Abstract
Fish–mammal genomic comparisons have proved powerful in identifying conserved noncoding elements likely to be cis-regulatory in nature, and the majority of those tested in vivo have been shown to act as tissue-specific enhancers associated with genes involved in transcriptional regulation of development. Although most of these elements share little sequence identity to each other, a small number are remarkably similar and appear to be the product of duplication events. Here, we searched for duplicated conserved noncoding elements in the human genome, using comparisons with Fugu to select putative cis-regulatory sequences. We identified 124 families of duplicated elements, each containing between two and five members, that are highly conserved within and between vertebrate genomes. In 74% of cases, we were able to assign a specific set of paralogous genes with annotation relating to transcriptional regulation and/or development to each family, thus removing much of the ambiguity in identifying associated genes. We find that duplicate elements have the potential to up-regulate reporter gene expression in a tissue-specific manner and that expression domains often overlap, but are not necessarily identical, between family members. Over two thirds of the families are conserved in duplicate in fish and appear to predate the large-scale duplication events thought to have occurred at the origin of vertebrates. We propose a model whereby gene duplication and the evolution of cis-regulatory elements can be considered in the context of increased morphological diversity and the emergence of the modern vertebrate body plan.
Regulation of gene expression in a spatial and temporal manner is crucial during vertebrate development. Such complex transcriptional regulation is thought to be mediated by the coordinated binding of transcription factors to discrete, typically noncoding DNA sequences, allowing the integration of multiple signals to regulate the expression of specific genes. These sequences, known as cis-regulatory modules (CRMs), can be up to several hundred bases in length (Arnone and Davidson 1997) and may be located at distances of several hundred kilobases to over a megabase in either direction from the genes on which they act (Bishop et al. 2000; Jamieson et al. 2002; Lettice et al. 2003). Moreover, CRMs may not act on the closest gene but can act across intervening genes (Spitz et al. 2003) and can also be located within the introns of neighboring genes (Lettice et al. 2003). Study of these elements may have medical implications as disruption to element function by mutations or by chromosomal rearrangements removing the proximity to their relevant transcriptional unit has been shown to cause disease (Kleinjan and van Heyningen 2005).
Identifying putative CRMs computationally relies on phylogenetic footprinting (for overview, see Ureta-Vidal et al. 2003) with sequence conservation implying functional constraint, although the confidence of such predictions depends on the evolutionary distance between the selected species. Recent large-scale computational comparative studies have resulted in the identification of hundreds of vertebrate conserved noncoding sequences, from those conserved between mammals (Dermitzakis et al. 2003; Margulies et al. 2003; Bejerano et al. 2004a) to those that show a high degree of conservation across larger evolutionary distances (Sandelin et al. 2004; Woolfe et al. 2005). These conserved noncoding sequences represent a diverse set of functional elements, a proportion of which are likely to act as CRMs.
A highly successful approach to filtering and prioritizing noncoding sequences most likely to be functional has been through fish–mammal comparisons using the compact genome of the pufferfish Fugu rubripes (Boffelli et al. 2004). Mammals and fish, being the most evolutionary distant extant vertebrates for which whole genome information is available, provide high stringency for the detection of vertebrate-specific regulatory elements. For example, in a previous study we identified ∼1400 highly conserved noncoding elements (CNEs) through fish–mammal comparisons that are likely to be cis-regulatory in nature (Woolfe et al. 2005). These CNEs represent a specific set of highly conserved sequences with an interesting evolutionary history. They have remained practically unchanged in the 450 million years (Myr) since the divergence of fish and mammals (sequence identity of >74% over at least 100 bases) but do not appear to be conserved in urochordates, such as Ciona intestinalis, or in other invertebrate genomes, despite the fact that these elements can exhibit a higher level of conservation than other functional sequences such as coding exons and noncoding RNAs. CNEs, and other similar highly conserved noncoding sequences, are found to cluster in the vicinity of genes implicated in transcriptional regulation and early development (Bejerano et al. 2004a; Sandelin et al. 2004; Woolfe et al. 2005) and the majority of those tested in vivo (5/7 in mice, Nobrega et al. 2003; 23/25 in fish, Woolfe et al. 2005) drive expression of reporter genes in a temporal and spatial specific manner during early development. Many other studies around specific developmental genes have also identified highly conserved noncoding sequences between humans and fish that have enhancer activity (Zerucha et al. 2000; Barton et al. 2001; Lien et al. 2002; Blader et al. 2003; Lettice et al. 2003; Dickmeis et al. 2004; Kimura-Yoshida et al. 2004; de la Calle-Mustienes et al. 2005; Goode et al. 2005). The association of these highly conserved sequences to genes implicated in the regulation in early development is most likely a result of the fundamental nature of the developmental process in vertebrates.
To date, confirmed and putative CRMs identified through comparative analysis appear to be distinct, single-copy elements within the human genome, with only a tiny proportion displaying local sequence similarity to each other (Margulies et al. 2003; Bejerano et al. 2004a, Bejerano et al. 2004a, b; Martin et al. 2004; Sandelin et al. 2004; Woolfe et al. 2005). These small numbers of nonunique sequences appear to be the product of duplication events and are found to be situated close to genes with clear paralogous relationships. This indicates a level of retention of regulatory elements between some gene duplicates over evolution. Comparisons with ancestral genes in the genomes of the urochordates C. intestalis (Dehal et al. 2002) and amphioxus (Branchiostoma floridae) (Panopoulou et al. 2003) have indicated that many vertebrate paralogs derive from large-scale duplications (including whole-genome duplications) thought to have occurred more than 500 million years ago (Mya) (Holland et al.1994; McLysaght et al. 2002), although contention still remains as to whether there were one or two rounds of polyploidy (the 1R vs. 2R hypothesis) (Seoighe 2003).
Here, we investigate more comprehensively the extent of the phenomenon of duplicated CNEs and their associated genes in vertebrate genomes. We identify families of duplicated CNEs in the human genome, concentrating on sequences likely to be cis-regulatory in nature by restricting the search to those which are conserved in Fugu and have little or no evidence of transcription. By assuming that CNEs are retained in proximity to the gene or genes on which they act following duplication, we are able to associate elements to nearby paralogous developmental genes and, through comparative analyses, study their evolution since duplication. Furthermore, we demonstrate that duplicated CNEs have the ability to up-regulate tissue-specific expression of a reporter gene in a manner that frequently reflects the endogenous expression pattern of their associated gene and that duplicated CNEs generally give overlapping, but not necessarily identical, temporal and spatial patterns of up-regulation.
Results
Detection and filtering of nonunique putative cis-regulatory elements within the human genome
We initially prioritized a set of potential regulatory elements through a comparative analysis of the human and Fugu genomes (see Methods). In a previous study, we identified 1373 CNEs using stringent search parameters (Woolfe et al. 2005). Here, by using more sensitive search parameters, we were able to identify a larger set of 2330 nonredundant human CNEs (mean percent identity = 85%, mean length = 145 bp) with no significant matches to known transcripts or noncoding RNAs. This new set overlaps ∼90% of the 1373 CNEs previously identified, as we now excluded sequences derived from untranslated regions (UTRs). This new set was compared back to the human genome to detect sequences that independently match the same CNE with significant sequence similarity. The resultant 349 sequences clustered into 169 groups of related sequences, which we refer to as duplicated CNE (dCNE) families. To focus on sequences likely to be CRMs, we removed 34 families that had EST evidence suggesting transcription, four families that were found to overlap small Ensembl (Hubbard et al. 2005) annotated exons, and one family with strong evidence for RNA secondary structure, leaving 130 families.
Conservation of dCNEs across vertebrates
Although we know that our set of dCNE families is duplicated within the human genome, it is of interest to ascertain whether they arose from recent (i.e., human or primate specific) or more ancient duplication event(s). We searched all dCNE families against the draft genome sequence assemblies of eight other vertebrate species, namely chimp, dog, mouse, rat, chicken, Xenopus, Tetraodon, and Fugu, and found that 94 families were conserved in duplicate across all vertebrates and 30 were duplicated only in tetrapods. This indicates that the majority of our dCNE families have an ancient origin that predates the fish–tetrapod split, and their wide-ranging conservation suggests they have an essential function in the vertebrate lineage. Six families were found to derive solely from a primate-specific duplication event; these were removed from further analysis as we wished to focus on potential regulatory sequences essential to the vertebrate lineage. The remaining 124 families, made up of 261 sequences, form the basis of this study. A total of 112 of the families contain two members, nine families contain three members, two families contain four members, and there is one five-member family. An example of a two-member dCNE family can be seen in Figure 1. Similarly to previous findings (Woolfe et al. 2005), none of the elements had significant matches to the closest nonvertebrate chordate genome, C. intestinalis, or to any cephalochordate or urochordate sequences. Less than 10% of these families have been previously documented (Bejerano et al. 2004a; Sandelin et al. 2004) and so this larger set of dCNEs provides data for a more in-depth analysis of their origin and evolution.
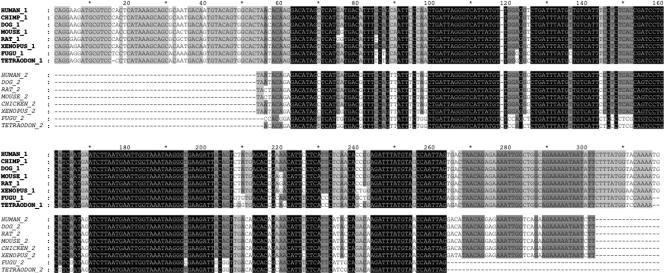
Figure 1.
A two-member dCNE family (#464) located within the introns of FOXP1 (464_1) and FOXP2 (464_2). Multiple alignment of sequences was carried out using CLUSTALW (v1.83) (Thompson et al. 1994). Element boundaries were defined by sequence conservation between human and Fugu for each family member. Human–Fugu orthologs of 464_1 are conserved at 92.7% identity over 316 bases while orthologs of 464_2 are conserved at 88.4% identity over 199 bases between these species. Conservation between human copies of 464_1 and 464_2 across the length of the smaller element (248 bp) was 83.5%, lower than that seen between orthologous copies but considerably higher than the average conservation between human dCNEs (Fig. 5). In addition, these elements have a length ratio (see Methods) of 0.78 indicating significant evolution of the elements at their edges. 464_1 was not detected in chicken and 464_2 was not detected in chimp, possibly because of missing sequence in these assemblies.
Association of dCNEs to paralogous genes
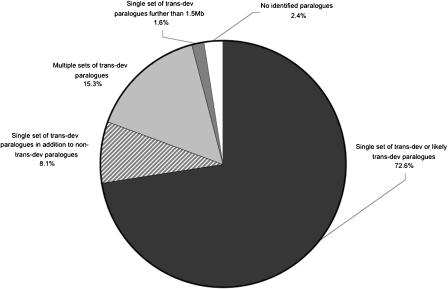
CRMs in vertebrates are often located large distances from the transcription start site of the genes upon which they act and, in some cases, in introns of neighboring genes (Aparicio et al. 2002; Lettice et al. 2003), making the association of genes to potential regulatory regions nontrivial. However, several studies have shown a distinct enrichment for genes involved in transcriptional regulation and/or development (which we term trans-dev genes) in the regions surrounding putative, highly conserved CRMs (Bejerano et al. 2004a; Boffelli et al. 2004; Sandelin et al. 2004; Woolfe et al. 2005). In addition, a small number of reported duplicated elements are found to be situated close to genes with clear paralogous relationships. (Bejerano et al. 2004a, Bejerano et al. 2004a; b; Martin et al. 2004; Sandelin et al. 2004; Woolfe et al. 2005), indicating that such elements are retained in the neighborhood of their target gene following a duplication event. Therefore, having identified families of duplicated CNEs, we searched for paralogous genes likely to be associated with each of these potential regulatory regions. In the genomic region 1.5 Mb upstream and downstream of each dCNE within a family, we compared genes to search for close paralogs common to all members of the family. The distance of 1.5 Mb exceeds that between all currently known CRMs and their associated genes, with the furthest separation being ∼1 Mb, for example, SHH (Lettice et al. 2003), Sox9 (Bishop et al. 2000), and MAF (Jamieson et al. 2002). Interestingly, close paralogs were found in the regions surrounding 119 of our 124 dCNE families, which is more than expected by chance (P < 0.001 based on 1000 randomizations), indicating that dCNEs are not independent of their genomic environment. The majority of dCNE families (90/124) were located in regions each containing just a single set of paralogous genes, 29 were located in regions containing multiple sets of paralogs, and five were located in regions in which no close paralogs could be identified.
As with CNEs in general, highly significant enrichment for Gene Ontology (GO) terms relating to transcriptional regulation and development has been found within the identified set of paralogs, reconfirming the likely association of such elements with genes of this type (Vavouri et al. 2005). Therefore, in regions in which close paralogs were detected, we identified whether or not each paralog could be considered trans-dev according to its functional annotation (see Methods). A summary of the results can be found in Figure 2. Of the 90 families located in regions containing just a single set of paralogous genes, 77 were located in regions containing trans-dev paralogs. These paralogs include some of the key regulators responsible for body patterning and morphogenesis in early vertebrate development, for example, members from the SOX, PAX, Forkhead, and DACH families. The remaining 13 families were located close to five single pairs of paralogous genes (NRXN1/NRXN3, ZNF521/ZNF423, ZNF503/ZNF703, ODZ3/ODZ4, and DLG1/DLG2) that were not considered trans-dev by our criteria. It is interesting to note, however, that orthologs of these genes have evidence to suggest they are implicated in development: NRXN1/NRXN3, ZNF521/ZNF423, and ZNF503/ZNF703 all have known mammalian developmental roles (Püschel and Betz 1995; Tsai and Reed 1998; Bond et al. 2004; Chang et al. 2004; Nakamura et al. 2004); the Drosophila odz gene, a homolog of mammalian ODZ3/ODZ4, is a pair-rule gene with many patterning roles throughout development (Ben-Zur et al. 2000); and DLG1 and DLG2, two relatively uncharacterized synapse-associated genes, have a close paralog, DLG3, that is known to be expressed in early brain development (Tarpey et al. 2004). Therefore, given their proven or probable developmental roles, as well as their status as the only close paralogs in the vicinity, we included these five pairs in our set of likely target genes for further analysis.
Figure 2.
Presence of trans-dev paralogs in the vicinity of the 124 dCNE families. For the majority of families, trans-dev paralogs were detected within 1.5 Mb, either upstream or downstream of dCNEs. In most cases, just a single set of paralogs was detected with annotation relating to trans-dev (black), with some regions containing additional non trans-dev paralogs (striped). Some regions contained multiple sets of trans-dev paralogs (light gray). For dCNEs located in gene deserts, a search region up to the next known gene was used (dark gray). A small proportion of dCNEs were located in regions with no functionally annotated paralogs (white).
Nineteen dCNE families were located in regions containing large clusters of related paralogous trans-dev genes (e.g., HOX and IRX clusters) or in regions containing several unrelated trans-dev paralogs (e.g., LMX1A and PBX1 on Chr1 and LMX1B and PBX3 on Chr9). A further 10 dCNE families were found in regions containing a single set of trans-dev paralogs in addition to other paralogous genes with no developmental or shared functional annotation (data not shown); in seven cases, each dCNE was located closest to the trans-dev paralog. In all cases containing multiple sets of paralogous genes, the closest trans-dev paralogs were selected as the most likely target genes for further analysis.
Five dCNE families were found to have no annotated paralogs in their vicinity. However, two of these families were located in gene deserts (Nobrega et al. 2003) and an additional search further up and downstream to the next gene regions revealed single sets of paralogous trans-dev genes (BCL11A/B and NR2F1/F2) located between 1.5 and 2.2 Mb from the dCNEs. No characterized CRMs are currently known to function at this distance, most likely because of a historical bias in functionally annotating CRMs that are located relatively close to a transcriptional unit. Nevertheless, the lack of alternative targets in these regions, as well as evidence that gene deserts harboring vertebrate-conserved elements are almost always adjacent to trans-dev genes (Ovcharenko et al. 2005), make it plausible that these elements and genes are indeed associated. Moreover, both sets of paralogs have other dCNE families within 1.5 Mb (Table 1, Supplemental Table S1). Although a functional survey of these distant elements is not undertaken here, it would be of interest to ascertain whether they act as long-range enhancer sequences to measure distance limits over which such elements operate. The remaining three families remain ambiguous in origin and function.
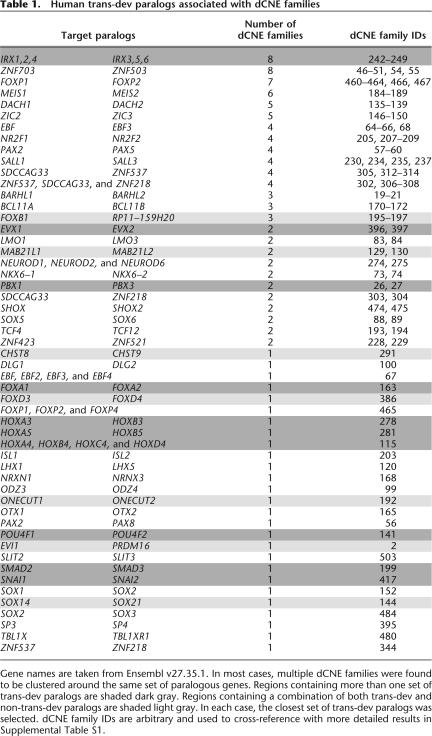
Table 1.
Human trans-dev paralogs associated with dCNE families
Gene names are taken from Ensembl v27.35.1. In most cases, multiple dCNE families were found to be clustered around the same set of paralogous genes. Regions containing more than one set of trans-dev paralogs are shaded dark gray. Regions containing a combination of both trans-dev and non-trans-dev paralogs are shaded light gray. In each case, the closest set of trans-dev paralogs was selected. dCNE family IDs are arbitrary and used to cross-reference with more detailed results in Supplemental Table S1.
Generally, in regions surrounding putative CNEs, there can be several potential candidate genes on which the elements may act, making predictions difficult. Here, however, the presence of dCNEs allowed us to resolve, in a number of cases, the likely target gene in regions containing several candidates. For example, five dCNE families have members in regions within chromosome 19 (q12-q13.11) and chromosome 20 (q13.2). Both regions contain a cluster of zinc-finger genes (ZNF536, ZNF537, and ZNF507 on Chr19, and SALL4, ZFP64, ZNF218, and ZNF217 on Chr20) of which only ZNF537 and ZNF218 are close paralogs, allowing a clear association of these dCNE families with these paralogs.
In total, 121 dCNE families were associated with 53 sets of trans-dev paralogs (as in many cases, several dCNE families cluster around the same set of paralogous genes) (Table 1). We therefore have identified a set of related sequences with high regulatory potential, the majority conserved across all vertebrates and all but three of which are located in the vicinity of one or more trans-dev paralogs. Consequently, through comparison within the human genome and across a range of other vertebrate genomes, we are able to undertake a more in-depth analysis of the evolution and constraints on genomic location and environment of dCNEs and their likely target genes.
Conservation of dCNEs and target genes across vertebrates
The proximity of CNEs to their target gene is constrained by their likely function as CRMs. Indeed, disruption of the proximity of CRMs from their target gene via chromosomal breakpoints has been shown to cause congenital disease in a number of cases (Kleinjan and van Heyningen 2005). Consequently, retention of the element and the gene in cis across vertebrate evolution increases confidence in their association. Therefore, for each of the 121 families (255 dCNEs) for which paralogous genes were identified, we searched for both the presence of the element and the ortholog of its human target gene in each of five vertebrate draft genome assemblies (mouse, rat, dog, chicken, and Tetraodon) for which chromosomal mapping exists. For 254 of the 255 dCNE-gene pairs, the dCNE was located in the vicinity of the orthologous gene in all organisms for which both a dCNE and orthologous gene could be retrieved (Supplemental Table S1). In only one case (associated with the NR2F2 ortholog in chicken) was a dCNE located on a different chromosome (ChrZ) from the gene (ChrW). However, a known assembly error on ChrW may mean that this annotation is incorrect (Ensembl Chicken, Release 29.1e).
Positional comparison of dCNE family members
As previously stated, it is known that regulatory elements reside in intergenic sequences (5′ or 3′ of genes), as well as within the introns of either the genes on which they act or the introns of neighboring genes. Assuming dCNE families and their target genes derive from a common ancestor, we investigated their position in relation to their target gene. For each of the 121 families for which we could assign a set of paralogous trans-dev genes, we looked at the relative genomic position (5′, 3′, or intronic) of each member with respect to its target gene within the human genome. In 110 cases, all dCNE family members were found to be in the same relative location with respect to the target gene (48% are 5′, 25% are 3′, and 27% intronic). In 10 cases, family members were found to be a mix of intergenic and intronic. However, for two of these (associated with EBF2 and NRXN1), transcript evidence suggests that Ensembl annotation may be missing one or more exons, thereby locating both dCNEs within an intron. If we assume annotation is correct for the other seven cases (where no additional transcript evidence exists), the change in genomic environment is most likely due to gene restructuring by exon gain or loss over evolution in one or more of the target paralogs rather than by chromosomal rearrangement. In all but one of the cases, we found the intergenic dCNE located 5′ of a trans-dev gene with a lower number of coding exons than its paralog, suggesting such a change in gene structure. We found no cases in which one member of a dCNE family was located in a 5′ position and the other in a 3′ position in relation to their target gene, although we found an intriguing exception of sorts in the case of a dCNE family associated with the zinc-finger paralogs SALL1 and SALL3 (Fig. 3A). Here, one member is located 5′ of SALL3, and two members are located both 5′ and 3′ of the SALL1 gene. Both SALL1 dCNEs are conserved in all vertebrates (Supplemental Table S1) and therefore may play functionally distinct roles in the regulation of SALL1, possibly related to their position around the gene. Although they differ substantially in length, they are 80% identical across the length of the smaller element, which comprises the “core” of the larger element. Additionally, the position of the dCNE in human with relation to its target gene (5′ upstream, intronic to target gene, or 3′ downstream) was found to be the same in five vertebrate genomes in 83% of families. Anomalies in individual genomes only occurred in situations in which a dCNE was intronic to the target gene in human but located outside of the target gene in one or more of the other genomes. However, in most cases, this appears to be due to the limitations of automated annotation in these genomes in Ensembl, as additional transcript evidence suggests that one or more coding exons have been missed, placing these dCNE within introns. Despite this, we cannot exclude the possibility that dCNEs move from intronic to intergenic positions (or vice versa) over evolution because of changes in gene structure between species as previously described.
Figure 3.
dCNE families with more than two members. Brown lines connect dCNEs within the same family. (A) An unusual three-member family is found around SALL1 and SALL3. Here, two of the members are found both 5′ and 3′ of SALL1, a feature not seen in any of the other families. (B) A three-member family of interest is located around EVX1 and EVX2. Here, the two members on Chr7 show significant similarity to different parts of the single element on Chr2 and are separated by a gap of 665 bp, little of which is conserved across orthologous regions in other vertebrates. The same region is only 150 bp on Chr2 and is conserved across vertebrates, indicating that this is likely to be the ancestral element. (C) dCNEs around NEUROD 1, 2, and 6 are retained in a similar manner to those in E although this set of paralogs contains no two-member families. (D) In contrast to dCNEs retained across three-member paralogous gene families as in C and E, PAX2, PAX5, and PAX8 retain only two-member dCNE families, connected by a central gene (PAX2). Blue boxes within the red dashed box represent dCNE located within the introns of these genes. (E) Four three-member families (yellow boxes) are located around three teashirt orthologs on human chromosomes 18, 19, and 20 that possess overlapping expression domains (Caubit et al. 2005). Additionally, seven two-member families (blue boxes) are retained between different pairs of these paralogs. Element lengths are represented relative to a 100-bp element shown in the key. Gene annotation was taken from Ensembl v27.35.1 for SDCCAG33 and ZNF537 and the Vertebrate Genome Annotation Database (http://vega.sanger.ac.uk/Homo_sapiens) for ZNF218. Distance of dCNEs from the presumed translation start site (TSS) in all three genes is fixed according to the lower scale. Different scales are used for the distance downstream of the TSS for ZNF218 (lower scale) and SDCCAG33 and ZNF537 (upper scale).
For dCNE families in which all members were located externally (i.e., 5′ or 3′) of the predicted target gene in human (77 families), we identified 27 cases in which one dCNE family member was located within the intron of an unrelated neighboring gene. Interestingly, the other dCNE member(s) was almost always located within a large intergenic region (an example of this can be seen in Fig. 4). The neighboring genes in which the elements were situated were found to have no paralogs in the region of the other dCNE family members, and interestingly most appear to have no paralogs at all in the human genome. We examined these cases in more detail by comparing the position of these dCNEs in the canine, rodent, and chicken genomes. We found that, in all but two cases (one in rat, one in chicken), the dCNE is situated within the ortholog of the human gene, indicating a high level of evolutionary constraint in the location of the dCNE (Supplemental Table S2). It is therefore likely that these dCNEs originated within the intron of these genes rather than being incorporated sometime after duplication and that their paralogs were lost through nonfunctionalization and subsequent neutral drift over evolution. In only two cases were all dCNE family members found to be located in the introns of paralogous genes (NBEA and LRBA) that were not the likely target genes. In these two specific cases the predicted target genes, MAB21L1 and MAB21L2, are also located in introns of NBEA and LRBA, respectively.
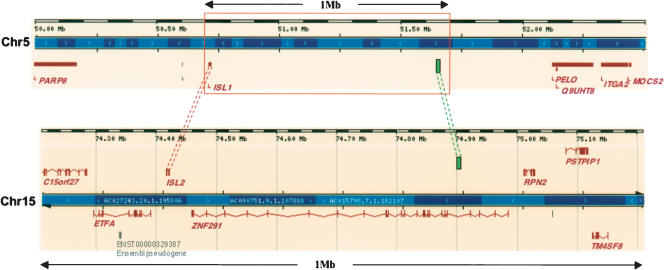
Figure 4.
Location of dCNEs in the vicinity of homeobox paralogs ISL1 (Chr5) and ISL2 (Chr15). ISL1 and ISL2 are the only paralogs within 1.5 Mb of the dCNEs (represented by green boxes) present in both regions (full extent not shown). The dCNE on Chr5 is located within a ‘gene desert’ and is ∼926 Kb 3′ of the ISL1 translation start site. In a similar manner to 27 other dCNE families (Supplemental Table S2), one dCNE is located within the intron of a gene (in this case ZNF291) while the other is located in a large intergenic region (spanning 1.39 Mb between ISL1 and PELO). In isolation, we would normally presume the dCNE on Chr15 to be associated with ZNF291, the closest trans-dev gene. However, as ZNF291 has no paralogs in the human genome, the ISL paralogs are far more likely to be the true associated genes of the dCNEs. In addition, this dCNE family has undergone an inversion event so that one dCNE is located in the same orientation to the target gene in one instance and the opposite orientation to the target gene in the other. Diagram adapted from the Ensembl Genome Browser (Hubbard et al. 2005).
CRMs, such as enhancers, have been shown to act both irrespective of orientation (e.g., Hill-Kapturczak et al. 2003) and in an orientation-dependent manner (e.g., Swamynathan and Piatigorsky 2002). Although it is not possible from our analysis to know the orientation of a dCNE, the known directionality of gene coding sequences allowed us to identify the relative orientation of each dCNE with respect to its putative target gene. Comparison of the relative orientation of orthologous dCNE-gene pairs across the five vertebrate genomes (see Methods) identified just four cases in which it appears the dCNE has undergone a local inversion since divergence (one in mouse, three in chicken). Similar comparisons between dCNE family members within the human genome also identified just four families in which dCNEs have undergone local inversion since duplication, three of which are located intergenically [SOX14/SOX21, ISL1/ISL2 (Fig. 4), two dCNEs located either side of SALL1 (Fig. 3A)] and one that is located within the introns of PBX1 and PBX3. This suggests that inversion events of dCNEs since duplication are relatively rare, but that such events are tolerated, possibly because of the orientation-independent nature of at least some enhancers.
Element evolution within dCNE families
dCNEs within a family have arisen through duplication events and share extensive sequence similarity within and between species. We investigated the extent to which dCNEs within the same organism have diverged compared with their orthologs across vertebrates by using the average percent identity (ignoring insertions/deletions) as a rough estimate of sequence divergence. For all two-member families, we compared the average pairwise sequence identities of human dCNEs with their orthologs in chicken and in Fugu and between dCNE copies within each of the organisms. Orthologous copies of the dCNEs were found to be, on average, more highly conserved than dCNE copies within each individual species (Figs. 1 and 5). Given the time scales involved, this indicates that dCNEs evolved rapidly after duplication but came under extreme evolutionary constraint sometime prior to the divergence of fish and tetrapods.
Figure 5.
Mean percent sequence identities of related dCNEs within and between species. “Between species” represents orthologous dCNEs; dCNEs from two-member families are extremely well conserved between human and chicken copies (Human1–Chick1, Human2–Chick2) with a lower level of conservation between human and Fugu copies (Human1–Fugu1, Human2–Fugu2), reflecting the longer phylogenetic branch length and higher rate of evolution in fish genomes (Jaillon et al. 2004). Error bars represent the standard error of the mean. “Within species” represents dCNEs within the same genome; mean conservation is much lower between dCNEs within the same species than between orthologs, indicating an increased rate of evolution following duplication followed by extreme evolutionary constraint sometime prior to the fish–tetrapod divergence. For >80% of families that contained at least two members in Fugu, phylogenetic trees constructed using maximum parsimony (with 1000 bootstrap replicates) fitted the expected topology, i.e., dCNE family members were more similar between genomes than within genomes.
Our set of dCNEs does not as a whole appear to be under greater evolutionary constraint than the remainder of unique CNEs (from our original set of 2330 Human–Fugu CNEs) with very similar mean percent identities (85.9 ± 0.49% and 84.5 ± 0.13% respectively, mean ±S.E.M). However, a subset of the elements does appear to be under extreme evolutionary constraint, as 32 dCNEs overlap with sequences previously identified as “ultraconserved” (100% identical over at least 200 bp between humans and rodent genomes, Bejerano et al. 2004a), which is a significant overrepresentation in this set (P ≤ 0.003). Indeed, by comparison with our set of dCNEs, more than 12% of noncoding ultraconserved elements (UCEs) have duplicates in the human genome. This is a higher proportion than reported (16/248) by Bejerano et al. (2004a) who searched for duplicated elements within their set of UCEs rather than by comparison with the human genome.
In addition to sequence divergence, the length of dCNEs can vary extensively between family members. The ratio of the length of the smaller element to the length of the larger element in all two-member families was found to vary between 0.23 and 1 (mean = 0.64). While a third of dCNE families had elements similar in size, over a third had a ratio below 0.5, indicating an extensive change in element length. An example can be seen in Figure 1 where evolution of sequence and length between related elements intronic of FOXP1 and FOXP2 is observed. Most of the elements that differed significantly in length can be attributed to loss of sequence similarity at the edges of the smaller element. We identified one exception in a dCNE family upstream of two homeobox paralogs EVX1 (Chr7) and EVX2 (Chr2), where substantial nucleotide changes have occurred at the core of one of the elements, essentially creating a split element upstream of EVX1 separated by 665 bp of nonconserved sequence, considerably larger than the 150-bp sequence that separates these sections in the element upstream of EVX2 (Fig. 3B). Comparison of the element on Chr7 with orthologous regions in other vertebrates reveals a similar pattern of nonconservation at the core of the sequence from rodents to fish, although the length of this nonconserved section ranges from 398 bp in rat to 168 bp in Fugu suggesting substantial insertions/deletions have occurred in this central section over evolution and it is no longer under functional constraint.
Families with more than two members
Although most of the dCNE families contain just two members located close to a pair of paralogs, a small number of dCNE families containing 3–5 members were also identified, suggesting these elements had been retained over two or more duplication events. This proved correct as the majority are located in the vicinity of genes from the same paralogous gene family [e.g., NEUROD1, 2, and 6 (Fig. 3C) and FOXP1, FOXP2 and FOXP4] with the largest number of examples located around and within three closely related but relatively uncharacterized homeobox genes SDCCAG33 (Chr18), ZNF537 (Chr19), and ZNF218 (Chr20). These genes are homologous to the Drosophila teashirt gene, and mouse orthologs have been shown to play critical roles in trunk, limb, and eye development (Caubit et al. 2000; Long et al. 2001; Manfroid et al. 2004). These paralogs exhibit a complex pattern of CNE retention, with four families retained around all three paralogs and several others retained between just two paralogs (Fig. 3E). In contrast, three paralogs of the PAX family of transcription factors PAX2, PAX5, and PAX8 have no related CNEs across all three genes, and two-member dCNE families are only retained between PAX2 and the other paralogs (Fig. 3D). One of the four-member dCNE families was found to be associated with all members of the EBF/Olf/Collier family of transcription factors (EBF, EBF2, EBF3, and EBF4) involved with differentiation of cells in early adipogenesis, as well as neuronal and B-cell development (Liberg et al. 2002). The dCNE associated with EBF4 either derives from a mammalian-specific duplication event or has been lost in birds and fish, as neither the EBF4 gene nor the dCNE is present in these lineages (Supplemental Table S1). The other four-member family is located within each of the four mammalian HOX clusters, closest to HOXA4, B4, C4, and D4. Although an enhancer has been identified that is conserved between the HOXA and HOXD clusters (Lehoczky et al. 2004), an element that shows sequence conservation across all four clusters has not previously been reported and may represent an element critical for expression of HOX genes to the same expression domain. The largest dCNE family contains five members that are located upstream of paralogs of the FOXD family of forkhead transcription factors. We can trace back two of the members to a tetrapod-specific duplication event that created FOXD3 (Chr1) and FOXD4 (Chr9). The remaining three members derive from primate-specific segmental duplications of the subtelomeric region of chromosome 9p around FOXD4 (Wong et al. 2004). Interestingly, all the dCNEs that derive from FOXD4 are within the introns of a neighboring gene, whereas the related dCNE upstream of FOXD3 is located in a large intergenic region, a feature common to a number of CNE families in our set (Supplemental Table S2). Similarly, lineage-specific duplication of elements was also seen in 15 of the families that had more members in the fish genomes than in tetrapods (Supplemental Table S1). These derive from an additional genome duplication event and subsequent retention of paralogs in the teleost lineage (Vandepoele et al. 2000; Christoffels et al. 2004). The remaining two multi-member families (Fig. 3A,B) are associated with just two paralogous genes as previously described.
Functional analysis of dCNEs
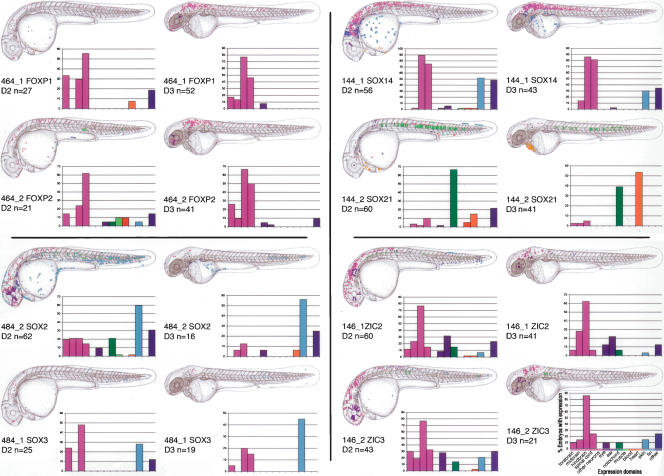
To assess whether the dCNEs are likely to have a regulatory role during development, we tested their ability to up-regulate a GFP reporter in zebrafish embryos, as described previously (Woolfe et al. 2005). We chose five two-member dCNE families that had just a single pair of trans-dev paralogs in their vicinity and in which both dCNEs and paralogs were conserved in mammals and fish: dCNEs associated with FOXP1/FOXP2 (two families), SOX14/SOX21, SOX2/SOX3, and ZIC2/ZIC3. For each element, we assayed the full-length dCNE as defined by sequence conservation between the human and Fugu genomes. In all cases this is larger than the region of conservation between the dCNEs (Supplemental Fig. S3). Eight of the ten dCNEs up-regulate GFP expression in a tissue-specific manner during day two and day three of zebrafish development (Fig. 6). Only one dCNE family (461_1 and 461_2), associated with the FOXP1 and FOXP2 genes, showed no expression in our assay. Of the four dCNE families that did up-regulate GFP, three families exhibit similar patterns of expression between members (FOXP1/FOXP2, SOX2/SOX3, and ZIC2/ZIC3), whereas the patterns exhibited by SOX14 and SOX21 are significantly different from each other, with very little overlap.
Figure 6.
dCNEs direct GFP reporter gene expression in specific tissues. For each dCNE, cumulative GFP expression data is pooled from a number of embryos (n ≥ 20 expressing embryos per dCNE on day 2 of development). Embryos are examined for GFP expression at ∼26–30 hpf and 50–54 hpf and schematically overlaid on camera lucida drawings of 2- and 3-day-old zebrafish embryos. Different cell types are color-coded, and the same key is used for all panels. Both the color code and the key are displayed under the day 3 chart for dCNE 146_2. Graphs encompass the same data set as the schematics and display the percentage of GFP-expressing embryos that show expression in each tissue category for a given dCNE. The total number of expressing embryos analyzed per CNE is displayed just below the schematic in each case. FOXP1/FOXP2 dCNEs 461_1 and 461_2 did not up-regulate GFP expression in this assay.
FOXP1 and FOXP2 are Winged helix/Forkhead domain transcription factors. Both are expressed in the developing brain, spinal cord, branchial arches, and eye (Tamura et al. 2003; Pohl et al. 2004; Bonkowsky and Chien 2005). All elements from dCNE families 461 and 464 are located in the introns of FOXP1 and FOXP2 in both human and Fugu. The GFP up-regulation profiles of dCNE elements 464_1 and 464_2 (Figs. 6 and 7A,B) are consistent with the known FOXP1/FOXP2 expression patterns. Both elements promote an increased level of expression particularly in day three embryos, in line with foxp2 expression patterns observed in zebrafish from day two through to day four (Bonkowsky and Chien 2005). In mouse, Foxp1 is expressed in the heart (Wang et al. 2004), so it is interesting that the FOXP1 dCNE 464_1 up-regulates GFP expression in the developing heart on day two, whereas no expression is seen in the heart with the FOXP2 dCNE 464_2. Both members of the 461 dCNE family were negative in our assay, suggesting that perhaps these elements are involved in repression or down-regulation of expression, rather than having enhancer function.
Figure 7.
Up-regulation of GFP expression by dCNEs. GFP expression is shown in live embryos as fluorescent images (A,B,C) or in fixed tissue following whole-mount anti-GFP immunostaining (D–H). All embryos are 48–54 hpf. Lateral views, anterior to the left, dorsal to the top. GFP expression is shown in the following tissue or cell types, indicated by arrowheads: (A) 464_1, FOXP1; hindbrain; (B) 464_2, FOXP2; hindbrain; (C) dCNE 144_1, SOX14; hindbrain; (D) 144_2, SOX21; heart; (E) 484_2, SOX2; epidermal cells; (F) 484_1, SOX3; epidermal cells; (G) 146_1, ZIC2; lens and various neurons in the fore-, mid-, and hindbrain; (H) 146_2, ZIC3; retina and various neurons in the fore- and hindbrain. Scale bar 50 μm (A–D,G,H) or 100 μm (E,F). (e) Eye; (f) fin; (fb) forebrain; (h) heart; (hb) hindbrain; (l) lens; (mb) midbrain; (ov) otic vesicle; (r), retina; (s) somite; (y) yolk.
SOX14 and SOX21 are members of the Sry-like Box gene family (Bowles et al. 2000). They are transcription factors containing the HMG (high mobility group) DNA binding domain. SOX14 and 21 belong to the B2 subgroup based on their repression domain at the C terminus (Uchikawa et al. 1999). Both genes are expressed in distinctive regions of the developing central nervous system (Rex et al. 1997; Rimini et al. 1999; Uchikawa et al. 1999; Hargrave et al. 2000). Whereas the profile of dCNE 144_1 reflects the endogenous pattern of expression of SOX14 (Figs. 6 and 7C), the profile of dCNE 144_2 is strikingly different and does not recapitulate any of the known zones of expression of SOX21 (Figs. 6 and 7D). GFP is most highly expressed on day two in notochord, and on day three in the heart, with over half of expressing embryos showing cardiac expression. These results are consistent, however, with previous assays using this element (element SOX21_1, Woolfe et al. 2005). The lack of any overlap in the expression patterns between these elements is surprising given that the dCNEs share 70% identity across 350 bp (Supplemental Fig. S3). However, it should be noted that there are nearly 140 bp of the SOX14 dCNE that are not present in the SOX21 dCNE, and 135 bp that are unique to the SOX21 dCNE (Supplemental Fig. S3), suggesting these extra sequences as well as nucleotide changes between elements play critical roles in directing expression to these domains.
SOX2 and SOX3 are also members of the Sry-like Box family of transcription factors, and are important embryonic regulators of organogenesis. Both genes are expressed in the early brain and play fundamental roles in placode formation in Xenopus (Wood and Episkopou 1999; Schlosser and Ahrens, 2004). In mouse, Sox2 is particularly associated with ear development (Kiernan et al. 2005) and in zebrafish it is expressed in the brain and spinal cord, eye, pharyngeal arches, and ventral mesoderm (ZFIN database). dCNE 484_2, associated with the SOX2 gene, appears to up-regulate GFP expression in a pattern consistent with the endogenous pattern of expression of SOX2. There is good expression in all regions of the brain and eye, and some of the more difficult to assign expression (labeled as ‘other’ in blue in Figure 6) is in the region of pharyngeal arch formation. SOX2 is also more highly expressed during the early stages of development, and this correlates with the fact that the profile of dCNE 484_2 shows much higher expression on day 2 than on day 3. SOX3 dCNE 484_1, although significantly longer, shows a similar yet more restricted pattern to 484_2, with CNS expression limited to the fore- and hindbrain. Once again, expression is much higher on day 2 than on day 3. Both dCNEs 484_1 and 484_2 also appear to up-regulate GFP expression in ventral and posterior epidermal cells (Figs. 6 and 7E,F).
The ZIC2 and ZIC3 genes are C2H2 zinc-finger domain genes, which are thought to play roles in embryonic pattern formation and early neurogenesis (Nagai et al. 1997). They are expressed widely in brain, spinal cord, and eye (Grinblat and Sive 2001; Warner et al. 2003; Toyama et al. 2004). In chick, Zic2 and Zic3 may also play a role in ear development (Warner et al. 2003). The GFP expression patterns for dCNEs 146_1 and 146_2 (Figs. 6 and 7G,H) are in good agreement with the endogenous zebrafish zic2 and zic3 patterns, with predominant expression in the brain, and with additional expression in the ear (ZIC2) and the eye (mostly ZIC3). Expression also appears stronger for both genes at day 2 compared with day 3.
Discussion
CRMs play a crucial role in the regulation of gene transcription, essential for the function and development of all organisms. To date, putative CRMs detected computationally through phylogenic footprinting appear to be unique within any one genome, lacking any close sequence similarity to one another, implying that they have evolved independently. However, a small number do exist that appear to have arisen from duplication events. The discovery of these dCNEs provides an opportunity to study their origin and evolution within the human and other vertebrate genomes.
In this study, we identified 124 families of dCNEs in the human genome that are likely to be cis-regulatory in nature. These families are all highly conserved both within and between vertebrate genomes and appear to have evolved remarkably slowly over the last 450 Myr. Their constrained evolution is more surprising because of their apparent absence in urochordates and cephalochordates, suggesting that they arose sometime near the beginning of the vertebrate lineage and play an essential functional role in vertebrates.
Under the assumption that these sequences have been retained after duplication with their associated genes and the genes they are likely to act on are annotated as trans-dev genes, we searched for paralogous relationships in the genomic environment around each member of a dCNE family. We found trans-dev paralogous genes in regions surrounding all but three of our dCNE families. In 74% of cases, just a single set of closely related paralogous genes was identified, all of which have proven or possible transcriptional or developmental roles, allowing us to associate the dCNEs and genes with confidence. Because CRMs do not necessarily act on the closest gene (Lettice et al. 2003; Spitz et al. 2003), this approach proved particularly powerful in regions containing clusters of unrelated trans-dev genes. Comparing the regions around all dCNE family members often indicates that the true association is actually with the only gene that has paralogs close to all members of that dCNE family. In contrast, for dCNE families with several trans-dev paralogs in their neighboring regions (e.g., clusters such as HOX and IRX) only provisional associations can be made. In these cases we selected the closest trans-dev paralogous set although, potentially, the dCNEs could have more distantly associated genes (e.g., Spitz et al. 2003) or be “shared” by more than one gene within the cluster (e.g., enhancers associated with Hoxb4 and Hoxb5; Sharpe et al. 1998). In total, 52 sets of paralogs were identified in the regions surrounding 121 dCNEs families, including many from the key regulatory gene families that orchestrate early development.
By confirming the retention of dCNEs in the vicinity of the same paralogs in five other vertebrate genomes, we were able to verify further the tight association of dCNE families with nearby paralogs over vertebrate evolution. Despite the existence of large regions of conserved synteny from humans to fish (McLysaght et al. 2000; Woods et al. 2000), gene order within syntenic chromosome segments is often rearranged (Woods et al. 2000). Here we find that changes in the relative position of dCNEs with respect to their paralogs (5′, intronic, or 3′) are rare, as are changes in orientation, suggesting that many of these elements may function in a position- and orientation-dependent manner. Indeed, the presence of CNEs interspersed across loci may play a role in conserving gene order in syntenic regions across species, for example, within a 4-Mb region around the SHH gene in human and Fugu (Goode et al. 2005).
For several of the dCNE families, the genomic environment surrounding each member can be very different (with one or more members located in the introns of a neighboring gene whilst the others are located intergenically), indicating that genomic environment may not be essential to element function. By comparing positions across vertebrate genomes, we identified several cases in which related dCNEs are found in an intergenic environment in one genome but are intronic of the target gene in another (and vice versa). These could be due either to limitations in accurate automated gene annotation (e.g., an exon has been missed, which would place an intergenic dCNE within an intron), or the loss or gain of an exon or exons within the target gene over evolution.
Although almost a quarter of the dCNEs found were not duplicated in fish genomes, most can still be dated to ancient, vertebrate-specific duplications, as their associated paralogous genes are present in all vertebrates. The fact that some dCNEs are only found in single copy in fish may be accounted for by fish-specific loss of elements over evolution or simply due to missing sequence as a result of incomplete nature of both Fugu and Tetraodon genomes. We can, in a number of cases, trace certain members of a dCNE family back to more recent duplication events, for example, the dCNE associated with EBF4 present only in the mammalian lineages and primate-specific duplications around FOXD4. A number of families also have more members in the Fugu and Tetraodon genomes because of an additional genome duplication event thought to have occurred sometime prior to the teleost radiation between 300 and 450 Mya (Vandepoele et al. 2000; Taylor et al. 2003). Fewer than 3% of the dCNE families are located in regions that do not contain any paralogs. These may constitute novel genomic elements of interest or indicate that novel, currently unannotated paralogs may exist in their vicinity. It is also possible that associated paralogs for these dCNE do exist but are located beyond the 1.5-Mb search boundaries used in this study.
Comparison of sequence divergence between related dCNEs within the human genome and their orthologous copies in the genomes of chicken and Fugu reveal an extraordinary evolutionary history. dCNEs within a genome have undergone greater evolutionary change in both nucleotide sequence and length than orthologous dCNEs between genomes. This suggests that across a period of 50–150 Myr following the duplication of these cis-regulatory elements and their associated genes, there was an increased rate of change within both the protein coding (Hughes and Friedman 2004) and regulatory sequences reflecting a possible relaxation of evolutionary constraint in one of the copies because of intergene redundancy (Fig. 8). Classical models predict the most likely fate of duplicated genes is the degeneration of one of the pair to a pseudogene (or lost from the genome altogether) or less frequently the acquisition of novel gene functions as a result of alterations in coding or regulatory sequences in a process known as neofunctionalization (Ohno 1970). Alternatively, a subfunctionalization model has been proposed in which duplicated genes undergo complementary loss-of-function mutations in independent subfunctions so that both genes are required to recapitulate the functions of the ancestral gene (Force et al. 1999). Here, it appears that, following duplication, paralogs evolved distinct and/or overlapping functions and expression domains and became effectively “fixed” in the ancestral genome to form the basis of early development in all subsequent vertebrates.
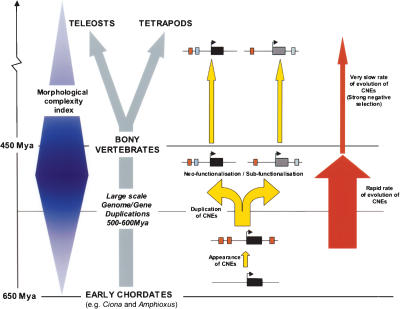
Figure 8.
Proposed model of CNE evolution in the context of other major genomic events during the early vertebrate radiation. Modern bony vertebrates evolved from the chordate lineage between 650 and 450 Mya, during a period of rapid morphological change (represented here in blue and based on the Morphological Complexity Index as described in Aburomia et al. 2003). It is now generally accepted that during this period an early ancestral vertebrate underwent one, or possibly two, whole-genome duplications, generating a greatly increased repertoire of genes, which in turn may have contributed to this increase in morphological complexity. The appearance of CNEs in vertebrate genomes (red boxes adjacent to gene loci, depicted as dark boxes) can be dated prior to these large-scale duplication events, as most of the dCNEs are associated with trans-dev paralogs that derive from these ancient duplications (yellow arrows). The duplication of gene loci together with associated cis-regulatory modules generates the plasticity for genes to develop new functions (neofunctionalization) and/or to perform a subset of the functions of the parent gene (subfunctionalization). This evolution must have occurred rapidly following duplication over a relatively short evolutionary period (∼50–150 Myr) during which time dCNEs evolved in length and sequence. In contrast, in the period since the teleost–tetrapod divergence (∼450 Mya), dCNEs have had a remarkably slow mutation rate and have remained practically unchanged.
One of the central tenants driving biological sequence analysis is the idea that sequences (whether DNA or protein) that show significant sequence similarity are likely to have the same or similar functions. Although this is known to be true (in general) when applied to coding-related sequences, it is unknown whether the same holds true for CRMs for which little is known about structure, language, or mode of action. A number of the paralogous genes in our set have been shown to have overlapping expression patterns (which may be driven by dCNEs) as well as distinct ones (possibly driven by CNEs unique to each gene), for example, PAX2, PAX5, and PAX8 (Heller and Brandli 1999) and Tsh1, Tsh2, and Tsh3 (mouse orthologs of human ZNF537, SDCCAG33, and ZNF218) (Caubit et al. 2005). Given the low rate of element retention between paralogs as evidenced by a much larger number of unique CNEs around the same genes (e.g., SOX21 and SOX14, Woolfe et al. 2005), we would assume dCNEs represent functional attributes (i.e., expression domains) that are shared by both paralogs. To test this assumption and further confirm the regulatory potential of our dCNE set, we tested a total of 10 duplicated elements in our zebrafish assay, representing five two-member dCNE families associated with eight genes, and found that all but one family up-regulated expression of GFP in a tissue-specific manner. Moreover, with the exception of the SOX14/SOX21 elements, the dCNEs exhibited expression profiles that not only recapitulated aspects of the endogenous pattern of the paralogs with which they were associated but also overlapped considerably between duplicate elements. These results therefore suggest a level of concurrence of sequence and functional homology. Indeed, recent functional studies on individual putative CRMs exhibiting sequence similarity report similar findings. In a functional analysis of a pair of duplicated UCEs within the introns of DACH1 and DACH2 (corresponding to dCNE family 136 from this study), both elements were shown to drive expression of a reporter gene within similar expression domains in mouse (Poulin et al. 2005). Similarly, duplicated CRMs conserved between the IrxA and IrxB clusters (de la Calle-Mustienes et al. 2005) and HoxA and HoxD clusters (Lehoczky et al. 2004) (not sufficiently conserved in Fugu to be identified by our study) were also shown to drive expression of genes within these clusters to similar expression domains.
In contrast, the stark differences in expression patterns observed for dCNEs around SOX14/SOX21 suggest that, as with protein sequences, sequence similarity may not always extend to functional similarity. However, although these elements share extensive sequence identity over the majority of their length, there is still considerable independent conservation not shared between dCNEs at their edges (Supplemental Fig. S3). This suggests that, in some cases, the function may require the complete dCNE for function, rather than being determined by the sum of smaller modules within the dCNE (i.e., multiple transcription factor binding sites, as proposed by current models; Davidson et al. 2002), which would presumably result in a heavily overlapping expression pattern for these two elements. However, the other six dCNE also have regions of independent conservation not shared by both family members but still exhibit highly overlapping expression patterns. In addition, it is important to note that the conserved elements are tested out of their genomic context (one of the main limitations of our assay) and that interaction between dCNEs and other CRMs in the vicinity may also be important in defining the precise function of each element. Without knowing the precise mechanism of action of these elements, it is difficult to speculate on the reason for the difference in expression patterns between the SOX14/SOX21 elements.
Whatever their mode of action, the ability to place the origin of these conserved elements at a specific evolutionary period has further implications. Between 450 and 600 Mya, the tremendous burst of gene duplication activity, as well as the appearance of a whole new repertoire of rapidly evolving cis-regulatory elements, coincides with fundamental and persistent changes in morphological complexity within the early vertebrate lineage (Aburomia et al. 2003) (Fig. 8). It is probable, therefore, that there is a direct connection between these events, given the association between CNEs and genes involved in developmental regulation. Gene paralogs identified in this study are some of the key regulators responsible for body patterning and morphogenesis in early vertebrate development. An increase in their copy number accompanied by the simultaneous evolution of a novel regulatory sequence network is likely to have played a major role in modeling these processes. Further studies are necessary to shed light on the function and mode of action of these elements. A key element of our studies will be to understand how evolutionary changes within members of dCNE families influence their regulatory potential and the consequence for the associated genes.
Methods
Detection of conserved noncoding sequences
To identify an initial set of CNEs between human and Fugu, the Fugu genome was masked for exons as described in Woolfe et al. (2005) and compared with human Ensembl v27.35.1 using MegaBLAST (Zhang et al. 2000) with a word size of 16 and an E-value cutoff of ≤10−4. Sequences with a significant similarity (E-value ≤10−4) to known expressed transcripts from SWISS-PROT/TrEMBL (http://us.expasy.org/sprot), EMBL mRNA (http://www.ebi.ac.uk/ embl), and Hs-UniGene (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene) as well as noncoding RNAs from miRNA Registry (Griffiths-Jones 2004) and Rfam (version 5.0) (Griffiths-Jones et al. 2003) were removed. Repeats and sequences dominated by low-complexity regions were detected using RepeatMasker and EntropyRep (v1.0, I. Abnizova, unpubl.), respectively, and removed.
Identification and filtering of human dCNE families
The human CNE set was made nonredundant by merging regions that overlapped. The resultant 2330 sequences were then compared back to the human genome using sensitive BLAST parameters (word size of nine, mismatch penalty of −1) (Altschul et al. 1997). An E-value cut-off of 5 × 10−4 was used. Sequences with more than one other sequence of significant similarity were grouped into families such that each sequence was similar (E-value ≤5 × 10−4) to at least one other in the family. Families with more than five members were regarded as likely to be repeat sequences and removed from the data set. Sequences showing similarity purely between human chromosomes X and Y were also ignored. All dCNE family members were BLAST searched against an EST database to see if they were likely to be transcribed. All families in which at least one member had more than three significant EST hits were removed. Families overlapping Ensembl annotated exons were also removed. Families were tested for significant RNA secondary structure using the program RNAfold from the Vienna RNA package (Hofacker et al. 1994). The minimum free energy was calculated for each family member along with 100 dinucleotide shuffled versions (Coward 1999) of that sequence. Z-scores were calculated for each sequence. dCNE families in which all members had Z-scores of less than −2 were considered to have significant RNA secondary structure.
Presence of dCNE families and copy numbers across vertebrate and chordate genomes
All human sequences from each dCNE family were BLAST searched with sensitive parameters against all vertebrate genome sequences from Ensembl [Chimp (v27.1a), Dog (v.27.1a), Mouse (v27.33.1), Rat (v.27.3e), Chicken (v.27.1e), Xenopus tropicalis (v.27.1), Tetraodon nigroviridis (v.27.1b), and Fugu rubripes (v27.3)] with the exception of zebrafish where sequence coverage is not reliable enough to make inferences. Families were considered (1) vertebrate-specific if conserved in at least one fish, one tetrapod, and one primate, (2) tetrapod-specific if not conserved in fish, and (3) primate-specific if conserved only in primates. Primate-specific dCNE families were not considered for further analysis. A similar BLAST search of all dCNE family members was carried out against the chordate genome C. intestinalis (JGI, v1.0), all urochordate and cephalochordate sequences from GenBank (Benson et al. 2005), and UCEs, as defined in Bejerano et al. 2004a. The expected number of UCEs within our dCNE set was calculated by choosing 261 CNEs at random from our original set of 2330 CNEs and calculating the mean number that overlapped UCEs in 1000 replicates. This was used to calculate a Z-score and probability that the observed proportion of UCEs within the dCNE set was significantly different to the expected value.
Finding associated genes
We defined a region of 1.5 Mb either side of each member of a dCNE family. Genes with paralogs within the regions of all family members were identified using paralogy assignments from Ensembl v27.35.1 (generated using all-against-all BLASTP sequence similarity search followed by a Markov Clustering algorithm. Eighty-seven percent of these paralogous families display full correspondence of domain structure across all annotated members [Enright et al. 2002]). To assess the likelihood of finding paralogs in these regions by chance we used the dCNE shuffling and family reassignment method as set out in Vavouri et al. (2005).
Previously (Woolfe et al. 2005), we reported that the genes found closest to CNEs are statistically overrepresented for GeneOntology (GO) annotations (Harris et al. 2004) relating to transcriptional regulation and/or development. Here, we defined paralogs as trans-dev if at least one member of the paralogous gene set had any of these 12 overrepresented GO ontologies (GO:0,003,700; 0,006,355; 0,006,351; 0,045,499; 0,019,219; 0,006,350; 0,006,366; 0,006,357; 0,007,399; 0,003,712; 0,003,714; 0,007,417). These GO ontologies encompass <8% of Ensembl human genes. In cases where paralogs were not identified and dCNEs were located in regions of low gene density (so-called ‘gene deserts’) we extended the region up to the next nearest gene.
Element evolution
Multiple alignments of CNE families were created using ClustalW (Thompson et al. 1994). Alignments were trimmed using the Gblocks program (Castresana 2000) and all columns containing no gaps were used to calculate the percent identities between pairs of sequences. The ratio (r) was calculated using r = s/l, where s is the length of the smallest element in the dCNE family and l is the length of the largest element in the family.
For those diverged genomes with chromosomal mapping and orthology data available (dog, mouse, rat, chicken, and Tetraodon) the location of the orthologous dCNE (obtained through a BLAST match) and orthologous human trans-dev gene (using Ensembl Compara 35.27.1) were compared. All pairs where both the dCNE and the orthologous gene were present and located within 2.5 Mb of each other were considered evidence of conserved association. Situations where the dCNE or gene was located on one of the assigned “random” chromosomes (i.e., sections of sequence that cannot yet be mapped to a specific chromosome) were ignored. dCNE gene sets were considered nonassociated if the dCNE was located on a different established chromosome to the orthologous gene or more than 2.5 Mb away on the same chromosome. Relative orientation of dCNEs in relation to their target gene was identified by using the orientation of each dCNE sequence in the genome and comparing it with that of the target gene. This was carried out for each individual dCNE identified in human and compared against the relative orientations of the orthologs in each of the genomes as specified above. Relative orientations were also compared between members of each dCNE family in human. dCNEs were considered to have undergone an inversion if relative orientations were different [e.g., orientation of one dCNE is (+) and its target gene is (+), but the other dCNE is (−) and its target gene is (+)].
Functional assaying of dCNE sequences.
In each case, dCNEs were PCR amplified from Fugu genomic DNA to encompass the region of sequence similarity between human and Fugu genomes (alignments can be found in Supplemental Fig. S3). Sequences used in the assays are also listed in Supplemental Figure S3 with primer sequences in upper case. PCR products were prepared and injected into 2–4 cells zebrafish embryos as described previously (Woolfe et al. 2005). Embryos were analyzed at ∼30 hours postfertilization (day 2) and 54 hours post-fertilization (day 3) and data processed as described (Woolfe et al. 2005).
Acknowledgments
We thank Walter Gilks and Sam Griffiths-Jones for insightful discussions and advice during the preparation of the manuscript, Laurent Fasano for useful correspondence regarding expression data of mouse teashirt genes, and two anonymous referees for their useful comments and suggestions. This work was funded by the Medical Research Council.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4143406
References
- Aburomia R., Khaner O., Sidow A., Khaner O., Sidow A., Sidow A. Functional evolution in the ancestral lineage of vertebrates or when genomic complexity was wagging its morphological tail. J. Struct. Funct. Genomics. 2003;3:45–52. [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang Z., Miller W., Lipman D.J., Miller W., Lipman D.J., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio S., Chapman J., Stupka E., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Chapman J., Stupka E., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Stupka E., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Christoffels A., Rash S., Hoon S., Smit A., Rash S., Hoon S., Smit A., Hoon S., Smit A., Smit A., et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Arnone M.I., Davidson E.H., Davidson E.H. The hardwiring of development: Organization and function of genomic regulatory systems. Development. 1997;124:1851–1864. doi: 10.1242/dev.124.10.1851. [DOI] [PubMed] [Google Scholar]
- Barton L.M., Gottgens B., Gering M., Gilbert J.G., Grafham D., Rogers J., Bentley D., Patient R., Green A.R., Gottgens B., Gering M., Gilbert J.G., Grafham D., Rogers J., Bentley D., Patient R., Green A.R., Gering M., Gilbert J.G., Grafham D., Rogers J., Bentley D., Patient R., Green A.R., Gilbert J.G., Grafham D., Rogers J., Bentley D., Patient R., Green A.R., Grafham D., Rogers J., Bentley D., Patient R., Green A.R., Rogers J., Bentley D., Patient R., Green A.R., Bentley D., Patient R., Green A.R., Patient R., Green A.R., Green A.R. Regulation of the stem cell leukemia (SCL) gene: A tale of two fishes. Proc. Natl. Acad. Sci. 2001;98:6747–6752. doi: 10.1073/pnas.101532998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Stephen S., Kent W.J., Mattick J.S., Haussler D., Kent W.J., Mattick J.S., Haussler D., Mattick J.S., Haussler D., Haussler D. Ultraconserved elements in the human genome. Science. 2004a;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Bejerano G., Haussler D., Blanchette M., Haussler D., Blanchette M., Blanchette M. Into the heart of darkness: Large-scale clustering of human noncoding DNA. Bioinformatics. 2004b;20:I40–I48. doi: 10.1093/bioinformatics/bth946. [DOI] [PubMed] [Google Scholar]
- Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L., Lipman D.J., Ostell J., Wheeler D.L., Ostell J., Wheeler D.L., Wheeler D.L. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Zur T., Feige E., Motro B., Wides R., Feige E., Motro B., Wides R., Motro B., Wides R., Wides R. The mammalian Odz gene family: Homologs of a Drosophila pair-rule gene with expression implying distinct yet overlapping developmental roles. Dev. Biol. 2000;217:107–120. doi: 10.1006/dbio.1999.9532. [DOI] [PubMed] [Google Scholar]
- Bishop C.E., Whitworth D.J., Qin Y., Agoulnik A.I., Agoulnik I.U., Harrison W.R., Behringer R.R., Overbeek P.A., Whitworth D.J., Qin Y., Agoulnik A.I., Agoulnik I.U., Harrison W.R., Behringer R.R., Overbeek P.A., Qin Y., Agoulnik A.I., Agoulnik I.U., Harrison W.R., Behringer R.R., Overbeek P.A., Agoulnik A.I., Agoulnik I.U., Harrison W.R., Behringer R.R., Overbeek P.A., Agoulnik I.U., Harrison W.R., Behringer R.R., Overbeek P.A., Harrison W.R., Behringer R.R., Overbeek P.A., Behringer R.R., Overbeek P.A., Overbeek P.A. A transgenic insertion upstream of sox9 is associated with dominant XX sex reversal in the mouse. Nat. Genet. 2000;26:490–494. doi: 10.1038/82652. [DOI] [PubMed] [Google Scholar]
- Blader P., Plessy C., Strahle U., Plessy C., Strahle U., Strahle U. Multiple regulatory elements with spatially and temporally distinct activities control neurogenin1 expression in primary neurons of the zebrafish embryo. Mech. Dev. 2003;120:211–218. doi: 10.1016/s0925-4773(02)00413-6. [DOI] [PubMed] [Google Scholar]
- Boffelli D., Nobrega M.A., Rubin E.M., Nobrega M.A., Rubin E.M., Rubin E.M. Comparative genomics at the vertebrate extremes. Nat. Rev. Genet. 2004;5:456–465. doi: 10.1038/nrg1350. [DOI] [PubMed] [Google Scholar]
- Bond H.M., Mesuraca M., Carbone E., Bonelli P., Agosti V., Amodio N., De Rosa G., Di Nicola M., Gianni A.M., Moore M.A., Mesuraca M., Carbone E., Bonelli P., Agosti V., Amodio N., De Rosa G., Di Nicola M., Gianni A.M., Moore M.A., Carbone E., Bonelli P., Agosti V., Amodio N., De Rosa G., Di Nicola M., Gianni A.M., Moore M.A., Bonelli P., Agosti V., Amodio N., De Rosa G., Di Nicola M., Gianni A.M., Moore M.A., Agosti V., Amodio N., De Rosa G., Di Nicola M., Gianni A.M., Moore M.A., Amodio N., De Rosa G., Di Nicola M., Gianni A.M., Moore M.A., De Rosa G., Di Nicola M., Gianni A.M., Moore M.A., Di Nicola M., Gianni A.M., Moore M.A., Gianni A.M., Moore M.A., Moore M.A., et al. Early hematopoietic zinc finger protein (EHZF), the human homolog to mouse Evi3, is highly expressed in primitive human hematopoietic cells. Blood. 2004;103:2062–2070. doi: 10.1182/blood-2003-07-2388. [DOI] [PubMed] [Google Scholar]
- Bonkowsky J.L., Chien C.B., Chien C.B. Molecular cloning and developmental expression of foxP2 in zebrafish. Dev. Dyn. 2005;234:740–746. doi: 10.1002/dvdy.20504. [DOI] [PubMed] [Google Scholar]
- Bowles J., Schepers G., Koopman P., Schepers G., Koopman P., Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Caubit X., Core N., Boned A., Kerridge S., Djabali M., Fasano L., Core N., Boned A., Kerridge S., Djabali M., Fasano L., Boned A., Kerridge S., Djabali M., Fasano L., Kerridge S., Djabali M., Fasano L., Djabali M., Fasano L., Fasano L. Vertebrate orthologues of the Drosophila region-specific patterning gene teashirt. Mech. Dev. 2000;91:445–448. doi: 10.1016/s0925-4773(99)00318-4. [DOI] [PubMed] [Google Scholar]
- Caubit X., Teviron M.C., Cremer H., Fasano L., Teviron M.C., Cremer H., Fasano L., Cremer H., Fasano L., Fasano L. Expression patterns of the three teashirt related genes define specific boundaries in the developing and postnatal mouse forebrain. J. Comp. Neurol. 2005;486:76–88. doi: 10.1002/cne.20500. [DOI] [PubMed] [Google Scholar]
- Chang C.W., Tsai C.W., Wang H.F., Tsai H.C., Chen H.Y., Tsai T.F., Takahashi H., Li H.Y., Fann M.J., Yang C.W., Tsai C.W., Wang H.F., Tsai H.C., Chen H.Y., Tsai T.F., Takahashi H., Li H.Y., Fann M.J., Yang C.W., Wang H.F., Tsai H.C., Chen H.Y., Tsai T.F., Takahashi H., Li H.Y., Fann M.J., Yang C.W., Tsai H.C., Chen H.Y., Tsai T.F., Takahashi H., Li H.Y., Fann M.J., Yang C.W., Chen H.Y., Tsai T.F., Takahashi H., Li H.Y., Fann M.J., Yang C.W., Tsai T.F., Takahashi H., Li H.Y., Fann M.J., Yang C.W., Takahashi H., Li H.Y., Fann M.J., Yang C.W., Li H.Y., Fann M.J., Yang C.W., Fann M.J., Yang C.W., Yang C.W., et al. Identification of a developmentally regulated striatum-enriched zinc-finger gene, Nolz-1, in the mammalian brain. Proc. Natl. Acad. Sci. 2004;101:2613–2618. doi: 10.1073/pnas.0308645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels A., Koh E.G., Chia J.M., Brenner S., Aparicio S., Venkatesh B., Koh E.G., Chia J.M., Brenner S., Aparicio S., Venkatesh B., Chia J.M., Brenner S., Aparicio S., Venkatesh B., Brenner S., Aparicio S., Venkatesh B., Aparicio S., Venkatesh B., Venkatesh B. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol. Biol. Evol. 2004;21:1146–1151. doi: 10.1093/molbev/msh114. [DOI] [PubMed] [Google Scholar]
- Coward E. Shufflet: Shuffling sequences while conserving the k-let counts. Bioinformatics. 1999;15:1058–1059. doi: 10.1093/bioinformatics/15.12.1058. [DOI] [PubMed] [Google Scholar]
- Davidson E.H., Rast J.P., Oliveri P., Ransick A., Calestani C., Rast J.P., Oliveri P., Ransick A., Calestani C., Oliveri P., Ransick A., Calestani C., Ransick A., Calestani C., Calestani C. A genomic gene regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Dehal P., Satou Y., Campbell R.K., Chapman J., Degnan B., De Tomaso A., Davidson B., Di Gregorio A., Gelpke M., Goodstein D.M., Satou Y., Campbell R.K., Chapman J., Degnan B., De Tomaso A., Davidson B., Di Gregorio A., Gelpke M., Goodstein D.M., Campbell R.K., Chapman J., Degnan B., De Tomaso A., Davidson B., Di Gregorio A., Gelpke M., Goodstein D.M., Chapman J., Degnan B., De Tomaso A., Davidson B., Di Gregorio A., Gelpke M., Goodstein D.M., Degnan B., De Tomaso A., Davidson B., Di Gregorio A., Gelpke M., Goodstein D.M., De Tomaso A., Davidson B., Di Gregorio A., Gelpke M., Goodstein D.M., Davidson B., Di Gregorio A., Gelpke M., Goodstein D.M., Di Gregorio A., Gelpke M., Goodstein D.M., Gelpke M., Goodstein D.M., Goodstein D.M., et al. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- de la Calle-Mustienes E., Feijoo C.G., Manzanares M., Tena J.J., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Feijoo C.G., Manzanares M., Tena J.J., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Manzanares M., Tena J.J., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Tena J.J., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Allende M.L., Gomez-Skarmeta J.L., Gomez-Skarmeta J.L. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res. 2005;15:1061–1072. doi: 10.1101/gr.4004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis E.T., Reymond A., Scamuffa N., Ucla C., Kirkness E., Rossier C., Antonarakis S.E., Reymond A., Scamuffa N., Ucla C., Kirkness E., Rossier C., Antonarakis S.E., Scamuffa N., Ucla C., Kirkness E., Rossier C., Antonarakis S.E., Ucla C., Kirkness E., Rossier C., Antonarakis S.E., Kirkness E., Rossier C., Antonarakis S.E., Rossier C., Antonarakis S.E., Antonarakis S.E. Evolutionary discrimination of mammalian conserved non-genic sequences (CNGs). Science. 2003;302:1033–1035. doi: 10.1126/science.1087047. [DOI] [PubMed] [Google Scholar]
- Dickmeis T., Plessy C., Rastegar S., Aanstad P., Herwig R., Chalmel F., Fischer N., Strahle U., Plessy C., Rastegar S., Aanstad P., Herwig R., Chalmel F., Fischer N., Strahle U., Rastegar S., Aanstad P., Herwig R., Chalmel F., Fischer N., Strahle U., Aanstad P., Herwig R., Chalmel F., Fischer N., Strahle U., Herwig R., Chalmel F., Fischer N., Strahle U., Chalmel F., Fischer N., Strahle U., Fischer N., Strahle U., Strahle U. Expression profiling and comparative genomics identify a conserved regulatory region controlling midline expression in the zebrafish embryo. Genome Res. 2004;14:228–238. doi: 10.1101/gr.1819204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A.J., Van Dongen S., Ouzounis C.A., Van Dongen S., Ouzounis C.A., Ouzounis C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F.B., Amores A., Yan Y.L., Lynch M., Pickett F.B., Amores A., Yan Y.L., Pickett F.B., Amores A., Yan Y.L., Amores A., Yan Y.L., Yan Y.L. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode D.K., Snell P., Smith S.F., Cooke J.E., Elgar G., Snell P., Smith S.F., Cooke J.E., Elgar G., Smith S.F., Cooke J.E., Elgar G., Cooke J.E., Elgar G., Elgar G. Highly conserved regulatory elements around the SHH gene may contribute to the maintenance of conserved synteny across human chromosome 7q36.3. Genomics. 2005;86:172–181. doi: 10.1016/j.ygeno.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Bateman A., Marshall M., Khanna A., Eddy S.R., Bateman A., Marshall M., Khanna A., Eddy S.R., Marshall M., Khanna A., Eddy S.R., Khanna A., Eddy S.R., Eddy S.R. Rfam: An RNA family database. Nucleic Acids Res. 2003;33:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinblat Y., Sive H., Sive H. zic gene expression marks anteroposterior pattern in the presumptive neurectoderm of the zebrafish gastrula. Dev. Dyn. 2001;222:688–689. doi: 10.1002/dvdy.1221. [DOI] [PubMed] [Google Scholar]
- Hargrave M., Karunaratne A., Cox L., Wood S., Koopman P., Yamada T., Karunaratne A., Cox L., Wood S., Koopman P., Yamada T., Cox L., Wood S., Koopman P., Yamada T., Wood S., Koopman P., Yamada T., Koopman P., Yamada T., Yamada T. The HMG box transcription factor gene Sox14 marks a novel subset of ventral interneurons and is regulated by sonic hedgehog. Dev. Biol. 2000;219:142. doi: 10.1006/dbio.1999.9581. [DOI] [PubMed] [Google Scholar]
- Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Eilbeck K., Lewis S., Marshall B., Mungall C., Lewis S., Marshall B., Mungall C., Marshall B., Mungall C., Mungall C., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller N., Brandli A.W., Brandli A.W. XenopusPax-2/5/8 orthologues: Novel insights into Pax gene evolution and identification of Pax-8 as the earliest marker for otic and pronephric cell lineages. Dev. Genet. 1999;24:208–219. doi: 10.1002/(SICI)1520-6408(1999)24:3/4<208::AID-DVG4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Hill-Kapturczak N., Sikorski E., Voakes C., Garcia J., Nick H.S., Agarwal A., Sikorski E., Voakes C., Garcia J., Nick H.S., Agarwal A., Voakes C., Garcia J., Nick H.S., Agarwal A., Garcia J., Nick H.S., Agarwal A., Nick H.S., Agarwal A., Agarwal A. An internal enhancer regulates heme- and cadmium-mediated induction of human heme oxygenase-1. Am. J. Physiol. Renal Physiol. 2003;285:F515–F523. doi: 10.1152/ajprenal.00137.2003. [DOI] [PubMed] [Google Scholar]
- Hofacker I.L., Fontana W., Stadler P.F., Bonhoeffer S., Tacker M., Schuster P., Fontana W., Stadler P.F., Bonhoeffer S., Tacker M., Schuster P., Stadler P.F., Bonhoeffer S., Tacker M., Schuster P., Bonhoeffer S., Tacker M., Schuster P., Tacker M., Schuster P., Schuster P. Fast folding and comparison of RNA secondary structures. Monatshefte f. Chemie. 1994;125:167–188. [Google Scholar]
- Holland P.W., Garcia-Fernandez J., Williams N.A., Sidow A., Garcia-Fernandez J., Williams N.A., Sidow A., Williams N.A., Sidow A., Sidow A. Gene duplications and the origins of vertebrate development. Dev. Suppl. 1994:125–133. [PubMed] [Google Scholar]
- Hubbard T., Andrews D., Caccamo M., Cameron G., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., Andrews D., Caccamo M., Cameron G., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., Caccamo M., Cameron G., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., Cameron G., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., Clarke L., Coates G., Cox T., Cunningham F., Coates G., Cox T., Cunningham F., Cox T., Cunningham F., Cunningham F., et al. Ensembl 2005. Nucleic Acids Res. 2005;33:D447–D453. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A.L., Friedman R., Friedman R. Pattern of divergence of amino acid sequences encoded by paralogous genes in human and pufferfish. Mol. Phylogenet. Evol. 2004;32:337–343. doi: 10.1016/j.ympev.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Jaillon O., Aury J.M., Brunet F., Petit J.L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Aury J.M., Brunet F., Petit J.L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Brunet F., Petit J.L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Petit J.L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Fischer C., Ozouf-Costaz C., Bernot A., Ozouf-Costaz C., Bernot A., Bernot A., et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Jamieson R.V., Perveen R., Kerr B., Carette M., Yardley J., Heon E., Wirth M.G., van Heyningen V., Donnai D., Munier F., Perveen R., Kerr B., Carette M., Yardley J., Heon E., Wirth M.G., van Heyningen V., Donnai D., Munier F., Kerr B., Carette M., Yardley J., Heon E., Wirth M.G., van Heyningen V., Donnai D., Munier F., Carette M., Yardley J., Heon E., Wirth M.G., van Heyningen V., Donnai D., Munier F., Yardley J., Heon E., Wirth M.G., van Heyningen V., Donnai D., Munier F., Heon E., Wirth M.G., van Heyningen V., Donnai D., Munier F., Wirth M.G., van Heyningen V., Donnai D., Munier F., van Heyningen V., Donnai D., Munier F., Donnai D., Munier F., Munier F., et al. Domain disruption and mutation of the bZIP transcription factor, MAF, associated with cataract, ocular anterior segment dysgenesis and coloboma. Hum. Mol. Genet. 2002;11:33–42. doi: 10.1093/hmg/11.1.33. [DOI] [PubMed] [Google Scholar]
- Kiernan A.E., Pelling A.L., Leung K.K., Tang A.S., Bell D.M., Tease C., Lovell-Badge R., Steel K.P., Cheah K.S., Pelling A.L., Leung K.K., Tang A.S., Bell D.M., Tease C., Lovell-Badge R., Steel K.P., Cheah K.S., Leung K.K., Tang A.S., Bell D.M., Tease C., Lovell-Badge R., Steel K.P., Cheah K.S., Tang A.S., Bell D.M., Tease C., Lovell-Badge R., Steel K.P., Cheah K.S., Bell D.M., Tease C., Lovell-Badge R., Steel K.P., Cheah K.S., Tease C., Lovell-Badge R., Steel K.P., Cheah K.S., Lovell-Badge R., Steel K.P., Cheah K.S., Steel K.P., Cheah K.S., Cheah K.S. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434:1031–1035. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- Kimura-Yoshida C., Kitajima K., Oda-Ishii I., Tian E., Suzuki M., Yamamoto M., Suzuki T., Kobayashi M., Aizawa S., Matsuo I., Kitajima K., Oda-Ishii I., Tian E., Suzuki M., Yamamoto M., Suzuki T., Kobayashi M., Aizawa S., Matsuo I., Oda-Ishii I., Tian E., Suzuki M., Yamamoto M., Suzuki T., Kobayashi M., Aizawa S., Matsuo I., Tian E., Suzuki M., Yamamoto M., Suzuki T., Kobayashi M., Aizawa S., Matsuo I., Suzuki M., Yamamoto M., Suzuki T., Kobayashi M., Aizawa S., Matsuo I., Yamamoto M., Suzuki T., Kobayashi M., Aizawa S., Matsuo I., Suzuki T., Kobayashi M., Aizawa S., Matsuo I., Kobayashi M., Aizawa S., Matsuo I., Aizawa S., Matsuo I., Matsuo I. Characterization of the pufferfish Otx2 cis-regulators reveals evolutionarily conserved genetic mechanisms for vertebrate head specification. Development. 2004;131:57–71. doi: 10.1242/dev.00877. [DOI] [PubMed] [Google Scholar]
- Kleinjan D.A., van Heyningen V., van Heyningen V. Long-range control of gene expression: Emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehoczky J.A., Williams M.E., Innis J.W., Williams M.E., Innis J.W., Innis J.W. Conserved expression domains for genes upstream and within the HoxA and HoxD clusters suggests a long-range enhancer existed before cluster duplication. Evol. Dev. 2004;6:423–430. doi: 10.1111/j.1525-142X.2004.04050.x. [DOI] [PubMed] [Google Scholar]
- Lettice L.A., Heaney S.J., Purdie L.A., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E., Heaney S.J., Purdie L.A., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E., Purdie L.A., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E., Goode D., Elgar G., Hill R.E., de Graaff E., Elgar G., Hill R.E., de Graaff E., Hill R.E., de Graaff E., de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- Liberg D., Sigvardsson M., Akerblad P., Sigvardsson M., Akerblad P., Akerblad P. The EBF/Olf/Collier family of transcription factors: Regulators of differentiation in cells originating from all three embryonal germ layers. Mol. Cell. Biol. 2002;22:8389–8397. doi: 10.1128/MCB.22.24.8389-8397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien C.L., McAnally J., Richardson J.A., Olson E.N., McAnally J., Richardson J.A., Olson E.N., Richardson J.A., Olson E.N., Olson E.N. Cardiac-specific activity of an Nkx2-5 enhancer requires an evolutionarily conserved Smad binding site. Dev. Biol. 2002;244:257–266. doi: 10.1006/dbio.2002.0603. [DOI] [PubMed] [Google Scholar]
- Long Q., Park B.K., Ekker M., Park B.K., Ekker M., Ekker M. Expression and regulation of mouse Mtsh1 during limb and branchial arch development. Dev. Dyn. 2001;222:308–312. doi: 10.1002/dvdy.1176. [DOI] [PubMed] [Google Scholar]
- Manfroid I., Caubit X., Kerridge S., Fasano L., Caubit X., Kerridge S., Fasano L., Kerridge S., Fasano L., Fasano L. Three putative murine Teashirt orthologues specify trunk structures in Drosophila in the same way as the Drosophila teashirt gene. Development. 2004;131:1065–1073. doi: 10.1242/dev.00977. [DOI] [PubMed] [Google Scholar]
- Margulies E.H., Blanchette M., Haussler D., Green E.D., Blanchette M., Haussler D., Green E.D., Haussler D., Green E.D., Green E.D. NISC Comparative Sequencing Program. Identification and characterization of multi-species conserved sequences. Genome Res. 2003;13:2507–2518. doi: 10.1101/gr.1602203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J., Han C., Gordon L.A., Terry A., Prabhakar S., She X., Xie G., Hellsten U., Chan Y.M., Altherr M., Han C., Gordon L.A., Terry A., Prabhakar S., She X., Xie G., Hellsten U., Chan Y.M., Altherr M., Gordon L.A., Terry A., Prabhakar S., She X., Xie G., Hellsten U., Chan Y.M., Altherr M., Terry A., Prabhakar S., She X., Xie G., Hellsten U., Chan Y.M., Altherr M., Prabhakar S., She X., Xie G., Hellsten U., Chan Y.M., Altherr M., She X., Xie G., Hellsten U., Chan Y.M., Altherr M., Xie G., Hellsten U., Chan Y.M., Altherr M., Hellsten U., Chan Y.M., Altherr M., Chan Y.M., Altherr M., Altherr M., et al. The sequence and analysis of duplication-rich human chromosome 16. Nature. 2004;432:988–994. doi: 10.1038/nature03187. [DOI] [PubMed] [Google Scholar]
- McLysaght A., Enright A.J., Skrabanek L., Wolfe K.H., Enright A.J., Skrabanek L., Wolfe K.H., Skrabanek L., Wolfe K.H., Wolfe K.H. Estimation of synteny conservation and genome compaction between pufferfish (Fugu) and human. Yeast. 2000;17:22–36. doi: 10.1002/(SICI)1097-0061(200004)17:1<22::AID-YEA5>3.0.CO;2-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A., Hokamp K., Wolfe K.H., Hokamp K., Wolfe K.H., Wolfe K.H. Extensive genomic duplication during early chordate evolution. Nat. Genet. 2002;31:200–204. doi: 10.1038/ng884. [DOI] [PubMed] [Google Scholar]
- Nagai T., Aruga J., Takada S., Gunther T., Sporle R., Schughart K., Mikoshiba K., Aruga J., Takada S., Gunther T., Sporle R., Schughart K., Mikoshiba K., Takada S., Gunther T., Sporle R., Schughart K., Mikoshiba K., Gunther T., Sporle R., Schughart K., Mikoshiba K., Sporle R., Schughart K., Mikoshiba K., Schughart K., Mikoshiba K., Mikoshiba K. The expression of the mouse Zic1, Zic2, and Zic3 gene suggests an essential role for Zic genes in body pattern formation. Dev. Biol. 1997;182:299–313. doi: 10.1006/dbio.1996.8449. [DOI] [PubMed] [Google Scholar]
- Nakamura M., Runko A.P., Sagerstrom C.G., Runko A.P., Sagerstrom C.G., Sagerstrom C.G. A novel subfamily of zinc finger genes involved in embryonic development. J. Cell. Biochem. 2004;93:887–895. doi: 10.1002/jcb.20255. [DOI] [PubMed] [Google Scholar]
- Nobrega M.A., Ovcharenko I., Afzal V., Rubin E.M., Ovcharenko I., Afzal V., Rubin E.M., Afzal V., Rubin E.M., Rubin E.M. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by Gene Duplication. Springer Verlag; New York.: 1970. [Google Scholar]
- Ovcharenko I., Loots G.G., Nobrega M.A., Hardison R.C., Miller W., Stubbs L., Loots G.G., Nobrega M.A., Hardison R.C., Miller W., Stubbs L., Nobrega M.A., Hardison R.C., Miller W., Stubbs L., Hardison R.C., Miller W., Stubbs L., Miller W., Stubbs L., Stubbs L. Evolution and functional classification of vertebrate gene deserts. Genome Res. 2005;15:137–145. doi: 10.1101/gr.3015505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulou G., Hennig S., Groth D., Krause A., Poustka A.J., Herwig R., Vingron M., Lehrach H., Hennig S., Groth D., Krause A., Poustka A.J., Herwig R., Vingron M., Lehrach H., Groth D., Krause A., Poustka A.J., Herwig R., Vingron M., Lehrach H., Krause A., Poustka A.J., Herwig R., Vingron M., Lehrach H., Poustka A.J., Herwig R., Vingron M., Lehrach H., Herwig R., Vingron M., Lehrach H., Vingron M., Lehrach H., Lehrach H. New evidence for genome-wide duplications at the origin of vertebrates using an amphioxus gene set and completed animal genomes. Genome Res. 2003;13:1056–1066. doi: 10.1101/gr.874803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl B.S., Schon C., Rossner A., Knochel W., Schon C., Rossner A., Knochel W., Rossner A., Knochel W., Knochel W. The FoxO-subclass in Xenopus laevisdevelopment. Gene Expr. Patterns. 2004;5:187–192. doi: 10.1016/j.modgep.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Poulin F., Nobrega M.A., Plajzer-Frick I., Holt A., Afzal V., Rubin E.M., Pennacchio L.A., Nobrega M.A., Plajzer-Frick I., Holt A., Afzal V., Rubin E.M., Pennacchio L.A., Plajzer-Frick I., Holt A., Afzal V., Rubin E.M., Pennacchio L.A., Holt A., Afzal V., Rubin E.M., Pennacchio L.A., Afzal V., Rubin E.M., Pennacchio L.A., Rubin E.M., Pennacchio L.A., Pennacchio L.A. In vivo characterization of a vertebrate ultraconserved enhancer. Genomics. 2005;85:774–781. doi: 10.1016/j.ygeno.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Püschel A.W., Betz H., Betz H. Neurexins are differentially expressed in the embryonic nervous system of mice. J. Neurosci. 1995;15:2849–2856. doi: 10.1523/JNEUROSCI.15-04-02849.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex M., Orme A., Uwanogho D., Tointon K., Wigmore P.M., Sharpe P.T., Scotting P.J., Orme A., Uwanogho D., Tointon K., Wigmore P.M., Sharpe P.T., Scotting P.J., Uwanogho D., Tointon K., Wigmore P.M., Sharpe P.T., Scotting P.J., Tointon K., Wigmore P.M., Sharpe P.T., Scotting P.J., Wigmore P.M., Sharpe P.T., Scotting P.J., Sharpe P.T., Scotting P.J., Scotting P.J. Dynamic expression of chicken Sox2 and Sox3 genes in ectoderm induced to form neural tissue. Dev. Dyn. 1997;209:323–332. doi: 10.1002/(SICI)1097-0177(199707)209:3<323::AID-AJA7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Rimini R., Beltrame M., Argenton F., Szymczak D., Cotelli F., Bianchi M.E., Beltrame M., Argenton F., Szymczak D., Cotelli F., Bianchi M.E., Argenton F., Szymczak D., Cotelli F., Bianchi M.E., Szymczak D., Cotelli F., Bianchi M.E., Cotelli F., Bianchi M.E., Bianchi M.E. Expression patterns of zebrafish sox11A, sox11B and sox21. Mech. Dev. 1999;89:167–171. doi: 10.1016/s0925-4773(99)00199-9. [DOI] [PubMed] [Google Scholar]
- Sandelin A., Bailey P., Bruce S., Engstrom P.G., Klos J.M., Wasserman W.W., Ericson J., Lenhard B., Bailey P., Bruce S., Engstrom P.G., Klos J.M., Wasserman W.W., Ericson J., Lenhard B., Bruce S., Engstrom P.G., Klos J.M., Wasserman W.W., Ericson J., Lenhard B., Engstrom P.G., Klos J.M., Wasserman W.W., Ericson J., Lenhard B., Klos J.M., Wasserman W.W., Ericson J., Lenhard B., Wasserman W.W., Ericson J., Lenhard B., Ericson J., Lenhard B., Lenhard B. Arrays of ultraconserved noncoding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics. 2004;5:99. doi: 10.1186/1471-2164-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser G., Ahrens K., Ahrens K. Molecular anatomy of placode development in Xenopus laevis. Dev. Biol. 2004;271:439–466. doi: 10.1016/j.ydbio.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Seoighe C. Turning the clock back on ancient genome duplication. Curr. Opin. Genet. Dev. 2003;13:636–643. doi: 10.1016/j.gde.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Sharpe J., Nonchev S., Gould A., Whiting J., Krumlauf R., Nonchev S., Gould A., Whiting J., Krumlauf R., Gould A., Whiting J., Krumlauf R., Whiting J., Krumlauf R., Krumlauf R. Selectivity, sharing and competitive interactions in the regulation of Hoxb genes. EMBO J. 1998;17:1788–1798. doi: 10.1093/emboj/17.6.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F., Gonzalez F., Duboule D., Gonzalez F., Duboule D., Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- Swamynathan S.K., Piatigorsky J., Piatigorsky J. Orientation-dependent influence of an intergenic enhancer on the promoter activity of the divergently transcribed mouse Shsp/α B-crystallin and Mkbp/HspB2 genes. J. Biol. Chem. 2002;277:49700–49706. doi: 10.1074/jbc.M209700200. [DOI] [PubMed] [Google Scholar]
- Tamura S., Morikawa Y., Iwanishi H., Hisaoka T., Senba E., Morikawa Y., Iwanishi H., Hisaoka T., Senba E., Iwanishi H., Hisaoka T., Senba E., Hisaoka T., Senba E., Senba E. Expression pattern of the winged-helix/forkhead transcription factor Foxp1 in the developing central nervous system. Gene Expr. Patterns. 2003;3:193–197. doi: 10.1016/s1567-133x(03)00003-6. [DOI] [PubMed] [Google Scholar]
- Tarpey P., Parnau J., Blow M., Woffendin H., Bignell G., Cox C., Cox J., Davies H., Edkins S., Holden S., Parnau J., Blow M., Woffendin H., Bignell G., Cox C., Cox J., Davies H., Edkins S., Holden S., Blow M., Woffendin H., Bignell G., Cox C., Cox J., Davies H., Edkins S., Holden S., Woffendin H., Bignell G., Cox C., Cox J., Davies H., Edkins S., Holden S., Bignell G., Cox C., Cox J., Davies H., Edkins S., Holden S., Cox C., Cox J., Davies H., Edkins S., Holden S., Cox J., Davies H., Edkins S., Holden S., Davies H., Edkins S., Holden S., Edkins S., Holden S., Holden S., et al. Mutations in the DLG3 gene cause nonsyndromic X-linked mental retardation. Am. J. Hum. Genet. 2004;75:318–324. doi: 10.1086/422703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.S., Braasch I., Frickey T., Meyer A., de Van Peer Y., Braasch I., Frickey T., Meyer A., de Van Peer Y., Frickey T., Meyer A., de Van Peer Y., Meyer A., de Van Peer Y., de Van Peer Y. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J., Higgins D.G., Gibson T.J., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama R., Gomez D.M., Mana M.D., Dawid I.B., Gomez D.M., Mana M.D., Dawid I.B., Mana M.D., Dawid I.B., Dawid I.B. Sequence relationships and expression patterns of zebrafish zic2 and zic5 genes. Gene Expr. Patterns. 2004;4:345–350. doi: 10.1016/j.modgep.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Tsai R.Y., Reed R.R., Reed R.R. Identification of DNA recognition sequences and protein interaction domains of the multiple-Zn-finger protein Roaz. Mol. Cell. Biol. 1998;18:6447–6456. doi: 10.1128/mcb.18.11.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchikawa M., Kamachi Y., Kondoh H., Kamachi Y., Kondoh H., Kondoh H. Two distinct subgroups of Group B Sox genes for transcriptional activators and repressors: Their expression during embryonic organogenesis of the chicken. Mech. Dev. 1999;84:103–120. doi: 10.1016/s0925-4773(99)00083-0. [DOI] [PubMed] [Google Scholar]
- Ureta-Vidal A., Ettwiller L., Birney E., Ettwiller L., Birney E., Birney E. Comparative genomics: Genome-wide analysis in metazoan eukaryotes. Nat. Rev. Genet. 2003;4:251–262. doi: 10.1038/nrg1043. [DOI] [PubMed] [Google Scholar]
- Vandepoele K., De Vos W., Taylor J.S., Meyer A., de Van Peer Y., De Vos W., Taylor J.S., Meyer A., de Van Peer Y., Taylor J.S., Meyer A., de Van Peer Y., Meyer A., de Van Peer Y., de Van Peer Y. Major events in the genome evolution of vertebrates: Paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc. Natl. Acad. Sci. 2000;101:1638–1643. doi: 10.1073/pnas.0307968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavouri T., McEwen G.K., Woolfe A., Gilks W.R., Elgar G., McEwen G.K., Woolfe A., Gilks W.R., Elgar G., Woolfe A., Gilks W.R., Elgar G., Gilks W.R., Elgar G., Elgar G. Defining a genomic radius for long-range enhancer action: Duplicated conserved non-coding elements hold the key. Trends Genet. 2006;22:5–10. doi: 10.1016/j.tig.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Wang B., Weidenfeld J., Lu M.M., Maika S., Kuziel W.A., Morrisey E.E., Tucker P.W., Weidenfeld J., Lu M.M., Maika S., Kuziel W.A., Morrisey E.E., Tucker P.W., Lu M.M., Maika S., Kuziel W.A., Morrisey E.E., Tucker P.W., Maika S., Kuziel W.A., Morrisey E.E., Tucker P.W., Kuziel W.A., Morrisey E.E., Tucker P.W., Morrisey E.E., Tucker P.W., Tucker P.W. Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocyte proliferation and maturation. Development. 2004;131:4477–4487. doi: 10.1242/dev.01287. [DOI] [PubMed] [Google Scholar]
- Warner S.J., Hutson M.R., Oh S.H., Gerlach-Bank L.M., Lomax M.I., Barald K.F., Hutson M.R., Oh S.H., Gerlach-Bank L.M., Lomax M.I., Barald K.F., Oh S.H., Gerlach-Bank L.M., Lomax M.I., Barald K.F., Gerlach-Bank L.M., Lomax M.I., Barald K.F., Lomax M.I., Barald K.F., Barald K.F. Expression of ZIC genes in the development of the chick inner ear and nervous system. Dev. Dyn. 2003;226:702–712. doi: 10.1002/dvdy.10262. [DOI] [PubMed] [Google Scholar]
- Wong A., Vallender E.J., Heretis K., Ilkin Y., Lahn B.T., Martin C.L., Ledbetter D.H., Vallender E.J., Heretis K., Ilkin Y., Lahn B.T., Martin C.L., Ledbetter D.H., Heretis K., Ilkin Y., Lahn B.T., Martin C.L., Ledbetter D.H., Ilkin Y., Lahn B.T., Martin C.L., Ledbetter D.H., Lahn B.T., Martin C.L., Ledbetter D.H., Martin C.L., Ledbetter D.H., Ledbetter D.H. Diverse fates of paralogs following segmental duplication of telomeric genes. Genomics. 2004;84:239–247. doi: 10.1016/j.ygeno.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Wood H.B., Episkopou V., Episkopou V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech. Dev. 1999;86:197–201. doi: 10.1016/s0925-4773(99)00116-1. [DOI] [PubMed] [Google Scholar]
- Woods I.G., Kelly P.D., Chu F., Ngo-Hazelett P., Yan Y.L., Huang H., Postlewait J.H., Talbot W.S., Kelly P.D., Chu F., Ngo-Hazelett P., Yan Y.L., Huang H., Postlewait J.H., Talbot W.S., Chu F., Ngo-Hazelett P., Yan Y.L., Huang H., Postlewait J.H., Talbot W.S., Ngo-Hazelett P., Yan Y.L., Huang H., Postlewait J.H., Talbot W.S., Yan Y.L., Huang H., Postlewait J.H., Talbot W.S., Huang H., Postlewait J.H., Talbot W.S., Postlewait J.H., Talbot W.S., Talbot W.S. A comparative map of the zebrafish genome. Genome Res. 2000;10:1903–1914. doi: 10.1101/gr.10.12.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A., Goodson M., Goode K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Goodson M., Goode K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Goode K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Smith S.F., North P., Callaway H., Kelly K., North P., Callaway H., Kelly K., Callaway H., Kelly K., Kelly K., et al. Highly conserved noncoding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerucha T., Stuhmer T., Hatch G., Park B.K., Long Q., Yu G., Gambarotta A., Schultz J.R., Rubenstein J.L., Ekker M., Stuhmer T., Hatch G., Park B.K., Long Q., Yu G., Gambarotta A., Schultz J.R., Rubenstein J.L., Ekker M., Hatch G., Park B.K., Long Q., Yu G., Gambarotta A., Schultz J.R., Rubenstein J.L., Ekker M., Park B.K., Long Q., Yu G., Gambarotta A., Schultz J.R., Rubenstein J.L., Ekker M., Long Q., Yu G., Gambarotta A., Schultz J.R., Rubenstein J.L., Ekker M., Yu G., Gambarotta A., Schultz J.R., Rubenstein J.L., Ekker M., Gambarotta A., Schultz J.R., Rubenstein J.L., Ekker M., Schultz J.R., Rubenstein J.L., Ekker M., Rubenstein J.L., Ekker M., Ekker M. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J. Neurosci. 2000;20:709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Schwartz S., Wagner L., Miller W., Schwartz S., Wagner L., Miller W., Wagner L., Miller W., Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]