Abstract
The exponential growth of pathogen nucleic acid sequences available in public domain databases has invited their direct use in pathogen detection, identification, and surveillance strategies. DNA microarray technology has offered the potential for the direct DNA sequence analysis of a broad spectrum of pathogens of interest. However, to achieve the practical attainment of this potential, numerous technical issues, especially nucleic acid amplification, probe specificity, and interpretation strategies of sequence detection, need to be addressed. In this report, we demonstrate an approach that combines the use of a custom-designed Affymetrix resequencing Respiratory Pathogen Microarray (RPM v.1) with methods for microbial nucleic acid enrichment, random nucleic acid amplification, and automated sequence similarity searching for broad-spectrum respiratory pathogen surveillance. Successful proof-of-concept experiments, utilizing clinical samples obtained from patients presenting adenovirus or influenza virus-induced febrile respiratory illness (FRI), demonstrate the ability of this approach for correct species- and strain-level identification with unambiguous statistical interpretation at clinically relevant sensitivity levels. Our results underscore the feasibility of using this approach to expedite the early surveillance of diseases, and provide new information on the incidence of multiple pathogens.
The critical need for advanced infectious diagnostic and surveillance systems has taken on a new urgency with increased concerns over bioterrorism agents as well as natural pathogens (e.g., Bacillus anthracis, coronavirus, avian influenza virus). A DNA microarray platform that can simultaneously detect and characterize many different types of human pathogens that cause similar symptoms provides considerable potential for both medical use and national defense purposes (Bodrossy and Sessitsch 2004; Cleland et al. 2004). DNA microarrays do this by simultaneously interrogating hundreds to thousands of immobilized probe DNA oligonucleotides, where each probe provides a single query for a known sequence that is unique for an organism or trait. Using DNA microarrays for pathogen detection has gained prominence leading to an explosive growth of research (Bryant et al. 2004).
The effective use of microarrays for pathogen detection requires the optimization of several factors, such as sample amplification, probe specificity, and interpretation strategy in order to obtain unambiguous and reproducible results (Striebel et al. 2003). A major technical hurdle that limits the straightforward application of DNA microarrays to broad-spectrum pathogen diagnostics has been the requirement of specific amplification reagents and protocols (primarily PCR) to amplify chosen targets prior to microarray hybridization (López et al. 2003; Striebel et al. 2003). A few random amplification strategies in conjunction with spotted microarrays have been developed using multiple rounds of amplification to detect a broad spectrum of pathogens in complex biological samples (Wang et al. 2002; Vora et al. 2004).
Another hurdle to using spotted microarrays is that the design of specific oligonucleotide probes for pathogen identification is dependent on assumptions regarding target sequence composition. Long (50–70mer) oligonucleotide probes used in most prior studies have the disadvantage of decreased specificity (threshold for differentiation at 75%–87% sequence similarity), making it necessary to target multiple markers and rely on hybridization patterns for pathogen identification, which can lead to unquantifiable errors (Bodrossy and Sessitsch 2004). Nevertheless, these microarrays have provided a successful platform for screening a large number of pathogens at a viral family level via the use of highly conserved and hybridization mismatch-tolerant 70mer oligonucleotides (Wang et al. 2002, 2003). An additional problem with this format is that cross-hybridization occurs when two sequences share a high degree of similarity (Kothapalli et al. 2002). Careful data interpretation is needed to differentiate subtypes of pathogens using spotted microarrays and hybridization patterns. This approach does not produce direct genomic sequence as an output, but requires manual isolation and conventional DNA sequencing of captured pathogen targets (Wang et al. 2003). Thus, it is obvious that any incorporation of these concepts into a broad-spectrum diagnostic device for hundreds of pathogenic microorganisms and their variants will require a significant reduction in design, processing, and analysis steps.
The exponentially increasing availability of microbial sequences makes it possible to envision the use of direct sequence for routine pathogen diagnostics and surveillance; however, this requires that pathogen sequence information be rapidly obtained. “Resequencing” microarrays use “tiled” sets of 105 to 106 probes of either 25mers or 29mers, containing one perfectly matched and three mismatched probes per base for both strands of target genes (Hacia 1999). This array-based format, combined with specific PCR, has proven ideal for single nucleotide polymorphism (SNP) genotyping and phylogenetic analysis (Kozal et al. 1996; Gingeras et al. 1998; K. Wilson et al. 2002; W. Wilson et al. 2002; Wong et al. 2004). Because several types of variations (especially insertion/deletion or frequent multiple substitutions) in pathogen sequence can perturb hybridization patterns, these approaches used differential measures of specific pathogen hybridization patterns to identify individual sequence variants. That is, identifications require a priori knowledge of a differential hybridization pattern that is empirically determined in control experiments. Even when control experiments are carried out, these characteristic and conserved hybridization patterns do not always occur with highly diverse pathogen targets obtained from clinical specimens.
In this study, our overall objective was to demonstrate the utility of a resequencing microarray approach for simultaneous detection of respiratory pathogens in a format that can be used in a clinical environment without requiring the design of pathogen-specific PCR primers (W. Wilson et al. 2002) or fixed hybridization patterns (Gingeras et al. 1998). We chose to use a custom-designed Affymetrix resequencing Respiratory Pathogen Microarray (RPM v.1). Furthermore, we developed a method for automatic assembly of incomplete and disconnected pathogen sequence data into cumulative sequences amenable for similarity-based (e.g., Basic Local Alignment Search Tool-BLAST, Altschul et al. 1990) identification. The combination of a resequencing microarray with the application of statistical metrics to the raw output of the assay can allow unambiguous and reproducible sequence-based pathogen identification from clinical specimens. Our results demonstrate the feasibility of this approach for correct species- and strain-level identification with unambiguous statistical interpretation of adenovirus and influenza A strains at clinically relevant sensitivity levels. This report further suggests the feasibility of using this technology for broad-spectrum surveillance of respiratory pathogens, while providing new information on the incidence of pathogen coinfection.
Results
Specificity of the RPM v.1 chip
The accuracy of RPM v.1 chips for sequence-specific pathogen detection was validated using clinical and/or controlled laboratory samples. Samples were amplified with either pathogen-specific PCR or random amplification strategies and then hybridized to the arrays. To test whether prototype tile regions could be used for the identification of a broad number of variants without relying on predetermined hybridization patterns, we used febrile respiratory illness (FRI)-causing adenoviruses (HAdV) as our model system. The capability of the RPM v.1 to discriminate HAdV serotypes was tested by interrogating degenerate PCR amplicons (Lin et al. 2004) from different HAdV strains that were fully sequenced by members of the Epidemic Outbreak Surveillance (EOS) Consortium. The amplicons were hybridized to the chip, and GDAS software (Affymetrix Inc.) was used to generate sequence calls by comparing the respective hybridization intensities. The parameters of the nucleotide base call algorithm (Cutler et al. 2001) within GDAS were set to allow the maximum number of base calls (Permissive Base Calling Algorithm Setting, Supplemental data). Once generated, the primary nucleotide sequence produced was filtered and subjected to sequence similarity searching using the Respiratory Pathogen Identifier (REPI) software developed by our group. For this report, we examined the results from REPI and counted a sample positive for a specific pathogen if at least one subsequence from the pathogen’s prototype tile region produced BLAST returns where the return with lowest expected (E) value was a match for the specific pathogen. E-values from multiple records that have the same score but indicate different strains generate ambiguous strain identification, but could still be unambiguous at the serotype level. This approach allows one or more individual subsequences to be used for pathogen identification without relying on the design of strain-specific probes and hybridization pattern recognition (Gingeras et al. 1998), and permits highly variable hybridization patterns to produce the same order of sequence rankings.
Our results demonstrate that the tile regions of HAdV-4, HAdV-5, and HAdV-7 of RPM v.1 can differentiate various FRI-associated HAdV strains (Table 1), and prove that prototype tile regions can be used for identifying a broad range of variants. Strain-level identification was obtained in all cases except for subgroup B2 strains that were identified only as belonging to that subgroup. In a similar fashion, the remaining tiled regions on the RPM v.1 were successfully validated with the exception of those for West Nile virus. For these validation tests, control laboratory strains (Table 2; Supplemental data) were used, except in the case of influenza A virus H5N1 tiled regions. Instead, total RNA obtained from a patient infected with influenza A-H5N1 in Southeast Asia was used for validating the H5N1 tile regions (Table 1). Each pathogen was validated with at least three independent amplifications. Our results reproducibly revealed that prototype reference regions exhibited little or no discernible cross-hybridization, and interference from one pathogen with one of the others never caused an erroneous identification. Similar results were obtained with either specific or random amplification (data not shown). No false positives were obtained due to microarray base call or analysis errors.
Table 1.
Differentiation of various FRI-causing pathogens with RPM v.1
aGenBank accession numbers obtained from BLAST results are shown in parentheses. Each sample was validated with at least three independent amplifications.
bSample showed the following multiple accession numbers, BK001454, AJ250783, AJ250786, AY271307, tied with highest probability score. All the found matches in each sample indicate subgroup B2 adenovirus members.
Table 2.
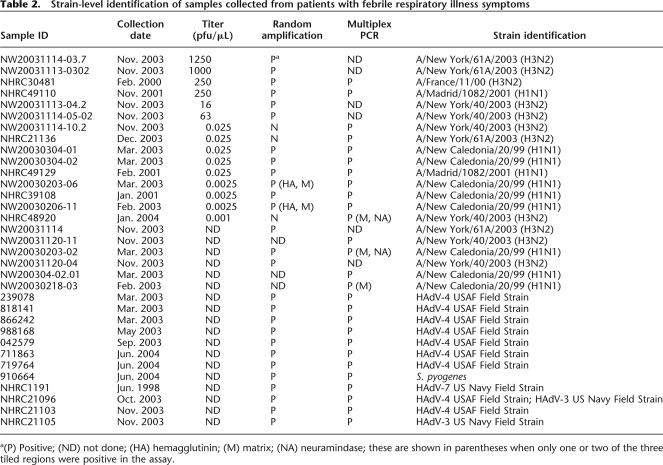
Strain-level identification of samples collected from patients with febrile respiratory illness symptoms
a(P) Positive; (ND) not done; (HA) hemagglutinin; (M) matrix; (NA) neuramindase; these are shown in parentheses when only one or two of the three tiled regions were positive in the assay.
RPM v.1 process development
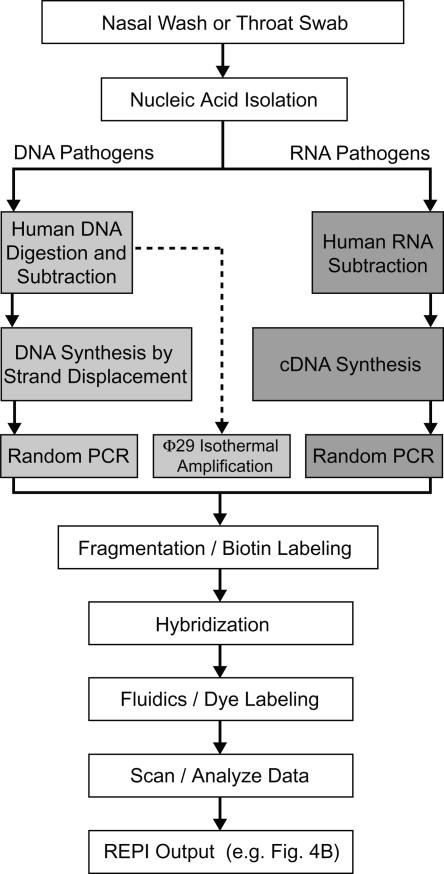
For resequencing (i.e., genotyping) applications, the Affymetrix GeneChip system and attendant protocols were optimized for highly accurate detection of SNPs by using specific PCR amplification. In order to achieve unbiased pathogen detection with RPM v.1, a random amplification strategy was developed. Rather than the random amplification protocols previously developed that require multiple amplification steps (Wang et al. 2002; Vora et al. 2004), a protocol that removes human nucleic acid (NA) first and then uses a single amplification step was developed. This protocol was combined with back-end automated sequence similarity searching to further simplify the microarray-based diagnostic strategy. This process allows for sensitive and unbiased microarray-based identification of respiratory tract pathogens (Fig. 1). Two common respiratory viral pathogens known to cause FRI outbreaks at military training facilities, HAdV-4 (DNA virus) and influenza A virus (negative-strand RNA virus), were used to test this process in a clinically relevant system. Assay sensitivity and specificity assessments were based on clinical nasal washes and throat swab samples obtained from the Naval Health Research Center (NHRC), San Diego, CA, or Lackland Air Force Base, San Antonio, TX.
Figure 1.
RPM v.1 process diagram. This diagram shows the process for each step when using RPM v.1 with a random amplification strategy for clinical samples.
To enhance detection sensitivity, separate human DNA and RNA subtraction pathways coupled with random NA amplification were developed. For DNA targets (e.g., HAdV-4), the isolated total NA from nasal wash or throat swab specimens was first subjected to McrBC enzymatic digestion at methylated CpG sites (Panne et al. 1999), reducing the human genomic DNA to a manageable size (≤10 kb), followed by the subtraction of repetitive sequences using Cot-1 human DNA (Fig. 1). The remaining DNA was subjected to whole-genome amplification (Lovmar et al. 2003) and routinely allowed full or partial detection of the E1A, hexon, and fiber genes at target concentrations of 103 copies/μL of the starting clinical sample. For RNA pathogen targets, non-human RNA species were enriched by the capture and removal of human 18S rRNA, 28S rRNA, and polyadenylatedmRNAs (MICROBEnrich, Ambion Inc.). Subsequent amplification of the enriched RNA, using a modified random reverse transcription PCR from a previously described method (Wang et al. 2002, 2003; Kessler et al. 2004) led to reproducible detection sensitivities of 2.5 × 10−3 plaque-forming units/μL of influenza A virus in previously frozen nasal wash specimens. In comparison to the detection sensitivity without subtraction (adenovirus, 106 copies/μL; influenza A virus, 250 plaque-forming units/μL of the starting clinical sample), the combination of human NA background subtraction and random amplification of the remaining NA in clinical samples greatly increased the detection sensitivity without multiple amplification steps.
Capability of RPM v.1 for multiple pathogen detection
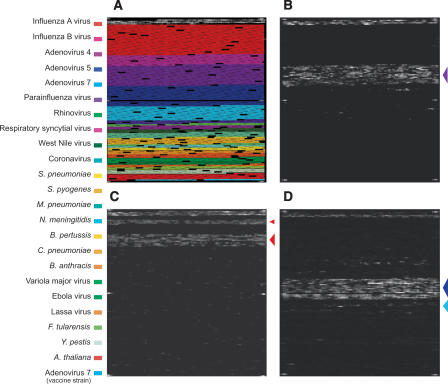
After successful proof-of-concept experiments, we tested the capability of RPM v.1 to discriminate pathogens. Figure 2B shows a raw data image generated by the hybridization of HAdV-4 (prototype strain RI-67) whole-genome random amplicons to the RPM v.1. It is of significance that no cross-hybridization resulted in base calls outside of the HAdV-4 tiled region. Similarly, a raw data image generated by the hybridization of randomly amplified NA from a clinical nasal wash sample was identified as influenza A virus (A/Fujian/411/2002) type H3N2 (Fig. 2C).
Figure 2.
Random amplification and resequencing microarray-based identification of two common respiratory tract viral pathogens. (A) RPM v.1 design overview. The tiled sequence regions for each of the targeted respiratory tract pathogens have been color-coded (left). (B) Hybridization profile of the HAdV-4 prototype strain RI-67. (C) Identification of an H3N2 influenza A virus (A/Fujian/411/2002) from Lackland AFB clinical nasal wash sample #NW20031114–03–7. (Upper arrow) Tile region for hemagglutinin (H3), (lower arrow) tile regions for neuraminidase (N2) and matrix. The black region interspersed between the two arrows constitutes the tile regions for hemagglutinin (H5) and neuraminidase (N1). (D) Identification of an HAdV-5/HAdV-21 coinfection in NHRC clinical throat swab sample #7151. The arrows on the right of each image are color coded according to the legend on the far left.
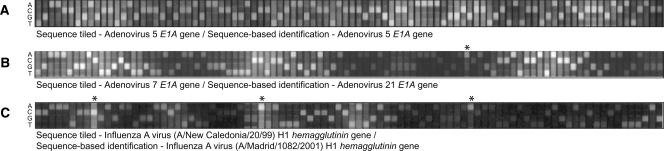
In addition to accurately identifying single pathogenic species, Figure 2D provides an example of another benefit of using this protocol for pathogen detection—the ability to detect co-infections without using reference control hybridizations for comparison. A throat swab specimen collected from a symptomatic patient previously vaccinated against HAdV-4 and HAdV-7 was shown to harbor adenovirus-specific DNA that hybridized specifically to the HAdV-5 and HAdV-7 prototype regions. The result of our analysis for subsequences from the HAdV-5 E1A gene (Fig. 3A) indicated the presence of HAdV-5 as expected. However, our analysis of the subsequences from the HAdV-7 E1A prototype region revealed that the subsequences were a match to HAdV-21 and not HAdV-7 (Fig. 3B; annotated genome sequence of HAdV-21, GenBank accession number AY601633). Similar results were also obtained from the HAdV-7 hexon and fiber gene prototype regions and strongly suggested the presence of two adenoviral species, HAdV-5 and HAdV-21. This finding was verified by several independent conventional and molecular adenovirus identification methods (G.J. Vora, B. Lin, K. Gratwick, C.E. Meador, C. Hansen, C. Tibbetts, D.A. Stenger, M. Irvine, D. Seto, A. Purkayastha, et al., in prep.).
Figure 3.
Examples of primary sequence data generated by the hybridization of randomly amplified targets to RPM v.1 tiled probe sets. Clinical throat swab sample NHRC #7151 was found to contain an adenoviral coinfection (Fig. 2D), as hybridization to the HAdV-5 E1A tiled prototype sequence region resulted in the identification of HAdV-5 E1A (A), whereas the sequence generated via hybridization to the HAdV-7 E1A tiled prototype sequence region suggested the presence of HAdV-21 (B). (C) Amplicons generated from clinical throat swab sample NHRC #49110 hybridized to the influenza A virus (A/New Caledonia/20/99) hemagglutinin (H1) gene prototype sequence but were identified (via REPI analysis) as influenza A virus (A/Madrid/1082/2001) H1N1. (*) Single nucleotides that differentiate the experimental sequence from the tiled prototype sequence.
For influenza virus strains, it is important not only to distinguish subtypes but also to identify the differences associated with significant shifts of the subtype from year to year. The accuracy of our microarray to identify these variations was demonstrated for an influenza A virus (Fig. 3C). Visual examination of the hybridization of amplicons from a clinical sample on the influenza A virus hemagglutinin (H1) gene prototype sequence correctly identified the presence of an influenza A virus. Sequence-based REPI analysis revealed the identity of the virus to be most nearly identical to subtype A/Madrid/1082/2001, another H1N1 strain that had been circulating during the same flu season as the A/New Caledonia/20/99 vaccine strain. This identification corresponded to identification made based on the sequence obtained using the conventional DNA sequencing. For every other clinical sample identified as influenza A virus H3 or H1 whose sequence was obtained using conventional DNA sequencing methods, the two methods identified strains that corresponded with each other (data not shown). The accuracy of the sequence information produced by the RPM v.1 for typing-level identification has been established and was not affected by either the reduced stringency settings of the Affymetrix base-calling algorithm or by the methods used to randomly amplify the pathogen targets from clinical specimens. A more detailed analysis of the accuracy of this microarray for specific strain identification compared with conventional sequencing for several influenza A and B strains is covered in a separate paper (Wang et al. 2006).
Preliminary clinical study
A preliminary clinical study was performed with samples collected from the NHRC and EOS team at Lackland Air Force Base. For comparison, two amplification strategies, random amplification and multiplex PCR, were employed with the microarray for the same set of clinical samples. As shown in Table 2, 21 influenza A virus culture-positive (15 nasal wash and six throat swab) samples were tested using both random and multiplex PCR amplification methods. Using multiplex PCR, 13 out of 13 samples were correctly diagnosed in comparison to the culture method. While using random amplification, 15 out of 18 samples were identified. RPM v.1 not only identified the samples as H3N2 and H1N1 subtype but also differentiated these samples, demonstrating the potential of the resequencing microarray. Using the hemagglutinin gene sequence as an example, sample NW20031114–03.7 collected in November 2003 was identified as A/New York/61A/2003, a relative of the dominant strain (A/Fujian/411/2002) for the 2003–2004 flu season, while sample NHRC30481, collected in February 2000, was identified as A/France/11/00 (H3N2) strain, a relative of the dominant strain (A/Panama/2007/99) for the 1999–2000 flu season. It is not surprising to see that samples collected from the same geographic region in the same season usually contained similar strains. Similar BLAST search results were observed from sequences generated for samples in which both random amplification and multiplex PCR were done, further suggesting that random amplification methods correspond well with multiplex PCR (Table 2). In addition, our assay correctly identified and typed 11 clinical samples (9-HAdV-4, 1-HAdV-3/HAdV-4 coinfection, and 1-Streptococcus pyogenes) when compared with traditional culture detection methods (Table 2). Two clinical samples that tested negative using conventional methods also did not have any pathogens detected when tested with our microarray (data not shown). These results demonstrate the ability of our microarray-based diagnostic to correctly identify and type clinically relevant HAdV and influenza A strains in a manner consistent with conventional culture detection.
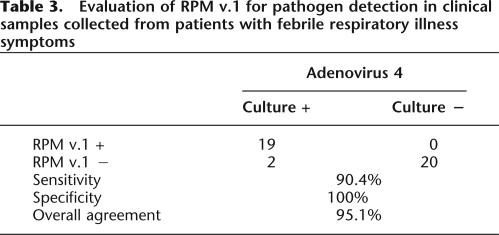
An additional study was carried out with clinical samples collected from the NHRC to further assess the utility of the microarray-based diagnostic for respiratory pathogen detection. The samples (n = 41) consisted of negative and positive throat swabs in viral transport medium from subjects with clinically documented respiratory illness. At the sites of collection, the samples were tested using conventional methods and sent to us in a coded fashion for testing. The experiments were conducted by two independent investigators, and the sample identities were revealed only after the resulting assessments had been finalized. The comparison demonstrated a complete concordance between our method and conventional methods for 19 of 21 samples (Table 3) for HAdV-4–positive clinical samples, and 20 of 20 negative samples. The failure to detect pathogens present in two HAdV-4 samples in this test and three influenza A samples in the first study, and the five false negatives, was probably due to the insufficient sensitivity of the current method. However, no false–positive results were obtained due to microarray base call or analysis errors. Future efforts will focus on improving assay sensitivity.
Table 3.
Evaluation of RPM v.1 for pathogen detection in clinical samples collected from patients with febrile respiratory illness symptoms
Discussion
We have demonstrated a straightforward approach that capitalizes on the ever-increasing availability of pathogen nucleic acid sequences that will provide strain-level information without relying on fixed hybridization patterns and does not rely on specific oligonucleotide sequences to amplify for specific targets. This approach not only allows simultaneous detection and differentiation of common circulating respiratory pathogens at clinically relevant sensitivity levels, but also enables us to identify coinfections and rarely encountered or typically unexpected pathogens. This technology does not require a priori knowledge of a differential hybridization pattern for pathogen identification. Thus, it is not necessary to build up a database of reference hybridization patterns through control experiments for differentiating subtypes of pathogens. Also, the false–positive rate caused by cross-hybridization when two sequences share a high degree of similarity was greatly reduced through this approach. In >300 experiments for all validated tiled sequences on the RPM v.1, no false–positive results were obtained when using clinical samples, and no misidentifications were made when using controlled laboratory samples (Table 1; Supplemental Fig. 1). Another salient benefit of this technique is demonstrated in the case of complex mixture samples, i.e., HAdV coinfection and Flu Vaccine (data not shown); the microarray not only distinguished the presence of two or more coinfectants, but also identified them correctly at the strain level. In all cases, the interpretation of a pathogen’s identity is much more straightforward.
While this system demonstrates remarkably low false–positive rates (a high specificity), the system remains somewhat limited in sensitivity as indicated by the false negatives. The sensitivity of the current system can detect adenovirus at target concentrations of 103 copies/μL of the starting clinical sample and influenza A virus at 2.5 × 10−3 plaque-forming units/μL using a combination of human NA background subtraction and random amplification of the remaining NA. Due to the use of specific primers and the exponential amplification of PCR, it is not surprising to see that the multiplex PCR is more sensitive than random amplification. Limitations in sensitivity can result in false negatives, especially if a patient is tested early or late in the infection or the pathogen of interest does not typically shed in high titer, such as HAdV or influenza A virus. Our future work will focus on improving assay sensitivity using more complete subtraction of background human DNA and RNA. Nevertheless, this assay may still be useful for some applications. This assay is not yet optimized for detailed strain-level identification, speed (∼12–16 h), or cost ($400 per chip, ∼$20 per pathogen species) relative to that desired for rapid infectious disease diagnostics. Improvements to processing methods (Lin et al. 2004) and the use of the next generation of resequencing chips will increase sequence content while substantially decreasing cost and time.
In comparison to the current state of the art, which would require multiple diagnostic tests to discern the offending agent, our assay is able not only to look for the most commonly occurring infectious agents, but also to survey for less common pathogens in a single test. The ability to order a single assay for effective differential diagnosis among the majority of pathogens causing FRI syndrome will increase the number of diagnoses made with far fewer assays. In a public health or an epidemic outbreak scenario, the ability to rapidly identify less common pathogens among a background of seasonal FRI will enable more effective identifications, leading to a better response to a naturally occurring epidemic outbreak or even a bioterrorism event. This information, combined with clinical symptom data and confirmatory laboratory tests, will result in more accurate disease reporting, decreased disease exposure, and improved outcomes for individuals and public health. Because viral agents cause most infections of the respiratory tract, it is satisfying that we could detect and type both DNA and RNA viruses from clinical samples using random amplification methods at clinically relevant sensitivity levels (Couch et al. 1966; Boivin et al. 2003). The success of the RPM v.1 has already led to the development and initial testing of a RPM v.2 that now includes 54 bacterial and viral species. Our future work will focus on improving sensitivity and assay speed in order to achieve high-throughput capability, which will provide a cost-effective diagnostic platform for pathogen detection and epidemic surveillance.
Methods
RPM v.1 design
The RPM v.1 (Fig. 2A) was designed primarily to test the hypothesis that a single tiled region could act as a prototype for the identification of a broad number of variants without relying on predetermined hybridization patterns. Prototype regions were selected to allow for both efficient hybridization and unique identification of most or all of a subtype of pathogenic species. (For probe tiling information of RPM v.1, see Supplemental Table 1). Two pathogens, HAdV and influenza A virus (H1N1, H3N2 and H5N1), were treated in much more detail. They were selected based upon recent outbreak information (Erdman et al. 2002; Kolavic-Gray et al. 2002; Thompson et al. 2003). Based on this, partial sequences from the E1A, hexon, and fiber genes containing diagnostic regions of adenovirus serotypes HAdV-4, HAdV-5, and HAdV-7 were tiled for the detection of all FRI-causing human adenoviruses. Similarly, tiled regions for influenza A virus detection were comprised of partial sequences from the hemagglutinin gene (subtypes H1, H3, and H5), the neuraminidase gene (subtypes N1 and N2), and the matrix gene. In addition to HAdV and influenza A virus, the current RPM design permits discrimination of 12 other common respiratory pathogens, and six Centers for Disease Control and Prevention category A bio-terrorism pathogens (Table 1) known to cause FRI, i.e., “flu-like” symptoms at early stage of infection.
Prototype strains
Detailed descriptions of all prototype and field strains used in this study and their sources are listed in Supplemental Table 2.
Clinical samples
Throat swabs were collected at the Molecular Biology Laboratory–NHRC (San Diego, CA) from patients with FRI symptoms and immediately placed in 2-mL cryogenic vials containing 1.5 mL of viral transport medium (VTM) to maintain the viral particles during transport. Nasal washes from the EOS team at Lackland Air Force Base were collected from basic military trainees with FRI symptoms. In both instances, samples were tested at the site of collection using conventional detection techniques and submitted for microarray-based detection in a masked fashion. The collection and transport of all clinical samples complied with the Wilford Hall Medical Center protocol for clinical investigation (FWH20020124H). Nucleic acid were extracted from clinical samples using the MasterPure DNA purification kit (Epicentre Technologies) following the manufacturer’s recommended protocol with slight modification; we omitted the RNase digestion step. Informed consents were obtained from all participants after the nature and possible consequences of the studies were explained.
Subtractive random amplification strategy
Bead-based subtraction steps were carried out to remove human genomic DNA and RNA from clinical samples. For human DNA removal, Cot Human DNA (Roche Applied Science), consisting largely of rapidly annealing repetitive elements, was labeled at the 3′ end with biotin-N6-ddATP (PerkinElmer Life Science, Inc.) with Terminal Transferase (New England Biolabs Inc.). Extracted NA from clinical samples was digested with 10 U of McrBC (New England Biolabs Inc.) for 20 min, then mixed with biotinylated Cot Human DNA. The reaction mixtures were brought to a final volume of 75 μL with a final concentration of 4× SSC and 0.2% SDS. The reaction mixtures were incubated for 10 min at 95°C, then slowly cooled to 65°C and incubated for 1 h. Streptavidin-coated magnetic beads (Bioclone Inc.) were then added to the mixture to capture hybridized human DNA. The enriched DNA in the supernatant was transferred to a fresh tube. Finally, DNA was precipitated with ethanol and subjected to a random amplification procedure (see below). For human RNA background subtraction, the MICROBEnrich (Ambion Inc.) kit was used following the manufacturer’s recommended protocol. RNA was precipitated with ethanol, then subjected to the random amplification procedure.
Random amplification for DNA samples was carried out with either bacteriophage ϕ29 DNA polymerase or the modified random amplification protocol from previously published papers (Wang et al. 2002, 2003). Briefly, DNA amplification utilizing bacteriophage ϕ29 DNA polymerase with random hexamers was performed according to the instructions of the GenomiPhi DNA Amplification Kit (Amersham Biosciences Corp.). DNA amplification utilizing modified random amplification was performed with an initial round of DNA synthesis using Sequenase version 2.0 DNA polymerase (United States Biochemical) and primer D, followed by PCR amplification with primer E. For RNA amplification, viral samples were amplified by a modified version of a random PCR protocol (Wang et al. 2002, 2003; Kessler et al. 2004). Briefly, 10 μL of total RNA was reverse transcribed by using primer D and superscript III reverse transcriptase (Invitrogen Corp.) and was then amplified by PCR with primer E. The random PCR reaction was carried out in a Peltier Thermal Cycler-PTC225 (MJ Research Inc.) with 40 cycles of: 30 sec at 94°C, 30 sec at 40°C, 30 sec at 50°C, 120 sec at 72°C; and a final extension for 7 min at 72°C.
For analysis of clinical specimens, samples were subjected to both DNA and RNA subtraction and amplification, then the amplified products were combined and subjected to purification and processing prior to hybridizing to the RPM v.1.
Multiplex RT–PCR
For influenza A viruses multiplex RT–PCR, the hemagglutinin, neuraminidase, and matrix genes were amplified with three sets of segment-specific primers (Bm-HA-1/Bm-NS-890R, Ba-Na-1/Ba-Na-1413R, and Bm-M-1/Bm-M-1027R; Hoffmann et al. 2001) and the HotStarTaq Multiplex PCR kit (Qiagen, Inc.). The amplification reaction was carried out in a Peltier Thermal Cycler-PTC225 (MJ Research Inc.) with an initial activation step for 15 min at 95°C, followed by 40 cycles of: 30 sec at 94°C, 90 sec at 58°C, 90 sec at 72°C; and a final extension for 10 min at 72°C. For adenovirus multiplex PCR, the amplification was carried out as previously described (Lin et al. 2004).
Microarray hybridization and processing
Microarray hybridization and processing were carried out according to the manufacturer’s recommended protocol (Affymetrix Inc.). After scanning, GCOS software is used to reduce the raw image (.DAT) file to a simplified file format (.CEL file) with intensities assigned to each of the corresponding probe positions. Finally, GDAS software is used to apply an embedded version of the ABACUS (Cutler et al. 2001) algorithm to produce an estimate of the correct base calls, comparing the respective intensities for the sense and antisense probe sets. To increase the percentage of base calls, we adjusted the parameters to allow the most permissive base calling (permissive setting, Supplemental data). The sequences from base calls made for each tiled region of the resequencing array then were exported from GDAS as the FASTA-formatted files.
Resequencing Pathogen Identifier (REPI)
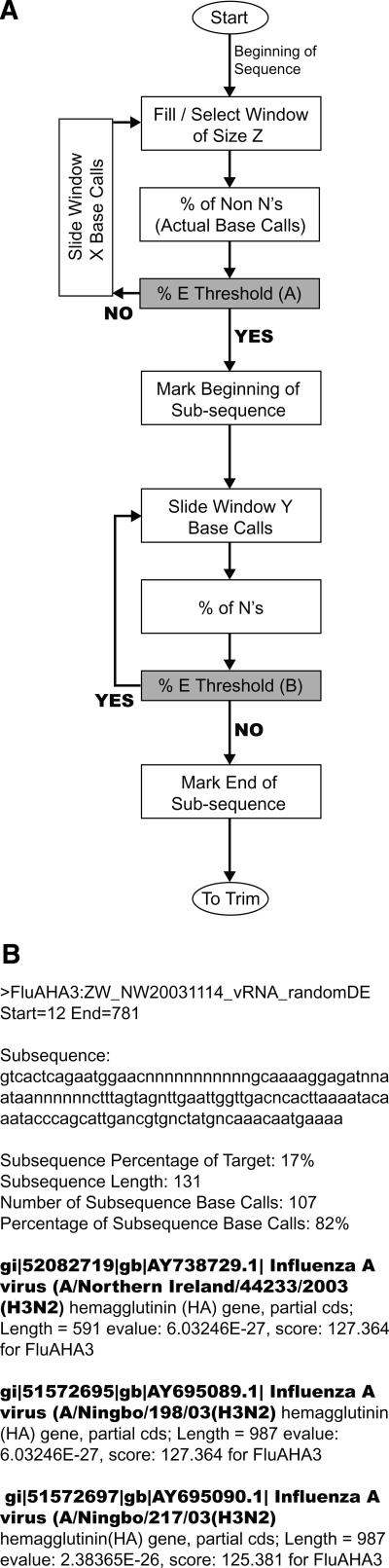
The Resequencing Pathogen Identification (REPI) software was developed and designed to filter the output of the FASTA file and perform sequence similarity searches using the NCBI BLASTN algorithm. The algorithm first removes control sequences, since they are intended only to indicate correct operation and are specifically designed by Affymetrix to be nonsense sequence; therefore, they will never have relevant returns. Next the sequence is evaluated for subsequences of usable data that will return a significant return from BLAST. A prototype tiled sequence is evaluated starting from the first base call. A window, m bases long, slides along the sequence searching for the first area that scores 25% or better, which is computed by dividing the total of valid bases in the window by the window’s length, m (Fig. 4A). Now that the start of usable data is determined, the program moves the window forward, searching for the location at which the window score is <25%. Once the end of the subsequence is found, beginning and trailing n’s are trimmed. Any subsequence <20 nucleotides (nt) is discarded. Subsequences >50 nt are accepted. Subsequences 20–50 nt in length are accepted if the total number of n’s in the sequence is ≤60% of the total subsequence length. The evaluation of the sequence continues in this manner, generating as many subsequences as needed to represent all usable data of the sequence. All accepted subsequences are queried against a public database (e.g., GenBank) and other nonpublic sequences using the BLAST algorithm. The output of this program discards subsequences that only have returns with an expected (E) value >1.0 × E−9 and only displays for each subsequence records having an E-value <1.0 × E−9. Raw BLAST outputs (subsequence length, database identifier, bit score, and E-value) for each parsed subsequence were saved and ranked in order of descending bit scores (Fig. 4B). In addition, a number of statistics on the subsequence are computed for user analysis, including the subsequence percentage of the target sequence, the subsequence length, the number of subsequence base calls, and the percentage of subsequence base calls. The REPI Java program is included in the Supplemental data.12
Figure 4.
(A) REPI logic diagram. The algorithm expands subsequences to the maximum length within the constraints of the allowable no-call (N) percentage (here, 25%). Each subsequence was submitted for BLAST analysis. (B) A sample REPI output (truncated) for Lackland AFB clinical nasal wash sample #NW20031114 that unambiguously identified influenza A virus type H3N2.
Quantification of HAdV-4 and influenza A viruses
For sensitivity assessments, real-time PCR assays were conducted on an iCycler instrument (Bio-Rad Laboratories) or R.A.P.I.D. LightCycler (Idaho Technology Inc.) to determine the number of adenovirus genomes in each sample. The findings for the samples were compared with those for 10-fold serial dilution of HAdV-4 prototype genomic DNA templates of known copy number (101 to 106 copies) by using Ad4hexon-F5, Ad4hexon-R4 with TaqMan probe (Ad4P2). HAdV-4 genomic copy number was calculated by measuring the DNA concentration from purified viral DNA and using the following conversion factor: 0.384 fg = a single adenoviral genome of ∼35 kb (Saitoh-Inagawa et al. 1996).
Similar assays were carried out to determine the plaque-forming units/μL of influenza A virus in each sample by using primers AMP-For and AMP-Rev (Stone et al. 2004) in the manner described in the publication.
Sequencing confirmation
Conventional sequencing result of influenza strains were provided by Luke T. Daum at the Air Force Institute for Operational Health ([AFIOH] San Antonio, TX).
Acknowledgments
Support for this research was provided by the Defense Threat Reduction Agency, the United States Army Medical Material Research Command, the Air Force Medical Service (Office of HQ USAF Surgeon General), and the Office of Naval Research. The help and constructive suggestions from members of the Epidemic Outbreak Surveillance Consortium were gratefully acknowledged. Dr. Klaus Schafer’s constructive advice is gratefully appreciated. We thank Margaret Ryan, Kevin Russell, and Christopher Barrozo at NHRC, Linda Canas at AFIOH, and Ted Hadfield at AFIP for kindly providing samples used in this study. This research has been conducted in compliance with all applicable federal and international regulations governing the protection of human subject in research, as documented in DoD protocol NHRC 1999.0002. The opinions and assertions contained herein are those of the authors and are not to be construed as official or reflecting the views of the Department of Defense or the U.S. Government.
Footnotes
[Supplemental material is available online at www.genome.org. REPI software is freely available at http://nrlbio.nrl.navy.mil/downloads/repi.zip.]
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4337206
Patent pending. This software embodies subject matter that is or may be claimed in one or more patent applications and/or issued patents. Please contact the Technology Transfer Office at the U.S. Naval Research Laboratory if you are interested in obtaining a license.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bodrossy L., Sessitsch A., Sessitsch A. Oligonucleotide microarrays in microbial diagnostics. Curr. Opin. Microbiol. 2004;7:1–10. doi: 10.1016/j.mib.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Boivin G., Coulombe Z., Wat C., Coulombe Z., Wat C., Wat C. Quantification of the influenza virus load by real-time polymerase chain reaction in nasopharyngeal swabs of patients treated with oseltamivir. J. Infect. Dis. 2003;188:578–580. doi: 10.1086/377046. [DOI] [PubMed] [Google Scholar]
- Bryant P.A., Venter D., Robins-Browne R., Curtis N., Venter D., Robins-Browne R., Curtis N., Robins-Browne R., Curtis N., Curtis N. Chips with everything: DNA microarrays in infectious diseases. Lancet Infect. Dis. 2004;4:100–111. doi: 10.1016/S1473-3099(04)00930-2. [DOI] [PubMed] [Google Scholar]
- Cleland C.A., White P.S., Deshpande A., Wolinsky M., Song J., Nolan J.P., White P.S., Deshpande A., Wolinsky M., Song J., Nolan J.P., Deshpande A., Wolinsky M., Song J., Nolan J.P., Wolinsky M., Song J., Nolan J.P., Song J., Nolan J.P., Nolan J.P. Development of rationally designed nucleic acid signatures for microbial pathogens. Expert Rev. Mol. Diagn. 2004;4:303–315. doi: 10.1586/14737159.4.3.303. [DOI] [PubMed] [Google Scholar]
- Couch R.B., Cate T.R., Fleet W.F., Gerone P.J., Knight V., Cate T.R., Fleet W.F., Gerone P.J., Knight V., Fleet W.F., Gerone P.J., Knight V., Gerone P.J., Knight V., Knight V. Aerosol-induced adenoviral illness resembling the naturally occurring illness in military recruits. Am. Rev. Respir. Dis. 1966;93:529–535. doi: 10.1164/arrd.1966.93.4.529. [DOI] [PubMed] [Google Scholar]
- Cutler D.J., Zwick M.E., Carrasquillo M.M., Yohn C.T., Tobin K.P., Kashuk C., Mathews D.J., Shah N.A., Eichler E.E., Warrington J.A., Zwick M.E., Carrasquillo M.M., Yohn C.T., Tobin K.P., Kashuk C., Mathews D.J., Shah N.A., Eichler E.E., Warrington J.A., Carrasquillo M.M., Yohn C.T., Tobin K.P., Kashuk C., Mathews D.J., Shah N.A., Eichler E.E., Warrington J.A., Yohn C.T., Tobin K.P., Kashuk C., Mathews D.J., Shah N.A., Eichler E.E., Warrington J.A., Tobin K.P., Kashuk C., Mathews D.J., Shah N.A., Eichler E.E., Warrington J.A., Kashuk C., Mathews D.J., Shah N.A., Eichler E.E., Warrington J.A., Mathews D.J., Shah N.A., Eichler E.E., Warrington J.A., Shah N.A., Eichler E.E., Warrington J.A., Eichler E.E., Warrington J.A., Warrington J.A. High-throughput variation detection and genotyping using microarrays. Genome Res. 2001;11:1913–1925. doi: 10.1101/gr.197201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman D.D., Xu W., Gerber S.I., Gray G.C., Schnurr D., Kajon A.E., Anderson L.J., Xu W., Gerber S.I., Gray G.C., Schnurr D., Kajon A.E., Anderson L.J., Gerber S.I., Gray G.C., Schnurr D., Kajon A.E., Anderson L.J., Gray G.C., Schnurr D., Kajon A.E., Anderson L.J., Schnurr D., Kajon A.E., Anderson L.J., Kajon A.E., Anderson L.J., Anderson L.J. Molecular epidemiology of adenovirus type 7 in the United States, 1966–2000. Emerg. Infect. Dis. 2002;8:269–277. doi: 10.3201/eid0803.010190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras T.R., Ghandour G., Wang E., Berno A., Small P.M., Drobniewski F., Alland D., Desmond E., Holodniy M., Drenkow J., Ghandour G., Wang E., Berno A., Small P.M., Drobniewski F., Alland D., Desmond E., Holodniy M., Drenkow J., Wang E., Berno A., Small P.M., Drobniewski F., Alland D., Desmond E., Holodniy M., Drenkow J., Berno A., Small P.M., Drobniewski F., Alland D., Desmond E., Holodniy M., Drenkow J., Small P.M., Drobniewski F., Alland D., Desmond E., Holodniy M., Drenkow J., Drobniewski F., Alland D., Desmond E., Holodniy M., Drenkow J., Alland D., Desmond E., Holodniy M., Drenkow J., Desmond E., Holodniy M., Drenkow J., Holodniy M., Drenkow J., Drenkow J. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Res. 1998;8:435–448. doi: 10.1101/gr.8.5.435. [DOI] [PubMed] [Google Scholar]
- Hacia J.G. Resequencing and mutational analysis using oligonucleotide microarrays. Nat. Genet. 1999;22:164–167. doi: 10.1038/4469. [DOI] [PubMed] [Google Scholar]
- Hoffmann E., Stech J., Guan Y., Webster R.G., Perez D.R., Stech J., Guan Y., Webster R.G., Perez D.R., Guan Y., Webster R.G., Perez D.R., Webster R.G., Perez D.R., Perez D.R. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- Kessler N., Ferraris O., Palmer K., Marsh W., Steel A., Ferraris O., Palmer K., Marsh W., Steel A., Palmer K., Marsh W., Steel A., Marsh W., Steel A., Steel A. Use of the DNA flow-thru chip, a three-dimensional biochip, for typing and subtyping of influenza viruses. J. Clin. Microbiol. 2004;42:2173–2185. doi: 10.1128/JCM.42.5.2173-2185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolavic-Gray S.A., Binn L.N., Sanchez J.L., Cersovsky S.B., Polyak C.S., Mitchell-Raymundo F., Asher L.V., Vaughn D.W., Feighner B.H., Innis B.L., Binn L.N., Sanchez J.L., Cersovsky S.B., Polyak C.S., Mitchell-Raymundo F., Asher L.V., Vaughn D.W., Feighner B.H., Innis B.L., Sanchez J.L., Cersovsky S.B., Polyak C.S., Mitchell-Raymundo F., Asher L.V., Vaughn D.W., Feighner B.H., Innis B.L., Cersovsky S.B., Polyak C.S., Mitchell-Raymundo F., Asher L.V., Vaughn D.W., Feighner B.H., Innis B.L., Polyak C.S., Mitchell-Raymundo F., Asher L.V., Vaughn D.W., Feighner B.H., Innis B.L., Mitchell-Raymundo F., Asher L.V., Vaughn D.W., Feighner B.H., Innis B.L., Asher L.V., Vaughn D.W., Feighner B.H., Innis B.L., Vaughn D.W., Feighner B.H., Innis B.L., Feighner B.H., Innis B.L., Innis B.L. Large epidemic of adenovirus type 4 infection among military trainees: Epidemiological, clinical, and laboratory studies. Clin. Infect. Dis. 2002;35:808–818. doi: 10.1086/342573. [DOI] [PubMed] [Google Scholar]
- Kothapalli R., Yoder S.J., Mane S., Loughran Jr , T P , Yoder S.J., Mane S., Loughran Jr , T P , Mane S., Loughran Jr , T P , Loughran Jr , T P , T P Microarray results: How accurate are they? BMC Bioinformatics. 2002;3:22. doi: 10.1186/1471-2105-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozal M.J., Shah N., Shen N., Yang R., Fucini R., Merigan T.C., Richman D.D., Morris D., Hubbell E., Chee M., Shah N., Shen N., Yang R., Fucini R., Merigan T.C., Richman D.D., Morris D., Hubbell E., Chee M., Shen N., Yang R., Fucini R., Merigan T.C., Richman D.D., Morris D., Hubbell E., Chee M., Yang R., Fucini R., Merigan T.C., Richman D.D., Morris D., Hubbell E., Chee M., Fucini R., Merigan T.C., Richman D.D., Morris D., Hubbell E., Chee M., Merigan T.C., Richman D.D., Morris D., Hubbell E., Chee M., Richman D.D., Morris D., Hubbell E., Chee M., Morris D., Hubbell E., Chee M., Hubbell E., Chee M., Chee M. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat. Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- Lin B., Vora G.J., Thach D., Walter E., Metzgar D., Tibbetts C., Stenger D.A., Vora G.J., Thach D., Walter E., Metzgar D., Tibbetts C., Stenger D.A., Thach D., Walter E., Metzgar D., Tibbetts C., Stenger D.A., Walter E., Metzgar D., Tibbetts C., Stenger D.A., Metzgar D., Tibbetts C., Stenger D.A., Tibbetts C., Stenger D.A., Stenger D.A. Use of oligonucleotide microarrays for rapid detection and serotyping of acute respiratory disease-associated adenoviruses. J. Clin. Microbiol. 2004;42:3232–3239. doi: 10.1128/JCM.42.7.3232-3239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López M.M., Bertolini E., Olmos A., Caruso P., Gorris M.T., Llop P., Penyalver R., Cambra M., Bertolini E., Olmos A., Caruso P., Gorris M.T., Llop P., Penyalver R., Cambra M., Olmos A., Caruso P., Gorris M.T., Llop P., Penyalver R., Cambra M., Caruso P., Gorris M.T., Llop P., Penyalver R., Cambra M., Gorris M.T., Llop P., Penyalver R., Cambra M., Llop P., Penyalver R., Cambra M., Penyalver R., Cambra M., Cambra M. Innovative tools for detection of plant pathogenic viruses and bacteria. Int. Microbiol. 2003;6:233–243. doi: 10.1007/s10123-003-0143-y. [DOI] [PubMed] [Google Scholar]
- Lovmar L., Fredriksson M., Liljedahl U., Sigurdsson S., Syvanen A.C., Fredriksson M., Liljedahl U., Sigurdsson S., Syvanen A.C., Liljedahl U., Sigurdsson S., Syvanen A.C., Sigurdsson S., Syvanen A.C., Syvanen A.C. Quantitative evaluation by minisequencing and microarrays reveals accurate multiplexed SNP genotyping of whole genome amplified DNA. Nucleic Acids Res. 2003;31:e129. doi: 10.1093/nar/gng129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panne D., Raleigh E.A., Bickle T.A., Raleigh E.A., Bickle T.A., Bickle T.A. The McrBC endonuclease translocates DNA in a reaction dependent on GTP hydrolysis. J. Mol. Biol. 1999;290:49–60. doi: 10.1006/jmbi.1999.2894. [DOI] [PubMed] [Google Scholar]
- Saitoh-Inagawa W., Oshima A., Aoki K., Itoh N., Isobe K., Uchio E., Ohno S., Nakajima H., Hata K., Ishiko H., Oshima A., Aoki K., Itoh N., Isobe K., Uchio E., Ohno S., Nakajima H., Hata K., Ishiko H., Aoki K., Itoh N., Isobe K., Uchio E., Ohno S., Nakajima H., Hata K., Ishiko H., Itoh N., Isobe K., Uchio E., Ohno S., Nakajima H., Hata K., Ishiko H., Isobe K., Uchio E., Ohno S., Nakajima H., Hata K., Ishiko H., Uchio E., Ohno S., Nakajima H., Hata K., Ishiko H., Ohno S., Nakajima H., Hata K., Ishiko H., Nakajima H., Hata K., Ishiko H., Hata K., Ishiko H., Ishiko H. Rapid diagnosis of adenoviral conjunctivitis by PCR and restriction fragment length polymorphism analysis. J. Clin. Microbiol. 1996;34:2113–2116. doi: 10.1128/jcm.34.9.2113-2116.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone B., Burrows J., Schepetiuk S., Higgins G., Hampson A., Shaw R., Kok T., Burrows J., Schepetiuk S., Higgins G., Hampson A., Shaw R., Kok T., Schepetiuk S., Higgins G., Hampson A., Shaw R., Kok T., Higgins G., Hampson A., Shaw R., Kok T., Hampson A., Shaw R., Kok T., Shaw R., Kok T., Kok T. Rapid detection and simultaneous subtype differentiation of influenza A viruses by real time PCR. J. Virol. Methods. 2004;117:103–112. doi: 10.1016/j.jviromet.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Striebel H.M., Birch-Hirschfeld E., Egerer R., Foldes-Papp Z., Birch-Hirschfeld E., Egerer R., Foldes-Papp Z., Egerer R., Foldes-Papp Z., Foldes-Papp Z. Virus diagnostics on microarrays. Curr. Pharm. Biotechnol. 2003;4:401–415. doi: 10.2174/1389201033377274. [DOI] [PubMed] [Google Scholar]
- Thompson W.W., Shay D.K., Weintraub E., Brammer L., Cox N., Anderson L.J., Fukuda K., Shay D.K., Weintraub E., Brammer L., Cox N., Anderson L.J., Fukuda K., Weintraub E., Brammer L., Cox N., Anderson L.J., Fukuda K., Brammer L., Cox N., Anderson L.J., Fukuda K., Cox N., Anderson L.J., Fukuda K., Anderson L.J., Fukuda K., Fukuda K. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- Vora G.J., Meador C.E., Stenger D.A., Andreadis J.D., Meador C.E., Stenger D.A., Andreadis J.D., Stenger D.A., Andreadis J.D., Andreadis J.D. Nucleic acid amplification strategies for DNA microarray-based pathogen detection. Appl. Environ. Microbiol. 2004;70:3047–3054. doi: 10.1128/AEM.70.5.3047-3054.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Coscoy L., Zylberberg M., Avila P.C., Boushey H.A., Ganem D., DeRisi J.L., Coscoy L., Zylberberg M., Avila P.C., Boushey H.A., Ganem D., DeRisi J.L., Zylberberg M., Avila P.C., Boushey H.A., Ganem D., DeRisi J.L., Avila P.C., Boushey H.A., Ganem D., DeRisi J.L., Boushey H.A., Ganem D., DeRisi J.L., Ganem D., DeRisi J.L., DeRisi J.L. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. 2002;99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Urisman A., Liu Y.T., Springer M., Ksiazek T.G., Erdman D.D., Mardis E.R., Hickenbotham M., Magrini V., Eldred J., Urisman A., Liu Y.T., Springer M., Ksiazek T.G., Erdman D.D., Mardis E.R., Hickenbotham M., Magrini V., Eldred J., Liu Y.T., Springer M., Ksiazek T.G., Erdman D.D., Mardis E.R., Hickenbotham M., Magrini V., Eldred J., Springer M., Ksiazek T.G., Erdman D.D., Mardis E.R., Hickenbotham M., Magrini V., Eldred J., Ksiazek T.G., Erdman D.D., Mardis E.R., Hickenbotham M., Magrini V., Eldred J., Erdman D.D., Mardis E.R., Hickenbotham M., Magrini V., Eldred J., Mardis E.R., Hickenbotham M., Magrini V., Eldred J., Hickenbotham M., Magrini V., Eldred J., Magrini V., Eldred J., Eldred J. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1:257–260. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Daum L.T., Vora G.J., Metzgar D., Walter E.A., Canas L.C., Malanoski A.P., Lin B., Stenger D.A. on behalf of the Epidemic Outbreak Surveillance Consortium., Daum L.T., Vora G.J., Metzgar D., Walter E.A., Canas L.C., Malanoski A.P., Lin B., Stenger D.A. on behalf of the Epidemic Outbreak Surveillance Consortium., Vora G.J., Metzgar D., Walter E.A., Canas L.C., Malanoski A.P., Lin B., Stenger D.A. on behalf of the Epidemic Outbreak Surveillance Consortium., Metzgar D., Walter E.A., Canas L.C., Malanoski A.P., Lin B., Stenger D.A. on behalf of the Epidemic Outbreak Surveillance Consortium., Walter E.A., Canas L.C., Malanoski A.P., Lin B., Stenger D.A. on behalf of the Epidemic Outbreak Surveillance Consortium., Canas L.C., Malanoski A.P., Lin B., Stenger D.A. on behalf of the Epidemic Outbreak Surveillance Consortium., Malanoski A.P., Lin B., Stenger D.A. on behalf of the Epidemic Outbreak Surveillance Consortium., Lin B., Stenger D.A. on behalf of the Epidemic Outbreak Surveillance Consortium., Stenger D.A. on behalf of the Epidemic Outbreak Surveillance Consortium.2006Rapid, broad-spectrum identification of influenza viruses by resequencing microarrays. Emerg. Infect. Dis. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W.J., Strout C.L., DeSantis T.Z., Stilwell J.L., Carrano A.V., Andersen G.L., Strout C.L., DeSantis T.Z., Stilwell J.L., Carrano A.V., Andersen G.L., DeSantis T.Z., Stilwell J.L., Carrano A.V., Andersen G.L., Stilwell J.L., Carrano A.V., Andersen G.L., Carrano A.V., Andersen G.L., Andersen G.L. Sequence-specific identification of 18 pathogenic microorganisms using microarray technology. Mol. Cell. Probes. 2002;16:119–127. doi: 10.1006/mcpr.2001.0397. [DOI] [PubMed] [Google Scholar]
- Wilson K.H., Wilson W.J., Radosevich J.L., DeSantis T.Z., Viswanathan V.S., Kuczmarski T.A., Andersen G.L., Wilson W.J., Radosevich J.L., DeSantis T.Z., Viswanathan V.S., Kuczmarski T.A., Andersen G.L., Radosevich J.L., DeSantis T.Z., Viswanathan V.S., Kuczmarski T.A., Andersen G.L., DeSantis T.Z., Viswanathan V.S., Kuczmarski T.A., Andersen G.L., Viswanathan V.S., Kuczmarski T.A., Andersen G.L., Kuczmarski T.A., Andersen G.L., Andersen G.L. High-density microarray of small-subunit ribosomal DNA probes. Appl. Environ. Microbiol. 2002;68:2535–2541. doi: 10.1128/AEM.68.5.2535-2541.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C.W., Albert T.J., Vega V.B., Norton J.E., Cutler D.J., Richmond T.A., Stanton L.W., Liu E.T., Miller L.D., Albert T.J., Vega V.B., Norton J.E., Cutler D.J., Richmond T.A., Stanton L.W., Liu E.T., Miller L.D., Vega V.B., Norton J.E., Cutler D.J., Richmond T.A., Stanton L.W., Liu E.T., Miller L.D., Norton J.E., Cutler D.J., Richmond T.A., Stanton L.W., Liu E.T., Miller L.D., Cutler D.J., Richmond T.A., Stanton L.W., Liu E.T., Miller L.D., Richmond T.A., Stanton L.W., Liu E.T., Miller L.D., Stanton L.W., Liu E.T., Miller L.D., Liu E.T., Miller L.D., Miller L.D. Tracking the evolution of the SARS coronavirus using high-throughput, high-density resequencing arrays. Genome Res. 2004;14:398–405. doi: 10.1101/gr.2141004. [DOI] [PMC free article] [PubMed] [Google Scholar]