Abstract
Compared with other sequenced animal genomes, human segmental duplications appear larger, more interspersed, and disproportionately represented as high-sequence identity alignments. Global sequence divergence estimates of human duplications have suggested an expansion relatively recently during hominoid evolution. Based on primate comparative sequence analysis of 37 unique duplication–transition regions, we establish a molecular clock for their divergence that shows a significant increase in their effective substitution rate when compared with unique genomic sequence. Fluorescent in situ hybridization (FISH) analyses from 1053 random nonhuman primate BACs indicate that great-ape species have been enriched for interspersed segmental duplications compared with representative Old World and New World monkeys. These findings support computational analyses that show a 12-fold excess of recent (>98%) intrachromosomal duplications when compared with duplications between nonhomologous chromosomes. These architectural shifts in genomic structure and elevated substitution rates have important implications for the emergence of new genes, gene-expression differences, and structural variation among humans and great apes.
Duplications play a pivotal role in disease process, gene evolution, and genome rearrangement. Structurally, these sequences have been linked to an increasing number of human genomic disorders within humans (Stankiewicz et al. 2004) as well as evolutionary breakpoints of conserved synteny between humans and other mammals (Armengol et al. 2003; Bailey et al. 2004a; Murphy et al. 2005). Segmental duplications have contributed significantly to large-scale copy number variation within the human population (Iafrate et al. 2004; Sebat et al. 2004; Sharp et al. 2005) and have contributed more to the genetic difference between chimpanzee and human than single-base mutations (Cheng et al. 2005). Importantly, several hominoid-specific genes have been uncovered within these dynamic regions (Johnson et al. 2001; Courseaux et al. 2003; Paulding et al. 2003), due either to fusions or adaptive evolution.
Previous analyses confirm that ∼5% (154.0 Mb) of the human genome is composed of duplications that are greater than 90% identical at the sequence level and greater than 1 kb in length (Bailey et al. 2002; Cheung et al. 2003a; International Human Genome Sequencing Consortium [IHGSC] 2004; She et al. 2004b; Zhang et al. 2005). Intrachromosomal duplications have occurred more frequently (3.97%, 113.66 Mb) compared with duplications between nonhomologous chromosomes (2.37%, 67.86 Mb). In addition, the sequence identity of intrachromosomal duplications is, on average, greater than that of interchromosomal duplications (She et al. 2004b). These properties have prompted speculation that the human and great-ape genomes have experienced a surge of intrachromosomal duplication activity (Samonte and Eichler 2002) or, alternatively, large-scale gene conversion or selection to maintain high-sequence identity within intrachromosomal duplications (Zhang et al. 2005). Such computational inferences, however, have two serious limitations. First, there is an assumption that neutral rates of unique genomic DNA divergence may be applied to duplicated DNA. Gene conversion, if rampant, may complicate extrapolations of neutral substitution rates, rendering such an assumption about a molecular clock invalid. Second, there is an ascertainment bias, in that most studies of primate segmental duplication are based on the human genome reference sequence. Considering the dynamic nature of these regions among closely related species such as chimpanzees and humans (Cheng et al. 2005), an unbiased view of genetic and genomic change is warranted.
In this study, we attempt to provide a preliminary, unbiased assessment of rates of substitution and changes in duplication architecture based on genomic comparisons with nonhuman primates. We begin by summarizing the apparent unique properties of human segmental duplications compared with other sequenced vertebrate genomes. We then establish a molecular clock for single-base divergence based on the analysis of orthologous primate sequence located at the transition regions between unique and duplicated sequence, and directly estimate the frequency of segmental duplication among other species based on FISH analysis of random genomic clones. Our data demonstrate a proclivity toward expansion of interspersed duplications during the emergence of humans and the great apes.
Results and Discussion
Properties of human segmental duplications
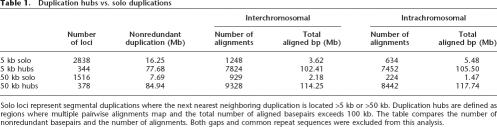
Segmental duplications are distributed nonrandomly across the human genome. We identified 378 regions in excess of 100 kb in length where duplications have accumulated—this includes 98 regions within 2 Mb of centromere and telomere positions (She et al. 2004a; Linardopoulou et al. 2005). The others map within euchromatic regions of the human genome, many of which are sites of instability associated with genomic disorders, cancer, and evolutionary rearrangement. We termed these regions “duplication hubs,” defined as regions where each neighboring pairwise alignment maps within a 5 or 50-kb genomic distance and the total aligned basepairs exceeds 100 kb (Table 1), to distinguish them from areas of the human genome where duplications occur sporadically and most often represented as a single pairwise alignment. Among these duplication hubs, the number of underlying pairwise alignments ranges from 2 to 676 (mean = 67, median = 41), suggesting that these regions have been bombarded by multiple rounds of duplication during the course of evolution.
Table 1.
Duplication hubs vs. solo duplications
Solo loci represent segmental duplications where the next nearest neighboring duplication is located >5 kb or >50 kb. Duplication hubs are defined as regions where multiple pairwise alignments map and the total number of aligned basepairs exceeds 100 kb. The table compares the number of nonredundant basepairs and the number of alignments. Both gaps and common repeat sequences were excluded from this analysis.
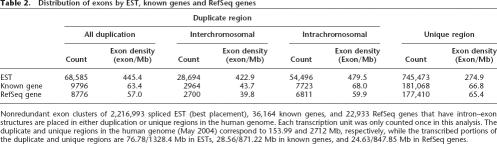
Interestingly, duplicated regions are particularly rich in transcripts. Overall, when the best-placement of spliced transcripts was considered, we found a higher exon density (62%) in duplicated regions when compared with unique regions of the human genome (Table 2), consistent with earlier findings of the draft genome sequence (Bailey et al. 2002). This effect is only observed for spliced ESTs, as opposed to transcripts identified by RefGene or “known gene” within the UCSC Genome Browser, suggesting either incomplete annotation or a higher frequency of transcribed pseudogenes. Details concerning the structure, composition and organization of the segmental duplications (>90% sequence identity and >1 kb in length) within the finished human genome may be found at http://humanparalogy.gs. washington.edu and as a complementary track on http://genome.ucsc.edu.
Table 2.
Distribution of exons by EST, known genes and RefSeq genes
Nonredundant exon clusters of 2,216,993 spliced EST (best placement), 36,164 known genes, and 22,933 RefSeq genes that have intron–exon structures are placed in either duplication or unique regions in the human genome. Each transcription unit was only counted once in this analysis. The duplicate and unique regions in the human genome (May 2004) correspond to 153.99 and 2712 Mb, respectively, while the transcribed portions of the duplicate and unique regions are 76.78/1328.4 Mb in ESTs, 28.56/871.22 Mb in known genes, and 24.63/847.85 Mb in RefSeq genes.
Compared with other sequenced vertebrate genomes, three properties of human segmental duplications emerge (Table 3; Methods). Human segmental duplications are larger, more interspersed, and show a high degree of sequence identity. Based on the analysis of 25,318 pairwise alignments, we determined that 86.5% of all duplicated bases are part of alignments that exceed 10 kb in length. A total of 55% of human segmental duplications are distributed in an interspersed fashion, where the paralogous pairs are separated by more than 1 Mb or map to nonhomologous chromosomes (Table 3). More than 77% (119/154 Mb) of duplicated bases are part of alignments with >95% sequence identity. These properties contrast sharply with other sequenced vertebrate genomes (Table 3). One caveat to this analysis is that the quality of the various genome sequences differ substantially. Two observations suggest that the observed differences are biological and not an artifact of assembly. First, assembly of the human genome based strictly on whole-genome shotgun sequence (Istrail et al. 2004) shows a similar distribution of interspersed and tandem duplicates. Second, recent experimental analyses (Bailey et al. 2004b) and finishing efforts of the mouse genome assembly (E.E. Eichler, unpubl.) confirm the disparity in the relative distribution of interspersed and tandem duplications seen between human and mouse. It should also be noted that structural variation has been shown to be significantly enriched within regions of recent segmental duplication (Iafrate et al. 2004; Sharp et al. 2005; Tuzun et al. 2005). It has been estimated that ∼20% of segmental duplications are polymorphic within the human and chimpanzee populations (Cheng et al. 2005; Sharp et al. 2005). In the case of human, however, this effect on our analysis was limited because we observed a similar pattern from two independent measures derived from different human DNA sources (Celera and public assembly) (She et al. 2004b). In the case of model organisms, species are highly inbred and it is expected that most duplications would have been fixed.
Table 3.
Segmental duplication detected in vertebrate genome sequence assemblies
WGAC (whole genome assembly comparison) analyses were applied to the updated genome assemblies of human (May 2004), mouse (May 2004), rat (June 2003), and chicken (February 2004). Pairwise alignments (>90%) were categorized based on length. It is important to note that the different genome assemblies represent various degrees of finishing and the true estimate of duplication content is still not known. Mouse and human represent the highest quality assemblies in this analysis. As a control, we also analyzed a human genome assembly based strictly on whole-genome shotgun sequence (Istrail et al. 2004; She et al. 2004b). In this assembly, we found a comparable level of tandem duplication (40%–45%) as the finished human genome assembly. The amount of duplication is computed as the nonredundant duplicated portion of the genome. The proportion is based on estimated size of the euchromatin, excluding gap sizes.
aThe percentage of tandem duplication is defined by the relative proportion of duplicated bases where the duplication alignments map within 1 Mb of one another. Pairwise alignments to unknown or random chromosomes are not included in this analysis.
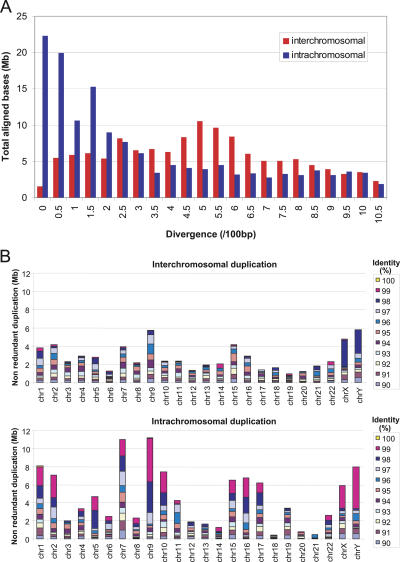
We analyzed the divergence of interchromosomal and intrachromosomal alignments and plotted the fraction of duplicated bases as a function of the total number of aligned bases (Fig. 1). Several trends emerge. First, there is mode at 0.05 substitutions per site for interchromosomal duplications, and this dramatically decreases by count and by base-pair representation at lower divergences (Supplemental Fig. S1). Most of the increase in higher sequence identity duplications is due to an expansion of intrachromosomal duplications. The majority of intrachromosomally duplicated bases show <0.03 substitutions per site. These high-identity duplications significantly outnumber interchromosomal duplications by count and by total base pairs (4:1 and 12:1, respectively) at comparable levels of divergence (Supplemental Fig. S1; Fig. 1A). The expansion increases until 0.005 substitutions per site, at which point the number of intrachromosomal alignments reduce. This intrachromosomal expansion of duplications is nonuniformly distributed among human chromosomes, largely restricted to nine autosomes and the sex chromosomes (Fig. 1B). In some cases, as much as 9.4% or 32.1% of the chromosomes total base pairs (chromosome 9 and Y, respectively) arose as a consequence of either recent intrachromosomal duplication or gene conversion events.
Figure 1.
Distribution of recent segmental duplications in the human genome. (A) Sequence identity. Interchromosomal (red) and intrachromomal (blue) segmental duplications in the human genome sequence (May 2004 build) were binned according to their divergence, and the total number of aligned basepairs was determined. Divergence (K) is calculated as the number of substitutions per site between the two duplication alignments. (B) Chromosomal distribution. The distribution of recent segmental duplications is depicted with the color bars representing different percent identities. The distribution within each chromosome was calculated as the proportion of pairwise alignments at each percent identity.
A molecular clock for primate segmental duplications
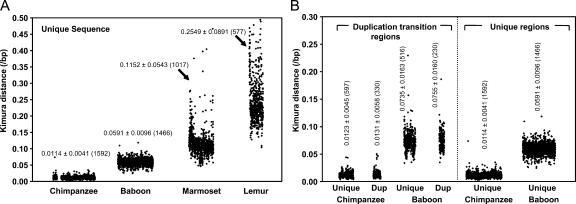
We sought to establish a molecular clock to determine the evolutionary age of segmental duplications within the human genome. We first aligned 16.78 Mb of unique noncoding genomic DNA between human and nonhuman primates. Four different nonhuman primate species (chimpanzee, macaque, marmoset, and lemur) were selected, representing different divergence branch points from the human lineage. We limited our analysis to high-quality (i.e., finished) sequences derived from bacterial artificial chromosome (BAC) clones; such sequences were associated with a known error rate. Using these sequences, we calculated the genetic distance (substitutions per base pair) from the human sequence (Fig. 2A; Supplemental Table S1a). Based on estimated divergence times of each primate (Goodman 1999), we calculated substitution rates ranging from 1.034 ± 0.04 × 10−9 for human–chimpanzee comparisons to 2.276 × 10−9 substitutions per bp/Mya for lemur–human comparisons. The reduction in the substitution rate among species more closely related to human has been noted before and may reflect an increase in hominoid generation time (Chen and Li 2001; Liu et al. 2003). These data served as a baseline to approximate the neutral genomic DNA as a function of divergence. A slower molecular clock in the humans may also contribute partially to the increase in the abundance of highly homologous segmental duplications observed for the human/ape genomes (see below).
Figure 2.
Single nucleotide substitution in unique and duplicated genomic regions. (A) Divergence of unique genomic regions. A scatter plot of genetic distances (changes/basepair, Kimura two-parameter model) determined from nonoverlapping 3-kb sliding windows for human–chimpanzee (5.0 Mb), human–baboon (5.0 Mb), human–marmoset (4.0 Mb), and human–lemur (2.8 Mb) sequence alignments. A total of 56 marmoset windows and 182 lemur windows were >0.50 in Kimura distance and thus not shown. The mean ± standard deviation (from the number of windows) are shown for each comparison based on the number of windows assessed (see Supplemental Table S1a for more details). (B) Divergence of duplicated genomic regions. Genomic sequence alignments that contain segmental duplications and that transition into unique sequence were examined for divergence between humans and nonhuman primates (baboon and chimpanzee). Duplicated and unique portions were considered separately in this analysis and compared with unique regions in A. Duplicated regions show an increase in substitution irrespective of their duplication or unique status based on alignment to the human genome.
Estimating the age of duplication events, however, is confounded by the propensity for these sequences to undergo gene conversion (Hurles 2001; Skaletsky et al. 2003; Jackson et al. 2005; Pavliček et al. 2005). Such homogenization events might erase patterns of divergence and lead to an underestimation of the true evolutionary age of the duplication. Alternatively, post-speciation gene conversion events might serve as a reservoir to increase substitution rates among orthologous copies. Indeed, there are several notable examples where the degree of sequence divergence is incompatible with the estimated timing of the duplication based on comparative analyses (Orti et al. 1998; DeSilva et al. 1999; Shaikh et al. 2001). To address this issue, we selected 17 different duplicons (99 copies throughout the genome) and assessed whether their divergence (K = 0.018 to K = 0.043) was consistent with their estimated time of expansion based on comparative FISH and/or hybridization data. Of the duplications that we investigated, 14/17 showed levels of sequence divergence consistent with the emergence, as predicted by comparative primate sequence data (Supplemental Table S2). In one case on chromosome 15q11, the higher degree of sequence identity was consistent with apparent large-scale gene conversion. In another case on chromosome LCR16a, duplications of the same locus had occurred independently within multiple lineages (E.E. Eichler, unpubl.). We conclude that gene conversion, while an important consideration, unlikely accounts for the bulk of high-sequence identity duplications and does not significantly complicate the estimated timing of events based on comparative FISH and sequence data.
To quantify the substitution rate for duplicated sequence more precisely, we specifically compared 37 duplicated regions between human and nonhuman primates (chimpanzee and baboon). We selected BACs containing duplications that were completely anchored within unique regions of the genome, allowing for unambiguous determination of orthologous relationships (Methods). Compared with strictly unique genome sequences, duplicated regions are significantly more diverged (Fig. 2B; Supplemental Table S1b). Between chimpanzee and human, we estimated a 10% increase in the rate of mutation, while an ∼25% increase was observed for orthologous sequence comparisons between human and baboon.
Several possible explanations might account for the increased substitution rate of duplicated DNA, including CpG bias, gene conversion, and/or relaxed selective constraint (Chen and Li 2001). Duplicated regions are known to be generally more Alu-repeat and GC-rich (Bailey et al. 2003; Jurka 2004). When we corrected for CpG-bias, the increase in substitution rate was reduced by more than one-half for chimpanzee–human comparisons. The effect was not as dramatic for more distant baboon–human comparisons, where it accounted for only 30% of the increase. We computed the genetic distance for both unique and duplicated portions of the transition regions. Since duplicated regions, in theory, could be targets for large-scale conversion events where paralogous sequence variants could replace orthologous variants, thereby creating the appearance of an increased substitution rate, we would expect to see a clear distinction between unique and duplicated sequences. Surprisingly, only a slight difference in the substitution was observed. If gene conversion is responsible for the increase in the observed substitution rate, the boundaries between unique and duplicated DNA would have had to have shifted among humans and nonhuman primates. Such a phenomenon, termed “duplication shadowing” was recently described based on comparisons of the human and chimpanzee genome (Cheng et al. 2005). Alternatively, a combination of CpG mutation and relaxed selective constraint may contribute to the overall hypermutability of these regions of the genome.
The human great-ape expansion of segmental duplications
Based on our adjusted molecular clock for duplicated DNA, we propose the following model for evolutionary expansion of segmental duplications. Our analyses support a model wherein interchromosomal duplication activity reached its peak during or after the separation of Old World monkeys and hominoid lineage (∼25 million years [Myr]). During this time, many of the duplicative transpositions of euchromatic DNA to pericentromeric and subtelomeric regions occurred, leading to the complex mosaic organization of euchromatic duplicons now found near centromeres and telomeres. Intrachromosomal duplications occurred at a relatively constant rate during this period, but were typically larger in size than their interchromosomal counterparts and subsequently occupied a slightly greater fraction of the genome. We predict that ∼10 Mya, the ancestral hominid genome experienced a sudden surge in the number and size of intrachromosomal duplications. In humans, this expansion was restricted to 72 gene-rich regions and primarily involved 11 chromosomes. We estimate that ∼2 Mya, the number of intrachromosomal duplication events began to decline at least in the human lineage.
The single most important caveat of this model is that our predictions are based on the human reference sequence and infer history based solely on that evolutionary trajectory. Based simply on the sequence, we cannot, for example, exclude the possibility that other nonhuman primate species have similarly undergone independent intrachromosomal expansions. Based on our model, such expansions are expected to be observed among great-ape species, but be less common among Old World and New World monkey species. To estimate the frequency of segmental duplications more directly, we performed FISH analyses with three nonhuman primates (chimpanzee, macaque, and marmoset). We randomly selected 384 BACs from each species and counted the number of clones displaying a multi-site distribution pattern, thereby indirectly providing an estimate of segmental duplication content (Table 4; Supplemental Table S3). The map position of each locus was determined based on matching the end-sequences of each BAC to positions along the human reference sequence. Previous cytogenetic and in silico estimates of segmental duplication in humans revealed that in situ estimates are a remarkably accurate indicator of recent duplication content (Cheung et al. 2001; Bailey et al. 2002).
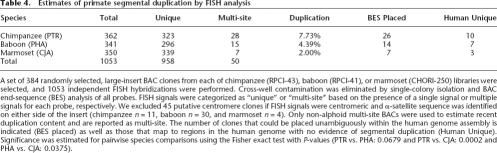
Table 4.
Estimates o primate segmental duplication by FISH analysis
A set of 384 randomly selected, large-insert BAC clones from each of chimpanzee (RPCI-43), baboon (RPCI-41), or marmoset (CHORI-250) libraries were selected, and 1053 independent FISH hybridizations were performed. Cross-well contamination was eliminated by single-colony isolation and BAC end-sequence (BES) analysis of all probes. FISH signals were categorized as “unique” or “multi-site” based on the presence of a single signal or multiple signals for each probe, respectively. We excluded 45 putative centromere clones if FISH signals were centromeric and α-satellite sequence was identified on either side of the insert (chimpanzee n = 11, baboon n = 30, and marmoset n = 4). Only non-alphoid multi-site BACs were used to estimate recent duplication content and are reported as multi-site. The number of clones that could be placed unambiguously within the human genome assembly is indicated (BES placed) as well as those that map to regions in the human genome with no evidence of segmental duplication (Human Unique). Significance was estimated for pairwise species comparisons using the Fisher exact test with P-values (PTR vs. PHA: 0.0679 and PTR vs. CJA: 0.0002 and PHA vs. CJA: 0.0375).
These FISH analyses augment two important aspects of our model. First, we observed an increase in the number of segmental duplications among chimpanzees compared with either baboon (P = 0.0679, Fisher exact test) or marmoset (P = 0.0002) (Table 4). In fact, the marmoset estimate for segmental duplications (∼2%) is similar to experimental and computational predictions for other mammals, such as the rat and mouse (Table 3) (Cheung et al. 2003b; Bailey et al. 2004b; Tuzun et al. 2004). Additional mammalian genomes of higher quality sequence, however, will need to be analyzed to definitively assess the significance of this primate expansion. Second, an examination of the mapped locations of the chimpanzee segmental duplications revealed that ∼30% (10/26) of the duplicated BACs map to corresponding unique regions in the human genome. This suggests that both great apes and humans have been predisposed to expansions of interspersed segmental duplications, and that a significant number of these will have occurred within different regions of the genome. These findings of extensive de novo duplication in each lineage are consistent with the recent analysis of the chimpanzee genome (Cheng et al. 2005). In addition, both analyses suggest a trend for increased segmental duplication in the chimpanzee lineage when compared with human.
Our studies establish a baseline for estimating the age of segmental duplication and predict an elevated primate substitution rate for duplicated DNA compared with unique noncoding sequence. Surprisingly, our analyses also show that unique sequence-flanking segmental duplications experienced comparable increases in substitution rate. Evolutionary variability in the boundaries between unique and duplicated DNA (i.e., duplication shadowing) may account for this property. We tested for this effect based on our knowledge of the duplication map of the chimpanzee genome (Cheng et al. 2005). The increased substitution rate did not significantly change if we excluded regions in chimpanzee that showed evidence of duplication shadowing, a phenomena that genomic sequence adjacent to duplication is predisposed to new duplication (Cheng et al. 2005). This suggests that the effect is particular to the region as opposed to being directly related to duplicated sequence as might be expected if gene conversion were responsible for the effect (Jackson et al. 2005; Pavliček et al. 2005).
Our assessment of segmental duplication among three nonhuman primates provides the first experimental evidence that humans and great apes are enriched for interspersed segmental duplications compared with other primate lineages. The evolutionary basis for this predilection is unknown, but may be related to smaller effective population size, adaptation, a slowdown in the molecular clock, and/or relaxed selective constraints (Li and Tanimura 1987; Wall et al. 2002; Keightley et al. 2005) during hominoid evolution. Analysis of duplication content in other large-bodied mammals with long generation times would help to address issues regarding population size, and a slowdown in the molecular clock. A systematic assessment of genes embedded within duplication regions should shed insight into the role of adaptive evolution. It will also be worthwhile to investigate the pattern of single nucleotide and structural polymorphism in the vicinity of these regions when compared with other less-variable areas of the genome. Comparisons of patterns of within-species and between-species variation among different primates will be essential in distinguishing effects due to population size and relaxed selective constraint.
Methods
Segmental duplication analysis
We used a BLAST-based detection scheme (WGAC) (Bailey et al. 2001) to identify all pairwise similarities representing duplicated regions within the NCBI genome assemblies of human (May 2004), mouse (May 2004), rat (June 2003) and chicken (February 2004). As a control for heterogeneity in the quality of genome assembly, we previously analyzed the Celera WGSA assembly (Istrail et al. 2004; She et al. 2004b), which was assembled using only whole-genome shotgun sequence. All duplications are longer than 1 kb with sequence identity >90%, except for rat and mouse (>5 kb and >90%). Duplications in the human genome were also identified by another approach using whole-genome shotgun sequence detection (WSSD) strategy (Bailey et al. 2002). Divergence estimates, the number of substitutions per site between the two sequences, were calculated using Kimura’s two-parameter method, which corrects for multiple events and transversion/transition mutational biases (Kimura 1980). Transcript content of unique and duplicated sequence was compared by computing the number of exons in each portion of the genome. Three sets of data were considered, i.e., human ESTs with intron/exon structure (4.56 million), RefSeq annotated genes (22,933), and known annotated genes (36,164). Both best-placement and tied genes/transcripts were distinguished based on BLAT score criteria (www.genome.ucsc.edu). In cases where a transcript could be mapped to two or more duplicated locations with equal score, one location was selected randomly. Exon density was defined as the number of nonoverlapping exons identified within the region.
Substitution rates
We optimally aligned a total of 16.8 Mb of unique nonhuman primate genomic sequence and the orthologous human sequence, and then calculated the (Kimura) genetic distance in nonoverlapping 3-kb sliding windows as described (Liu et al. 2003). We examined 51 loci (5.0 Mb/1592 windows) for human–chimpanzee, 42 loci (5.0 Mb, 1466 windows) for human–baboon, 45 loci (4.0 Mb, 1017 windows) for human–marmoset, and 29 loci (2.8 Mb, 577 windows) for human–lemur genomic sequence alignments. For duplication boundary regions, we selected 37 finished nonhuman primate BACs (20 for chimpanzee and 17 for baboon) containing at least 20 kb for both duplication and unique regions. Alignments were completely anchored within unique regions of the human sequence to allow for unambiguous determination of orthologous relationship with the nonhuman primate sequences.
FISH analysis
A random set of BAC clones (384) was selected from each of chimpanzee (RP43), baboon (RP41), and marmoset (CH250) genomic libraries (www.bacpac.chori.org). Isolated single colonies from each clone were end-sequenced and hybridized to metaphase preparations from two unrelated individuals of each species. Each clone was then classified as generating unique or multi-site signals. Multi-site signals were categorized as alphoid if α-satellite sequences were identified on either side of the hybridizing signal. Non-alphoid multi-site BACs were used to estimate recent duplication content. The BAC-end sequences were used to establish the in silico best placement of each clone in the human genome sequence (build35). In total, 50 BACs yielded multiple FISH signals (chimpanzee 28, baboon 15, marmoset 7) (Supplemental Table S3). Only those BACs where the underlying sequence was determined to be duplicated in human were considered to be concordant. The discordant BACs were further confirmed by FISH using human chromosomal spreads.
Acknowledgments
We thank Devin Locke and Matthew Johnson for technical assistance. This work was supported, in part, by NIH grants GM58815 to E.E.E and by funds provided through the NHGRI Intramural Program of the NIH to E.D.G. E.E.E. is an investigator of the Howard Hughes Medical Institute. In addition, the authors gratefully acknowledge CEGBA (Centro di Eccellenza Geni in campo Biosanitario e Agroalimentare), MIUR (Ministero Italiano della Università e della Ricerca; Cluster C03, Prog. L.488/92), the European Commission (INPRIMAT, QLRI-CT-2002-01325), and the BMBF (Bundesministerium für Bildung und Forschung) for financial support.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4949406
References
- Armengol L., Pujana M.A., Cheung J., Scherer S.W., Estivill X., Pujana M.A., Cheung J., Scherer S.W., Estivill X., Cheung J., Scherer S.W., Estivill X., Scherer S.W., Estivill X., Estivill X. Enrichment of segmental duplications in regions of breaks of synteny between the human and mouse genomes suggest their involvement in evolutionary rearrangements. Hum. Mol. Genet. 2003;12:2201–2208. doi: 10.1093/hmg/ddg223. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Yavor A.M., Massa H.F., Trask B.J., Eichler E.E., Yavor A.M., Massa H.F., Trask B.J., Eichler E.E., Massa H.F., Trask B.J., Eichler E.E., Trask B.J., Eichler E.E., Eichler E.E. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Myers E.W., Li P.W., Eichler E.E., Li P.W., Eichler E.E., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Liu G., Eichler E.E., Liu G., Eichler E.E., Eichler E.E. An Alu transposition model for the origin and expansion of human segmental duplications. Am. J. Hum. Genet. 2003;73:823–834. doi: 10.1086/378594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A., Baertsch R., Kent W.J., Haussler D., Eichler E.E., Baertsch R., Kent W.J., Haussler D., Eichler E.E., Kent W.J., Haussler D., Eichler E.E., Haussler D., Eichler E.E., Eichler E.E. Hotspots of mammalian chromosomal evolution. Genome Biol. 2004a;5:R23. doi: 10.1186/gb-2004-5-4-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A., Church D.M., Ventura M., Rocchi M., Eichler E.E., Church D.M., Ventura M., Rocchi M., Eichler E.E., Ventura M., Rocchi M., Eichler E.E., Rocchi M., Eichler E.E., Eichler E.E. Analysis of segmental duplications and genome assembly in the mouse. Genome Res. 2004b;14:789–801. doi: 10.1101/gr.2238404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F.C., Li W.H., Li W.H. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Ventura M., She X., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Pääbo S., Ventura M., She X., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Pääbo S., She X., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Pääbo S., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Pääbo S., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Pääbo S., Osoegawa K., Church D., DeJong P., Wilson R.K., Pääbo S., Church D., DeJong P., Wilson R.K., Pääbo S., DeJong P., Wilson R.K., Pääbo S., Wilson R.K., Pääbo S., Pääbo S., et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- Cheung V.G., Nowak N., Jang W., Kirsch I.R., Zhao S., Chen X.N., Furey T.S., Kim U.J., Kuo W.L., Olivier M., Nowak N., Jang W., Kirsch I.R., Zhao S., Chen X.N., Furey T.S., Kim U.J., Kuo W.L., Olivier M., Jang W., Kirsch I.R., Zhao S., Chen X.N., Furey T.S., Kim U.J., Kuo W.L., Olivier M., Kirsch I.R., Zhao S., Chen X.N., Furey T.S., Kim U.J., Kuo W.L., Olivier M., Zhao S., Chen X.N., Furey T.S., Kim U.J., Kuo W.L., Olivier M., Chen X.N., Furey T.S., Kim U.J., Kuo W.L., Olivier M., Furey T.S., Kim U.J., Kuo W.L., Olivier M., Kim U.J., Kuo W.L., Olivier M., Kuo W.L., Olivier M., Olivier M., et al. Integration of cytogenetic landmarks into the draft sequence of the human genome. The BAC Resource Consortium. Nature. 2001;409:953–958. doi: 10.1038/35057192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J., Estivill X., Khaja R., MacDonald J.R., Lau K., Tsui L.C., Scherer S.W., Estivill X., Khaja R., MacDonald J.R., Lau K., Tsui L.C., Scherer S.W., Khaja R., MacDonald J.R., Lau K., Tsui L.C., Scherer S.W., MacDonald J.R., Lau K., Tsui L.C., Scherer S.W., Lau K., Tsui L.C., Scherer S.W., Tsui L.C., Scherer S.W., Scherer S.W. Genome-wide detection of segmental duplications and potential assembly errors in the human genome sequence. Genome Biol. 2003a;4:R25. doi: 10.1186/gb-2003-4-4-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J., Wilson M.D., Zhang J., Khaja R., MacDonald J.R., Heng H.H., Koop B.F., Scherer S.W., Wilson M.D., Zhang J., Khaja R., MacDonald J.R., Heng H.H., Koop B.F., Scherer S.W., Zhang J., Khaja R., MacDonald J.R., Heng H.H., Koop B.F., Scherer S.W., Khaja R., MacDonald J.R., Heng H.H., Koop B.F., Scherer S.W., MacDonald J.R., Heng H.H., Koop B.F., Scherer S.W., Heng H.H., Koop B.F., Scherer S.W., Koop B.F., Scherer S.W., Scherer S.W. Recent segmental and gene duplications in the mouse genome. Genome Biol. 2003b;4:R47. doi: 10.1186/gb-2003-4-8-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courseaux A., Richard F., Grosgeorge J., Ortola C., Viale A., Turc-Carel C., Dutrillaux B., Gaudray P., Nahon J.L., Richard F., Grosgeorge J., Ortola C., Viale A., Turc-Carel C., Dutrillaux B., Gaudray P., Nahon J.L., Grosgeorge J., Ortola C., Viale A., Turc-Carel C., Dutrillaux B., Gaudray P., Nahon J.L., Ortola C., Viale A., Turc-Carel C., Dutrillaux B., Gaudray P., Nahon J.L., Viale A., Turc-Carel C., Dutrillaux B., Gaudray P., Nahon J.L., Turc-Carel C., Dutrillaux B., Gaudray P., Nahon J.L., Dutrillaux B., Gaudray P., Nahon J.L., Gaudray P., Nahon J.L., Nahon J.L. Segmental duplications in euchromatic regions of human chromosome 5: A source of evolutionary instability and transcriptional innovation. Genome Res. 2003;13:369–381. doi: 10.1101/gr.490303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSilva U., Massa H., Trask B.J., Green E.D., Massa H., Trask B.J., Green E.D., Trask B.J., Green E.D., Green E.D. Comparative mapping of the region of human chromosome 7 deleted in williams syndrome. Genome Res. 1999;9:428–436. [PMC free article] [PubMed] [Google Scholar]
- Goodman M. The genomic record of Humankind’s evolutionary roots. Am. J. Hum. Genet. 1999;64:31–39. doi: 10.1086/302218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles M.E. Gene conversion homogenizes the CMT1A paralogous repeats. BMC Genomics. 2001;2:11. doi: 10.1186/1471-2164-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Qi Y., Scherer S.W., Lee C., Scherer S.W., Lee C., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (IHGSC) Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Istrail S., Sutton G.G., Florea L., Halpern A.L., Mobarry C.M., Lippert R., Walenz B., Shatkay H., Dew I., Miller J.R., Sutton G.G., Florea L., Halpern A.L., Mobarry C.M., Lippert R., Walenz B., Shatkay H., Dew I., Miller J.R., Florea L., Halpern A.L., Mobarry C.M., Lippert R., Walenz B., Shatkay H., Dew I., Miller J.R., Halpern A.L., Mobarry C.M., Lippert R., Walenz B., Shatkay H., Dew I., Miller J.R., Mobarry C.M., Lippert R., Walenz B., Shatkay H., Dew I., Miller J.R., Lippert R., Walenz B., Shatkay H., Dew I., Miller J.R., Walenz B., Shatkay H., Dew I., Miller J.R., Shatkay H., Dew I., Miller J.R., Dew I., Miller J.R., Miller J.R., et al. Whole-genome shotgun assembly and comparison of human genome assemblies. Proc. Natl. Acad. Sci. 2004;101:1916–1921. doi: 10.1073/pnas.0307971100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M.S., Oliver K., Loveland J., Humphray S., Dunham I., Rocchi M., Viggiano L., Park J.P., Hurles M.E., Santibanez-Koref M., Oliver K., Loveland J., Humphray S., Dunham I., Rocchi M., Viggiano L., Park J.P., Hurles M.E., Santibanez-Koref M., Loveland J., Humphray S., Dunham I., Rocchi M., Viggiano L., Park J.P., Hurles M.E., Santibanez-Koref M., Humphray S., Dunham I., Rocchi M., Viggiano L., Park J.P., Hurles M.E., Santibanez-Koref M., Dunham I., Rocchi M., Viggiano L., Park J.P., Hurles M.E., Santibanez-Koref M., Rocchi M., Viggiano L., Park J.P., Hurles M.E., Santibanez-Koref M., Viggiano L., Park J.P., Hurles M.E., Santibanez-Koref M., Park J.P., Hurles M.E., Santibanez-Koref M., Hurles M.E., Santibanez-Koref M., Santibanez-Koref M. Evidence for widespread reticulate evolution within human duplicons. Am. J. Hum. Genet. 2005;77:824–840. doi: 10.1086/497704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.E., Viggiano L., Bailey J.A., Abdul-Rauf M., Goodwin G., Rocchi M., Eichler E.E., Viggiano L., Bailey J.A., Abdul-Rauf M., Goodwin G., Rocchi M., Eichler E.E., Bailey J.A., Abdul-Rauf M., Goodwin G., Rocchi M., Eichler E.E., Abdul-Rauf M., Goodwin G., Rocchi M., Eichler E.E., Goodwin G., Rocchi M., Eichler E.E., Rocchi M., Eichler E.E., Eichler E.E. Positive selection of a gene family during the emergence of humans and African apes. Nature. 2001;413:514–519. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- Jurka J. Evolutionary impact of human Alu repetitive elements. Curr. Opin. Genet. Dev. 2004;14:603–608. doi: 10.1016/j.gde.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Keightley P.D., Kryukov G.V., Sunyaev S., Halligan D.L., Gaffney D.J., Kryukov G.V., Sunyaev S., Halligan D.L., Gaffney D.J., Sunyaev S., Halligan D.L., Gaffney D.J., Halligan D.L., Gaffney D.J., Gaffney D.J. Evolutionary constraints in conserved nongenic sequences of mammals. Genome Res. 2005;15:1373–1378. doi: 10.1101/gr.3942005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Li W.H., Tanimura M., Tanimura M. The molecular clock runs more slowly in man than in apes and monkeys. Nature. 1987;326:93–96. doi: 10.1038/326093a0. [DOI] [PubMed] [Google Scholar]
- Linardopoulou E.V., Williams E.M., Fan Y., Friedman C., Young J.M., Trask B.J., Williams E.M., Fan Y., Friedman C., Young J.M., Trask B.J., Fan Y., Friedman C., Young J.M., Trask B.J., Friedman C., Young J.M., Trask B.J., Young J.M., Trask B.J., Trask B.J. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005;437:94–100. doi: 10.1038/nature04029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Zhao S., Bailey J.A., Sahinalp S.C., Alkan C., Tuzun E., Green E.D., Eichler E.E., Zhao S., Bailey J.A., Sahinalp S.C., Alkan C., Tuzun E., Green E.D., Eichler E.E., Bailey J.A., Sahinalp S.C., Alkan C., Tuzun E., Green E.D., Eichler E.E., Sahinalp S.C., Alkan C., Tuzun E., Green E.D., Eichler E.E., Alkan C., Tuzun E., Green E.D., Eichler E.E., Tuzun E., Green E.D., Eichler E.E., Green E.D., Eichler E.E., Eichler E.E. Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res. 2003;13:358–368. doi: 10.1101/gr.923303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy W.J., Larkin D.M., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Larkin D.M., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Chowdhary B.P., Galibert F., Gatzke L., Galibert F., Gatzke L., Gatzke L., et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- Orti R., Potier M.C., Maunoury C., Prieur M., Creau N., Delabar J.M., Potier M.C., Maunoury C., Prieur M., Creau N., Delabar J.M., Maunoury C., Prieur M., Creau N., Delabar J.M., Prieur M., Creau N., Delabar J.M., Creau N., Delabar J.M., Delabar J.M. Conservation of pericentromeric duplications of a 200-kb part of the human 21q22.1 region in primates. Cytogenet. Cell Genet. 1998;83:262–265. doi: 10.1159/000015201. [DOI] [PubMed] [Google Scholar]
- Paulding C.A., Ruvolo M., Haber D.A., Ruvolo M., Haber D.A., Haber D.A. The Tre2 (USP6) oncogene is a hominoid-specific gene. Proc. Natl. Acad. Sci. 2003;100:2507–2511. doi: 10.1073/pnas.0437015100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavliček A., House R., Gentles A.J., Jurka J., Morrow B.E., House R., Gentles A.J., Jurka J., Morrow B.E., Gentles A.J., Jurka J., Morrow B.E., Jurka J., Morrow B.E., Morrow B.E. Traffic of genetic information between segmental duplications flanking the typical 22q11.2 deletion in velo-cardio-facial syndrome/DiGeorge syndrome. Genome Res. 2005;15:1487–1495. doi: 10.1101/gr.4281205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samonte R.V., Eichler E.E., Eichler E.E. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 2002;3:65–72. doi: 10.1038/nrg705. [DOI] [PubMed] [Google Scholar]
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Lundin P., Maner S., Massa H., Walker M., Chi M., Maner S., Massa H., Walker M., Chi M., Massa H., Walker M., Chi M., Walker M., Chi M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Shaikh T.H., Kurahashi H., Emanuel B.S., Kurahashi H., Emanuel B.S., Emanuel B.S. Evolutionarily conserved low copy repeats (LCRs) in 22q11 mediate deletions, duplications, translocations, and genomic instability: An update and literature review. Genet. Med. 2001;3:6–13. doi: 10.1097/00125817-200101000-00003. [DOI] [PubMed] [Google Scholar]
- Sharp A.J., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Clark R.A., Schwartz S., Segraves R., Schwartz S., Segraves R., Segraves R., et al. Segmental duplications and copy number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X., Horvath J.E., Jiang Z., Liu G., Furey T.S., Christ L., Clark R., Graves T., Gulden C.L., Alkan C., Horvath J.E., Jiang Z., Liu G., Furey T.S., Christ L., Clark R., Graves T., Gulden C.L., Alkan C., Jiang Z., Liu G., Furey T.S., Christ L., Clark R., Graves T., Gulden C.L., Alkan C., Liu G., Furey T.S., Christ L., Clark R., Graves T., Gulden C.L., Alkan C., Furey T.S., Christ L., Clark R., Graves T., Gulden C.L., Alkan C., Christ L., Clark R., Graves T., Gulden C.L., Alkan C., Clark R., Graves T., Gulden C.L., Alkan C., Graves T., Gulden C.L., Alkan C., Gulden C.L., Alkan C., Alkan C., et al. The structure and evolution of centromeric transition regions within the human genome. Nature. 2004a;430:857–864. doi: 10.1038/nature02806. [DOI] [PubMed] [Google Scholar]
- She X., Jiang Z., Clark R.A., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Jiang Z., Clark R.A., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Clark R.A., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Sutton G., Halpern A.L., Eichler E.E., Halpern A.L., Eichler E.E., Eichler E.E. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature. 2004b;431:927–930. doi: 10.1038/nature03062. [DOI] [PubMed] [Google Scholar]
- Skaletsky H., Kuroda-Kawaguchi T., Minx P.J., Cordum H.S., Hillier L.W., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Kuroda-Kawaguchi T., Minx P.J., Cordum H.S., Hillier L.W., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Minx P.J., Cordum H.S., Hillier L.W., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Cordum H.S., Hillier L.W., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Hillier L.W., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Repping S., Pyntikova T., Ali J., Bieri T., Pyntikova T., Ali J., Bieri T., Ali J., Bieri T., Bieri T., et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;433:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P., Shaw C.J., Withers M., Inoue K., Lupski J.R., Shaw C.J., Withers M., Inoue K., Lupski J.R., Withers M., Inoue K., Lupski J.R., Inoue K., Lupski J.R., Lupski J.R. Serial segmental duplications during primate evolution result in complex human genome architecture. Genome Res. 2004;14:2209–2220. doi: 10.1101/gr.2746604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun E., Bailey J.A., Eichler E.E., Bailey J.A., Eichler E.E., Eichler E.E. Recent segmental duplications in the working draft assembly of the brown Norway rat. Genome Res. 2004;14:493–506. doi: 10.1101/gr.1907504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun E., Sharp A.J., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., Sharp A.J., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., Haugen E., Hayden H., Albertson D., Pinkel D., Hayden H., Albertson D., Pinkel D., Albertson D., Pinkel D., Pinkel D., et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- Wall J.D., Andolfatto P., Przeworski M., Andolfatto P., Przeworski M., Przeworski M. Testing models of selection and demography in Drosophila simulans. Genetics. 2002;162:203–216. doi: 10.1093/genetics/162.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lu H.H., Chung W.Y., Yang J., Li W.H., Lu H.H., Chung W.Y., Yang J., Li W.H., Chung W.Y., Yang J., Li W.H., Yang J., Li W.H., Li W.H. Patterns of segmental duplication in the human genome. Mol. Biol. Evol. 2005;22:135–141. doi: 10.1093/molbev/msh262. [DOI] [PubMed] [Google Scholar]