Abstract
Recently there has been a great interest in the identification of microRNAs and their targets as well as understanding the spatial and temporal regulation of microRNA genes. To understand how microRNA genes evolve, we looked at several rapidly evolving families in Arabidopsis thaliana, and found that they arose from a process of genome-wide duplication, tandem duplication, and segmental duplication followed by dispersal and diversification, similar to the processes that drive the evolution of protein gene families. Using multiple expression data sets to examine the transcription patterns of different members of the microRNA families, we find the sequence diversification of duplicated microRNA genes to be accompanied by a change in spatial and temporal expression patterns, suggesting that duplicated copies acquire new functionality as they evolve.
It has been suggested that microRNAs, or miRNAs, play a central role in regulating basic developmental processes, such as meristem cell identity, organ polarity, and timing of developmental events, by interfering with the expression of targeted messenger RNAs (mRNAs) (Emery et al. 2003; Palatnik et al. 2003; Bartel 2004). Understanding the role of miRNAs could help answer fundamental biological questions while also enhancing the ability to precisely engineer plants for improved crop yields, increased resistance to disease, and adaptation to environmental extremes.
miRNAs are a class of small single-stranded non-coding RNAs that range in length from roughly 20 to 24 nucleotides (nt) (Bartel and Bartel 2003; Bartel 2004). The biogenesis of miRNAs differs between plants and animals. Within plants, it is believed that polymerase II transcribes miRNAs into a primary miRNA transcript (pri-miRNA). In the nucleus, a ribonuclease III-like nuclease, DICER-LIKE 1 (DCL1) (Papp et al. 2003), then processes the pri-miRNA, potentially with the assistance of one or more unknown enzymes. This process yields the precursor miRNA (pre-miRNA) and ultimately the mature miRNA:miRNA* duplex (Bartel 2004). The mature miRNA duplex is exported to the cytoplasm, where it is unwound and incorporated into the RISC complex (Bartel 2004). The miRNA then guides the complex to its specific protein-coding gene target mRNA, partially or completely silencing the transcript by either degrading it or by inhibiting its translation into a protein (Llave et al. 2002).
Plant miRNAs can be grouped into distinct families of one or more precursors. Each precursor within the family produces similar, if not identical, mature miRNA products. Within a family, the greatest sequence conservation occurs in the stem that becomes the mature miRNA product, followed by the stem that opposes the mature miRNA in the precursor. Within both plants and animals, the unpaired loop regions are the most variable parts of the precursor despite the characteristically smaller loop lengths found in animal hairpins (Lai et al. 2003; Maher et al. 2004). High levels of sequence similarity among loop regions of Arabidopsis precursors appear only in tandemly duplicated precursors (Maher et al. 2004). In most cases, there is no obvious sequence similarity among the loop regions of members of the same miRNA family.
Direct evidence pertaining to the mechanism of miRNA transcription has only recently been published (Lee et al. 2004; Xie et al. 2005). Currently, the majority of plant miRNAs reside within intergenic regions or in the opposite strand of annotated genes. miRNAs, like mRNAs, are transcribed by polymerase II. We therefore expect miRNA sequences to be found in collections of Pol II-transcribed RNAs, such as Massively Parallel Signature Sequencing (MPSS) collections.
In Arabidopsis, protein-coding gene families arise by a process of gene duplication and diversification (The Arabidopsis Genome Initiative 2000; Prince and Pickett 2002; Cannon et al. 2004). The processes driving gene duplication are whole-genome duplication (polyploidization), duplications of subchromosomal-length regions known as segmental duplications, and local duplications that involve one or two genes known as tandem duplications (Bowers et al. 2003; Lawton-Rauh 2003; Blanc and Wolfe 2004b). Gene- and chromosomal-level rearrangements increase the difficulty of numbering and dating polyploidy events (Lawton-Rauh 2003; Blanc and Wolfe 2004a; Adams and Wendel 2005)
The goal of this study was to ask whether this model of protein-coding gene family evolution applies to the miRNA gene families as well, and, if so, whether there exists an association between the evolution of miRNA genes and changes in expression patterns that might indicate diversification of function.
Results
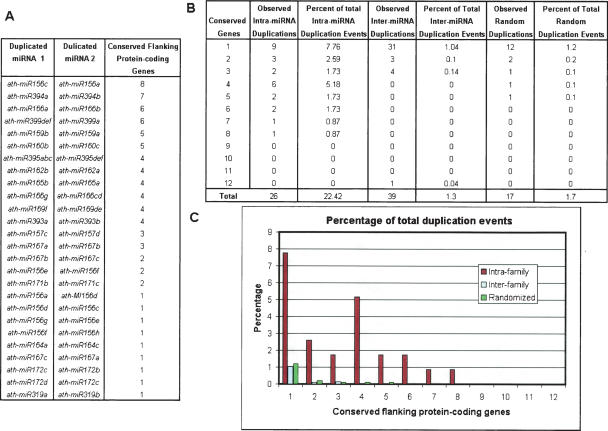
The haploid genome of Arabidopsis consists of five chromosomes containing many internally duplicated regions. To begin this work, we obtained all 92 Arabidopsis miRNA precursor gene sequences and coordinates from the miRNA Registry (http://microrna.sanger.ac.uk/) (Ambros et al. 2003; Griffiths-Jones 2004). The miRNA genes were grouped into 26 families based on the similarity of the mature miRNA product (Ambros et al. 2003). Of the 26 families, 22 (84.6%) contain more than one miRNA gene and six families (25%) contain five or more miRNA genes (Supplemental Table 1). Given the large number of miRNA families with multiple genes, it is reasonable to hypothesize that they have undergone a history of expansion events similar to those that underlie the amplification and diversification of families of protein-coding genes. Therefore, we expect to see different members from the same miRNA family residing within duplicated regions of the genome.
Tandem duplications
We first identified apparent tandem duplications among the miRNA gene families. We did so by looking for contiguous miRNAs in the same intergenic region, or in neighboring intergenic regions, and found 23 genes from six gene families that met these criteria. The longest run of miRNA genes arising from an apparent tandem duplication was six, while the remainder occurred in arrays of two or three miRNA genes. Of the 23 tandemly duplicated miRNAs, if each miRNA is paired with the nearest downstream tandemly duplicated miRNA, two-thirds are on the same strand and the average distance between tandemly duplicated miRNAs is 1987 nt (data not shown).
Large duplication events
We next wished to test the hypothesis that large-scale duplication events play a role in the evolution of miRNA gene families. We reasoned that, if this were the case, then the protein-coding genes flanking members of the same miRNA family would be more similar to each other than protein-coding genes flanking randomly selected genes, because the protein-coding genes would also be involved in the duplications. The alternative hypothesis is that miRNAs are not evolving through duplication events, but rather via random translocations and insertional events. To identify large duplication events, we chose to align protein-coding genes rather than non-coding nucleotide sequence because of the low level of nucleotide sequence conservation among non-coding regions in Arabidopsis duplicated regions (Vision et al. 2000). We therefore consider miRNAs to originate from a duplication event if they reside within a region of conserved protein-coding genes. Two chromosomal regions, containing one or more miRNAs, were classified as residing within such a duplicated block if one or more of the 10 upstream or 10 downstream protein-coding genes flanking the miRNA were found to have a best non-self match to a protein-coding gene flanking another miRNA according to BLASTP (E-value < 0.001). Only the best match was used for this analysis so that tandemly duplicated genes did not enrich the number of conserved genes flanking a miRNA. In addition, using the best match selects for paralogs that are more likely to be recently duplicated from one another over less conserved genes from the same family. As a control, we generated a simulated data set in which we selected random genomic locations and aligned their flanking protein-coding genes.
Our approach excluded miRNA families containing a single gene and therefore leaves us with 88 miRNA precursors from 22 distinct miRNA families. Since we are aligning the flanking protein-coding genes for a miRNA, tandemly duplicated miRNAs were counted only once. Therefore, our 88 miRNA precursors were located within 73 chromosomal regions.
To characterize the pattern of miRNA duplication, we compared the rates of duplicated blocks surrounding miRNAs within the same family (intrafamily), between families (interfamily), and randomly selected locations (Fig. 1). In our analysis we found that there are 26 duplicated chromosomal regions containing miRNAs from the same family that have conservation between their flanking protein-coding genes out of the 116 total possible miRNA pairs (22.42%) as opposed to 1.3% of interfamily miRNA pairs and 1.94% of randomly selected genomic locations. Together, these data suggest that large-scale duplication plays a major role in miRNA evolution and are inconsistent with the random insertion hypothesis. Our procedure may misclassify duplicated blocks at a rate of ∼2%.
Figure 1.
Percentage of intrafamily, interfamily, and randomly duplicated blocks. The total number of duplication events is the sum of all possible miRNA pairs within the set. Therefore, the percentage equals the total number of duplication events observed compared to the total number of possible duplication events. (A) Duplicated miRNAs from within the same family with the number of conserved protein-coding genes flanking the miRNAs. (B) Percentages of observed duplicated blocks against the total number of potential duplicated blocks and the number of flanking genes that are conserved within each block. (C) Plot comparing the percentage of observed duplication events against the total number of potential duplications for interfamily miRNAs, intrafamily miRNAs, and the randomized simulation.
While the randomized set represents the upper bound of our false-positive rate, we also observed that the randomized and interfamily duplicated blocks tend to have fewer conserved protein-coding regions than the intrafamily duplicated blocks. In fact, interfamily duplicated blocks all occur with three or fewer conserved flanking genes, with the exception of the miR169a–miR158b pair, which has 12 conserved flanking genes. Therefore, we believe our classification system is more likely to fail when applied to duplicated blocks having three or fewer conserved flanking protein-coding genes. Almost half of the putative intrafamily duplicated blocks that we have identified have at least three conserved flanking genes. We therefore defined all predicted duplicated blocks as our “loose” set and the duplicated blocks containing four or more conserved flanking protein-coding genes as our “strict” set.
While our previous methodology analyzed 10 upstream and downstream protein-coding genes in order to identify duplicated blocks, these duplicated regions can span much larger regions, which we will refer to as extended duplicated blocks. To enable a more detailed analysis of miRNA families, we wanted to provide a broad overview of each duplicated region and therefore extracted 200 protein-coding genes flanking each miRNA. We then plotted these protein-coding genes surrounding the miRNAs to highlight our previously identified duplicated blocks, but in addition show the varying degrees of chromosomal rearrangements, if any, within the extended duplicated block. This enables us to establish relationships between miRNAs that are more closely related to one another within a particular family. In addition, we incorporate expression data to further support the diversification of miRNAs.
Table 1 summarizes the number of segmental and tandem duplications for each miRNA family according to our definitions. It would appear that 18 of the 22 families (81.8%) arise from either a segmental or tandem duplication, or a combination of the two processes. Of these 18 families, six were involved in tandem duplications, and 17 were involved in segmental duplications. In total, 23 (26.1%) miRNAs are involved in tandem duplications, while 51 miRNAs (57.9%) are involved in large-scale duplication events. A more conservative estimate of segmental duplications, which would discard all miRNAs that have three or fewer conserved flanking protein-coding genes, predicts that 32 miRNAs (36.3%) would be involved in duplicated blocks. This suggests that miRNA genes are evolving by segmental duplications and tandem duplications, just as protein-coding genes have evolved.
Table 1.
Duplication events of miRNAs in multigene families
This table indicates the number of loci within a family found to be tandemly duplicated or within a duplicated block, along with the target mRNA. The number of segmental duplications is shown under both a loose and a strict definition. The loose definition shows all possible miRNAs that reside in duplicated blocks, while the strict definition shows the number of miRNAs in duplicated blocks with four or more conserved flanking genes.
Dating duplication events
Under the assumption that synonymous silent substitutions per site (Ks) occur with a constant rate over time, we can use the conserved flanking protein-coding genes to estimate the dates of the large-scale duplication events. For this analysis, we used duplicated blocks in our strict set only. Each pair of proteins in the duplicated block was aligned at the amino acid level, and then codons from gapless aligned regions were used to calculate Ks values using codeml (Yang 1997). We discarded any Ks values >2.0 because of the risk of saturation (Blanc and Wolfe 2004b). The approximate date of the duplication event was then calculated using the mean Ks and an estimated rate of silent-site substitutions of 1.5 × 10−8 substitutions/synonymous site/year (Koch et al. 2000; Blanc and Wolfe 2004b). Table 2 shows the mean Ks values for each duplication event and the estimated date. We conclude that the large-scale duplication events involving miRNAs have all occurred within the last 28–39 million years (Myr). Given that traces of duplication events erode with time, we believe our approach may be limited to duplication events that have occurred within the last 39 Myr. Thus, miRNAs lacking conserved flanking protein-coding genes, which nevertheless maintain sequence conservation across both stems of their precursor, may have evolved prior to the events we have detected.
Table 2.
Estimation of the absolute date for large-scale duplication events
For each duplicated region containing miRNAs, we indicate the number of protein-coding genes, n, used for the Ks estimation. Only duplication events containing four or more conserved protein-coding genes were used to calculate the duplication event date. The events range from 28 to 39 million years ago (Mya), with the average date occurring around 33.5 Myr.
Relationship of miRNAs and their targets
For multigene miRNA families that target multiple mRNAs, with similar or identical target sites, we were interested to see whether there was a correlation in the physical locations of known miRNAs and their targets. If so, miRNAs in close proximity to their respective target mRNAs could be indicative of a regulatory relationship. Previous studies have identified potential miRNA targets based on a predetermined set of rules for base-pairing between a miRNA and its target mRNA (Rhoades et al. 2002; Jones-Rhoades and Bartel 2004; Schwab et al. 2005). Using these predicted targets, we observed that precursors from within the same and different families are scattered physically throughout the genome and that there is no apparent correlation between miRNA genes and their protein-coding targets (Supplemental Fig. 1).
Expansion of miRNA families in conjunction with expression data
When protein-coding genes duplicate and diverge, they can lose function (become pseudogenes), maintain their current function (redundant function), acquire new functions (neofunctionalization), or take on more specialized functions (subfunctionalization). We next asked whether the same processes apply to the miRNA genes. To answer this question, we obtained spatial- and temporal-specific expression pattern data from Massively Parallel Signature Sequencing (MPSS) collections (Meyers et al. 2004a).
MPSS is a large-scale expression resource capturing transcript expression levels within 17 different libraries. The MPSS signatures are derived from the 3′-end of the mRNA molecule (Meyers et al. 2004a). Therefore, mapping the signature relative to the miRNA should show a higher density of signatures downstream of the miRNA (Meyers et al. 2004b). It is a possibility that some of the miRNAs are alternatively spliced, given the close proximity of multiple signatures downstream of the miRNA, but for our purposes we were interested in the minimum distance of downstream signatures. Therefore, for each miRNA, we record only the first occurring significantly expressed signature downstream of the miRNA yet upstream of the adjacent protein-coding gene, as shown in Table 3. We observed a greater density of expressed class 4 (intergenic) signatures located slightly downstream of the known miRNAs within the first 400 nt (data not shown).
Table 3.
Class 4, intergenic MPSS signatures for known miRNAs
For each miRNA with an associated MPSS signature downstream, the neighboring protein-coding genes, the 17-nt MPSS signature, nucleotide distance downstream of the precursor 3′-end, and expression level are shown. The expression level is normalized to show how many transcripts, containing the signature, occurred for every million different transcripts captured within the library. Tandemly duplicated miRNAs were kept in this table to show the specific small RNA, 5′-RACE, and 3′-RACE results for each gene despite having the same MPSS signature. 5′-RACE and 3′-RACE values are indicated as Yes, No, or NT (Not Tested) (Xie et al. 2005). The small RNAs in ASRP represent the number of clones related to that particular miRNA gene (Gustafson et al. 2005).
We analyzed the 92 miRNAs from 26 different families and merged the 19 tandemly duplicated miRNAs that reside within the same intergenic region since it is not known whether they are expressed as one large transcriptional unit or as two separate primary transcripts. Overall, 32 of the 92 miRNAs (34.8%) have an associated class 4 signature that is expressed at significant levels, as shown in Table 3, assuming the two tandemly duplicated miRNAs are polycistronic. The average expression level is 26 transcripts per million (TPM), with a range of 4–173. We then correlated the tissue distribution of the MPSS signatures associated with each known miRNA (Supplemental Table 2).
For those miRNAs that did not have an MPSS signature, it is possible that their expression patterns are specific to tissues not sampled by the MPSS libraries. This is consistent with a recent analysis of Arabidopsis miRNA gene expression, in which 47 out of 99 (47.4%) miRNAs failed to produce a detectable signal using 5′-RACE or 3′-RACE (Xie et al. 2005). Of the 52 miRNAs detected by RACE, 25 (48.1%) miRNAs have an associated MPSS signature (Table 3). Of 47 miRNAs not detected by RACE, nine (14.9%) miRNAs have an associated MPSS signature. Overall, the miRNAs for which we failed to find MPSS signatures were more likely to be undetectable by RACE.
In the following sections, we describe specific examples of how the miR156, miR159, and miR166 families seem to evolve and take on new functionality through duplication events.
miR159 family evolution
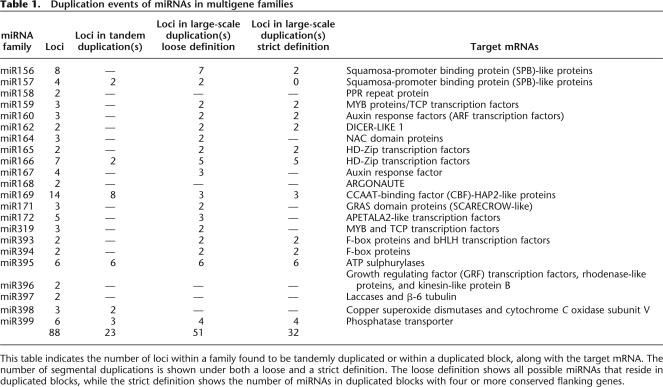
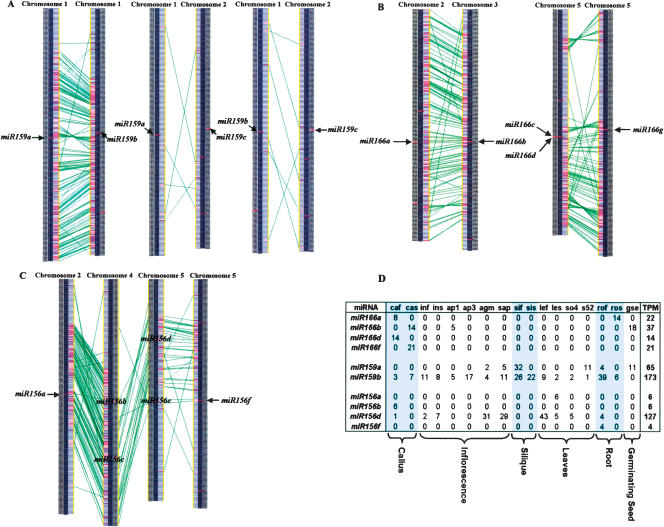
The three precursors within the miR159 family target mRNAs coding for MYB proteins, which are known to bind to the promoter of the floral meristem identity gene LEAFY and have varying degrees of conservation in their surrounding regions (Reinhart et al. 2002; Rhoades et al. 2002; Achard et al. 2004). Using the extended duplicated blocks identified earlier, we find that miR159a and miR159b reside within an intrachromosomal duplication within chromosome 1 (Fig. 2A). While many of the conserved genes within the duplicated block appear to maintain their order between the two chromosomal segments, the inversion within the middle of the duplicated region indicates it has undergone an additional rearrangement event. However, the origin of miR159c is more mysterious. There are very few conserved genes surrounding miR159a and miR159c or miR159b and miR159c, indicating that either miR159c arose via a small duplication that did not involve flanking protein-coding genes, that the duplication is ancient and cannot be detected by our methods, or that miR159c arose via an unknown mechanism. Overall, we believe that miR156a and miR156b evolved from a duplication event within the last 30 Myr, while miR159c existed prior to this duplication event, as shown in Figure 3A.
Figure 2.
Conserved protein-coding genes surround flanking miRNA genes. The chromosomal regions surrounding two miRNAs are displayed as vertical yellow lines. Each of the protein-coding genes nearby are shown as black horizontal lines, while the miRNA is displayed as a red horizontal line, and indicated by the arrow because of the resolution of the images. The green lines represent genes that are conserved according to BLASTN analysis. (A) miR159 family. (B) miR166 family. (C) miR156 family. (D) MPSS tissue expression for miRNA genes from miR166, miR159, and miR156.
Figure 3.
Reconstruction of miRNA family evolution. These phylogenetic trees were generated to demonstrate the order of duplication events for three miRNA families. Circles indicate duplication events for which we have supporting evidence. Red circles indicate duplication events supported by conserved protein-coding genes flanking two miRNA genes. Green circles represent duplication events supported by conserved non-coding sequence flanking two miRNA genes. Blue circles indicate tandemly duplicated miRNAs. A combination of circles indicates that it is supported by multiple methods. The branch lengths are of uniform length and are not meant to indicate time since each duplication event. The connection between two green circles indicates that it is the same duplication event. Arrows establish which two miRNAs were found to be involved in a specific duplication event. (A) miR159 family; (B) miR166 family; (C) miR156 family.
The closest downstream MPSS tags for miR159a and miR159b show slight variations in their tissue expression profiles (Fig. 2D). Under identical conditions, each miRNA demonstrates expression within inflorescence, leaves, root, and silique. However, miR159a is expressed in germinating seed, and only miR159b is expressed in callus tissue. This example suggests that the duplicated copies exhibit both redundancy of function and diversification. These miRNAs have a wide range of tissue expression and are expressed at low levels; however, there remains the possibility that the MPSS technique failed to detect low levels of expression in callus and seed. Regardless, this does demonstrate the high level of redundant function between the two miRNAs.
miR166 family evolution
Class III HD-ZIP genes are predicted transcription factors that are involved in the adaxial identity of lateral organs and meristem development in Arabidopsis (Engstrom et al. 2004; Juarez et al. 2004). The putative binding site, as determined by sequence identity, overlaps with a gain-of-function mutation, suggesting that members of the miRNA166 family regulate these transcription factors (Reinhart et al. 2002; Rhoades et al. 2002). Figure 2B shows two duplicated blocks containing five of the seven miRNA genes within the miR166 family. The first example is of a duplicated block between miR166a and miR166b, located on chromosomes 2 and 3, respectively. The second example shows the duplicated region between the tandem duplication of miR166c and miR166d to the chromosomal region surrounding miR166g. Within the highly conserved regions of this intrachromosomal duplication on chromosome 5, some duplicated blocks have undergone smaller inversions and rearrangements.
Differential gene loss after a genome-wide duplication could contribute to a number of miRNA genes that are not visible in the analysis (Paterson et al. 2004). For instance, miR166c and miR166d are tandemly duplicated, yet there is only one corresponding miRNA, miR166g, residing within the duplicated region. The first explanation is that the tandem duplication occurred before the larger duplication event and was followed by differential gene loss near miR166g. Alternatively, this could be due to a tandem duplication occurring after a genome-wide duplication event.
To help resolve the evolutionary history of the miR166 family, we looked for conservation in the non-coding flanking regions of the miRNAs. We aligned flanking regions using Dotmatcher from the EMBOSS analysis package (Rice et al. 2000). This demonstrated that there are conserved non-coding regions flanking miR166b and miR166e, but no regions of conservation between miR166a and miR166e outside of the conserved stems (Fig. 4A,B,C). The number of conserved regions flanking miR166b and miR166e is less than the number of regions with sequence similarity between miR166a and miR166b (Supplemental Fig. 2). This supports the model that the duplication event between miR166b and miR166e predates the duplication event between miR166a and miR166b.
Figure 4.
Dotmatcher results for miR166 family. These three plots highlight non-coding flanking regions that are conserved between miRNAs. The red boxes highlight the conserved stems between the two miRNAs. (A) miR166a and miR166b. (B) miR166a and miR166e. (C) miR166b and miR166e.
The overall evolutionary model we propose for the miR166 family is shown in Figure 3B. miR166f lacks any relation, other than having a similar mature miRNA sequence, to all miR166 genes except for miR166a with which it has conservation in the opposing stem of the precursor; therefore, we place miR166f closest to miR166a. We believe miR166a and miR166b evolved from a recent large-scale duplication event. miR166b and miR166e have conserved non-coding flanking sequences, while miR166a and miR166e lack this conservation, indicating that miR166b and miR166e most likely evolved from a duplication prior to the large-scale duplication event between miR166a and miR166b. The best explanation is that miR166e is anciently related to miR166g. miR166g resides within a duplicated block with the tandem duplication containing miR166c and miR166d.
All miR166 family members with an associated MPSS signature demonstrate expression in callus, indicating substantial redundancy of function (Fig. 2D). However, in addition, miR166a is expressed in root, and miR166b is expressed in germinating seed and inflorescence tissue. miR166a and miR166b have demonstrated redundant and diversified expression following duplication. Within another duplicated region, miR166d (and potentially miR166c, depending on whether it resides in the same transcription unit as miR166d) has a significant expression level, while its duplicated counterpart, miR166g, lacks any detectable level of expression. This either represents the loss of miR166g functionality, or indicates that it is transcribed at very low levels indistinguishable from background levels.
The functional implications based on the expression profiles of two tandemly duplicated miRNAs that are located on the same strand is challenging since in many instances only the 3′-miRNA has an associated MPSS signature. For instance, the tandem duplication between miR166c and miR166d, which resides within the intergenic region between At5g08710 and At5g08720, has one MPSS signature located downstream of the 3′-miRNA, implying that they may be transcribed as one transcriptional unit.
miR156 family evolution
The miR156 family has been demonstrated to target proteins resembling the Squamosa-promoter-binding proteins (SPB). SPB proteins are a plant-specific group of transcription factors involved in plant development (Yamasaki et al. 2004). The complementary target sites for the miRNAs within this family do not reside in the conserved domain defining SPB-like proteins but instead fall within a region weakly conserved among the target family (Bartel 2004).
Figure 2C shows an overview of the relationships between the different members of the family. The different members reside within both inter- and intrachromosomal duplications and appear to occur in pairs (miR156b/miR156c and miR156d/miR156e). These closely related pairs are located many genes apart, whereas most pairs that we have characterized as tandem duplications occur within the same intergenic region.
Our overall evolutionary reconstruction (Fig. 3C) shows miR156g as an outlier since it has a low level of conservation in the flanking protein-coding genes with miR156e, but lacks any other relationship within this family indicating its ancient origins (Supplemental Fig. 2). miR156h and miR156d have conservation in their flanking non-coding sequence, indicating they have evolved from a duplication event. miR156b has conservation across both stems of the precursor with miR156f, whereas it only shares similarity in its mature miRNA product with the remainder of the miRNA genes in the family. This suggests an ancient relationship between miR156b and miR156f. We observed an apparent large-scale duplication involving miR156e and miR156f. The protein-coding genes conserved in this duplicated block span the region containing miR156d (Fig. 2C), yet there isn’t a known miRNA in the corresponding region of the duplicated block. We used Patscan to search the region for a miRNA sequence with up to five mismatches that could form a hairpin structure representing a potentially undetected member of the miR156 family but failed to find such a candidate. We therefore believe a gene loss occurred within this region after the duplication event. miR156d and miR156c were then duplicated from one another based on their conserved flanking protein-coding genes. The most recent duplication event occurred between miR156a and miR156c as determined by the high level of conserved flanking protein-coding genes. In addition, we think that an ancestor of miR156b originally resided within this duplicated block, but once again there were no remnants of a corresponding miRNA within the duplicated block, indicating that the duplication of miR156b was again followed by gene loss.
While we lack MPSS expression data for any two miRNAs that are directly involved within a large-scale duplicated region, we do have two miRNAs that are indirectly related according to our evolutionary reconstruction. miR156c was involved in a duplication event with miR156d prior to its recent duplication with miR156a. Interestingly, we do not have an MPSS signature for miR156c, but we do have a signature for miR156a and miR156d (Fig. 3D). We observed a broad expression profile (callus, inflorescence, leaves, and root) for the more divergent miR156d and a very specific expression profile (leaves) for miR156a. This suggests that miR156a is providing redundant functionality with miR156d, while miR156c may have lost some functionality following its duplication with miR156a.
miR395 family evolution
The miR395 family, predicted to target mRNAs coding for ATP sulphurylases, can be broken into two groups of tandem duplications (Bartel 2004; Jones-Rhoades and Bartel 2004). Each group of tandemly duplicated miRNAs has two miRNAs on the same strand and another on the opposite strand. Our previous expansion analysis indicated that seven protein-coding genes flanking both sets of tandemly duplicated miR395 genes were conserved. This suggests that an intrachromosomal duplication event occurred after the tandem duplication events, thereby conserving the orientation of the miR395 genes (Fig. 5).
Figure 5.
Schematic representation of intrachromosomal duplication within the miR395 family.
Within both sets of tandem duplications, we observed a high sequence complementarity within the loop regions, providing further support that each set arose from tandem duplication events. One example of two highly conserved precursors within the miR395 family is between miR395b and miR395c. Both miRNAs are in the same orientation and have an identical precursor length of 100 nt, yet only two nucleotides within their loop regions are different.
In the other set of tandem duplications among miR395d, miR395e, and miR395f, the two miRNAs on the same strand also have a higher level of similarity in their loop region than they do with the miRNA on the opposing strand. Regardless, they all have a high level of sequence conservation, being tandem duplications of one another.
Only miR395e has an associated MPSS signature, making it difficult to draw any conclusions about potential diversification. The expression of miR395, which depends on environmental stress, increases during sulfate starvation (Bartel 2004; Jones-Rhoades and Bartel 2004). The specificity of this condition makes miR395 less likely to appear in tissue libraries tested with MPSS. According to the MPSS data, in the instance of miR395e, the tandem duplications do not appear to be transcribed as one transcriptional unit, given that the signature is located downstream of miR395e, which was not the 3′ member of the pair.
Discussion
The evolution of protein-coding genes arises from genome-wide duplication events, large-scale chromosomal duplication, and local rearrangements. Recent efforts in miRNA predictions provide a solid foundation for analyzing the evolution of miRNAs. By analyzing the genomic position of known miRNA families, we demonstrate that miRNAs evolve through segmental duplications and tandem duplications in the same manner as protein-coding genes.
Five of the six sets of tandemly duplicated miRNAs that we observed are in arrays or two or three miRNAs, which is in agreement with the observation that 87% of all tandemly duplicated Arabidopsis protein-coding genes occur in arrays or two or three genes (Zhang and Gaut 2003) due to shrinkage of the genome over the last 50 Myr. In addition, ∼17% of all Arabidopsis protein-coding genes reside within tandemly repeated segments (Vision et al. 2000), which is slightly lower than that of miRNAs at 25%.
For large-scale duplications, we observed a higher rate of intrafamily duplicated blocks than we did for randomly selected locations or for miRNAs from different families. In addition to seeing a higher rate of apparent duplicated blocks surrounding miRNAs from the same families, the level of conservation of the flanking proteins was generally higher within miRNA families than duplicated blocks surrounding randomly selected locations and miRNAs from different families. Two of the duplications having at least four or more conserved flanking protein-coding genes (miR159a/miR159b and miR166a/miR166b) were also found in the initial study of large-scale duplications conducted by the Arabidopsis Genome Initiative (2000). This demonstrates that duplication events have caused miRNA family expansion just as they have for protein-coding genes.
A total of 59 (67%) multifamily miRNA genes were within either a tandem or large-scale duplication. We believe that miRNAs not occurring within duplicated regions are the result of older, less detectable, duplication events, rather than random insertions. The accumulation of chromosomal rearrangements over time, in addition to events such as gene loss, are some of the more well-known hindrances to detecting older duplications, and therefore may limit our findings to more recently duplicated miRNAs (∼39 Myr).
Our understanding of miRNA evolution serves as a starting point for elucidating their complex regulatory roles. Expression data provide some insight into the functional divergence of duplicated miRNAs by capturing differences in specific tissue samples. We chose to use the MPSS expression data set because it can distinguish between different miRNA loci and has 17 different tissue-specific libraries for comparing expression profiles. Additional large-scale expression data sets such as ESTs or cloned libraries were too limiting to incorporate into our analysis. Only two miRNAs were captured via ESTs. The ASRP data set (Gustafson et al. 2005) is highly sensitive, but fails to distinguish among family members.
The cutoff that we used to determine whether a downstream MPSS signature should be associated with a miRNA is arbitrary. Supporting our cutoff choice, we were able to observe the characteristically higher density of signatures slightly downstream, ∼400 nt, of the miRNA precursor. This observation is consistent with previous work done on public and private MPSS sets in which the majority of miRNAs had a signature within 500 nt downstream of the miRNA (Wang et al. 2004). To provide further evidence that these downstream signatures are in fact representing the miRNA transcript, we looked at ESTs. Due to the lack of Arabidopsis ESTs containing miRNAs, available ESTs from other plant species provide evidence for expression downstream of the precursor (Bonnet et al. 2004; Xie et al. 2005; Zhang et al. 2005). Therefore, the downstream MPSS signatures are associated with the miRNA transcript.
Data from MPSS detect just over a third of all known miRNAs. While this number may appear low, it still provides locus-specific expression data. Many of the miRNA loci that were not captured by MPSS were also missed by a combination, if not all, of experimental methods such as cloning, 5′-RACE, and 3′-RACE. This indicates that many of these miRNA genes have low or very cell-specific expression.
Using expression data beyond validating miRNA existence is a challenging task and is limited by the sampling of the tissues at specific points in time. While presence of miRNA expression is informative, absence of expression must be interpreted cautiously. The tissues that lack expression may result from low expression levels, sensitivity, or limitations of the assay. Therefore, the expression data serve as a good starting point for understanding the expression patterns within miRNA families, but will need to be expanded on to have a true understanding of the temporal and spatial patterns of miRNA genes.
Within animal species, miRNAs are commonly found in clusters in which multiple miRNAs are transcribed at the same time in one large polycistronic unit. Consistent with this is our observation that for three tandem duplications in the Arabidopsis genome in which the miRNA is found in the same orientation, there is a single associated MPSS signature downstream of the 3′-miRNA. In these instances, the 5′-miRNA lacked a signature with a significant level of expression. An alternative explanation is simply the lack of expression of the 5′-miRNA.
Overall, we have demonstrated that plant miRNAs families are evolving through duplication events similar to those that drive the evolution of protein-coding genes, and that the duplicated copies take on new expression patterns potentially resulting in neo- and subfunctionalization. The evolutionary relationships within a miRNA family in conjunction with public data enable us to explore the subsequent functional divergence of duplicated genes and can be used for further experimental analysis of their interactions with target mRNA and resulting regulatory effects in plant development. While we have documented specific examples of divergent expression profiles following a duplication event, a more comprehensive understanding will become clear as more expression data become available within Arabidopsis. Our procedures can also be applied in other cereal species, which contain similar families to Arabidopsis, and some monocot-specific families. On a more practical note, our understanding and ability to control gene expression during plant development have the potential to improve crop yields, increase resistance to disease, and increase the adaptability of the plant to its environment. The ability to understand the evolution of plant miRNAs will enable us to understand the complexities of miRNA-based regulation.
Methods
Identification of miRNA genes
To determine the genomic locations of miRNA genes, we downloaded miRNA sequences from the miRNA Registry version 5.0 (http://www.sanger.ac.uk/cgi-bin/Rfam/mirna/), a database of published miRNAs (Griffiths-Jones 2004), and aligned them against the TIGR Arabidopsis genome version 5.0. The protein-coding genes flanking each miRNA were then extracted for our miRNA family expansion analysis.
Categorization of miRNA expansions
For this analysis, we focused on the processes of segmental and tandem duplication, using the similarity among sets of protein-coding genes as markers for regions involved in such duplications (Vision et al. 2000). To categorize apparent expansions of miRNA gene families, we looked at the physical locations of all the members of a family. Tandem duplications are characterized as multiple members occurring within the same intergenic region, or within neighboring intergenic regions.
In order to classify two miRNAs as residing within a duplicated block, their neighboring protein-coding genes must have high similarity to one another at the amino acid level. Therefore, to identify segmental duplications, we developed Perl scripts that extract 10 protein-coding genes upstream and downstream of each miRNA, or tandemly duplicated miRNAs since their flanking protein-coding genes would be the same. The protein-coding genes flanking each miRNA were aligned against a set of 29,161 Arabidopsis peptide sequences (http://www.arabidopsis.org) at the amino acid level, using BLASTP, to retrieve the best non-self matches (assuming E-value < 0.001). For each miRNA, we tallied the number of flanking protein-coding genes with a best non-self match to a protein-coding gene neighboring a miRNA from the same family (i.e., miR156a and miR156b).
Simulation of miRNA expansions
We generated a simulation to determine the random likelihood of a protein-coding gene flanking a miRNA to have a best match to a protein-coding gene neighboring a related miRNA. The simulation randomly selected two protein-coding genes as anchors, representing two related miRNAs from the same family, and then aligned the 10 flanking protein-coding genes against all Arabidopsis genes using BLASTP. It then tallied the total number of protein-coding genes from the first anchor that had a best non-self match (E-value < 0.001) with a protein-coding gene neighboring the other anchor point. We repeated this process 1000 times to recreate the frequency of observing a duplication event between two genomic regions.
Estimation of synonymous substitutions and duplication event dating
A Perl script parsed the peptide alignment, from BLASTP, for each pair of conserved flanking protein-coding genes within each miRNA duplicated region to obtain a high quality alignment. Using the protein alignment as our guide, the codons were extracted for each amino acid that was aligned between genes, excluding regions containing gaps. The level of synonymous substitution for these nucleotide sequences was calculated with codeml (Yang 1997), which uses a maximum likelihood method under the F3×4 model (Goldman and Yang 1994). The mean Ks value was calculated for each pair of protein-coding genes within a duplicated block and then used for determining the approximate date of divergence, D, with the equation: D = Ks/2E. We assumed a constant rate of synonymous substitution for dicots, E, as 1.5 × 10−8 substitutions/synonymous site/year (Koch et al. 2000).
Expression analysis
We obtained MPSS signatures from the Delaware Biotechnology Institute (http://mpss.udel.edu/at/). All of the MPSS signatures were loaded into a custom MySQL database designed for this task. The intergenic region downstream of each miRNA was extracted and then scanned for dpn-II restriction sites, used by the MPSS technology. For each dpn-II site, the 20-mer signature was extracted and queried against our database to filter out all signatures lacking reliability, uniqueness, or a significant expression level. Each signature is grouped into a class indicating the signature position relative to the genome annotation. Only class 4 signatures, indicating transcript expression within an intergenic region, were extracted. We associated the first downstream signature meeting these criteria with the miRNA.
Acknowledgments
We thank T. Kellog and N. Chen for their critical reading of the manuscript and K. Nabuta and B. Meyers for the MPSS data. This work was supported by the National Science Foundation (grants #0321685 and #27870201) and USDA ARS CRIS project 1907-21000-014.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4680506
References
- Achard P., Herr A., Baulcombe D.C., Harberd N.P., Herr A., Baulcombe D.C., Harberd N.P., Baulcombe D.C., Harberd N.P., Harberd N.P. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;131:3357–3365. doi: 10.1242/dev.01206. [DOI] [PubMed] [Google Scholar]
- Adams K.L., Wendel J.F., Wendel J.F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Ambros V., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Eddy S.R., Griffiths-Jones S., Marshall M., Griffiths-Jones S., Marshall M., Marshall M., et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative Analysis of the genome sequenceof the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel B., Bartel D.P., Bartel D.P. MicroRNAs: At the root of plant development? Plant Physiol. 2003;132:709–717. doi: 10.1104/pp.103.023630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Wolfe K.H., Wolfe K.H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004a;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Wolfe K.H., Wolfe K.H. Widespread paleopolyploidy in model plant species inferredfrom age distributions of duplicate genes. Plant Cell. 2004b;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E., Wuyts J., Rouze P., de Van Peer Y., Wuyts J., Rouze P., de Van Peer Y., Rouze P., de Van Peer Y., de Van Peer Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. 2004;101:11511–11516. doi: 10.1073/pnas.0404025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers J.E., Chapman B.A., Rong J., Paterson A.H., Chapman B.A., Rong J., Paterson A.H., Rong J., Paterson A.H., Paterson A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Cannon S.B., Mitra A., Baumgarten A., Young N.D., May G., Mitra A., Baumgarten A., Young N.D., May G., Baumgarten A., Young N.D., May G., Young N.D., May G., May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;2004:4–10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L., Izhaki A., Baum S.F., Bowman J.L., Baum S.F., Bowman J.L., Bowman J.L. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Engstrom E.M., Izhaki A., Bowman J.L., Izhaki A., Bowman J.L., Bowman J.L. Promoter bashing, microRNAs, and Knox genes. New insights, regulators, and targets-of-regulation in the establishment of lateral organ polarity in Arabidopsis. Plant Physiol. 2004;135:685–694. doi: 10.1104/pp.104.040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman N., Yang Z., Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson A.M., Allen E., Givan S., Smith D., Carrington J.C., Kasschau K.D., Allen E., Givan S., Smith D., Carrington J.C., Kasschau K.D., Givan S., Smith D., Carrington J.C., Kasschau K.D., Smith D., Carrington J.C., Kasschau K.D., Carrington J.C., Kasschau K.D., Kasschau K.D. ASRP: The Arabidopsis Small RNA Project Database. Nucleic Acids Res. 2005;33:D637–D640. doi: 10.1093/nar/gki127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Juarez M.T., Kui J.S., Thomas J., Heller B.A., Timmermans M.C., Kui J.S., Thomas J., Heller B.A., Timmermans M.C., Thomas J., Heller B.A., Timmermans M.C., Heller B.A., Timmermans M.C., Timmermans M.C. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature. 2004;428:84–88. doi: 10.1038/nature02363. [DOI] [PubMed] [Google Scholar]
- Koch M.A., Haubold B., Mitchell-Olds T., Haubold B., Mitchell-Olds T., Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- Lai E.C., Tomancak P., Williams R.W., Rubin G.M., Tomancak P., Williams R.W., Rubin G.M., Williams R.W., Rubin G.M., Rubin G.M. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton-Rauh A. Evolutionary dynamics of duplicated genes in plants. Mol. Phylogenet. Evol. 2003;29:396–409. doi: 10.1016/j.ympev.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N., Yeom K.H., Lee S., Baek S.H., Kim V.N., Lee S., Baek S.H., Kim V.N., Baek S.H., Kim V.N., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C., Kasschau K.D., Rector M.A., Carrington J.C., Kasschau K.D., Rector M.A., Carrington J.C., Rector M.A., Carrington J.C., Carrington J.C. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher C., Timmermans M., Stein L., Ware D., Timmermans M., Stein L., Ware D., Stein L., Ware D., Ware D.2004. Identifying microRNAs in plant genomes. In Computational systems bioinformatics (ed. IEEE), (ed. F. Titsworth), pp. 718–723. IEEE; Stanford, CA [Google Scholar]
- Meyers B.C., Tej S.S., Vu T.H., Haudenschild C.D., Agrawal V., Edberg S.B., Ghazal H., Decola S., Tej S.S., Vu T.H., Haudenschild C.D., Agrawal V., Edberg S.B., Ghazal H., Decola S., Vu T.H., Haudenschild C.D., Agrawal V., Edberg S.B., Ghazal H., Decola S., Haudenschild C.D., Agrawal V., Edberg S.B., Ghazal H., Decola S., Agrawal V., Edberg S.B., Ghazal H., Decola S., Edberg S.B., Ghazal H., Decola S., Ghazal H., Decola S., Decola S. The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res. 2004a;14:1641–1653. doi: 10.1101/gr.2275604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., Vu T.H., Tej S.S., Ghazal H., Matvienko M., Agrawal V., Ning J., Haudenschild C.D., Vu T.H., Tej S.S., Ghazal H., Matvienko M., Agrawal V., Ning J., Haudenschild C.D., Tej S.S., Ghazal H., Matvienko M., Agrawal V., Ning J., Haudenschild C.D., Ghazal H., Matvienko M., Agrawal V., Ning J., Haudenschild C.D., Matvienko M., Agrawal V., Ning J., Haudenschild C.D., Agrawal V., Ning J., Haudenschild C.D., Ning J., Haudenschild C.D., Haudenschild C.D. Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat. Biotechnol. 2004b;22:1006–1011. doi: 10.1038/nbt992. [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D., Schommer C., Schwab R., Carrington J.C., Weigel D., Schwab R., Carrington J.C., Weigel D., Carrington J.C., Weigel D., Weigel D. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- Papp I., Mette M.F., Aufsatz W., Daxinger L., Schauer S.E., Ray A., van der Winden J., Matzke M., Matzke A.J., Mette M.F., Aufsatz W., Daxinger L., Schauer S.E., Ray A., van der Winden J., Matzke M., Matzke A.J., Aufsatz W., Daxinger L., Schauer S.E., Ray A., van der Winden J., Matzke M., Matzke A.J., Daxinger L., Schauer S.E., Ray A., van der Winden J., Matzke M., Matzke A.J., Schauer S.E., Ray A., van der Winden J., Matzke M., Matzke A.J., Ray A., van der Winden J., Matzke M., Matzke A.J., van der Winden J., Matzke M., Matzke A.J., Matzke M., Matzke A.J., Matzke A.J. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 2003;132:1382–1390. doi: 10.1104/pp.103.021980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A.H., Bowers J.E., Chapman B.A., Bowers J.E., Chapman B.A., Chapman B.A. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince V.E., Pickett F.B., Pickett F.B. Splitting pairs: The diverging fates of duplicated genes. Nat. Rev. Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P., Rhoades M.W., Bartel B., Bartel D.P., Bartel B., Bartel D.P., Bartel D.P. MicroRNAs in plants. Genes & Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D P , Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D P , Lim L.P., Burge C.B., Bartel B., Bartel D P , Burge C.B., Bartel B., Bartel D P , Bartel B., Bartel D P , Bartel D P Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A., Longden I., Bleasby A., Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D., Riester M., Schommer C., Schmid M., Weigel D., Schommer C., Schmid M., Weigel D., Schmid M., Weigel D., Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Vision T.J., Brown D.G., Tanksley S.D., Brown D.G., Tanksley S.D., Tanksley S.D. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- Wang X.J., Reyes J.L., Chua N.H., Gaasterland T., Reyes J.L., Chua N.H., Gaasterland T., Chua N.H., Gaasterland T., Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5:R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Allen E., Fahlgren N., Calamar A., Givan S.A., Carrington J.C., Allen E., Fahlgren N., Calamar A., Givan S.A., Carrington J.C., Fahlgren N., Calamar A., Givan S.A., Carrington J.C., Calamar A., Givan S.A., Carrington J.C., Givan S.A., Carrington J.C., Carrington J.C. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005;138:2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K., Kigawa T., Inoue M., Tateno M., Yamasaki T., Yabuki T., Aoki M., Seki E., Matsuda T., Nunokawa E., Kigawa T., Inoue M., Tateno M., Yamasaki T., Yabuki T., Aoki M., Seki E., Matsuda T., Nunokawa E., Inoue M., Tateno M., Yamasaki T., Yabuki T., Aoki M., Seki E., Matsuda T., Nunokawa E., Tateno M., Yamasaki T., Yabuki T., Aoki M., Seki E., Matsuda T., Nunokawa E., Yamasaki T., Yabuki T., Aoki M., Seki E., Matsuda T., Nunokawa E., Yabuki T., Aoki M., Seki E., Matsuda T., Nunokawa E., Aoki M., Seki E., Matsuda T., Nunokawa E., Seki E., Matsuda T., Nunokawa E., Matsuda T., Nunokawa E., Nunokawa E., et al. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 2004;337:49–63. doi: 10.1016/j.jmb.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Zhang L., Gaut B.S., Gaut B.S. Does recombination shape the distribution and evolution of tandemly arrayed genes (TAGs) in the Arabidopsis thalianagenome? Genome Res. 2003;13:2533–2540. doi: 10.1101/gr.1318503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.H., Pan X.P., Wang Q.L., Cobb G.P., Anderson T.A., Pan X.P., Wang Q.L., Cobb G.P., Anderson T.A., Wang Q.L., Cobb G.P., Anderson T.A., Cobb G.P., Anderson T.A., Anderson T.A. Identification and characterization of new plant microRNAs using EST analysis. Cell Res. 2005;15:336–360. doi: 10.1038/sj.cr.7290302. [DOI] [PubMed] [Google Scholar]