Abstract
8-Oxoguanine (8-oxoG), a major spontaneous form of oxidative DNA damage, is considered to be a natural cause of genomic diversity in organisms because of its mutagenic potential. The steady-state level of 8-oxoG in the nuclear genome of a human cell has been estimated to be several residues per 106 guanines. In the present study, to clarify the genome-wide distribution of 8-oxoG in the steady state, we performed fluorescence in situ detection of 8-oxoG on human metaphase chromosomes using a monoclonal antibody. Multiple dot-like signals were observed on each metaphase chromosome. We then mapped the position of the signal at megabase resolution referring to the cytogenetically identified chromosomal band, and demonstrated that 8-oxoG is unevenly distributed in the normal human genome and that the distribution pattern is conserved among different individuals. Moreover, we found that regions with a high frequency of recombination and single nucleotide polymorphisms (SNPs) are preferentially located within chromosomal regions with a high density of 8-oxoG. Our findings suggest that 8-oxoG is one of the main causes of frequent recombinations and SNPs in the human genome, which largely contribute to the genomic diversity in human beings.
Mutations are sequence variations in the genome caused by base substitutions, insertions/deletions, duplications, and recombinations, which largely contribute to genomic diversity in living organisms (Aquadro et al. 2001; Nevo 2001). Mutagenesis is known to be a biological outcome of processing damaged DNA during genome replication, transcription, repair, and recombination, and enzymes that act upon DNA in the transactions have been well documented (Hanawalt 1998; Maki 2002). Analyses of spontaneous mutagenesis in various organisms revealed that these mutations do not occur randomly in the genome sequence and that their distribution and mutation spectrum exhibit strong bias in the genome (Drake 1991; Maki 2002). Recent progress in human genome projects demonstrated that human genome sequence variation, such as single nucleotide polymorphisms (SNPs), differs greatly in extent across the genome (Sachidanandam et al. 2001; Lercher and Hurst 2002; Malcom et al. 2003). This may be a result of the nonrandom distribution of premutagenic lesions; however, how and where DNA is damaged in the genome remains to be elucidated.
Amongst the various forms of DNA damage, oxidative DNA lesions caused by reactive oxygen species (ROS) that are generated both as a byproduct of oxidative metabolism and as a consequence of exposure to ionizing radiation and other environmental factors are considered to be a major threat to the genome (Hanawalt 1998; Barnes and Lindahl 2004). Among the four bases, guanine is known to be the most susceptible to oxidation, and its simple oxidized form, 8-oxoguanine (8-oxoG), is one of the major oxidation products in DNA or nucleotides (Kasai and Nishimura 1984). 8-oxoG is known as a potent premutagenic lesion, because it can pair with adenine as well as cytosine during DNA replication and cause a G:C to T:A transversion mutation (Shibutani et al. 1991). To counteract the mutagenic potential of 8-oxoG, organisms are equipped with elaborate error-avoiding mechanisms. In Escherichia coli, MutT hydrolyzes 8-oxo-dGTP, thus avoiding its incorporation into DNA (Maki and Sekiguchi 1992), MutM is an 8-oxoG DNA glycosylase that excises the 8-oxoG opposite cytosine (Tchou et al. 1991), and MutY with adenine DNA glycosylase activity excises adenine misincorporated opposite 8-oxoG (Michaels et al. 1992). In humans, homologs of MutT such as MTH1 (Sakumi et al. 1993; Furuichi et al. 1994), MTH2 (Cai et al. 2003), and NUDT5 (Ishibashi et al. 2003), OGG1, a functional homolog of MutM (Aburatani et al. 1997; Boiteux and Radicella 1999; Nishioka et al. 1999), and a MutY homolog, MUTYH (MYH) (Slupska et al. 1996; Ohtsubo et al. 2000), are likely to suppress spontaneous mutagenesis initiated by 8-oxoG. Mutant bacteria or mice lacking one of these genes exhibit a mutator phenotype with an increased spontaneous mutation rate or an increased susceptibility to carcinogenesis (Miller 1996; Sekiguchi and Tsuzuki 2002; Nakabeppu et al. 2004). Evolutionary conservation of error-avoiding mechanisms suggests that the frequent occurrence of 8-oxoG in the genome is associated with genomic diversity. However, the effects of 8-oxoG on nucleotide diversity such as due to SNPs as well as mutagenesis in humans remain to be elucidated.
It has been reported that there are constitutively several thousand residues of 8-oxoG in the nuclear genome of normal human tissues or cultured cells (Gedik and Collins 2005); however, their genomic distribution has not yet been intensively characterized. In the present study, we quantitatively examined the distribution of 8-oxoG in the human genome in order to delineate the genetic influence of 8-oxoG. In situ detection of 8-oxoG using a monoclonal antibody on a human metaphase spread prepared from peripheral lymphocytes revealed that the genome-wide distribution of chromosomal regions with a high density of 8-oxoG is highly conserved among individuals. Moreover, we found that chromosomal regions with a high density of 8-oxoG coincide with regions exhibiting a high meiotic recombination rate as well as with those with a high density of single nucleotide polymorphisms (SNPs).
Results
In situ detection of 8-oxoG in human chromosomes
Using HPLC-MS/MS, we determined the contents of 2′-deoxy-8-oxoguanosine (8-oxo-dG) in human nuclear DNA prepared from freshly isolated peripheral lymphocytes or from those cultured after PHA stimulation. The former contained 2.66 (SEM = 0.05) residues of 8-oxo-dG per 106 residues of deoxyguanosine (dG), and the latter, 3.07 (SEM = 0.17) residues of 8-oxo-dG per 106 residues of dG. These numbers correspond to ∼10,000 8-oxoG residues per single nucleus of a human cell (2n: 6.4 × 109 bp = 12.8 × 109 bases, which contain ∼3.2 × 109 guanine residues), and this represents the steady-state level of 8-oxoG in the human genome.
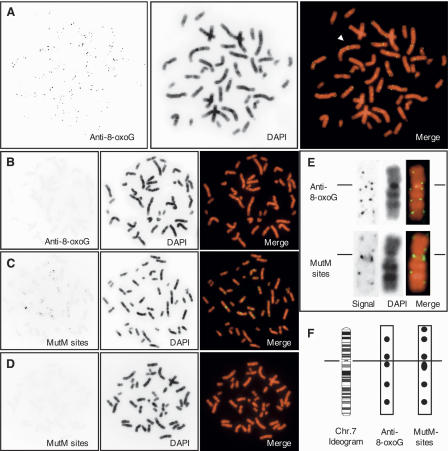
Next, we performed immunofluorescence microscopy using anti-8-oxo-dG monoclonal antibody (N45.1) for in situ detection of 8-oxoG in a human metaphase spread prepared from PHA-stimulated peripheral lymphocytes. The preparation was pretreated with RNase A to eliminate fluorescent signals derived from 8-oxoG in RNA and with HCl to denature the double-stranded DNA. In addition to diffused fine signals with a weak fluorescence intensity, multiple dot-like signals with a strong fluorescence intensity were observed in all interphase nuclei (data not shown) or metaphase chromosomes (Fig. 1A). Since each dot-like signal exhibited a different fluorescence intensity, each single fluorescent dot was likely derived from multiple 8-oxoG residues in the same region. Thus, a chromosomal region with a strong fluorescent signal was considered to have a high density of 8-oxoG, in contrast to a chromosomal region without such a signal. Twin dots of fluorescent signals were often found at the same region in sister chromatids (Fig. 1A, arrowhead).
Figure 1.
In situ detection of 8-oxoG on human metaphase chromosomes. (A) Immunological detection of 8-oxoG. The arrowhead indicates one example of a twin dot on sister chromatids. (B) Immunological detection of 8-oxoG after treatment with MutM. (C) MutM-dependent in situ end-labeling. (D) MutM (−) negative control for in situ end-labeling. (E) Signal patterns on chromosome 7: comparison between immunological detection (top) and MutM-sensitive sites (bottom). Lines indicate centromere. The monochrome images of signal (left) and DAPI (middle) are digitally enhanced and positively inverted. For a merged image, signal and DAPI images are pseudo-colored with green and red, respectively (right). (F) Signal positions on chromosome 7 are indicated as filled circles on corresponding regions in the ideogram of the G-banding pattern at the 850-band resolution obtained from the NCBI ftp site. The line indicates centromeres.
When a specimen was sequentially treated with RNase A and DNase I prior to the first antibody reaction, no fluorescent signal was observed, while pretreatment with RNase A and RNase H, instead of with DNase I, altered neither its intensity nor pattern (data not shown). Pretreatment of a specimen with a bacterial enzyme, MutM, which excises 8-oxoG opposite cytosine, completely abolished the fluorescent signal (Fig. 1B). Furthermore, pre-adsorption of the anti-8-oxo-dG with free 8-oxo-dG also diminished the immunoreactivity to the level obtained without the primary antibody (data not shown). We thus established a method for in situ detection of 8-oxoG in human chromosomes.
We next performed a MutM-dependent in situ end-labeling assay to detect MutM-sensitive sites, and multiple dot-like signals were again detected on each chromosome (Fig. 1C). Without the pretreatment with MutM, no signal was detected (Fig. 1D). By comparing the distribution of fluorescent signals on chromosome 7 obtained with the two approaches (Fig. 1E,F), we found that five of six regions found to be MutM sensitive coincided with 8-oxoG-containing regions detected by immunofluorescence microscopy. From these results, we concluded that the fluorescent signals obtained with the two approaches represent 8-oxoG present in the human genome, thus indicating that the fluorescent dots represent chromosomal regions with a high density of steady-state 8-oxoG. Minor differences between the sites identified by the anti-8-oxoG antibody and those found to be MutM sensitive may reflect the fact that MutM recognizes formamidepyrimidine derivatives as well as 8-oxoG (Boiteux et al. 1987, 1992; Tchou et al. 1991). In subsequent analyses, we therefore applied in situ immunodetection to map the location of 8-oxoG on human chromosomes.
Genome-wide distribution of 8-oxoG in the human genome
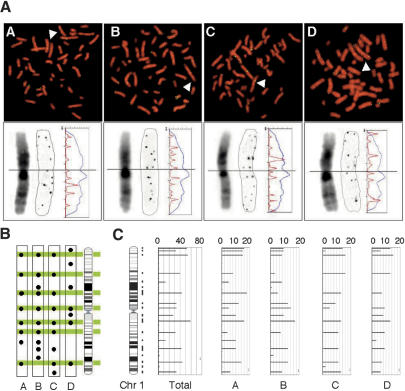
To examine whether chromosomal regions with a high density of 8-oxoG that we observed in one human subject are common among different individuals, we compared the distribution of 8-oxoG on the chromosome of peripheral lymphocytes prepared from four healthy volunteers. As shown in Figure 2A (top), the number and intensity of 8-oxoG immunofluorescent signals detected in a single cell were essentially the same among the four individuals. In order to compare the intrachromosomal distribution of 8-oxoG among the four, we next analyzed one copy of chromosome 1 in a single cell from each individual at higher magnification (Fig. 2A, bottom). Based on chromosomal bands visualized by DAPI and on an intensity plot of the immunofluorescent signals detected along chromosome 1, we mapped each signal position on an ideogram of this chromosome (Fig. 2A, bottom), and found that the distribution pattern of 8-oxoG signals on chromosome 1 was highly conserved among the four volunteers (Fig. 2B). To carry out genome-wide mapping of chromosomal regions with a high density of 8-oxoG, we next analyzed the distribution of immunofluorescent signals on all chromosomes of the four subjects (Fig. 2C; Supplemental Fig. 1). We identified the position of each immunofluorescent dot at megabase resolution referring to the cytogenetically identified chromosomal band. A single immunofluorescent dot was counted as one in 8-oxoG scoring (See Methods and Supplemental Table 1a); thus, the 8-oxoG score for a metaphase (4n) was determined to be 306 (SD = 22, n = 4 individuals), while the number of chromosomal regions with a high density of 8-oxoG was 190 (SD = 8, n = 4 individuals, Supplemental Table 1b) on average. There was no marked difference in these values among the four individuals. The total number of chromosomal regions with a high density of 8-oxoG was 237 in the four subjects, and among them, four regions were mapped on the Y chromosome. Of 233 regions (excluding the four on the Y chromosome), 147 regions (63%) were detected in all four subjects, 37 (16%) were detected in three, and 30 regions (13%) were detected only in one subject (Supplemental Table 1c).
Figure 2.
Genome-wide distribution of 8-oxoG is conserved among individuals. (A, top) Immunological detection of 8-oxoG on metaphase chromosomes from four individuals (A–D, green: immuno-fluorescence signal, red: DAPI). Arrowheads indicate a single chromosome 1. (A, bottom) High-magnification image of chromosome 1 (arrow, top). A positively inverted monochrome image of DAPI (left) and signal (middle) is presented. The fluorescence intensity of DAPI (blue) and signal (red) are plotted on the profile (vertical axis: chromosome length, horizontal axis: fluorescence intensity). (B) The position of each signal detected on chromosome 1 is indicated by a filled circle on the corresponding region in the ideogram. The regions detected in more than three chromosomal copies of the four individuals are colored. (C) Total scoring for 8-oxoG from 10 copies of chromosome 1 of the four subjects are represented as bar plots with the chromosome length on the y-axis and the 8-oxoG score on the x-axis, max = 20 points, see Methods). The vertical lines on the right of the ideogram indicate mapped regions. A bar indicating frequency per region was plotted at the center of the corresponding region.
An averaged 8-oxoG score per megabase in a given region of each chromosome except for the Y chromosome was calculated and its distribution was mapped on a connected physical map of chromosomes 1–22 and X (Supplemental Fig. 2a). The distribution of chromosomal regions with a high density of 8-oxoG across the entire genome was statistically compared among four subjects. The Kruskal-Wallis test revealed that there was no significant difference among the four subjects (P = 0.54, Supplemental Fig. 2a), and moreover, there was a strong correlation between any pair of the four subjects (coefficient >0.7, P < 0.0001, Spearman’s rank correlation test, Supplemental Fig. 2b). These analyses revealed that the genome-wide distribution of 8-oxoG, in terms of its chromosomal region and frequency, was highly similar among the four individuals, suggesting that 8-oxoG at steady state is generally distributed in distinct regions of human chromosomes. Thus, results from the four subjects were combined to obtain a comprehensive map of chromosomal regions with a high density of 8-oxoG (Fig. 3). These high-density regions were distributed throughout the genome, and at least a single region was mapped on each arm of all chromosomes. The high densities of 8-oxoG were found in regions corresponding to GC-rich R-bands and GC-very-rich T-bands or near boundaries between R and G bands more frequently than in those corresponding to AT-rich G-bands. The total 8-oxoG score per chromosome increased with increasing chromosomal length (Supplemental Table 1a). However, the 8-oxoG density in each chromosome (total 8-oxoG score per megabase) varied among the chromosomes, and the shorter chromosomes tended to exhibit a higher 8-oxoG density (Supplemental Table 1a). Chromosomes 18 and 19, whose sizes are very close to each other, exhibit a similar level of 8-oxoG density, regardless of the difference in their GC content; i.e., 40% in chromosome 18 and 48% in chromosome 19, suggesting that another factor(s) in addition to the GC content may have control over the distribution of 8-oxoG in the human genome.
Figure 3.
Comprehensive mapping of 8-oxoG. The positions at which 8-oxoG was detected are indicated as vertical lines on the right of the ideogram. The total 8-oxoG score for each region was graded from zero to five and indicated as filled circles.
8-oxoG and meiotic recombination rate in the human genome
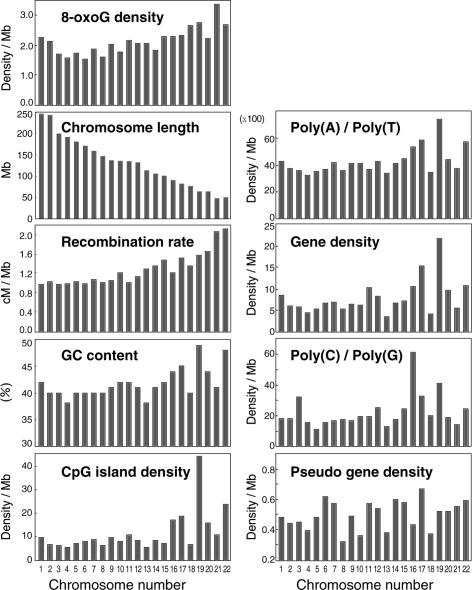
In recent years, it has been shown that each human chromosome has different features in terms of certain sequence parameters revealed by the human genome project (Venter et al. 2001). To determine the factors involved in the distinct distribution of 8-oxoG in human chromosomes, we first performed simple regression analyses considering the 8-oxoG density as the response and eight different genomic parameters as the predictor (Fig. 4; Table 1). The strongest correlation was found between the 8-oxoG density and the meiotic recombination rate (data from Kong et al. 2002) (coefficient = 0.79, R2 = 0.6, P < 0.0001, Table 1). We also noticed its significant positive correlation with GC content, CpG-island density, poly(A) or poly(T) fraction and gene density (coefficient = 0.53, 0.51, 0.45, 0.39, respectively, Table 1). Since it is well known that CpG-island density, the poly(A) or poly(T) fraction and gene density strongly correlate with the GC content, the GC content can be considered as one of the factors determining the distribution of 8-oxoG in the human genome. There is a significant negative correlation between the 8-oxoG density and chromosomal length as we noted above. On the other hand, the multiple regression analysis with the six predictors, which showed a significant correlation with the 8-oxoG density in simple regression analyses, indicated that only the recombination rate has a significant correlation with the 8-oxoG density (P = 0.0127, Table 1).
Figure 4.
Bar plots of 8-oxoG density and eight predictors. The recombination rate (sex-averaged) was obtained from the deCODE study (Kong et al. 2002). The GC content and CpG island density were obtained from Venter et al. (2001). Gene density, pseudo gene density, and chromosomal length were obtained from the Ensembl Genome browser (http://www.ensembl.org/Homo_sapiens/). Poly (A or T) and (C or G) fractions were obtained from Rohrer et al. (2002).
Table 1.
Results of regression analyses with 8-oxoG density as the response
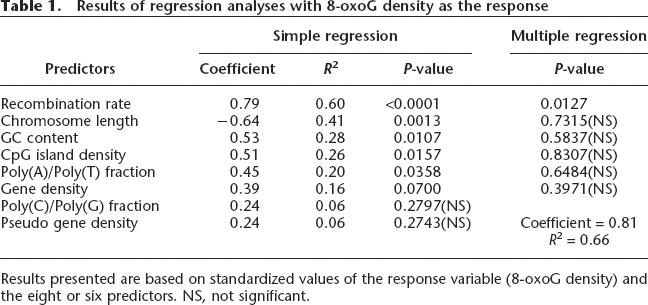
Results presented are based on standardized values of the response variable (8-oxoG density) and the eight or six predictors. NS, not significant.
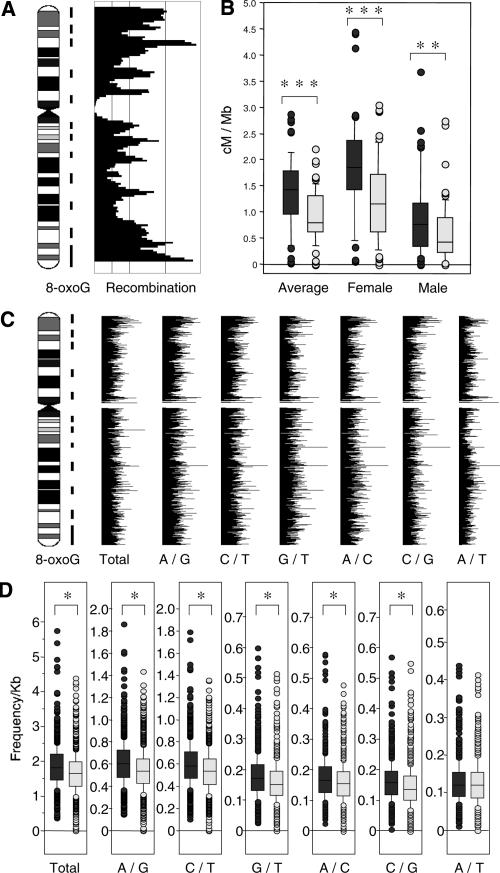
To further examine whether the intrachromosomal distribution of 8-oxoG has any correlation with that of recombination-prone sites, we compared locations of regions with a high density of 8-oxoG and regions with a high frequency of recombination on chromosome 11. As shown in Figure 5A, most of the recombination peaks were located within regions with a high density of 8-oxoG. Statistical analysis revealed that regions with the high 8-oxoG density exhibited a significantly higher recombination rate in comparison with regions lacking this high density (Fig. 5B). The same results were obtained from a genome-wide analysis using data from all autosomes (Supplemental Table 2).
Figure 5.
Comparison of 8-oxoG distribution on chromosome 11 and recombination rate or SNP frequency. (A) Regions with a high density of 8-oxoG are indicated by vertical lines on the right of the ideogram. The sex-averaged recombination rate (deCODE) was calculated with a 3-Mb window in 1-Mb steps and indicated as a bar plot with the chromosome lengths on the y-axis and recombination rates on the x-axis. (B) Box-whisker plot of recombination rates in chromosome 11 within regions with a high density of 8-oxoG (gray) and regions without a high density of 8-oxoG (light gray). (Mann-Whitney U-test, **P < 0.01, ***P < 0.0001). The center line within the box gives the median. The top and bottom boxes show the 75 and 25 percentiles, respectively. The top and bottom of the whisker show the 5 and 95 percentiles, respectively. (C) Bar plots of SNP frequencies. The SNP frequency for each base substitution was calculated with a 200-Kb window and in 50-Kb steps. The substitution spectra are indicated by two letters separated with a slash. For example, A/G means that A and G were recorded in a single position, and does not suggest the direction of the substitution. The regions with a high density of 8-oxoG are indicated by vertical lines on the right of the ideogram of chromosome 11. (D) Box-whisker plots of SNP frequencies in chromosome 11 regions with a high density of 8-oxoG (gray) and in regions without a high density of 8-oxoG (light gray) (Mann-Whitney U-test, *P < 0.05). Centrometic regions were excluded from the calculation.
8-oxoG and SNPs in the human genome
Since some of the base substitutions that occurred in the human genome in the past are preserved as SNPs in the current population, we hypothesized that regions with a high density of 8-oxoG may be colocalized with a high density of SNPs. To evaluate our hypothesis, we compared both the distributions and frequencies across the entire genome. From the SNP database at NCBI (build 33), we selected SNPs with only two reported differences in bases at the same position (A/G, C/T, G/T, A/C, C/G, A/T), and frequencies of the six types of substitutions were calculated. The chromosomal position and frequency of each type of substitution were expressed on an ideogram as bar plots (Fig. 5C for chromosome 11, Supplemental Fig. 3 for all autosomes). All types of substitutions were distributed throughout the genome, and their frequencies varied from place to place. All chromosomes possess multiple regions with a significantly higher frequency of each base substitution spectrum compared with surrounding regions. In general, regions with a high density of 8-oxoG tend to exhibit a higher frequency of SNPs (Supplemental Fig. 3). Statistical analyses clearly showed that the frequencies of all substitutions except the A/T substitution were significantly higher in regions with a high density of 8-oxoG in chromosome 11 than in the regions without this high density (Fig. 5D). The same results were obtained from genome-wide analysis using data from all autosomes (Supplemental Table 3). We also performed the same analysis against the SNP data set obtained from shotgun sequencing, and the results were similar (Supplemental Table 4).
Discussion
In the present study, we detected and mapped positions of 8-oxoG in human chromosomes by means of in situ immunodetection to clarify its genome-wide distribution. The chromosomal distribution of 8-oxoG, detected as clusters of several residues at steady state, is uneven in the total genome of a single human cell in terms of position and frequency, while there are many common regions with a high density of 8-oxoG in the human genome, even in cells of different individuals. Chromosomal regions with a high density of 8-oxoG contain peaks of recombination, as do those with a high density of SNPs.
To elucidate the underlying cause for genomic diversity in humans, it is essential to determine why 8-oxoG preferentially locates within distinct regions of the human genome. In the in situ immunodetection, MutM, which excises 8-oxoG opposite cytosine, largely abolished the immune signals for 8-oxoG, thus suggesting that most 8-oxoG present in the normal human genome is a product of oxidation of a G:C pair. Our results clearly indicate that the GC content or GC content-related factors are one of the determinants for 8-oxoG density in autosomes. However, we are still speculating as to whether there is another factor(s) that more strongly influences the local 8-oxoG density. The steady-state level of 8-oxoG in the normal human genome reflects its generation and repair; thus, in regions with a high density of 8-oxoG, the generation of this lesion exceeds its removal by repair. The results of in situ detection of 8-oxoG indicate that the genome may not be a homogeneous target for oxidation and repair. It has been pointed out that various factors contribute to the outcome of oxidative damage to DNA (Beckman and Ames 1997; Evans and Cooke 2004). The first of these, guanine, possesses the lowest oxidation potential of all DNA bases, and 5′-guanine in sequences GG and GGG is the selective target for oxidation mediated by long-distance electron transport along the double helix (Hall et al. 1996; Steenken 1997; Burrows and Muller 1998). Secondly, internucleosomal DNA appears to be more susceptible to oxidation by physiological iron chelates, such as iron-ADP, while histone-associated nucleosomal DNA is protected (Enright et al. 1996). Moreover, there is accumulating evidence indicating that the local chromatin structure and/or transcription status alter the repair efficiency, thus contributing to the regional differences in damage repair (Livingstone-Zatchej et al. 1997; Wellinger and Thoma 1997; Suter et al. 2000; Sanders et al. 2004). Recently, Ikemura and coworkers have reported that boundary regions of cytogenetically identified chromosomal bands, which are also known to be a transition zone of DNA replication timing and GC content (Tenzen et al. 1997), are likely to be highly mutagenic judging from the higher density of SNPs in these regions (Watanabe et al. 2002). The regions with a high density of 8-oxoG in chromosome 11 apparently correspond to the transition zones, as we noted that the 8-oxoG immunoreactivities are often located in boundary regions of R and/or G bands. We therefore suggest that several structural factors in the genome such as sequence contexts and chromatin structures as well as functional factors such as replication/transcription and repair itself, cooperatively determine the local density of 8-oxoG in a particular genomic region together with the cellular environment that is affected by such factors as redox status.
We showed that chromosomal regions associated with peaks in meiotic recombination are preferentially located within regions with a high density of 8-oxoG. In the present study, we compared the chromosomal distribution of 8-oxoG observed in somatic lymphocytes with that of meiotic recombination, which occurs in meiotic germ cells. In meiotic cells in mouse testis as well as in cultured human cell lines, we observed a similar number of chromosomal regions with a high density of 8-oxoG (M. Ohno and Y. Nakabeppu, unpubl.). Thus, we may assume that there should be the same chromosomal distribution of 8-oxoG in germ cells as observed in somatic lymphocytes.
One can argue that there must be another factor that coincidentally increases the recombination rate in addition to the accumulation of 8-oxoG in a particular region of chromosome, implying that there is no causality between the two events, and we do not have any data at present to deny this possibility. On the contrary, it is also possible that there is causality between the two events. However, we can eliminate the possibility that meiotic recombination itself causes an increased accumulation of 8-oxoG within the recombinogenic site, because we detected 8-oxoG accumulation in the long arms of both X chromosomes in females, which undergoes meiotic recombination only during oogenesis, as well as in the long arm of the Y chromosome which has no counterpart for homologous recombination (see Supplemental Fig. 1) (Burgoyne 1982). Furthermore, the chromosomal distribution of 8-oxoG was observed in somatic lymphocytes, which never undergo meiotic recombination. We thus suggest that 8-oxoG accumulated in a particular region of a chromosome causes meiotic recombination, based on supporting data published by others, as discussed below.
As in other eukaryotes, meiotically programmed double-strand breaks by the enzyme SPO11, a member of the type II-like topoisomerase family of proteins, are essential for initiating meiotic recombination in the mouse (Romanienko and Camerini-Otero 2000). However, meiotic arrest in Spo11-deficient mice was partially rescued by generating DNA damage with irradiation or cisplatin treatment (Romanienko and Camerini-Otero 2000), thus indicating that the double-strand breaks in spermatocytes introduced by DNA damage or its repair resulted in the meiotic recombination. Recently, it was shown that nitric oxide-induced homologous recombination in E. coli is promoted by DNA glycosylases, such as uracil DNA glycosylase or MutM (Spek et al. 2002), suggesting that homologous recombination is induced by strand breaks accumulated during the repair reaction. It is reasonable to assume that excision repair of 8-oxoG or misincorporation of an adenine opposite 8-oxoG increases the chance of strand breaks in the genome when these events are clustered within a limited region. OGG1, an enzyme that excises 8-oxoG opposite cytosine, and MUTYH which excises adenine opposite 8-oxoG, are therefore important for maintaining informative 8-oxoG in the genome, and both are highly expressed in ovary and testis, the reproductive organs (Nishioka et al. 1999; Ohtsubo et al. 2000). Thus, base excision repair of 8-oxoG or adenine opposite 8-oxoG in germ cells may increase recombination, especially in regions containing clusters of 8-oxoG. It has been reported that the meiotic recombination rate in women is higher than that in men, indicating that there is another factor(s), which increases the recombination rate other than solely the genomic sequence context (Kong et al. 2002). In females, environmental factor(s) to which oocytes at the prophase of meiosis I are exposed for many years may increase the recombination rate. Thus, one can speculate that 8-oxoG, which accumulates in oocytes during aging, may result in the increase in the recombination rate seen in females. However, further analysis is required to prove the causal relationship between 8-oxoG and recombination.
Areas with a higher density of SNPs are predominantly distributed within regions with a high density of 8-oxoG, suggesting that 8-oxoG may contribute to the generation of SNPs because of its mutagenic potential. SNPs are unevenly distributed across the human genome; however, the cause(s) of these base substitutions and the factor(s) determining their distribution are still largely unknown. The transition from C:G to T:A, for example, is believed to be largely induced by deamination of 5-methylcytosine at the CpG sequence, but this accounts for only part of the substitution (Venter et al. 2001). A:T to T:A or T:A to A:T substitutions are distinctly independent of the regions with a high density of 8-oxoG (Supplemental Table 2), suggesting that 8-oxoG is somehow involved in base substitutions from or toward a G:C base pair. The accumulation of 8-oxoG in DNA is a result of the incorporation of 8-oxo-dGTP generated in nucleotide pools as well as of the direct oxidation of guanine in DNA, thus primarily causing A:T to C:G or G:C to T:A transversions (Egashira et al. 2002; Arai et al. 2003; Nakabeppu et al. 2006). Recently, it has been shown that 8-oxoG in DNA induces various substitutions not only at the 8-oxoG site, but also in the neighboring regions through error-prone DNA replication in addition to the typical transversions (Efrati et al. 1999). Furthermore, it has been noted that there are correlations between the recombination rate and SNPs or base substitutions (Lercher and Hurst 2002; Hellmann et al. 2003). Thus, we suggest that 8-oxoG is one of the major causes of SNPs as well as recombination, and that its generation and repair are responsible for their uneven genomic distribution, as discussed above. In conclusion, we propose that the spontaneously oxidized base, 8-oxoG, contributes to genomic diversity in human beings.
Methods
Blood samples and cell culture
Peripheral blood samples were obtained from healthy adult volunteers (four unrelated Japanese, two males and two females). We obtained informed consent from all volunteers. Lymphocytes were cultured in RPMI1640 (Gibco-BRL) medium supplemented with 10% FBS, antibiotics and PHA-M (Sigma) for 72 h under normal conditions according to a standard cytogenetic method (Barch 1991).
HPLC–MS/MS analysis of 8-oxoG
Nuclear DNA was prepared from the uncultured or cultured lymphocytes and were subjected to HPLC-MS/MS analysis for 8-oxoG, according to a previously described method (Tsuruya et al. 2003).
In situ immunodetection of 8-oxoG
Toyokuni et al. (1997, 1999) previously described immunofluorescence staining with anti-8-oxo-dG monoclonal antibody (N45.1) to study changes in the intranuclear distribution of 8-oxoG in renal cells of male Wistar rats. We therefore used this monoclonal antibody to detect 8-oxoG in human chromosomes with modifications to the method, as described below. Metaphase chromosome spreads on glass slides were prepared from cultured lymphocytes according to a standard cytogenetic method (Barch 1991). The slides were incubated with 1 mg/mL of RNase A (Sigma) in PBS at 37°C for 1h. Chromosomal DNA was denatured by treating with 2N HCl for 7 min, and was neutralized in 50 mM Tris-base and with PBS. The slides were then incubated with anti-8-oxo-dG mouse monoclonal antibody (JaICA, N45.1, 1:10 dilution) for 2 h at room temperature, and washed three times with PBD (0.1% TritonX-100 in PBS). Then, the slides were incubated with anti-mouse IgG conjugated to Alexa-488 (Molecular Probes Inc., 1:300 dilution) for 40 min at room temperature, and washed and mounted with anti-fade mounting medium containing DAPI (Vector). To verify the specificity of the immunoreactivity, some slides were treated with either 0.1 μg/mL of MutM protein (Sigma) in nicking buffer (10 mM Tris-HCl, 5 μM ZnCl2, 0.5 mM DTT, 0.5 mM EDTA, 1.5% glycerol, 100 μg/mL bovine serum albumin) or 1000 U/mL of DNase I (Sigma) in reaction buffer (50 mM Tris-HCl [pH 7.5], 0.1 mM MgCl2) at 37°C for 1 h after RNase A treatment.
MutM-dependent in situ end-labeling assay
Chromosome slide preparations were incubated with 0.1 μg/mL of BSA or MutM protein and 0.02 μg/mL of purified mAPEX1 protein (Tominaga et al. 2004) in nicking buffer at 37°C for 1 h after RNase A treatment. mAPEX1 was used to convert abasic sites or nicks with 3′-phosphate, which were produced by MutM, to nicks with a 3′-hydroxy group (Madhusudan et al. 2005). The 3′-hydroxy ends were labeled with fluorescein-dUTP using terminal deoxynucleotidyl transferase (Roche Diagnostics) according to the manufacturer’s instructions. The slides were then washed and mounted with anti-fade mounting medium.
Image analysis
Digital images were acquired using Axioskop2 plus equipped with a CCD camera, AxioCam and AxioVision software (Carl Zeiss). Captured images were processed for publication with Adobe Photoshop 5.1 software (Adobe Systems, Inc.). The intensity of each fluorescence was quantified using ImageGage 4.0 software (Fujifilm).
Chromosomal mapping of 8-oxoG
Digital images of well-spread metaphases with clear DAPI-banding patterns were independently acquired for each of the four subjects. The number and positions of 8-oxoG signals on 10 copies of each chromosome from each individual were determined from the images. The chromosomal position of the signal was determined by the DAPI-banding pattern with megabase (Mb) resolution. A single fluorescent dot on one of the sister chromatids of each chromosome was given an 8-oxoG score of 1; thus, twin dots observed in the same region of both sister chromatids were counted as 2. The maximum score was 20 at any given chromosomal region calculated from 20 sister chromatids of each chromosome. When two sister chromatids were not separated at the centromeric region, only a single fluorescence dot or no dot at all was observed; therefore, these signals were excluded from the analysis. This analysis was performed by an individual unaware of the source of each sample.
Calculation of the recombination rate and SNP frequency
All data including results of deCODE high-resolution genetic mapping, human genome sequence data (build33), uniSTS, dbSNPs (build116), and ideogram data were obtained from the NCBI ftp site (ftp://ftp.ncbi.nlm.nih.gov/). The physical positions associated with each of the deCODE genetic markers were obtained by comparing the uniSTS IDs in the databases. The recombination rates at each deCODE genetic marker were calculated with the genetic position and the corresponding physical position according to Kong et al. (2002). We calculated the moving average of the recombination rate for each chromosome with a 3-Mb window and in 1-Mb steps. To determine the base-substitution frequency across each chromosome, we extracted single nucleotide substitutions from the dbSNP database discounting insertions and deletions. We selected data on one-base substitutions having only two different bases recorded in a single position using the refSNP in dbSNP build 116 (http://www.ncbi.nlm.nih.gov/projects/SNP/). Multispectrum data were excluded from the calculation. The moving average of the SNP frequency of all chromosomes was calculated with a 200-Kb window and in 50-Kb steps.
Acknowledgments
We are grateful to Drs. Toshimichi Ikemura and Kenshi Hayashi for valuable discussions. We thank N. Adachi for HPLC MS/MS analysis and A. Matsuyama and S. Kitamura for technical support. We also acknowledge laboratory coworkers for their involvement in many discussions. This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (grant nos. 16012248 and 17014070), and the Japan Society for the Promotion of Science (grant no. 16390119).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.4769606
References
- Aburatani H., Hippo Y., Ishida T., Takashima R., Matsuba C., Kodama T., Takao M., Yasui A., Yamamoto K., Asano M., Hippo Y., Ishida T., Takashima R., Matsuba C., Kodama T., Takao M., Yasui A., Yamamoto K., Asano M., Ishida T., Takashima R., Matsuba C., Kodama T., Takao M., Yasui A., Yamamoto K., Asano M., Takashima R., Matsuba C., Kodama T., Takao M., Yasui A., Yamamoto K., Asano M., Matsuba C., Kodama T., Takao M., Yasui A., Yamamoto K., Asano M., Kodama T., Takao M., Yasui A., Yamamoto K., Asano M., Takao M., Yasui A., Yamamoto K., Asano M., Yasui A., Yamamoto K., Asano M., Yamamoto K., Asano M., Asano M. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997;57:2151–2156. [PubMed] [Google Scholar]
- Aquadro C.F., Bauer Dumont V., Reed F.A., Bauer Dumont V., Reed F.A., Reed F.A. Genome-wide variation in the human and fruitfly: A comparison. Curr. Opin. Genet. Dev. 2001;11:627–634. doi: 10.1016/s0959-437x(00)00245-8. [DOI] [PubMed] [Google Scholar]
- Arai T., Kelly V.P., Komoro K., Minowa O., Noda T., Nishimura S., Kelly V.P., Komoro K., Minowa O., Noda T., Nishimura S., Komoro K., Minowa O., Noda T., Nishimura S., Minowa O., Noda T., Nishimura S., Noda T., Nishimura S., Nishimura S. Cell proliferation in liver of Mmh/Ogg1-deficient mice enhances mutation frequency because of the presence of 8-hydroxyguanine in DNA. Cancer Res. 2003;63:4287–4292. [PubMed] [Google Scholar]
- Barch M.J. The ACT cytogenetics laboratory manual. Raven Press; New York.: 1991. [Google Scholar]
- Barnes D.E., Lindahl T., Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- Beckman K.B., Ames B.N., Ames B.N. Oxidative decay of DNA. J. Biol. Chem. 1997;272:19633–19636. doi: 10.1074/jbc.272.32.19633. [DOI] [PubMed] [Google Scholar]
- Boiteux S., Radicella J.P., Radicella J.P. Base excision repair of 8-hydroxyguanine protects DNA from endogenous oxidative stress. Biochimie. 1999;81:59–67. doi: 10.1016/s0300-9084(99)80039-x. [DOI] [PubMed] [Google Scholar]
- Boiteux S., O’Connor T.R., Laval J., O’Connor T.R., Laval J., Laval J. Formamidopyrimidine-DNA glycosylase of Escherichia coli: Cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 1987;6:3177–3183. doi: 10.1002/j.1460-2075.1987.tb02629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S., Gajewski E., Laval J., Dizdaroglu M., Gajewski E., Laval J., Dizdaroglu M., Laval J., Dizdaroglu M., Dizdaroglu M. Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): Excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry. 1992;31:106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- Burgoyne P.S. Genetic homology and crossing over in the X and Y chromosomes of mammals. Hum. Genet. 1982;61:85–90. doi: 10.1007/BF00274192. [DOI] [PubMed] [Google Scholar]
- Burrows C.J., Muller J.G., Muller J.G. Oxidative nucleobase modifications leading to strand scission. Chem. Rev. 1998;98:1109–1152. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- Cai J.P., Ishibashi T., Takagi Y., Hayakawa H., Sekiguchi M., Ishibashi T., Takagi Y., Hayakawa H., Sekiguchi M., Takagi Y., Hayakawa H., Sekiguchi M., Hayakawa H., Sekiguchi M., Sekiguchi M. Mouse MTH2 protein which prevents mutations caused by 8-oxoguanine nucleotides. Biochem. Biophys. Res. Commun. 2003;305:1073–1077. doi: 10.1016/s0006-291x(03)00864-7. [DOI] [PubMed] [Google Scholar]
- Drake J.W. Spontaneous mutation. Annu. Rev. Genet. 1991;25:125–146. doi: 10.1146/annurev.ge.25.120191.001013. [DOI] [PubMed] [Google Scholar]
- Efrati E., Tocco G., Eritja R., Wilson S.H., Goodman M.F., Tocco G., Eritja R., Wilson S.H., Goodman M.F., Eritja R., Wilson S.H., Goodman M.F., Wilson S.H., Goodman M.F., Goodman M.F. “Action-at-a-distance” mutagenesis. 8-oxo-7, 8-dihydro-2′-deoxyguanosine causes base substitution errors at neighboring template sites when copied by DNA polymerase β. J. Biol. Chem. 1999;274:15920–15926. doi: 10.1074/jbc.274.22.15920. [DOI] [PubMed] [Google Scholar]
- Egashira A., Yamauchi K., Yoshiyama K., Kawate H., Katsuki M., Sekiguchi M., Sugimachi K., Maki H., Tsuzuki T., Yamauchi K., Yoshiyama K., Kawate H., Katsuki M., Sekiguchi M., Sugimachi K., Maki H., Tsuzuki T., Yoshiyama K., Kawate H., Katsuki M., Sekiguchi M., Sugimachi K., Maki H., Tsuzuki T., Kawate H., Katsuki M., Sekiguchi M., Sugimachi K., Maki H., Tsuzuki T., Katsuki M., Sekiguchi M., Sugimachi K., Maki H., Tsuzuki T., Sekiguchi M., Sugimachi K., Maki H., Tsuzuki T., Sugimachi K., Maki H., Tsuzuki T., Maki H., Tsuzuki T., Tsuzuki T. Mutational specificity of mice defective in the MTH1 and/or the MSH2genes. DNA Repair (Amst.) 2002;1:881–893. doi: 10.1016/s1568-7864(02)00113-1. [DOI] [PubMed] [Google Scholar]
- Enright H., Miller W.J., Hays R., Floyd R.A., Hebbel R.P., Miller W.J., Hays R., Floyd R.A., Hebbel R.P., Hays R., Floyd R.A., Hebbel R.P., Floyd R.A., Hebbel R.P., Hebbel R.P. Preferential targeting of oxidative base damage to internucleosomal DNA. Carcinogenesis. 1996;17:1175–1177. doi: 10.1093/carcin/17.5.1175. [DOI] [PubMed] [Google Scholar]
- Evans M.D., Cooke M.S., Cooke M.S. Factors contributing to the outcome of oxidative damage to nucleic acids. Bioessays. 2004;26:533–542. doi: 10.1002/bies.20027. [DOI] [PubMed] [Google Scholar]
- Furuichi M., Yoshida M.C., Oda H., Tajiri T., Nakabeppu Y., Tsuzuki T., Sekiguchi M., Yoshida M.C., Oda H., Tajiri T., Nakabeppu Y., Tsuzuki T., Sekiguchi M., Oda H., Tajiri T., Nakabeppu Y., Tsuzuki T., Sekiguchi M., Tajiri T., Nakabeppu Y., Tsuzuki T., Sekiguchi M., Nakabeppu Y., Tsuzuki T., Sekiguchi M., Tsuzuki T., Sekiguchi M., Sekiguchi M. Genomic structure and chromosome location of the human mutT homologue gene MTH1 encoding 8-oxo-dGTPase for prevention of A:T to C:G transversion. Genomics. 1994;24:485–490. doi: 10.1006/geno.1994.1657. [DOI] [PubMed] [Google Scholar]
- Gedik C.M., Collins A., Collins A. Establishing the background level of base oxidation in human lymphocyte DNA: Results of an interlaboratory validation study. FASEB J. 2005;19:82–84. doi: 10.1096/fj.04-1767fje. [DOI] [PubMed] [Google Scholar]
- Hall D.B., Holmlin R.E., Barton J.K., Holmlin R.E., Barton J.K., Barton J.K. Oxidative DNA damage through long-range electron transfer. Nature. 1996;382:731–735. doi: 10.1038/382731a0. [DOI] [PubMed] [Google Scholar]
- Hanawalt P.C. Genomic instability: Environmental invasion and the enemies within. Mutat. Res. 1998;400:117–125. doi: 10.1016/s0027-5107(98)00084-0. [DOI] [PubMed] [Google Scholar]
- Hellmann I., Ebersberger I., Ptak S.E., Pääbo S., Przeworski M., Ebersberger I., Ptak S.E., Pääbo S., Przeworski M., Ptak S.E., Pääbo S., Przeworski M., Pääbo S., Przeworski M., Przeworski M. A neutral explanation for the correlation of diversity with recombination rates in humans. Am. J. Hum. Genet. 2003;72:1527–1535. doi: 10.1086/375657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi T., Hayakawa H., Sekiguchi M., Hayakawa H., Sekiguchi M., Sekiguchi M. A novel mechanism for preventing mutations caused by oxidation of guanine nucleotides. EMBO Rep. 2003;4:479–483. doi: 10.1038/sj.embor.embor838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai H., Nishimura S., Nishimura S. Hydroxylation of deoxyguanosine at the C-8 position by ascorbic acid and other reducing agents. Nucleic Acids Res. 1984;12:2137–2145. doi: 10.1093/nar/12.4.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A., Gudbjartsson D.F., Sainz J., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Gudbjartsson D.F., Sainz J., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Sainz J., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Barnard J., Hallbeck B., Masson G., Hallbeck B., Masson G., Masson G., et al. A high-resolution recombination map of the human genome. Nat. Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Lercher M.J., Hurst L.D., Hurst L.D. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 2002;18:337–340. doi: 10.1016/s0168-9525(02)02669-0. [DOI] [PubMed] [Google Scholar]
- Livingstone-Zatchej M., Meier A., Suter B., Thoma F., Meier A., Suter B., Thoma F., Suter B., Thoma F., Thoma F. RNA polymerase II transcription inhibits DNA repair by photolyase in the transcribed strand of active yeast genes. Nucleic Acids Res. 1997;25:3795–3800. doi: 10.1093/nar/25.19.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhusudan S., Smart F., Shrimpton P., Parsons J.L., Gardiner L., Houlbrook S., Talbot D.C., Hammonds T., Freemont P.A., Sternberg M.J., Smart F., Shrimpton P., Parsons J.L., Gardiner L., Houlbrook S., Talbot D.C., Hammonds T., Freemont P.A., Sternberg M.J., Shrimpton P., Parsons J.L., Gardiner L., Houlbrook S., Talbot D.C., Hammonds T., Freemont P.A., Sternberg M.J., Parsons J.L., Gardiner L., Houlbrook S., Talbot D.C., Hammonds T., Freemont P.A., Sternberg M.J., Gardiner L., Houlbrook S., Talbot D.C., Hammonds T., Freemont P.A., Sternberg M.J., Houlbrook S., Talbot D.C., Hammonds T., Freemont P.A., Sternberg M.J., Talbot D.C., Hammonds T., Freemont P.A., Sternberg M.J., Hammonds T., Freemont P.A., Sternberg M.J., Freemont P.A., Sternberg M.J., Sternberg M.J., et al. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res. 2005;33:4711–4724. doi: 10.1093/nar/gki781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H. Origins of spontaneous mutations: Specificity and directionality of base-substitution, frameshift, and sequence-substitution mutageneses. Annu. Rev. Genet. 2002;36:279–303. doi: 10.1146/annurev.genet.36.042602.094806. [DOI] [PubMed] [Google Scholar]
- Maki H., Sekiguchi M., Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- Malcom C.M., Wyckoff G.J., Lahn B.T., Wyckoff G.J., Lahn B.T., Lahn B.T. Genic mutation rates in mammals: Local similarity, chromosomal heterogeneity, and X-versus-autosome disparity. Mol. Biol. Evol. 2003;20:1633–1641. doi: 10.1093/molbev/msg178. [DOI] [PubMed] [Google Scholar]
- Michaels M.L., Tchou J., Grollman A.P., Miller J.H., Tchou J., Grollman A.P., Miller J.H., Grollman A.P., Miller J.H., Miller J.H. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry. 1992;31:10964–10968. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- Miller J.H. Spontaneous mutators in bacteria: Insights into pathways of mutagenesis and repair. Annu. Rev. Microbiol. 1996;50:625–643. doi: 10.1146/annurev.micro.50.1.625. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y., Tsuchimoto D., Furuichi M., Sakumi K., Tsuchimoto D., Furuichi M., Sakumi K., Furuichi M., Sakumi K., Sakumi K. The defense mechanisms in mammalian cells against oxidative damage in nucleic acids and their involvement in the suppression of mutagenesis and cell death. Free Radic. Res. 2004;38:423–429. doi: 10.1080/10715760410001688348. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y., Behmanesh M., Yamaguchi H., Yoshimura D., Sakumi K., Behmanesh M., Yamaguchi H., Yoshimura D., Sakumi K., Yamaguchi H., Yoshimura D., Sakumi K., Yoshimura D., Sakumi K., Sakumi K.2006. Prevention of the mutagenecity and cytotoxicity of oxidized purine nucleotides. In Oxidative damage to nucleic acids (eds. M.D. Evans and M.S. Cooke). Landes Bioscience; Georgetown, TX.http://eurekah.com/abstract.php?chapid=2920&bookid=214&catid=78 [Google Scholar]
- Nevo E. Evolution of genome-phenome diversity under environmental stress. Proc. Natl. Acad. Sci. 2001;98:6233–6240. doi: 10.1073/pnas.101109298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K., Ohtsubo T., Oda H., Fujiwara T., Kang D., Sugimachi K., Nakabeppu Y., Ohtsubo T., Oda H., Fujiwara T., Kang D., Sugimachi K., Nakabeppu Y., Oda H., Fujiwara T., Kang D., Sugimachi K., Nakabeppu Y., Fujiwara T., Kang D., Sugimachi K., Nakabeppu Y., Kang D., Sugimachi K., Nakabeppu Y., Sugimachi K., Nakabeppu Y., Nakabeppu Y. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol. Biol. Cell. 1999;10:1637–1652. doi: 10.1091/mbc.10.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo T., Nishioka K., Imaiso Y., Iwai S., Shimokawa H., Oda H., Fujiwara T., Nakabeppu Y., Nishioka K., Imaiso Y., Iwai S., Shimokawa H., Oda H., Fujiwara T., Nakabeppu Y., Imaiso Y., Iwai S., Shimokawa H., Oda H., Fujiwara T., Nakabeppu Y., Iwai S., Shimokawa H., Oda H., Fujiwara T., Nakabeppu Y., Shimokawa H., Oda H., Fujiwara T., Nakabeppu Y., Oda H., Fujiwara T., Nakabeppu Y., Fujiwara T., Nakabeppu Y., Nakabeppu Y. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res. 2000;28:1355–1364. doi: 10.1093/nar/28.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer G.A., Fahrenkrug S.C., Nonneman D., Tao N., Warren W.C., Fahrenkrug S.C., Nonneman D., Tao N., Warren W.C., Nonneman D., Tao N., Warren W.C., Tao N., Warren W.C., Warren W.C. Mapping microsatellite markers identified in porcine EST sequences. Anim. Genet. 2002;33:372–376. doi: 10.1046/j.1365-2052.2002.00880.x. [DOI] [PubMed] [Google Scholar]
- Romanienko P.J., Camerini-Otero R.D., Camerini-Otero R.D. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- Sachidanandam R., Weissman D., Schmidt S.C., Kakol J.M., Stein L.D., Marth G., Sherry S., Mullikin J.C., Mortimore B.J., Willey D.L., Weissman D., Schmidt S.C., Kakol J.M., Stein L.D., Marth G., Sherry S., Mullikin J.C., Mortimore B.J., Willey D.L., Schmidt S.C., Kakol J.M., Stein L.D., Marth G., Sherry S., Mullikin J.C., Mortimore B.J., Willey D.L., Kakol J.M., Stein L.D., Marth G., Sherry S., Mullikin J.C., Mortimore B.J., Willey D.L., Stein L.D., Marth G., Sherry S., Mullikin J.C., Mortimore B.J., Willey D.L., Marth G., Sherry S., Mullikin J.C., Mortimore B.J., Willey D.L., Sherry S., Mullikin J.C., Mortimore B.J., Willey D.L., Mullikin J.C., Mortimore B.J., Willey D.L., Mortimore B.J., Willey D.L., Willey D.L., et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Sakumi K., Furuichi M., Tsuzuki T., Kakuma T., Kawabata S., Maki H., Sekiguchi M., Furuichi M., Tsuzuki T., Kakuma T., Kawabata S., Maki H., Sekiguchi M., Tsuzuki T., Kakuma T., Kawabata S., Maki H., Sekiguchi M., Kakuma T., Kawabata S., Maki H., Sekiguchi M., Kawabata S., Maki H., Sekiguchi M., Maki H., Sekiguchi M., Sekiguchi M. Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J. Biol. Chem. 1993;268:23524–23530. [PubMed] [Google Scholar]
- Sanders M.H., Bates S.E., Wilbur B.S., Holmquist G.P., Bates S.E., Wilbur B.S., Holmquist G.P., Wilbur B.S., Holmquist G.P., Holmquist G.P. Repair rates of R-band, G-band and C-band DNA in murine and human cultured cells. Cytogenet. Genome Res. 2004;104:35–45. doi: 10.1159/000077464. [DOI] [PubMed] [Google Scholar]
- Sekiguchi M., Tsuzuki T., Tsuzuki T. Oxidative nucleotide damage: Consequences and prevention. Oncogene. 2002;21:8895–8904. doi: 10.1038/sj.onc.1206023. [DOI] [PubMed] [Google Scholar]
- Shibutani S., Takeshita M., Grollman A.P., Takeshita M., Grollman A.P., Grollman A.P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- Slupska M.M., Baikalov C., Luther W.M., Chiang J.H., Wei Y.F., Miller J.H., Baikalov C., Luther W.M., Chiang J.H., Wei Y.F., Miller J.H., Luther W.M., Chiang J.H., Wei Y.F., Miller J.H., Chiang J.H., Wei Y.F., Miller J.H., Wei Y.F., Miller J.H., Miller J.H. Cloning and sequencing a human homolog (hMYH) of the Escherichia coli mutY gene whose function is required for the repair of oxidative DNA damage. J. Bacteriol. 1996;178:3385–3392. doi: 10.1128/jb.178.13.3885-3892.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spek E.J., Vuong L.N., Matsuguchi T., Marinus M.G., Engelward B.P., Vuong L.N., Matsuguchi T., Marinus M.G., Engelward B.P., Matsuguchi T., Marinus M.G., Engelward B.P., Marinus M.G., Engelward B.P., Engelward B.P. Nitric oxide-induced homologous recombination in Escherichia coli is promoted by DNA glycosylases. J. Bacteriol. 2002;184:3501–3507. doi: 10.1128/JB.184.13.3501-3507.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenken S. Electron transfer in DNA? Competition by ultra-fast proton transfer? Biol. Chem. 1997;378:1293–1297. [PubMed] [Google Scholar]
- Suter B., Wellinger R.E., Thoma F., Wellinger R.E., Thoma F., Thoma F. DNA repair in a yeast origin of replication: Contributions of photolyase and nucleotide excision repair. Nucleic Acids Res. 2000;28:2060–2068. doi: 10.1093/nar/28.10.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchou J., Kasai H., Shibutani S., Chung M.H., Laval J., Grollman A.P., Nishimura S., Kasai H., Shibutani S., Chung M.H., Laval J., Grollman A.P., Nishimura S., Shibutani S., Chung M.H., Laval J., Grollman A.P., Nishimura S., Chung M.H., Laval J., Grollman A.P., Nishimura S., Laval J., Grollman A.P., Nishimura S., Grollman A.P., Nishimura S., Nishimura S. 8-oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc. Natl. Acad. Sci. 1991;88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenzen T., Yamagata T., Fukagawa T., Sugaya K., Ando A., Inoko H., Gojobori T., Fujiyama A., Okumura K., Ikemura T., Yamagata T., Fukagawa T., Sugaya K., Ando A., Inoko H., Gojobori T., Fujiyama A., Okumura K., Ikemura T., Fukagawa T., Sugaya K., Ando A., Inoko H., Gojobori T., Fujiyama A., Okumura K., Ikemura T., Sugaya K., Ando A., Inoko H., Gojobori T., Fujiyama A., Okumura K., Ikemura T., Ando A., Inoko H., Gojobori T., Fujiyama A., Okumura K., Ikemura T., Inoko H., Gojobori T., Fujiyama A., Okumura K., Ikemura T., Gojobori T., Fujiyama A., Okumura K., Ikemura T., Fujiyama A., Okumura K., Ikemura T., Okumura K., Ikemura T., Ikemura T. Precise switching of DNA replication timing in the GC content transition area in the human major histocompatibility complex. Mol. Cell. Biol. 1997;17:4043–4050. doi: 10.1128/mcb.17.7.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga Y., Ushijima Y., Tsuchimoto D., Mishima M., Shirakawa M., Hirano S., Sakumi K., Nakabeppu Y., Ushijima Y., Tsuchimoto D., Mishima M., Shirakawa M., Hirano S., Sakumi K., Nakabeppu Y., Tsuchimoto D., Mishima M., Shirakawa M., Hirano S., Sakumi K., Nakabeppu Y., Mishima M., Shirakawa M., Hirano S., Sakumi K., Nakabeppu Y., Shirakawa M., Hirano S., Sakumi K., Nakabeppu Y., Hirano S., Sakumi K., Nakabeppu Y., Sakumi K., Nakabeppu Y., Nakabeppu Y. MUTYH prevents OGG1 or APEX1 from inappropriately processing its substrate or reaction product with its C-terminal domain. Nucleic Acids Res. 2004;32:3198–3211. doi: 10.1093/nar/gkh642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyokuni S., Tanaka T., Hattori Y., Nishiyama Y., Yoshida A., Uchida K., Hiai H., Ochi H., Osawa T., Tanaka T., Hattori Y., Nishiyama Y., Yoshida A., Uchida K., Hiai H., Ochi H., Osawa T., Hattori Y., Nishiyama Y., Yoshida A., Uchida K., Hiai H., Ochi H., Osawa T., Nishiyama Y., Yoshida A., Uchida K., Hiai H., Ochi H., Osawa T., Yoshida A., Uchida K., Hiai H., Ochi H., Osawa T., Uchida K., Hiai H., Ochi H., Osawa T., Hiai H., Ochi H., Osawa T., Ochi H., Osawa T., Osawa T. Quantitative immunohistochemical determination of 8-hydroxy-2'-deoxyguanosine by a monoclonal antibody N45.1: Its application to ferric nitrilotriacetate-induced renal carcinogenesis model. Lab. Invest. 1997;76:365–374. [PubMed] [Google Scholar]
- Toyokuni S., Iwasa Y., Kondo S., Tanaka T., Ochi H., Hiai H., Iwasa Y., Kondo S., Tanaka T., Ochi H., Hiai H., Kondo S., Tanaka T., Ochi H., Hiai H., Tanaka T., Ochi H., Hiai H., Ochi H., Hiai H., Hiai H. Intranuclear distribution of 8-hydroxy-2′-deoxyguanosine. An immunocytochemical study. J. Histochem. Cytochem. 1999;47:833–836. doi: 10.1177/002215549904700613. [DOI] [PubMed] [Google Scholar]
- Tsuruya K., Furuichi M., Tominaga Y., Shinozaki M., Tokumoto M., Yoshimitsu T., Fukuda K., Kanai H., Hirakata H., Iida M., Furuichi M., Tominaga Y., Shinozaki M., Tokumoto M., Yoshimitsu T., Fukuda K., Kanai H., Hirakata H., Iida M., Tominaga Y., Shinozaki M., Tokumoto M., Yoshimitsu T., Fukuda K., Kanai H., Hirakata H., Iida M., Shinozaki M., Tokumoto M., Yoshimitsu T., Fukuda K., Kanai H., Hirakata H., Iida M., Tokumoto M., Yoshimitsu T., Fukuda K., Kanai H., Hirakata H., Iida M., Yoshimitsu T., Fukuda K., Kanai H., Hirakata H., Iida M., Fukuda K., Kanai H., Hirakata H., Iida M., Kanai H., Hirakata H., Iida M., Hirakata H., Iida M., Iida M., et al. Accumulation of 8-oxoguanine in the cellular DNA and the alteration of the OGG1 expression during ischemia-reperfusion injury in the rat kidney. DNA Repair (Amst.) 2003;2:211–229. doi: 10.1016/s1568-7864(02)00214-8. [DOI] [PubMed] [Google Scholar]
- Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Smith H.O., Yandell M., Evans C.A., Holt R.A., Yandell M., Evans C.A., Holt R.A., Evans C.A., Holt R.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Fujiyama A., Ichiba Y., Hattori M., Yada T., Sakaki Y., Ikemura T., Fujiyama A., Ichiba Y., Hattori M., Yada T., Sakaki Y., Ikemura T., Ichiba Y., Hattori M., Yada T., Sakaki Y., Ikemura T., Hattori M., Yada T., Sakaki Y., Ikemura T., Yada T., Sakaki Y., Ikemura T., Sakaki Y., Ikemura T., Ikemura T. Chromosome-wide assessment of replication timing for human chromosomes 11q and 21q: Disease-related genes in timing-switch regions. Hum. Mol. Genet. 2002;11:13–21. doi: 10.1093/hmg/11.1.13. [DOI] [PubMed] [Google Scholar]
- Wellinger R.E., Thoma F., Thoma F. Nucleosome structure and positioning modulate nucleotide excision repair in the non-transcribed strand of an active gene. EMBO J. 1997;16:5046–5056. doi: 10.1093/emboj/16.16.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]