Abstract
The discovery of direct downstream targets of transcription factors (TFs) is necessary for understanding the genetic mechanisms underlying complex, highly regulated processes such as development. In this report, we have used a combinatorial strategy to conduct a genome-wide search for novel direct targets of Eyeless (Ey), a key transcription factor controlling early eye development in Drosophila. To overcome the lack of high-quality consensus binding site sequences, phylogenetic shadowing of known Ey binding sites in sine oculis (so) was used to construct a position weight matrix (PWM) of the Ey protein. This PWM was then used for in silico prediction of potential binding sites in the Drosophila melanogaster genome. To reduce the false positive rate, conservation of these potential binding sites was assessed by comparing the genomic sequences from seven Drosophila species. In parallel, microarray analysis of wild-type versus ectopic ey-expressing tissue, followed by microarray-based epistasis experiments in an atonal (ato) mutant background, identified 188 genes induced by ey. Intersection of in silico predicted conserved Ey binding sites with the candidate gene list produced through expression profiling yields a list of 20 putative ey-induced, eye-enriched, ato-independent, direct targets of Ey. The accuracy of this list of genes was confirmed using both in vitro and in vivo methods. Initial analysis reveals three genes, eyes absent, shifted, and Optix, as novel direct targets of Ey. These results suggest that the integrated strategy of computational biology, genomics, and genetics is a powerful approach to identify direct downstream targets for any transcription factor genome-wide.
Transcriptional regulation plays a key role in complex biological processes such as development. This occurs when combinations of nuclear transcription factors (TFs) acting with the nuclear effectors of signal transduction pathways bind DNA to activate and repress target genes. The structure of most gene regulatory networks is highly complex and often involves cooperative interactions and feedback regulation. While genetic dissection of such networks has been very productive, the discovery of direct targets of TFs remains largely underexplored, due mostly to technical limitations. Techniques to discover direct targets of TFs on a genomic scale include gene expression profiling using microarrays (Tavazoie et al. 1999), in silico prediction of binding sites (Halfon et al. 2002), and direct chromatin profiling (Ren et al. 2000). However, microarray analysis is limited by an inability to distinguish direct targets of TFs from indirect targets; in silico prediction of binding sites is limited by poor characterization of most TF binding sequences and by lack of experimental confirmation of most TF binding events in vivo; and direct chromatin profiling using techniques such as chromatin immunoprecipitation with microarray detection (ChIP-on-chip) is still technically challenging and cannot be easily applied to all TFs in all tissues.
A combination of microarrays and in silico binding site prediction is a very promising approach to identifying direct targets of TFs. For example, a previous study used a combination of these two techniques to perform a genome-wide prediction of direct targets of Drosophila Dorsal (Dl) (Stathopoulos and Levine 2002). However, as performed, such studies have been limited to a few well-characterized TFs, such as Dl, whose binding site consensus sequence is well defined with many targets previously identified (Stathopoulos and Levine 2002). Moreover, the structure of these regulatory modules is relatively clear with binding sites from multiple TFs positioned within a small window (Berman et al. 2002). In contrast, it remains a great challenge to identify direct targets of TFs for which binding site consensus sequences and cofactors are either not known or poorly defined. We report several improvements to this multi-pronged approach that now allows genome-wide discovery of potential direct targets for TFs with degenerate and/or poorly characterized binding sites. These improvements are demonstrated through a genome-wide discovery of the direct targets of Eyeless (Ey), whose direct downstream targets are largely unknown.
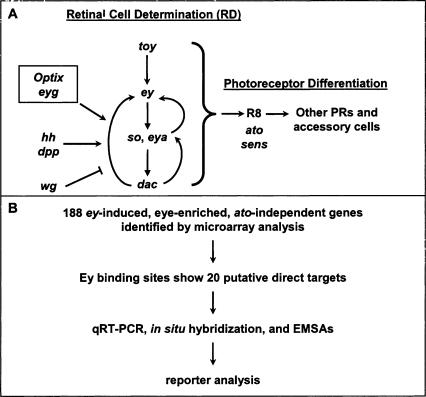
ey is a key regulator of Drosophila eye development from its earliest stages in embryogenesis, where a group of about 20 cells is partitioned into an eye primordium (Garcia-Bellido and Merriam 1969; Halder et al. 1995; Halder et al. 1995; 1998; Loosli et al. 1998). During larval development, these cells develop into an epithelial monolayer called the eye imaginal disc. After a proliferative stage in which the eye disc is undifferentiated and unpatterned, an indentation in the epithelium, the morphogenetic furrow (MF), sweeps anteriorly across the eye disc for two days during late larval and early pupal development, leaving differentiated ommatidial cells in its wake (Ready et al. 1976; Wolff and Ready 1993). Immediately anterior to the furrow, retinal cell fate is irreversibly established in a process termed retinal determination. Genes involved in retinal determination share many similar features: Loss-of-function mutations cause an early block in eye development; misexpression of these genes can induce the entire cascade of eye development in other imaginal discs; and these genes are expressed early and anterior to the MF in the eye imaginal disc (for review, see Pappu and Mardon 2004). So far, seven retinal determination (RD) genes have been identified: toy, ey, eyes absent (eya), sine oculis (so), dachshund (dac), Optix, and eye gone (eyg) (Bonini et al. 1993; Cheyette et al. 1994; Mardon et al. 1994; Loosli et al. 1998; Czerny et al. 1999; Seimiya and Gehring 2000; Jang et al. 2003). Encoding proteins that function in multiple complexes, these genes form a genetic network that controls the specification and determination stages of retinal development (Akimaru et al. 1997; Pignoni et al. 1997). At the top of this network are the two Pax6 homologs in Drosophila, toy and ey (Fig. 1A).
Figure 1.
Strategy to find direct targets of Ey. (A) The Drosophila retinal determination network. Retinal cell fate is established by an interregulatory network of conserved nuclear factors. toy induces ey, which induces so and eya, which induce dac. This pathway receives input from secreted signals from the hh and dpp pathways, which promote retinal determination, and the wg pathway that inhibits it. Several other genes are necessary and/or sufficient for eye development, including Optix and eyegone (eyg). These factors establish competency for the next stage of development, photoreceptor differentiation, whose first step, R8 recruitment, requires genes such as ato and sens. (B) Genes that upregulated by ey were identified by conducting microarray analysis of wild-type versus dpp-GAL4/UAS-ey discs. Quantitative real-time RT-PCR and hand curation through comparison to online databases were used to evaluate the results. We searched for possible Ey binding sites in this list of 188 genes, identifying 20 putative direct targets. Expression pattern and response to ey was examined by in situ hybridization and RT-PCR. In vitro Ey binding was tested by EMSAs. Reporter analysis of fragments containing identified Ey binding sites was used to test their function in vivo.
ey encodes a TF containing two conserved DNA binding domains, the homeodomain (HD) and the paired domain (PD) (Dahl et al. 1997). Despite its involvement in many stages of retinal development, the only direct target of Ey identified to date is so, which is coordinately regulated by both Ey and Toy (Czerny et al. 1999; Hauck et al. 1999; Niimi et al. 1999; Punzo et al. 2002, 2004). Since ey mutants cannot be rescued by so and ectopic so expression is not sufficient to induce ectopic eye formation or expression of other RD genes, additional targets of ey must exist (Pignoni et al. 1997; Halder et al. 1998). Thus, to gain a full understanding of ey function, it is essential to identify these additional targets. We have designed a novel approach to identify Ey targets genome-wide using a combination of gene expression profiling, comparative genomics, and in silico binding site prediction (Fig. 1B). First, genes that are up-regulated by ectopic expression of ey were identified using microarrays to profile gene expression in three different tissues. Only genes consistently up-regulated in all three tissues were selected as potential targets of ey. Second, microarray-based genetic epistasis experiments were conducted to identify genes immediately downstream of ey by repeating expression profiling in an atonal (ato) mutant background. Only those genes induced by ey even in the absence of ato function were retained. In parallel, potential Ey binding sites in the Drosophila genome were identified using in silico methods. To overcome the limitation of only three previously identified Ey binding sites being available, phylogenetic shadowing of these Ey binding sites in closely related Drosophila species was used to produce a Drosophila Ey position weight matrix (PWM) (Epstein et al. 1994; Niimi et al. 1999; Punzo et al. 2002). Based on this new PWM, potential Ey binding sites were identified genome-wide. By intersecting these data with potential targets identified through microarray analyses, we identified three novel direct targets of Ey: eya, Optix, and shf, which were confirmed using both in vitro and in vivo methods. Reiteration of this in silico prediction process together with comparative genomics of seven Drosophila species yields a final list of 20 genes that are likely to be true targets of Ey during retinal development in Drosophila. This rapid and efficient discovery of new direct targets of Ey genome-wide demonstrates the utility of this approach and adds significantly to our knowledge of early eye development. It also shows that a combinatorial approach using microarray epistasis, PWM construction through phylogenetic shadowing, and comparative genomics can allow circumvention of the problems inherent in searching for direct targets of a TF with a degenerate or poorly characterized binding site.
Results
Even though Eyeless has been known as a critical regulator of early retinal development for more than a decade, only one direct target, so, has been confirmed (Niimi et al. 1999). However, since so expression is not sufficient to rescue the ey mutant phenotype or to induce ectopic eye formation or expression of other RD genes, additional targets of ey must exist. One striking feature of ey is that it is sufficient to induce ectopic eye formation in other imaginal discs. Therefore, direct targets of Ey relevant to normal eye development should be ectopically induced where ey is misexpressed. Taking advantage of this feature, we first identified genes that are up-regulated by ey in other imaginal tissues using gene expression microarray technology.
Identification of 300 ey-induced, eye-enriched genesby microarray analysis
To identify genes responsive to ey, we determined the gene expression profiles in wild-type and ectopic dpp-GAL4/UAS-ey (dpp>ey) leg, wing, and antennal discs and identified genes that are consistently induced by ey in all three tissues. Intersecting the gene expression profiles of three distinct tissues reduces the false positive rate of our screen as compared with previous studies that examined only one tissue (Michaut et al. 2003). A total of 956 genes are identified when an average of twofold induction and a false discovery rate of 0.001 are used as cutoffs (Klipper-Aurbach et al. 1995; Storey and Tibshirani 2003). As in previous studies, to identify genes with specific roles during retinal development, we compared this list with genes identified by microarray analysis of wild-type eye discs (Michaut et al. 2003). Genes that are not expressed in wild-type eye discs or with a lower expression level in wild-type eye discs than in wild-type wing, leg, and antennal discs were also treated as false positives. As a result, we generated a list of 300 candidate genes that are likely to act downstream of ey and play specific roles during Drosophila retinal development (Supplemental Table 1). Both direct and indirect ey targets are expected to be included in this list.
Arranging potential ey targets by microarray epistasis experiments
Based on the current model of Drosophila retinal development (Fig. 1A), we reasoned that genes dependent on atonal (ato) for expression are involved in processes occurring after retinal specification and determination and are thus likely to be indirect targets. Indeed, RD genes are still expressed in ato discs, while many genes known to be associated with retinal differentiation including sevenless, glass, and senseless, are greatly reduced or absent (data not shown; Jarman et al. 1994; Frankfort et al. 2001). We therefore repeated our microarray studies in the absence of ato function, where all photoreceptor differentiation is blocked.
To identify genes upstream of ato but downstream of ey, we first compared the expression profiles of wild-type and ato mutant eye discs that were dissected and separated from antennal discs. Among the 300 ey-responsive, eye-enriched genes identified, expression of 178 genes is significantly reduced in ato mutant eye discs, suggesting that these genes are downstream of ato and are likely to be indirect targets of ey. Since genes functioning both anterior and posterior to the furrow may be reduced in ato mutant eye discs, we also examined the gene expression profiles of ato mutant leg discs containing ectopic ey driven by dpp-GAL4. ey induction of 129 genes in the leg disc is abolished in an ato mutant background. These two sets overlap by a total of 112 genes (P-value of 1e-12 by χ2 test), indicating changes in the gene expression profiles are very similar between ato mutant eye discs and leg discs with ectopic ey expression in an ato mutant background. Significant changes in expression of the known RD genes are not detected in these microarray experiments, consistent with a genetic model where these genes act upstream of ato. Thus, we filtered 112 of 300 genes, producing a list of 188 ey-induced, eye-enriched, ato-independent genes (Supplemental Table 1).
Two methods were used to validate the microarray results. First, manual inspection of the list of 188 genes identified was compared with the literature. Consistent with the current model of the retinal determination pathway, many early retinal development genes, such as eya, so, Optix, and ato, are significantly upregulated. Overall, 24 genes known to be required or expressed in the eye are found on our list of 188 genes. As an experimental confirmation, real-time RT-PCR was conducted to test induction by ey misexpression. We randomly selected 85 novel genes and tested their mRNA levels in wild-type and ectopic ey leg discs. Results consistent with microarray analysis were obtained for 74 of these genes, suggesting that at least 87% of the 188 ey-induced genes are actual direct or indirect targets (data not shown).
Construction of a Drosophila Ey PWM for in silico Ey binding site prediction
There are several reasons that identification of direct downstream targets of Ey via expression profiling alone is unlikely to succeed. These include nonspecific probe hybridization and ectopic expression-mediated changes in biological processes other than retinal development. We sought to overcome these limitations by employing in silico methods that use data sets entirely independent from those used for expression profiling. To do so, we searched for direct downstream targets of Ey in our list by identifying nearby binding site consensus sequences. While this approach is also limited by a high false-positive rate caused by binding site degeneracy and low information content of most binding sites, we found that the specificity of computational methods can be greatly improved by combining them with experimental expression profiling data.
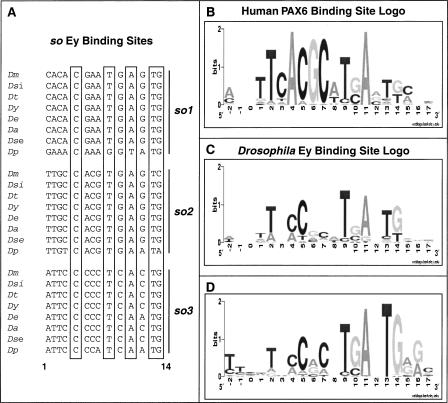
A well-established approach to identify TF binding sites is to scan the genome with a site represented by a PWM (Stormo et al. 1982). Our first task was to determine an appropriate PWM for the Ey binding site. Generally, a large collection of binding sites (20 or more) is necessary to obtain an appropriate PWM. Unfortunately, only three Ey binding sites have been discovered and validated in the Drosophila genome, all of which are in an eye-specific enhancer of so (Niimi et al. 1999; Punzo et al. 2002). As these three sites alone are not sufficient to construct a proper PWM, we turned to the PWM of the human PAX6 paired domain, which was obtained through in vitro oligonucleotide selection (Epstein et al. 1994). However, when the human PAX6 PWM (Fig. 2B) is used to screen the Drosophila genome, relatively low scores are obtained for all three Ey binding sites in the so eye-specific enhancer. This could reflect either amino acid differences between Drosophila Ey and human PAX6 or bias in the in vitro method used to construct the PWM for human PAX6.
Figure 2.
Sequence conservation of Ey binding sites in the so10 eye enhancer. (A) Sequence alignment of three Ey binding sites from eight Drosophila species. Positions 5, 9, 11, 13, and 14 (highlighted in boxes) are almost identical across all Drosophila species. (B) The human PAX6 position weight matrix (PWM) is represented by a logo, where nucleotide preference is represented by the height of the letter. This was combined with the so binding site data to build an Ey PWM, shown by the logo in C. (D) Optimized Ey PWM based on new Ey binding sites identified in this report. (Dm) D. melanogaster, (Dsi) D. simulans, (Dt) D. teisseri, (Dy) D. yakuba, (De) D. erecta, (Da) D. ananassae, (Dse) D. sechelia, (Dp) D. pseudoobscura.
To overcome this problem, we modified the human PAX6 PWM using the known Drosophila binding sites as a guide. Since bases that are critical for binding are under negative selection and change more slowly than other bases in a binding site, we used phylogenetic shadowing to refine the PWM (Boffelli et al. 2003). We determined the sequence of the so eye enhancer from seven additional Drosophila species that span an evolutionary distance of about 35 million years (Tamura et al. 2004). When aligned, we found that all three Ey binding sites are highly conserved (Fig. 2A and Supplemental data). Bases at positions 5, 9, 11, 13, and 14 are almost identical in all sites and all species and have similar base preferences as human PAX6 binding sites. In contrast, although positions 2, 4, 6, and 7 in the human PAX6 PWM are highly constrained, most or all bases are observed at these positions in Drosophila species (Fig. 2C). Thus, we changed the PAX6 matrix at these positions to reflect the reduced constraint. As expected, the resulting PWM produced greatly improved scores for all three Ey sites in the so enhancer.
Combining in silico and microarray data to identify novel direct Ey targets
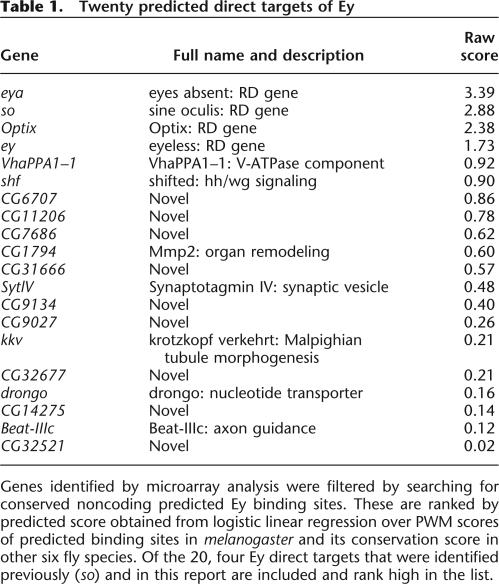
The PWM obtained above was used to score all 14 mers within 1 kb of each gene in the list of 188 genes produced by microarray analysis. To estimate the P-value of a score, a random sequence was generated using a 5th-order Markov Model and scanned with the same PWM (Lawrence 1989). Based on the score distribution of the random sequence, sites with a score of 9.6 have a P-value of 0.00005. Since one of the weakest Ey binding sites in the so eye-specific enhancer scores at 9.6, this cutoff was used to exclude other sites from further analysis. To improve the accuracy of this method, we first filtered out sites in repetitive regions or within exons based on the Drosophila genome annotation. Second, comparative genomics was used to assess conservation of putative binding sites, as true cis-regulatory elements may tend to be more conserved due to functional constraint. As shown in Figure 2A, all three Ey binding sites in the so eye enhancer are highly conserved from Drosophila melanogaster to pseudoobscura. We examined other putative Ey sites by comparing melanogaster with pseudoobscura using the BLASTZ program (Elnitski et al. 2003). Low-quality alignment blocks were excluded to identify the best matches in pseudoobscura using the melanogaster genome as a reference. The filtered alignment was then used to identify corresponding sequences for each putative melanogaster Ey binding site in pseudoobscura. Sites were considered conserved if the corresponding pseudoobscura site scored 9.6 or higher. Genes were ranked by summation of all conserved binding site scores in melanogaster and pseudoobscura (data not shown). Strikingly, four out of the seven known RD genes (toy, so, eya, and Optix) are among the top-ranked genes with the sole known target of ey, so, ranking highest. We chose to further test two genes with no characterized role in eye development, shf and VhaPPA1–1, and two known RD genes that had not previously been characterized as direct targets of Ey, eyes absent (eya) and Optix. shf encodes a Drosophila homolog of Wnt-inhibitory factor-1 (WIF-1) and has recently been shown to be necessary for extracellular transport of Hedgehog (Hh) (Hsieh et al. 1999; Glise et al. 2005; Gorfinkiel et al. 2005). VhaPPA1–1 is a member of a multigene family encoding a Drosophila vacuolar ATPase (Dow 1999). Optix is the Drosophila homolog of vertebrate Six3, encodes a Six-class homeodomain protein, and is a paralog of sine oculis (Toy et al. 1998). eya encodes the only member of a novel class of phosphatases and has been shown to bind to Dac and So (Chen et al. 1997; Pignoni et al. 1997; Rayapureddi et al. 2003; Tootle et al. 2003). These four genes were tested for direct Ey regulation by electrophoretic mobility shift assays (EMSAs) and reporter analysis.
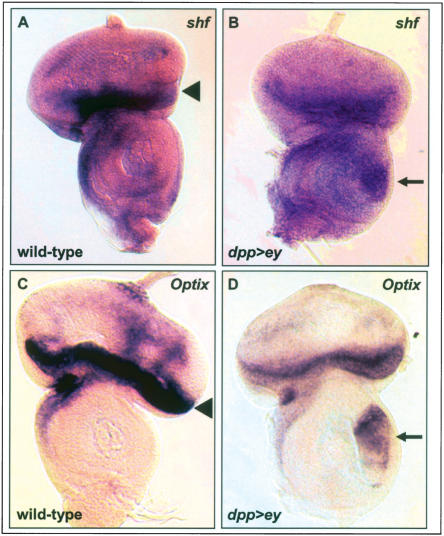
First, in situ hybridization was used to determine the expression patterns of shf and Optix in wild-type eye discs and in response to ectopic ey in dpp>ey antennal discs. shf is expressed at the anterior margin of the eye disc and in scattered cells posterior to the morphogenetic furrow (Fig. 3A). Optix shows strong expression anterior to the morphogenetic furrow (Fig. 3C). Both genes are markedly induced in the ventral antennal dpp expression domain in dpp>ey discs (Fig. 3B,D). These data indicate that shf and Optix are expressed in the wild-type eye and are responsive to ectopic ey. Previous studies have already shown eye disc expression and induction by ectopic ey for eya and eye disc expression for Optix (Bonini et al. 1993; Halder et al. 1998; Seimiya and Gehring 2000).
Figure 3.
In situ hybridizations on predicted direct targets of Ey. (A,B) shf probe on wild-type (A) and dpp-GAL4/UAS-ey (B) eye-antennal discs. shf is expressed at the anterior margin of the disc and in scattered cells posterior to the morphogenetic furrow. (C,D) Optix probe on wild-type (C) and dpp-GAL4/UAS-ey (D) eye-antennal discs. Optix is expressed anterior to the morphogenetic furrow. Arrowheads in A and C indicate endogenous expression; arrows in B and D indicate up-regulation of expression in the ventral dpp expression domain of the antennal disc. (dpp>ey) dpp-GAL4/UAS-ey.
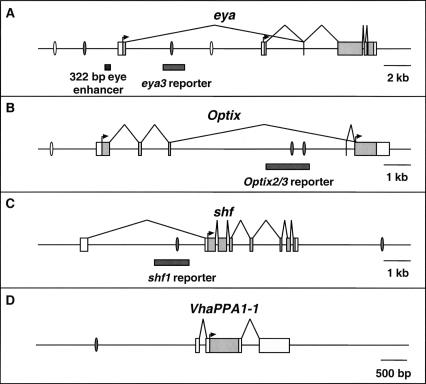
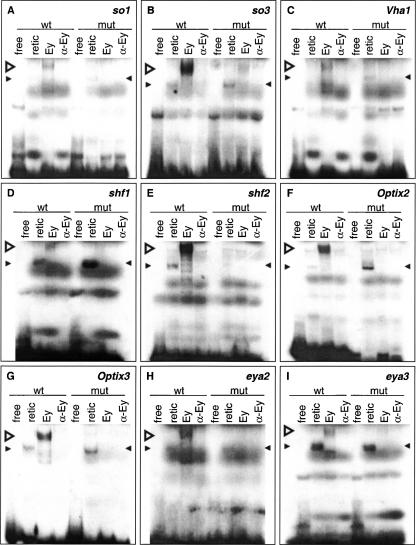
Second, we performed EMSAs to test whether the predicted Ey binding sites are bound by Ey in vitro. Four sites are predicted in eya, three in Optix, two in shf, and one in VhaPPA1–1 (Fig. 4). Seven of these ten sites showed a shift in EMSAs by reticulocyte-expressed full-length Ey: two eya sites, two Optix sites, the VhaPPA1–1 site, and both shifted sites (Fig. 5), while one predicted site in Optix (Optix1) and two predicted sites in eya do not shift (eya1 and eya4) (data not shown). Two sites in the so10 enhancer were also tested as positive controls and both show a shift by Ey in EMSAs (Niimi et al. 1999). The efficacy of binding among these nine sites differs, with eya2, shf2, Optix2, Optix3, and so3 shifting strongly and Vha1 (from VhaPPA1–1), shf1, so1, and eya3 shifting more weakly. Comparing sequences from sites that shifted by Ey, the only unchanged base is at position 13 (Supplemental data). In addition, bases at position 5, 9, 11 and 14 are almost identical with only one substitution observed. Base substitution at these positions abolishes Ey binding to the oligo, further supporting the importance of these bases.
Figure 4.
The genomic loci of eya (A), Optix (B), shf (C), and VhaPPA1–1 (D). Ey binding sites shown to bind Ey by EMSAs are shown as grey ovals, predicted binding sites that did not bind Ey as open ovals. Reporter fragments responsive to Ey are shown as gray bars. In the transcribed regions, untranslated sequence is shown as white boxes and the coding region is shown in gray. The shf1 and Optix2/3 reporters are expressed in the third instar eye disc and are induced by ectopic ey; the eya3 reporter is only induced by ectopic ey. In A, a previously described 322-bp eye enhancer is indicated by a black bar.
Figure 5.
Electromobility shift assays with reticulocyte-expressed Ey protein on 30-bp double-stranded oligonucleotides containing predicted wild-type or mutated Ey binding sites. Sites are from so (A,B), VhaPPA1–1 (C), shf (D,E), Optix (F,G), and eya (H,I). Both wild-type and mutated oligonucleotides were tested for binding to Ey. Four experiments were conducted with each probe with the first two lanes as negative controls. In vitro synthesized Ey was incubated with the probe as shown in the third lane. The binding is Ey specific since addition of polyclonal anti-Eyeless serum abolished the binding as shown in the fourth lane. Wild-type oligonucleotides were bound by Ey protein while mutated binding sites were not. (wt) Wild-type oligonucleotide, (mut) mutated oligonucleotide, (free) free probe alone, (retic) reticulocyte lysate with no plasmid, (Ey) reticulocyte-expressed Ey protein, (α-Ey) Ey protein with polyclonal anti-Eyeless serum added. Open arrowhead indicates specific gel shift; filled arrowheads indicate nonspecific binding.
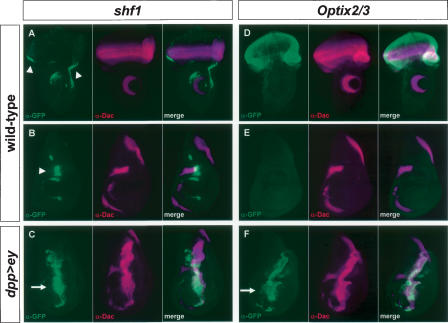
Finally, the ability of predicted Ey binding sites in these four genes to function as Ey-responsive enhancer elements in vivo was tested. If these Ey binding sites are active in vivo, DNA encompassing these sites should drive reporter gene expression in the eye disc and respond to ectopic ey expression. We designed ∼1.5 kb constructs containing the sites and nearby conserved noncoding sequences identified by comparison with the D. pseudoobscura genome. These sequences were inserted into reporter constructs and tested for their ability to drive gene expression in vivo. The Optix2 and Optix3 binding sites are close enough together to be included in one reporter construct. Significantly, constructs containing the shf1 or the Optix2/3 binding sites drive GFP expression in the third instar larval eye disc anterior to the furrow where ey is normally expressed and are strongly induced by ectopic ey expression (Fig. 6). The shf1 reporter drives GFP expression along the lateral margins of the wild-type eye disc anterior to the furrow (Fig. 6A). It also drives expression near the anterior/posterior (A/P) boundary of the wing disc and in the antennal and leg discs (Fig. 6A,B, and data not shown). To test if this expression is under the control of ey, GFP expression was also examined in a dpp>ey background. The shf1 reporter is strongly induced in the dpp expression domains of the wing, leg, haltere, and antennal discs (Fig. 6C and data not shown). Similarly, the Optix2/3 reporter is also sufficient to drive reporter gene expression anterior to the furrow where Optix is normally expressed (Fig. 6D). No expression is seen in any other larval tissue (a wing disc is shown in Fig. 6E). Like shf1, Optix2/3 is also strongly induced by ectopic ey expression in all imaginal discs (Fig. 6F; data not shown). For eya2, eya3, shf2, Vha1, and Optix1, no activity is seen in the wild-type eye disc. However, the eya3 reporter is induced in dpp>ey discs (data not shown), suggesting that this Ey binding site is functional as well. Consistent with the EMSA results, reporter constructs for shf1, eya3, and Optix2–3 with Ey binding sites mutated fail to respond to ey induction in vivo (data not shown). Therefore, our initial in vitro and in vivo analyses strongly suggest that at least three of the four predicted ey targets, eya, Optix, and shf, are under the direct control of Ey through the predicted Ey binding sites in vivo.
Figure 6.
In vivo reporter activity of Ey binding sites from shf and Optix. α-GFP is shown in green; α-Dac is shown in magenta in all panels. (A) A construct containing an Ey binding site in shf drives GFP in the eye disc anterior to the morphogenetic furrow. This construct also drives expression in the antennal disc, near the anterior/posterior (A/P) compartment boundary of the wing disc (B), and in the leg disc (not shown). A merge of A and B is shown in C. In a dpp-GAL4/UAS-ey background, the shf1 reporter is strongly induced throughout the dpp expression domain along the A/P boundary of the wing disc. (D) A construct containing the Optix2 and Optix3 binding sites (Optix2/3) expresses strongly throughout the eye disc and in no other imaginal tissues examined (a wing disc is shown in E). Optix2/3 is also strongly induced in the wing disc throughout the dpp expression domain of dpp-GAL4/UAS-ey animals. A merge of D and E is shown in F. Arrowheads in A and B indicate reporter activity in a wild-type background. Arrows in C and F indicate ectopic induction.
Genome-wide prediction of Ey direct targets
With the identification of six additional Ey binding sites based on EMSAs and in vivo reporter assays (eya3, Optix2/3, shf1/2, Vha1), we sought to optimize the PWM of Ey further. Corresponding binding site sequences of these confirmed Ey binding sites in melanogaster were identified in six other fly species using the genomic sequences generated from the Drosophila genome project. Sequences from a total of 66 binding sites were obtained to generate a new PWM (Fig. 2D). The new PWM was used to score binding sites that have been tested above and while the score of sites that are bound by Ey have been improved, all three sites that failed to be shifted by Ey have scores below the cutoff of 9.6 (data not shown), suggesting that the new PWM is indeed more optimal.
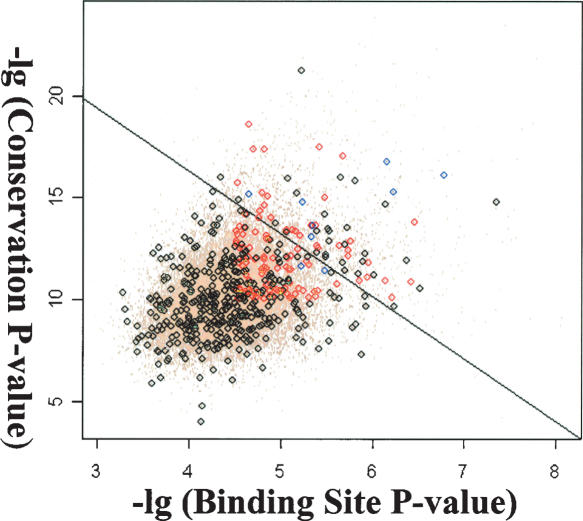
Using the new PWM, we scanned the entire Drosophila genome and identified 5773 sites that score 9.6 or higher. To reduce false positive predictions, conservation of these putative sites was examined in seven fly species and the conservation score of each site was calculated. To determine the cutoff value of the conservation score, we mapped all 5773 binding sites to individual genes and two types of putative sites were obtained. Sites mapped within genes that do not respond to ectopic ey are grouped into the negative set while sites mapped within the 188 genes that are induced by ey independent of ato are grouped into the positive set. Based on the hypothesis that true binding sites should closely resemble the new PWM and tend to be more conserved across the fly species, each putative binding site was classified based on the score of its melanogaster sequence and its conservation score. A linear classifier that maximizes the separation of the positive and negative training sets was obtained through logistic regression (Fig. 7) and sites that have positive predicted score are considered potential true binding sites. Among the 188 genes, a total of 20 genes containing 22 putative Ey binding sites were identified as potential Ey direct targets (Table 1). Among this list, all four targets of Ey are included and rank at the top. In addition, genes that are involved in axon guidance and synapse formation are identified. Finally, many novel genes are in this list and further studies of their function will likely provide further insights concerning the molecular mechanism of ey function during retinal development.
Figure 7.
Classification of putative Ey binding sites based on the score in melanogaster and their conservation score. Black circles represent sites that fall within genes not induced by ey (negative set) whereas red circles represent sites that fall within ey induced genes (positive set). Blue circles are known Ey binding sites. A line is obtained using logistic regression and sites above the line are considered true binding sites.
Table 1.
Twenty predicted direct targets of Ey
Genes identified by microarray analysis were filtered by searching for conserved noncoding predicted Ey binding sites. These are ranked by predicted score obtained from logistic linear regression over PWM scores of predicted binding sites in melanogaster and its conservation score in other six fly species. Of the 20, four Ey direct targets that were identified previously (so) and in this report are included and rank high in the list.
Discussion
We have used a combinatorial approach using microarrays and in silico binding site prediction to identify 20 putative direct targets of the Drosophila Pax6 homolog Ey. To increase specificity in our microarray analysis, we only considered genes induced by ey in multiple tissues and that are expressed independently of ato. For in silico prediction, an Ey PWM was constructed by phylogenetic shadowing and then used to identify three novel Ey direct targets, eyes absent, Optix, and shifted, which were confirmed using in situ hybridization, gel shift assays, and reporter analysis. Further optimization of the Ey PWM by phylogenetic shadowing on these novel Ey binding sites allows us to identify potential Ey binding sites genome-wide, whose specificity is further improved through comparative genomics. Intersection of the in silico prediction with ey downstream genes identified from microarray experiments reveals a short list of 20 genes as Ey direct targets, providing the basis for further functional studies.
Combination of microarray analysis and genetic epistasis yields a short list of downstream targets
Microarray analysis is a powerful method to obtain expression profiles of all genes in the genome in a single experiment. However, the major challenge of this approach is to distinguish genes that play specific roles in a process of interest versus background noise. To minimize nonspecific genes, we tested gene expression in three different tissues and only genes that are consistently induced by ey were considered further. As a result, our microarray experiments yielded a set of consistent, reproducible data in concurrence with previous knowledge of the retinal determination pathway. Our data set apparently does not share some of the inconsistencies found in a previous study (Michaut et al. 2003). Most significantly, our list contains the RD genes toy, eya, so, and Optix, where the previous study identified only eya (among RD genes) as up-regulated by ey. For novel genes identified from microarray experiments that have been tested, at least 87% could be independently verified using real-time quantitative RT-PCR. This estimation is likely to be conservative since RT-PCR verification was performed using only the leg disc sample pair. Among the 11 genes with negative results, three of them are relatively weakly induced in leg discs but are strongly induced in antennal and wing discs based on microarray data. In addition, five genes failed for the RT-PCR reaction probably due to primers. Additional RT-PCR tests and/or other methods such as disc in situs can be used to further examine these genes. While our analysis excludes genes with high levels of endogenous expression in all other imaginal discs, it is likely that our list of eye-enriched, ey-induced genes accounts for a significant fraction of genes important in early eye development.
The combination of microarrays and genetic epistasis analysis is another powerful approach that was used to rapidly refine our list using current models of eye development as a guide. Using ato mutant discs with and without ectopic ey, we selected genes that are active prior to photoreceptor differentiation, which reduced the number of potential ey targets to 188. These epistasis experiments represent a powerful tool for refining the vast amounts of data arising from microarray experiments by allowing a rapid subdivision of genetic pathways into distinct levels. This technique is widely applicable to many developmental systems and can be used to discover coregulated gene groups and interregulated genetic networks on a genomic scale rapidly.
Genome-wide identification of putative Ey binding site in Drosophila
To identify direct Ey targets, we used independent data from computational genomics to predict possible Ey binding sites in Drosophila genome. We first used phylogenetic shadowing to build an Ey PWM that was then used to identify putative Ey binding sites in the melanogaster genome. Although our PWM is based on only three sites found in the so10 enhancer, its accuracy is greatly improved using phylogenetic shadowing. ThroughEMSAs, we tested 10 predicted binding sites in four loci and found that seven of the 10 are indeed bound by Ey. Therefore, we believe that phylogenetic shadowing is a widely applicable technique for expanding a pool of TF binding sites, especially to facilitate rapid construction of PWMs for other TFs where only a few sites are known. The genomes of close relatives of many relevant model organisms are being sequenced and much of the data required to perform binding site shadowing is already publicly available. Using these data, robust PWMs for accurate TF binding site discovery can be readily constructed even if one starts with only a few characterized binding sites. Furthermore, since transcriptional control often depends on coordinate regulation by numerous TFs, in silico prediction can be adapted to find putative enhancers containing clusters of predicted binding sites. This approach has already been used to find enhancers in systems with well-characterized multi-factorial regulation (e.g., Berman et al. 2002; Markstein and Levine 2002; Zhu et al. 2002; Schroeder et al. 2004). Phylogenetic shadowing to build PWMs will improve this characterization and open new fields to these kinds of analyses. Finally, such in silico binding site identification can be iterated efficiently. Taking advantage of the newly identified Ey binding sites in our report, a revised Ey PWM was built. The specificity in predicting true Ey binding sites is clearly improved using the new PWM with scores of all four Ey false-positive sites predicted using the first Ey PWM falling below the new cutoff.
Comparative genomics is another powerful tool to refine binding site prediction further. Based on recently sequenced fly genomes, conservation scores of all 5773 predicted Ey binding sites were calculated. Using a linear classifier obtained from training sets, the total number of potential sites is reduced to 1265 without excluding most of the confirmed Ey binding sites. The accuracy may be further improved when sequences from additional fly species become available. One limitation of a comparative genomics approach is that many regions are conserved due to insufficient separation among species, resulting in over prediction. This limitation can be largely overcome by combining with other types of data, such as microarray data, as demonstrated in this report. Another limitation of a comparative genomics approach is the difficultly obtaining accurate sequence alignments, especially among highly diverged species. With the continuing reduction of sequencing costs, it is reasonable to expect that this problem could be overcome by sequencing many relatively closely related species in the near future. Therefore, we believe comparative genomics will play an increasingly important role in TF binding site prediction not only in Drosophila but in other model systems as well.
Combinatorial approach to generate a short list of interesting genes with high accuracy
The intersection of gene expression profiling and Ey binding site prediction yield a list of 20 potential Ey targets. This list is likely to be enriched by important primary targets crucial for specifying retinal cell fate and the first developmental steps associated with that decision. Consistent with this idea, among the list, eight putative direct targets of ey have previously characterized eye function or expression. In addition, EMSAs and reporter analysis based on predicted binding sites were used to test four candidate loci, including two RD genes (eya and optix) and two novel genes (shf and VhaPP1). We show that the three genes (eya, Optix, and shf) are direct targets of Ey. We also show that the Ey binding sites of Optix and shf are located in functional ey-responsive eye enhancers. Another construct that contains a putative Ey binding site in the eya locus, eya3, does not drive reporter expression in the eye disc but is induced by ectopic ey. Since the reporter constructs were designed based only on the location of the predicted Ey binding sites and any nearby conserved noncoding sequence, it is possible that other relevant regulatory regions were not included. This is likely to explain why the shf1 and Optix2/3 reporters do not completely recapitulate the expression patterns seen by in situ hybridization (Seimiya and Gehring 2000; Glise et al. 2005; Gorfinkiel et al. 2005). However, our current predicted list is unlikely to be perfect. For example, no enhancer activity has been observed with the Vhapp1 reporter, raising the possibility that it is a false prediction. In addition, it is also worth noting that the current list is unlikely to be complete due to the limited number of known Ey binding sites identified thus far and the arbitrary cutoff criteria we used in the screening process. Fortunately, the reiterating combinatorial approach described here will continue to optimize the Ey PWM as newly identified Ey binding sites and further accumulation of genomic sequences from additional closely related species are incorporated, resulting in an increasingly more accurate list of Ey targets.
Since ey is both necessary and sufficient for the formation of a complete and properly constructed eye, a process that requires as many as 3000 genes, it is likely that Ey has dozens of direct targets, as its expression initiates a cascade impacting virtually all areas of cellular function (Thaker and Kankel 1992). Indeed, based on the function of our candidate genes, it suggests possible roles of ey in morphogenesis (Mmp2), synapse formation and axon guidance (SytlV, BeatIII-C), and other signaling cascades (shf). These correspond well to the morphologic and cell-fate changes associated with retinal determination and entry into the morphogenetic furrow. The pleiotropy uncovered in this study for Drosophila Eyeless mirrors the diverse functions of the characterized targets of vertebrate PAX6, which include TFs, cytoskeletal components, and lens crystallins (Marquardt et al. 2001; Renaud and Simpson 2002). Further studies of these targets will shed light on the molecular mechanism of how ey regulates the RD network to establish retinal cell fate by regulating multiple processes necessary for creating an eye disc permissive for retinal cell differentiation.
In summary, our approach has wide implications for the discovery of direct downstream targets of TFs. Similar microarray experiments can be conducted with any TF for which it is possible to acquire mutant tissue or tissue in which it is overexpressed. For partially dissected genetic pathways, additional mutants can be combined with ectopic overexpression to conduct microarray epistasis experiments. For building a PWM using genomic shadowing, only a few binding sites are needed now that genome projects are contributing the sequence of many closely related species. Putative sites can be efficiently ranked by conservation by comparing the genomic sequences from a large number of closely related species, making this approach applicable even for TFs with degenerate binding sites that occur thousands of times throughout the genome. Therefore, this combination of approaches can reduce the high false positive rate inherent with individual techniques, producing a rapid and accurate methodology for discovery of direct targets of TFs.
Methods
RNA sample preparation and hybridization
Leg, wing, and antennal discs of Canton S, w; dpp-GAL4/UAS-ey, w; ato1, w; UAS-ey/+; ato1 dpp-GAL4/ato1 and eye discs of w; ato1 and Canton S were dissected in phosphate buffered saline (PBS) at pH 6.8 and placed directly into Trizol reagent (Invitrogen) on ice. Eye discs were separated from antennal discs using tungsten needles. Approximately 80 leg or eye discs, 100 antennal discs, or 60 wing discs were used per independent sample. Three independent samples were used for each genotype. Total RNA was extracted from the discs in Trizol using the manufacturer’s protocol and purified using the RNeasy MinElute kit (Qiagen). cDNA synthesis was carried out as described in the Expression Analysis Technical Manual (Affymetrix, two-cycle protocol) using 100 ng of total RNA for each sample. The cRNA reactions were carried out using the BioArray High-Yield Transcript Labeling kit (Enzo). Fifteen micrograms of labeled cRNA was fragmented and sequentially hybridized to the Affymetrix Drosophila Genome Array 2 following the manufacturer’s instructions.
Microarray data analysis
All arrays were normalized and analyzed using the PerfectMatch program (Zhang et al. 2003). A two-way ANOVA test was performed on wild-type and ectopic eyeless antennal, leg, and wing discs. To adjust for multiple testing, the false discovery rate (FDR) was calculated and a total of 956 genes with the cutoff set at 0.001 and at least twofold induction in one sample pair were identified. To further reduce false positives, genes that are not expressed in the top 40 percentile were considered absent and were excluded from further analysis. Furthermore, only genes that are expressed at a higher level in wild-type eye discs than in wild-type leg, wing, or antennal discs were included for further analysis.
Genomic shadowing and PWM assembly
D. sechelia (14021–0248.1), simulans (14021–0251.161), teisseri (14021–0257.0), yakuba (14021–0261.0), erecta (14021–0224.0), and ananassae (14024–0371.00) flies were ordered from the Tucson Stock Center. Genomic DNA was prepared using standard protocols. Genomic PCR primers were designed to regions of the sine oculis eye enhancer conserved between D. melanogaster and D. pseudoobscura. PCR products were sequenced directly using PCR primers. All primer sequences are available upon request.
In situ hybridizations
Approximately 1-kb fragments were amplified from the Drosophila Gene Collection (DGC) clones GH27042 (shf) and LD05472 (Optix) using Pfx polymerase (Invitrogen) followed by a post-incubation with Taq polymerase to add A-ends. The PCR products were then T/A cloned using an Invitrogen kit and sequenced. The inserts were excised with EcoRI and subcloned into pBluescript II (Stratagene). Digoxygenin-labeled RNA probes were made with T7 polymerase using a Roche kit. Imaginal discs were dissected and fixed in 4% paraformaldehyde for 30 min at room temperature, then washed four times in methanol and four times in ethanol at room temperature for ten minutes apiece. Discs were then permeabilized using xylene/ethanol for 1 h at room temperature, washed 2 times in ethanol and 2 times in methanol, and then rehydrated in a methanol/PBT (PBS + 0.1% Tween-20) series. Discs were then fixed again for 30 min at room temperature in 4% formaldehyde/PBT, washed, and treated for 3 min with 6 μg/mL Proteinase K, then washed in 3 changes of PBT. After post-fixing for 30 min at room temperature in 4% formaldehyde/PBT, washes in PBT and a PBT/hybridization buffer (50% formamide, 5X SSC, 2% Boehringer blocking powder, 0.1% Tween-20, 0.5% CHAPS, 1 mg/mL Yeast RNA, 5 mM EDTA, 50 μg/mL heparin) series were done at room temperature. Discs were prehybridized at 55°C in hybridization buffer for at least 2 h before probe was added and then hybridized at 55°C overnight. Washes were done through a hybridization buffer/PBT series, the discs were blocked in PBT + 5% normal goat serum. Alkaline phosphatase-conjugated anti-digoxygenin antibody (1:1000) (Roche Applied Science) preabsorbed against wild-type imaginal discs was used for detection at 4°C overnight. Staining with NBT/BCIP was performed by the manufacturer’s protocol (Roche Applied Science).
Gel shift assays
Gel shift assays were performed on 30-bp double-stranded oligonucleotides centered on the putative 14-bp Ey binding site using full-length Ey protein. Bases 2, 5, 9, 11, 13, and 14 of the oligonucleotide, which are predicted to be constrained nucleotides by the Ey PWM, were changed in the mutated binding site oligonucleotides. Oligonucleotides were end-labeled with α-32P-dATP using Klenow. The Ey ORF was amplified from a full-length cDNA (provided by Georg Halder) and cloned into the XhoI/XbaI site of the pLINK vector (Dalton and Treisman 1992). Ey protein was synthesized with the TNT reticulocyte lysate kit (Promega) using T7 polymerase according to the manufacturer’s protocol. Gel shift assays were performed as previously described (Niimi et al. 1999), with the addition of 0.5 μl of normal goat serum to the 20-μl binding reaction. All oligonucleotide sequences can be found in the Supplemental data.
Reporter assays
Fragments centered on putative Ey binding sites were amplified from D. melanogaster genomic DNA using Pfx polymerase (Invitrogen). A-ends were added with a post-incubation with Taq polymerase. The fragments were T/A cloned (Invitrogen) and sequenced. The fragments were then excised using EcoRI and cloned into the EcoRI site of pH-Stinger (Barolo et al. 2000). For Optix2/3, the primers 5′-CGTTTTCGGTGGCTTGTGAGAA-3′ and 5′−AGCCAAAGCCCACGATGCTGGAAT-3′ were used. For shf1, the primers used were 5′-CGACCTGATACGGTGTTTGATA-3′ and 5′-CGGTGTGAAGTTACCCAAGTCAA-3′. Insulated vectors were used to minimalize position effect. Expression was confirmed in 2 independent insertions of Optix2/3 and 3 independent insertions of shf1.
Binding site conservation assessment
Based on global alignment (http://hanuman.math.berkeley.edu/genomes/drosophila.html), 500-bp flanking sequences around the putative binding site in seven Drosophila species were extracted and local alignment was conducted to identify the best alignment for each putative binding site. This optimal alignment was then used to calculate the conservation score of the putative binding site using the program MONKEY (Moses et al. 2004). All sites were mapped to genes based on Flybase annotation, version 3.1, using an ad hoc Perl program. Logistic regression over the minus logarithm of the binding site P-value and its conservation P-value was conducted and plotted using the R program (http://www.r-project.org/).
Acknowledgments
We thank the Drosophila Comparative Genome Sequencing Consortium for providing the genomic sequences used in this study. We thank Georg Halder, Ben Frankfort, and Kartik Pappu for critical reading of the manuscript. This work was supported by the National Eye Institute, the Retina Research Foundation, and the Curtis & Doris K. Hankamer Foundation. E.J.O. is supported by a grant from The Robert and Janice McNair Foundation. Y.L. is supported by an NEI T32 training grant (EY07102).
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GEO under accession no. GSE4008.]
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4673006
References
- Akimaru H., Chen Y., Dai P., Hou D.X., Nonaka M., Smolik S.M., Armstrong S., Goodman R.H., Ishii S., Chen Y., Dai P., Hou D.X., Nonaka M., Smolik S.M., Armstrong S., Goodman R.H., Ishii S., Dai P., Hou D.X., Nonaka M., Smolik S.M., Armstrong S., Goodman R.H., Ishii S., Hou D.X., Nonaka M., Smolik S.M., Armstrong S., Goodman R.H., Ishii S., Nonaka M., Smolik S.M., Armstrong S., Goodman R.H., Ishii S., Smolik S.M., Armstrong S., Goodman R.H., Ishii S., Armstrong S., Goodman R.H., Ishii S., Goodman R.H., Ishii S., Ishii S. Drosophila Cbp is a co-activator of cubitus interruptus in hedgehog signalling. Nature. 1997;386:735–738. doi: 10.1038/386735a0. [DOI] [PubMed] [Google Scholar]
- Barolo S., Carver L.A., Posakony J.W., Carver L.A., Posakony J.W., Posakony J.W. GFP and β-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques. 2000;29:726–732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- Berman B.P., Nibu Y., Pfeiffer B.D., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Nibu Y., Pfeiffer B.D., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Pfeiffer B.D., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Levine M., Rubin G.M., Eisen M.B., Rubin G.M., Eisen M.B., Eisen M.B. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffelli D., McAuliffe J., Ovcharenko D., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., McAuliffe J., Ovcharenko D., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., Ovcharenko D., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., Ovcharenko I., Pachter L., Rubin E.M., Pachter L., Rubin E.M., Rubin E.M. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- Bonini N.M., Leiserson W.M., Benzer S., Leiserson W.M., Benzer S., Benzer S. The eyes absent gene: Genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- Chen R., Amoui M., Zhang Z.H., Mardon G., Amoui M., Zhang Z.H., Mardon G., Zhang Z.H., Mardon G., Mardon G. Dachshund and Eyes Absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997;91:893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- Cheyette B.N., Green P.J., Martin K., Garren H., Hartenstein V., Zipursky S.L., Green P.J., Martin K., Garren H., Hartenstein V., Zipursky S.L., Martin K., Garren H., Hartenstein V., Zipursky S.L., Garren H., Hartenstein V., Zipursky S.L., Hartenstein V., Zipursky S.L., Zipursky S.L. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- Czerny T., Halder G., Kloter U., Souabni A., Gehring W.J., Busslinger M., Halder G., Kloter U., Souabni A., Gehring W.J., Busslinger M., Kloter U., Souabni A., Gehring W.J., Busslinger M., Souabni A., Gehring W.J., Busslinger M., Gehring W.J., Busslinger M., Busslinger M. Twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol. Cell. 1999;3:297–307. doi: 10.1016/s1097-2765(00)80457-8. [DOI] [PubMed] [Google Scholar]
- Dahl E., Koseki H., Balling R., Koseki H., Balling R., Balling R. PAX genes and organogenesis [Review]. Bioessays. 1997;19:755–765. doi: 10.1002/bies.950190905. [DOI] [PubMed] [Google Scholar]
- Dalton S., Treisman R., Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68:597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- Dow J.A. The multifunctional Drosophila melanogaster V-ATPase is encoded by a multigene family. J. Bioenerg. Biomembr. 1999;31:75–83. doi: 10.1023/a:1005400731289. [DOI] [PubMed] [Google Scholar]
- Elnitski L., Hardison R.C., Li J., Yang S., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Hardison R.C., Li J., Yang S., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Li J., Yang S., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Yang S., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Schwartz S., Miller W., Chiaromonte F., Miller W., Chiaromonte F., Chiaromonte F. Distinguishing regulatory DNA from neutral sites. Genome Res. 2003;13:64–72. doi: 10.1101/gr.817703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein J., Cai J., Glaser T., Jepeal L., Maas R., Cai J., Glaser T., Jepeal L., Maas R., Glaser T., Jepeal L., Maas R., Jepeal L., Maas R., Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J. Biol. Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- Frankfort B.J., Nolo R., Zhang Z., Bellen H., Mardon G., Nolo R., Zhang Z., Bellen H., Mardon G., Zhang Z., Bellen H., Mardon G., Bellen H., Mardon G., Mardon G. Senseless repression of rough is required for R8 photoreceptor differentiation in the developing Drosophilaeye. Neuron. 2001;32:403–414. doi: 10.1016/s0896-6273(01)00480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bellido A., Merriam J.R., Merriam J.R. Cell lineage of the imaginal discs in Drosophila gynandromorphs. J. Exp. Zool. 1969;170:61–75. doi: 10.1002/jez.1401700106. [DOI] [PubMed] [Google Scholar]
- Glise B., Miller C.A., Crozatier M., Halbisen M.A., Wise S., Olson D.J., Vincent A., Blair S.S., Miller C.A., Crozatier M., Halbisen M.A., Wise S., Olson D.J., Vincent A., Blair S.S., Crozatier M., Halbisen M.A., Wise S., Olson D.J., Vincent A., Blair S.S., Halbisen M.A., Wise S., Olson D.J., Vincent A., Blair S.S., Wise S., Olson D.J., Vincent A., Blair S.S., Olson D.J., Vincent A., Blair S.S., Vincent A., Blair S.S., Blair S.S. Shifted, the Drosophila ortholog of Wnt inhibitory factor-1, controls the distribution and movement of Hedgehog. Dev. Cell. 2005;8:255–266. doi: 10.1016/j.devcel.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Gorfinkiel N., Sierra J., Callejo A., Ibanez C., Guerrero I., Sierra J., Callejo A., Ibanez C., Guerrero I., Callejo A., Ibanez C., Guerrero I., Ibanez C., Guerrero I., Guerrero I. The Drosophila ortholog of the human Wnt inhibitor factor Shifted controls the diffusion of lipid-modified Hedgehog. Dev. Cell. 2005;8:241–253. doi: 10.1016/j.devcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Halder G., Callaerts P., Gehring W.J., Callaerts P., Gehring W.J., Gehring W.J. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila [see comments]. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Halder G., Callaerts P., Flister S., Walldorf U., Kloter U., Gehring W.J., Callaerts P., Flister S., Walldorf U., Kloter U., Gehring W.J., Flister S., Walldorf U., Kloter U., Gehring W.J., Walldorf U., Kloter U., Gehring W.J., Kloter U., Gehring W.J., Gehring W.J. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998;125:2181–2191. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- Halfon M.S., Grad Y., Church G.M., Michelson A.M., Grad Y., Church G.M., Michelson A.M., Church G.M., Michelson A.M., Michelson A.M. Computation-based discovery of related transcriptional regulatory modules and motifs using an experimentally validated combinatorial model. Genome Res. 2002;12:1019–1028. doi: 10.1101/gr.228902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck B., Gehring W.J., Walldorf U., Gehring W.J., Walldorf U., Walldorf U. Functional analysis of an eye specific enhancer of the eyeless gene in Drosophila. Proc. Natl. Acad. Sci. 1999;96:564–569. doi: 10.1073/pnas.96.2.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J.C., Kodjabachian L., Rebbert M.L., Rattner A., Smallwood P.M., Samos C.H., Nusse R., Dawid I.B., Nathans J., Kodjabachian L., Rebbert M.L., Rattner A., Smallwood P.M., Samos C.H., Nusse R., Dawid I.B., Nathans J., Rebbert M.L., Rattner A., Smallwood P.M., Samos C.H., Nusse R., Dawid I.B., Nathans J., Rattner A., Smallwood P.M., Samos C.H., Nusse R., Dawid I.B., Nathans J., Smallwood P.M., Samos C.H., Nusse R., Dawid I.B., Nathans J., Samos C.H., Nusse R., Dawid I.B., Nathans J., Nusse R., Dawid I.B., Nathans J., Dawid I.B., Nathans J., Nathans J. A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature. 1999;398:431–436. doi: 10.1038/18899. [DOI] [PubMed] [Google Scholar]
- Jang C.C., Chao J.L., Jones N., Yao L.C., Bessarab D.A., Kuo Y.M., Jun S., Desplan C., Beckendorf S.K., Sun Y.H., Chao J.L., Jones N., Yao L.C., Bessarab D.A., Kuo Y.M., Jun S., Desplan C., Beckendorf S.K., Sun Y.H., Jones N., Yao L.C., Bessarab D.A., Kuo Y.M., Jun S., Desplan C., Beckendorf S.K., Sun Y.H., Yao L.C., Bessarab D.A., Kuo Y.M., Jun S., Desplan C., Beckendorf S.K., Sun Y.H., Bessarab D.A., Kuo Y.M., Jun S., Desplan C., Beckendorf S.K., Sun Y.H., Kuo Y.M., Jun S., Desplan C., Beckendorf S.K., Sun Y.H., Jun S., Desplan C., Beckendorf S.K., Sun Y.H., Desplan C., Beckendorf S.K., Sun Y.H., Beckendorf S.K., Sun Y.H., Sun Y.H. Two Pax genes, eye gone and eyeless, act cooperatively in promoting Drosophila eye development. Development. 2003;130:2939–2951. doi: 10.1242/dev.00522. [DOI] [PubMed] [Google Scholar]
- Jarman A.P., Grell E.H., Ackerman L., Jan L.Y., Jan Y.N., Grell E.H., Ackerman L., Jan L.Y., Jan Y.N., Ackerman L., Jan L.Y., Jan Y.N., Jan L.Y., Jan Y.N., Jan Y.N. Atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- Klipper-Aurbach Y., Wasserman M., Braunspiegel-Weintrob N., Borstein D., Peleg S., Assa S., Karp M., Benjamini Y., Hochberg Y., Laron Z., Wasserman M., Braunspiegel-Weintrob N., Borstein D., Peleg S., Assa S., Karp M., Benjamini Y., Hochberg Y., Laron Z., Braunspiegel-Weintrob N., Borstein D., Peleg S., Assa S., Karp M., Benjamini Y., Hochberg Y., Laron Z., Borstein D., Peleg S., Assa S., Karp M., Benjamini Y., Hochberg Y., Laron Z., Peleg S., Assa S., Karp M., Benjamini Y., Hochberg Y., Laron Z., Assa S., Karp M., Benjamini Y., Hochberg Y., Laron Z., Karp M., Benjamini Y., Hochberg Y., Laron Z., Benjamini Y., Hochberg Y., Laron Z., Hochberg Y., Laron Z., Laron Z. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med. Hypotheses. 1995;45:486–490. doi: 10.1016/0306-9877(95)90228-7. [DOI] [PubMed] [Google Scholar]
- Lawrence P.A.1989Cell lineage and cell states in the Drosophila embryo. Ciba Foundation Symposium. 144131–140. discussion 140–149, 150–155. [PubMed] [Google Scholar]
- Loosli F., Koster R.W., Carl M., Krone A., Wittbrodt J., Koster R.W., Carl M., Krone A., Wittbrodt J., Carl M., Krone A., Wittbrodt J., Krone A., Wittbrodt J., Wittbrodt J. Six3, a medaka homologue of the Drosophila homeobox gene sine oculis is expressed in the anterior embryonic shield and the developing eye. Mech. Dev. 1998;74:159–164. doi: 10.1016/s0925-4773(98)00055-0. [DOI] [PubMed] [Google Scholar]
- Mardon G., Solomon N.M., Rubin G.M., Solomon N.M., Rubin G.M., Rubin G.M. dachshund encodes a nuclear protein required for normal eye and leg development in. Drosophila. Development. 1994;120:3473–3486. doi: 10.1242/dev.120.12.3473. [DOI] [PubMed] [Google Scholar]
- Markstein M., Levine M., Levine M. Decoding cis-regulatory DNAs in the Drosophila genome. Curr. Opin. Genet. Dev. 2002;12:601. doi: 10.1016/s0959-437x(02)00345-3. [DOI] [PubMed] [Google Scholar]
- Marquardt T., Ashery-Padan R., Andrejewski N., Scardigli R., Guillemot F., Gruss P., Ashery-Padan R., Andrejewski N., Scardigli R., Guillemot F., Gruss P., Andrejewski N., Scardigli R., Guillemot F., Gruss P., Scardigli R., Guillemot F., Gruss P., Guillemot F., Gruss P., Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Michaut L., Flister S., Neeb M., White K.P., Certa U., Gehring W.J., Flister S., Neeb M., White K.P., Certa U., Gehring W.J., Neeb M., White K.P., Certa U., Gehring W.J., White K.P., Certa U., Gehring W.J., Certa U., Gehring W.J., Gehring W.J. Analysis of the eye developmental pathway in Drosophila using DNA microarrays. Proc. Natl. Acad. Sci. 2003;100:4024–4029. doi: 10.1073/pnas.0630561100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses A.M., Chiang D.Y., Pollard D.A., Iyer V.N., Eisen M.B., Chiang D.Y., Pollard D.A., Iyer V.N., Eisen M.B., Pollard D.A., Iyer V.N., Eisen M.B., Iyer V.N., Eisen M.B., Eisen M.B. MONKEY: Identifying conserved transcription-factor binding sites in multiple alignments using a binding site-specific evolutionary model. Genome Biol. 2004;5:R98. doi: 10.1186/gb-2004-5-12-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi T., Seimiya M., Kloter U., Flister S., Gehring W.J., Seimiya M., Kloter U., Flister S., Gehring W.J., Kloter U., Flister S., Gehring W.J., Flister S., Gehring W.J., Gehring W.J. Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development. 1999;126:2253–2260. doi: 10.1242/dev.126.10.2253. [DOI] [PubMed] [Google Scholar]
- Pappu K.S., Mardon G., Mardon G. Genetic control of retinal specification and determination in Drosophila. Int. J. Dev. Biol. 2004;48:913–924. doi: 10.1387/ijdb.041875kp. [DOI] [PubMed] [Google Scholar]
- Pignoni F., Hu B., Zavitz K.H., Xiao J., Garrity P.A., Zipursky S.L., Hu B., Zavitz K.H., Xiao J., Garrity P.A., Zipursky S.L., Zavitz K.H., Xiao J., Garrity P.A., Zipursky S.L., Xiao J., Garrity P.A., Zipursky S.L., Garrity P.A., Zipursky S.L., Zipursky S.L. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophilaeye development. Cell. 1997;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- Punzo C., Seimiya M., Flister S., Gehring W.J., Plaza S., Seimiya M., Flister S., Gehring W.J., Plaza S., Flister S., Gehring W.J., Plaza S., Gehring W.J., Plaza S., Plaza S. Differential interactions of eyeless and twin of eyeless with the sine oculis enhancer. Development. 2002;129:625–634. doi: 10.1242/dev.129.3.625. [DOI] [PubMed] [Google Scholar]
- Punzo C., Plaza S., Seimiya M., Schnupf P., Kurata S., Jaeger J., Gehring W.J., Plaza S., Seimiya M., Schnupf P., Kurata S., Jaeger J., Gehring W.J., Seimiya M., Schnupf P., Kurata S., Jaeger J., Gehring W.J., Schnupf P., Kurata S., Jaeger J., Gehring W.J., Kurata S., Jaeger J., Gehring W.J., Jaeger J., Gehring W.J., Gehring W.J. Functional divergence between eyeless and twin of eyeless in Drosophila melanogaster. Development. 2004;131:3943–3953. doi: 10.1242/dev.01278. [DOI] [PubMed] [Google Scholar]
- Rayapureddi J.P., Kattamuri C., Steinmetz B.D., Frankfort B.J., Ostrin E.J., Mardon G., Hegde R.S., Kattamuri C., Steinmetz B.D., Frankfort B.J., Ostrin E.J., Mardon G., Hegde R.S., Steinmetz B.D., Frankfort B.J., Ostrin E.J., Mardon G., Hegde R.S., Frankfort B.J., Ostrin E.J., Mardon G., Hegde R.S., Ostrin E.J., Mardon G., Hegde R.S., Mardon G., Hegde R.S., Hegde R.S. Eyes absent represents a class of protein tyrosine phosphatases. Nature. 2003;426:295–298. doi: 10.1038/nature02093. [DOI] [PubMed] [Google Scholar]
- Ready D.F., Hanson T.E., Benzer S., Hanson T.E., Benzer S., Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Ren B., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Schreiber J., Hannett N., Kanin E., Hannett N., Kanin E., Kanin E., et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Renaud O., Simpson P., Simpson P. Movement of bristle precursors contributes to the spacing pattern in Drosophila. Mech. Dev. 2002;119:201–211. doi: 10.1016/s0925-4773(02)00381-7. [DOI] [PubMed] [Google Scholar]
- Schroeder M.D., Pearce M., Fak J., Fan H., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Pearce M., Fak J., Fan H., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Fak J., Fan H., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Fan H., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Emberly E., Rajewsky N., Siggia E.D., Gaul U., Rajewsky N., Siggia E.D., Gaul U., Siggia E.D., Gaul U., Gaul U. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2:E271. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seimiya M., Gehring W.J., Gehring W.J. The Drosophila homeobox gene optix is capable of inducing ectopic eyes by an eyeless-independent mechanism. Development. 2000;127:1879–1886. doi: 10.1242/dev.127.9.1879. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A., Levine M., Levine M. Whole-genome expression profiles identify gene batteries in Drosophila. Dev. Cell. 2002;3:464–465. doi: 10.1016/s1534-5807(02)00300-3. [DOI] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G.D., Schneider T.D., Gold L., Ehrenfeucht A., Schneider T.D., Gold L., Ehrenfeucht A., Gold L., Ehrenfeucht A., Ehrenfeucht A. Use of the ‘Perceptron’ algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 1982;10:2997–3011. doi: 10.1093/nar/10.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Subramanian S., Kumar S., Subramanian S., Kumar S., Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- Tavazoie S., Hughes J.D., Campbell M.J., Cho R.J., Church G.M., Hughes J.D., Campbell M.J., Cho R.J., Church G.M., Campbell M.J., Cho R.J., Church G.M., Cho R.J., Church G.M., Church G.M. Systematic determination of genetic network architecture. Nat. Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- Thaker H.M., Kankel D.R., Kankel D.R. Mosaic analysis gives an estimate of the extent of genomic involvement in the development of the visual system in Drosophila melanogaster. Genetics. 1992;131:883–894. doi: 10.1093/genetics/131.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tootle T.L., Silver S.J., Davies E.L., Newman V., Latek R.R., Mills I.A., Selengut J.D., Parlikar B.E., Rebay I., Silver S.J., Davies E.L., Newman V., Latek R.R., Mills I.A., Selengut J.D., Parlikar B.E., Rebay I., Davies E.L., Newman V., Latek R.R., Mills I.A., Selengut J.D., Parlikar B.E., Rebay I., Newman V., Latek R.R., Mills I.A., Selengut J.D., Parlikar B.E., Rebay I., Latek R.R., Mills I.A., Selengut J.D., Parlikar B.E., Rebay I., Mills I.A., Selengut J.D., Parlikar B.E., Rebay I., Selengut J.D., Parlikar B.E., Rebay I., Parlikar B.E., Rebay I., Rebay I. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature. 2003;426:299–302. doi: 10.1038/nature02097. [DOI] [PubMed] [Google Scholar]
- Toy J., Yang J.M., Leppert G.S., Sundin O.H., Yang J.M., Leppert G.S., Sundin O.H., Leppert G.S., Sundin O.H., Sundin O.H. The optx2 homeobox gene is expressed in early precursors of the eye and activates retina-specific genes. Proc. Natl. Acad. Sci. 1998;95:10643–10648. doi: 10.1073/pnas.95.18.10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T., Ready D.F., Ready D.F.1993. Pattern formation in the Drosophilaretina. In The Development of Drosophila melanogaster (eds. M. Bate and A. Martinez Arias), pp. 1277–1326. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY [Google Scholar]
- Zhang L., Miles M.F., Aldape K.D., Miles M.F., Aldape K.D., Aldape K.D. A model of molecular interactions on short oligonucleotide microarrays. Nat. Biotechnol. 2003;21:818–821. doi: 10.1038/nbt836. [DOI] [PubMed] [Google Scholar]
- Zhu Z., Pilpel Y., Church G.M., Pilpel Y., Church G.M., Church G.M. Computational identification of transcription factor binding sites via a transcription-factor-centric clustering (TFCC) algorithm. J. Mol. Biol. 2002;318:71–81. doi: 10.1016/S0022-2836(02)00026-8. [DOI] [PubMed] [Google Scholar]