakuba for male- and female-biased genes on chromosome arms X and 3L. On the X chromosome, there is no significant difference in rates of evolution between male- and female-biased genes (pairwise Wilcoxon rank sum test, P = 0.17). However, male-biased genes on the autosomes evolve faster than female-biased, autosomal genes (pairwise Wilcoxon rank sum test, P = 5e − 06). Despite this interesting difference, sex linkage has no effect on the rates of evolution of male-biased genes in these data (pairwise Wilcoxon rank sum test, P = 0.79). Significantly faster evolution of male-biased genes on the autosomes, if anything, contradicts the predictions of the faster-X model.
Figure 3.
Boxplot of divergence between D. melanogaster and D. yakuba for genes that are significantly sex-biased in expression. The boxplots show the quartiles of the distribution of dN/dS, with a notch at the median value and whiskers extending to three times the interquartile distance. (M) Significantly male-biased; (F) significantly female-biased; (X) X-linked loci; (3L) loci on chromosome arm 3L.
Discussion
If beneficial amino acid substitutions are partially recessive on average and if selection acts primarily on newly arising mutations, then the rate of protein evolution should be higher for an X-linked gene compared to an autosomal one (Charlesworth et al. 1987). Here, we find no evidence that orthologous loci evolve faster when X-linked than when autosomal (Tables 1 and 2). This result may reflect on properties of beneficial mutations and adaptive evolution. First, adaptive protein evolution might not be common enough to be detectable using genomic data. If only a very small fraction of all amino acid changes observed between species is driven by positive selection, than the background noise introduced by neutral amino acid substitutions accumulating uniformly on different chromosomes might overshadow any possible faster-X effect. This scenario, however, is not supported by recent analysis from Drosophila (Bustamante et al. 2002; Fay et al. 2002; Smith and Eyre-Walker 2002; Sawyer et al. 2003; Bierne and Eyre-Walker 2004; Andolfatto 2005). By contrasting the amount of amino acid polymorphism to amino acid divergence, various studies have concluded that more than one-third (and up to 90%) of amino acid differences observed between species were, in fact, driven to fixation by positive selection between species of the D. melanogaster species group. Thus, adaptive protein evolution appears to be common enough to be detectable in genome-wide scans.
Secondly, adaptation may not often use newly arising mutations, but instead may act on standing genetic variation. The alleles selected may have been previously neutral (Dykhuizen and Hartl 1980; Innan and Kim 2004) or previously deleterious and maintained at mutation–selection equilibrium. For such alleles, Orr and Betancourt (2001) showed that evolution from mutation–selection balance always proceeds more slowly at X-linked than autosomal genes, independent of how dominant a mutation is. Thus, if adaptive evolution primarily occurs from amino acids that were previously deleterious and segregating at mutation–selection balance, the rate of protein evolution should actually be lower on the X chromosome compared to autosomes. Finally, beneficial amino acid mutations might not be recessive on average, but their average effect may be close to additive. In that case, no faster-X effect would be expected (Charlesworth et al. 1987).
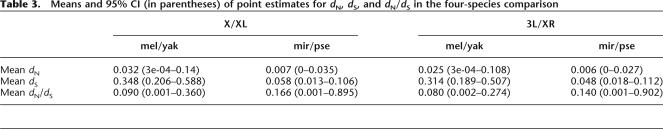
Differences in mutation rates between the X chromosome and autosomes affect the ability to detect a faster-X effect. In mammals, for example, the rate of substitution at nonsynonymous, synonymous, and non-coding sites is lower on X than on the autosomes (McVean and Hurst 1997; Malcolm et al. 2003; Lu and Wu 2005), but whether a faster-X effect is observed depends on whether synonymous or non-coding sites are used as the neutral benchmark (Lu and Wu 2005). In contrast to mammals, studies of different Drosophila species pairs suggest that there is no evidence that substitution rates differ significantly between the X chromosome and autosomes (Bauer and Aquadro 1997; Betancourt et al. 2002; Bartolome et al. 2005) (see comparison between D. melanogaster and D. yakuba in Table 3). Therefore, a difference in substitution rates among chromosomes is unlikely to account for the lack of a faster-X effect in our data. In Drosophila species, the X chromosome shows a higher degree of codon usage bias than the autosomes (Comeron et al. 1999; Singh et al. 2005), but codon bias does not appear to correlate with estimates of dS for the maximum likelihood method used here (Dunn et al. 2001). However, it is unlikely that differences in codon bias have had a significant impact on our analysis. First, the lack of evidence for differences in substitution rates between the X chromosome and autosomes suggests that the higher codon bias on X is not having a significant effect on long-term rates of divergence. Second, if higher codon bias leads to decreased rates of synonymous substitution (lower dS), then our analysis would have been biased toward finding a faster-X effect, because the reduction in dS inflates estimates of dN/dS, as is observed in mammals (Lu and Wu 2005).
Table 3.
Means and 95% CI (in parentheses) of point estimates for dN, dS, and dN/dS in the four-species comparison
The faster-X hypothesis was originally proposed to explain the disproportionately large effects of loci on the X chromosome to species differences. Such large X effects have been reported in interspecific crosses in studies of mating behavior, morphology, and hybrid sterility (Coyne and Orr 2004), suggesting a special role for X in species differentiation. Another evolutionary phenomenon connected with the sex chromosomes is Haldane’s rule (Haldane 1922), which states that it is nearly always the heterogametic sex that is inviable or infertile in interspecific hybrids. While the favored explanation of Haldane’s rule is based on the recessivity of genetic incompatibilities (Turelli and Orr 1995; Coyne and Orr 2004), the faster-X effect has been proposed as an additional contributing factor (Charlesworth et al. 1987; Naveira 2003; Tao and Hartl 2003; Coyne et al. 2004). The present analysis provides strong evidence that rates of adaptive evolution, as measured by dN/dS, do not differ substantially between X-linked and autosomal loci. This implies that the disproportionately large contribution of the X chromosome to species differences and Haldane’s rule is not adequately explained by the faster-X model (in the context of protein evolution).
In addition, several recent polymorphism studies in Drosophila (Begun and Whitley 2000; Andolfatto 2001), chickens (Sundström et al. 2004), and humans (Payseur et al. 2002) have found reduced levels of polymorphism on X (or Z) chromosomes relative to autosomes. This finding has often been interpreted as evidence for a higher rate of adaptive evolution on the X chromosome relative to the autosomes due to recessive beneficial mutations, wiping out linked variability more frequently on X (i.e., greater genetic hitchhiking on X). Again, the lack of evidence of a general faster-X effect for amino acid substitutions, at least in Drosophila, is inconsistent with this interpretation. Indeed, alternative explanations such as demographic effects (Wall et al. 2002) or other confounding factors such as chromosomal inversions and different effective population sizes in males and females (Andolfatto 2001; Charlesworth 2001) have been suggested as alternative explanations for patterns of variation on X versus autosomes, at least in Drosophila.
Methods
Primer design
Primers were designed using the annotation from the “freeze1” assembly of the D. pseudoobscura genome project (Richards et al. 2005). Loci were selected on the basis of having at least one exon of >1000 bp in length. Based on the exon annotations (B. Bettencourt, pers. comm.), we designed PCR and internal sequencing primers by an automated procedure using the primer3 software (Rozen and Skaletsky 2000) for every annotated exon fitting our length criterion. We chose 120 loci from both the XL and XR arms of D. pseudoobscura for sequencing in D. miranda without regard for any feature other than exon length (i.e., functional annotations, gene names, etc., were ignored).
PCR and sequencing
Genomic DNA was extracted from a single male of Drosophila miranda (line MSH22) using the Puregene DNA extraction Kit (Gentra). PCR reactions were performed in 50-μL volumes, purified using Exo-SAP treatment, and sequenced using ABI BigDye version 3 (Perkin Elmer). The phred/phrap/consed (Ewing and Green 1998; Ewing et al. 1998; Gordon et al. 1998) software suite was used to automate the base-calling and assembly, leading to a preliminary assembly of the data. We used BLASTN (Altschul et al. 1990) against our D. pseudoobscura coding sequence (CDS) database, to confirm that we had sequenced what we expected, and used the BLAST alignments as guides for manually confirming every base call in D. miranda that differed from D. pseudoobscura. Low-quality regions were trimmed manually. Sequences have been deposited in GenBank under accession nos. DQ334053–DQ33425.
Alignment, annotation, and analysis for paired comparisons
We annotated the coding regions of the finished contigs using the GeneWise tool (http://www.ebi.ac.uk/Wise2/), which is a comparative annotation program on which many of the D. pseudoobscura gene models are based. We used the peptide sequence from the D. pseudoobscura sequence to annotate the D. miranda coding sequence.
We obtained scaffolds from the April 2004 freeze of the D. yakuba genome sequencing project from the Washington University Genome Sequencing Center (http://genome.wustl.edu/projects/yakuba; R. Wilson pers. comm.). First, we generated melanogaster/miranda/pseudoobscura alignments using DIALIGN 2.2 (Morgenstern 1999), with the option to align based on CDS translations. We used the D. melanogaster CDS sequences (Adams et al. 2000) that were used to annotate the D. pseudoobscura genome (B. Bettencourt, pers. comm.). These alignments were manually edited and trimmed in order to find the portion of the melanogaster CDS homologous to the region sequenced in miranda. The resulting homologous fragments from melanogaster were used to search the D. yakuba contigs using BLASTN. While translated BLAST programs (i.e., TBLASTX) would be preferable, these species are closely related at the sequence level (Takano 1998), and obtaining nucleotide alignments directly from BLAST was useful for our purposes.
For both miranda/pseudoobscura and melanogaster/yakuba comparisons, final alignments were made using peptide alignments generated with DIALIGN 2.2, and the CDS sequences were aligned with the tranalign tool in the EMBOSS software suite (http://www.emboss.org), which uses the peptide alignments as guides.
Whole chromosome analysis
Using CDS sequences for D. melanogaster loci for which gene models exist in the annotation of the “freeze1” assembly of the D. pseudoobscura genome (Richards et al. 2005), we performed BLASTN searches against the D. yakuba contigs. Using the BLASTN results for all D. melanogaster CDS sequences mapping to X/XL and 3L/XR, we extracted the region of the best BLASTN hit in D. yakuba plus 6000 bp on either side. The D. melanogaster peptide was then used to predict the gene structure in D. yakuba using the GeneWise tool. We were able to obtain 1087 loci on X/XL and 1559 on 3L/XR for which we were able to align D. melanogaster, D. yakuba, and D. pseudoobscura. The three-species alignments were performed on the peptide alignments using DIALIGN and tranalign, as described above.
Estimation of divergence parameters
Estimates of the number of amino acid differences per site (dN) and the number of synonymous differences per site (dS) were calculated using the likelihood methods implemented in the PAML software package (Yang 1997). Estimates based on the three-species phylogeny were performed using codeml, allowing dN/dS to vary across branches (i.e., each lineage can have its own dN/dS value). For these analyses, it is important that we are comparing orthologous sequences rather than paralogs (e.g., gene duplications). In Drosophila, 90% of D. melanogaster gene models were successfully assigned orthologs in D. pseudoobscura (Richards et al. 2005). As our identification of genes in D. yakuba was based on the set of melanogaster/pseudoobscura orthologs, we have high confidence that we are, indeed, comparing orthologs. It is formally possible that some of the genes sequenced from D. miranda are recent duplications in that species, rather than true orthologs. However, recent duplications (as measured by dS) appear to be relatively rare in Drosophila species (e.g., Thornton and Long 2002). Furthermore, we observed no “heterozygous” base calls in our sequencing traces, which we would have expected to find if our PCR had amplified closely related sequences.
Expression data
Data of sex-biased gene expression were taken from the Supplemental information of Parisi et al. (2003). We used the GFF annotation files from Releases 3 and 4 of the D. melanogaster genome (http://www.flybase.org) in order to map the gene names and synonyms from Parisi et al. to the “CG” numbers that we used as locus labels in this study. We were able to map 7309 of the genes studied in Parisi et al. to CG numbers. We used the same measure of significant sex-biased expression as Parisi et al. to identify sex-biased genes on chromosomes X/XL and 3L/XR.
Acknowledgments
K.T. is supported by an A.P. Sloan Postdoctoral Fellowship. D.B. is supported by a Postdoctoral Fellowship from the Austrian Academy of Sciences. P.A. is supported by an A.P. Sloan Fellowship in Molecular and Computational Biology.
Footnotes
[The Drosophila three and four species alignments are available at http://www.molpopgen.org/krthornt/Data/ FastXGR. The sequence data from this study have been submitted to GenBank under accession nos. DQ334053– DQ33425.]
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4447906
References
- Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Li P.W., Hoskins R.A., Galle R.F., Hoskins R.A., Galle R.F., Galle R.F., et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Muller W., Myers E.W., Lipman D.J., Gish W., Muller W., Myers E.W., Lipman D.J., Muller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andolfatto P. Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 2001;18:279–290. doi: 10.1093/oxfordjournals.molbev.a003804. [DOI] [PubMed] [Google Scholar]
- Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1153. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- Barbash D.A., Awadalla P., Tarone A.M., Awadalla P., Tarone A.M., Tarone A.M. Functional divergence caused by ancient positive selection of a Drosophila hybrid incompatibility locus. PLoS Biol. 2004;2:839–848. doi: 10.1371/journal.pbio.0020142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolome C., Maside X., Yi S., Grant A.L., Charlesworth B., Maside X., Yi S., Grant A.L., Charlesworth B., Yi S., Grant A.L., Charlesworth B., Grant A.L., Charlesworth B., Charlesworth B. Patterns of selection on synonymous and nonsynonymous variants in Drosophila miranda. Genetics. 2005;169:1495–1507. doi: 10.1534/genetics.104.033068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer V.L., Aquadro C.F., Aquadro C.F. Rates of DNA sequence evolution are not sex-biased in Drosophila melanogaster and D. simulans. Mol. Biol. Evol. 1997;14:1252–1257. doi: 10.1093/oxfordjournals.molbev.a025734. [DOI] [PubMed] [Google Scholar]
- Begun D., Whitley P., Whitley P. Reduced X-linked nucleotide polymorphism in Drosophila simulans. Proc. Natl. Acad. Sci. 2000;97:5960–5965. doi: 10.1073/pnas.97.11.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman C.M., Pfeiffer B.D., Rincón-Limas D.E., Hoskins R.A., Gnirke A., Mungall C.J., Wang A.M., Kronmiller B., Pacleb J., Park S., Pfeiffer B.D., Rincón-Limas D.E., Hoskins R.A., Gnirke A., Mungall C.J., Wang A.M., Kronmiller B., Pacleb J., Park S., Rincón-Limas D.E., Hoskins R.A., Gnirke A., Mungall C.J., Wang A.M., Kronmiller B., Pacleb J., Park S., Hoskins R.A., Gnirke A., Mungall C.J., Wang A.M., Kronmiller B., Pacleb J., Park S., Gnirke A., Mungall C.J., Wang A.M., Kronmiller B., Pacleb J., Park S., Mungall C.J., Wang A.M., Kronmiller B., Pacleb J., Park S., Wang A.M., Kronmiller B., Pacleb J., Park S., Kronmiller B., Pacleb J., Park S., Pacleb J., Park S., Park S., et al. 2002Assessing the impact of comparative genomic sequence data on the functional annotation of the Drosophila genome. Genome Biol. 3r0086 0081–0086.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt A.J., Presgraves D.C., Swanson W.J., Presgraves D.C., Swanson W.J., Swanson W.J. A test for faster X evolution in Drosophila. Mol. Biol. Evol. 2002;19:1816–1819. doi: 10.1093/oxfordjournals.molbev.a004006. [DOI] [PubMed] [Google Scholar]
- Bierne N., Eyre-Walker A., Eyre-Walker A. The genomic rate of adaptive amino acid substitution in Drosophila. Mol. Biol. Evol. 2004;21:1350–1360. doi: 10.1093/molbev/msh134. [DOI] [PubMed] [Google Scholar]
- Braverman J.M., Hudson R.R., Kaplan N.L., Langley C.H., Stephan W., Hudson R.R., Kaplan N.L., Langley C.H., Stephan W., Kaplan N.L., Langley C.H., Stephan W., Langley C.H., Stephan W., Stephan W. The hitchhiking effect on the site frequency-spectrum of DNA polymorphisms. Genetics. 1995;140:783–796. doi: 10.1093/genetics/140.2.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C.D., Nielsen R., Sawyer S.A., Olsen K.M., Purugganan M.D., Hartl D.L., Nielsen R., Sawyer S.A., Olsen K.M., Purugganan M.D., Hartl D.L., Sawyer S.A., Olsen K.M., Purugganan M.D., Hartl D.L., Olsen K.M., Purugganan M.D., Hartl D.L., Purugganan M.D., Hartl D.L., Hartl D.L. The cost of inbreeding in Arabidopsis. Nature. 2002;416:531–534. doi: 10.1038/416531a. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. The effect of life-history and mode of inheritance on neutral genetic variability. Genet. Res. 2001;77:153–166. doi: 10.1017/s0016672301004979. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Coyne J.A., Barton N.H., Coyne J.A., Barton N.H., Barton N.H. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 1983;130:113–146. [Google Scholar]
- Comeron J.M., Kreitman M., Aguade M., Kreitman M., Aguade M., Aguade M. Natural selection on synonymous sites is correlated with gene length and recombination in Drosophila. Genetics. 1999;151:239–249. doi: 10.1093/genetics/151.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counterman B.A., Ortiz-Barrientos D., Noor M.A., Ortiz-Barrientos D., Noor M.A., Noor M.A. Using comparative genomic data to test for faster-X evolution. Evolution Int. J. Org. Evolution. 2004;58:656–660. [PubMed] [Google Scholar]
- Coyne J.A., Orr H.A., Orr H.A.2004Speciation. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Coyne J.A., Elwyn S., Kim S.Y., Llopart A., Elwyn S., Kim S.Y., Llopart A., Kim S.Y., Llopart A., Llopart A. Genetic studies of two sister species in the Drosophila melanogaster subgroup, D. yakuba and. D. santomea. Genet. Res. 2004;84:11–26. doi: 10.1017/s0016672304007013. [DOI] [PubMed] [Google Scholar]
- Dunn K.A., Bielawski J.P., Yang Z., Bielawski J.P., Yang Z., Yang Z. Substitution rates in Drosophila nuclear genomes: Implications for translational selection. Genetics. 2001;157:295–305. doi: 10.1093/genetics/157.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen D., Hartl D.L., Hartl D.L. Selective neutrality of 6PGD allozymes in E. coli and the effects of genetic background. Genetics. 1980;96:801–817. doi: 10.1093/genetics/96.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B., Green P., Green P. Basecalling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M., Green P., Hillier L., Wendl M., Green P., Wendl M., Green P., Green P. Basecalling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Fay J., Wyckoff G., Wu C., Wyckoff G., Wu C., Wu C. Testing the neutral theory of molecular evolution with genomic data from Drosophila. Nature. 2002;415:1024–1026. doi: 10.1038/4151024a. [DOI] [PubMed] [Google Scholar]
- Gordon G.C., Abajian C., Green P., Abajian C., Green P., Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Haldane J.B.S. Sex ratio and unisexual sterility in animal hybrids. J. Genet. 1922;12:101–109. [Google Scholar]
- Innan H., Kim Y., Kim Y. Pattern of polymorphism after strong artificial selection in a domestication event. Proc. Natl. Acad. Sci. 2004;101:10667–10672. doi: 10.1073/pnas.0401720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Wu C.I., Wu C.I. Weak selection revealed by the whole-genome comparison of the X chromosome and autosomes of human and chimpanzee. Proc. Natl. Acad. Sci. 2005;102:4063–4067. doi: 10.1073/pnas.0500436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcolm C.M., Wyckoff G.J., Lahn B.T., Wyckoff G.J., Lahn B.T., Lahn B.T. Genic mutation rates in mammals: Local similarity, chromosomal heterogeneity, and X-versus-autosome disparity. Mol. Biol. Evol. 2003;20:1633–1641. doi: 10.1093/molbev/msg178. [DOI] [PubMed] [Google Scholar]
- McVean G.T., Hurst L.D., Hurst L.D. Evidence for a selectively favourable reduction in the mutation rate of the X chromosome. Nature. 1997;386:388–392. doi: 10.1038/386388a0. [DOI] [PubMed] [Google Scholar]
- Morgenstern B. DIALIGN 2: Improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- Muller H.J.1940. Bearings of the Drosophila work on systematics. In The new systematics (ed. J.S. Huxley) pp. 185–268. Oxford University Press, London [Google Scholar]
- Naveira H.F. On the relative roles of faster-X evolution and dominance in the establishment of intrinsic postzygotic isolating barriers. Genetica. 2003;118:41–45. doi: 10.1023/a:1022978222021. [DOI] [PubMed] [Google Scholar]
- Orr H.A., Betancourt A.J., Betancourt A.J. Haldane’s sieve and adaptation from the standing genetic variation. Genetics. 2001;157:875–884. doi: 10.1093/genetics/157.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M., Nuttall R., Naiman D., Bouffard G., Malley J., Andrews J., Eastman S., Oliver B., Nuttall R., Naiman D., Bouffard G., Malley J., Andrews J., Eastman S., Oliver B., Naiman D., Bouffard G., Malley J., Andrews J., Eastman S., Oliver B., Bouffard G., Malley J., Andrews J., Eastman S., Oliver B., Malley J., Andrews J., Eastman S., Oliver B., Andrews J., Eastman S., Oliver B., Eastman S., Oliver B., Oliver B. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur B.A., Cutter A.D., Nachman M.W., Cutter A.D., Nachman M.W., Nachman M.W. Searching for evidence of positive selection in the human genome using patterns of microsatellite variability. Mol. Biol. Evol. 2002;19:1143–1153. doi: 10.1093/oxfordjournals.molbev.a004172. [DOI] [PubMed] [Google Scholar]
- Presgraves D.C., Balagopalan L., Abmayr S.M., Orr H.A., Balagopalan L., Abmayr S.M., Orr H.A., Abmayr S.M., Orr H.A., Orr H.A. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature. 2003;423:715–719. doi: 10.1038/nature01679. [DOI] [PubMed] [Google Scholar]
- Richards S., Liu Y., Bettencourt B.R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Liu Y., Bettencourt B.R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Bettencourt B.R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Hubisz M.J., Chen R., Meisel R.P., Chen R., Meisel R.P., Meisel R.P., et al. Comparative genome sequencing of Drosophila pseudoobscura: Chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H., Skaletsky H.2000. Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics methods and protocols: Methods in molecular biology (eds. S. Krawertz and S. Misener), pp. 365–386. Humana Press; Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- Sawyer S., Kulathinal R., Bustamante C., Hartl D., Kulathinal R., Bustamante C., Hartl D., Bustamante C., Hartl D., Hartl D. Bayesian analysis suggests that most amino acid replacements in Drosophila are driven by positive selection. J. Mol. Evol. 2003;57:S154–S164. doi: 10.1007/s00239-003-0022-3. [DOI] [PubMed] [Google Scholar]
- Singh N.D., Davis J.C., Petrov D.A., Davis J.C., Petrov D.A., Petrov D.A. X-linked genes evolve higher codon bias in Drosophila and Caenorhabditis. Genetics. 2005;169:709–722. doi: 10.1534/genetics.105.043497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N., Eyre-Walker A., Eyre-Walker A. Adaptive protein evolution in Drosophila. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- Sundström H., Webster M., Ellegren H., Webster M., Ellegren H., Ellegren H. Reduced variation on the chicken Z chromosome. Genetics. 2004;167:377–385. doi: 10.1534/genetics.167.1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T.S. Rate variation of DNA sequence evolution in the Drosophila lineages. Genetics. 1998;149:959–970. doi: 10.1093/genetics/149.2.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Hartl D.L., Hartl D.L. Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana.III. Heterogeneous accumulation of hybrid incompatibilities, degree of dominance, and implications for Haldane’s rule. Evolution. 2003;57:2580–2598. doi: 10.1111/j.0014-3820.2003.tb01501.x. [DOI] [PubMed] [Google Scholar]
- Thornton K., Long M., Long M. Rapid divergence of gene duplicates on the Drosophila melanogaster X chromosome. Mol. Biol. Evol. 2002;19:918–925. doi: 10.1093/oxfordjournals.molbev.a004149. [DOI] [PubMed] [Google Scholar]
- Ting C.-T., Tsaur S.-C., Wu M.-L., Wu C.I., Tsaur S.-C., Wu M.-L., Wu C.I., Wu M.-L., Wu C.I., Wu C.I. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science. 1998;282:1501–1504. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- Turelli M., Orr H.A., Orr H.A. The dominance theory of Haldane’s rule. Genetics. 1995;140:389–402. doi: 10.1093/genetics/140.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall J.D., Andolfatto P., Przeworski M., Andolfatto P., Przeworski M., Przeworski M. Testing models of selection and demography in Drosophila simulans. Genetics. 2002;162:203–216. doi: 10.1093/genetics/162.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.H. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Hambuch T.M., Parsch J., Hambuch T.M., Parsch J., Parsch J. Molecular evolution of sex-biased genes in Drosophila. Mol. Biol. Evol. 2004;21:2130–2139. doi: 10.1093/molbev/msh223. [DOI] [PubMed] [Google Scholar]