Abstract
Correct gene prediction is impaired by the presence of processed pseudogenes: nonfunctional, intronless copies of real genes found elsewhere in the genome. Gene prediction programs frequently mistake processed pseudogenes for real genes or exons, leading to biologically irrelevant gene predictions. While methods exist to identify processed pseudogenes in genomes, no attempt has been made to integrate pseudogene removal with gene prediction, or even to provide a freestanding tool that identifies such erroneous gene predictions. We have created PPFINDER (for Processed Pseudogene finder), a program that integrates several methods of processed pseudogene finding in mammalian gene annotations. We used PPFINDER to remove pseudogenes from N-SCAN gene predictions, and show that gene prediction improves substantially when gene prediction and pseudogene masking are interleaved. In addition, we used PPFINDER with gene predictions as a parent database, eliminating the need for libraries of known genes. This allows us to run the gene prediction/PPFINDER procedure on newly sequenced genomes for which few genes are known.
With the sequencing of more and more genomes, the need for accurate gene prediction is greater than ever. One of the key hurdles in mammalian genome annotation is the presence of large numbers of pseudogenes—copies of real genes that have lost their ability to encode a functional protein product (Zhang and Gerstein 2004). Pseudogenes are frequently predicted to be functional by de novo gene prediction programs such as N-SCAN (Gross and Brent 2006), TWINSCAN (Korf et al. 2001; Flicek et al. 2003), SGP2 (Parra et al. 2003), and SLAM (Alexandersson et al. 2003), as well as annotation programs that make use of transcript evidence such as Ensembl (Hubbard et al. 2005) and Acembly (Kim et al. 2004). Both kinds of gene predictors are attracted to pseudogenes because the sequences of pseudogenes are similar to those of their transcribed parents.
There are two classes of pseudogenes: nonprocessed and processed. Nonprocessed pseudogenes arise through segmental duplication, and hence, they typically retain at least part of the exon–intron structure of the parent gene. Processed pseudogenes arise through retrotransposition of a spliced mRNA and therefore do not contain introns (Vanin 1985). However, secondary integration events may occur within such pseudogenes, leading to intron-like interruptions. Typically, both kinds of pseudogenes accumulate mutations over time until they are indistinguishable from other sequences without any known function. In rare cases, however, they may be incorporated into other genes and thereby acquire new functions (Buzdin 2004).
Estimates of the total number of processed pseudogenes in the human genome vary. Zhang et al. (2003) put the number of processed pseudogenes in the human genome at ∼7800, while Torrents et al. (2003) predict ∼13,800. Ohshima et al. (2003) reported 3664 processed pseudogenes in the human genome and predict that the total number of human processed pseudogenes is ∼7000, based on an estimation of ∼35,000 human genes. The three groups use different thresholds for pseudogene completeness in their methods, and this is the most likely explanation for the difference in estimates (Zhang and Gerstein 2004).
Currently, none of these pseudogene detection methods is available as a standalone tool that can be used to screen genomes or gene sets. Furthermore, they have been optimized for finding as many pseudogenes as possible, rather than the younger pseudogenes that typically get incorporated into models of functional genes. We have created PPFINDER, a standalone tool that can be used to identify processed pseudogenes that have been incorporated into gene models in any mammalian genome annotation. PPFINDER is optimized for this purpose rather than for finding all processed pseudogenes in a genome. In this article, we show that it can be used to improve gene models by iteratively masking pseudogenes incorporated in models and rerunning a gene predictor until no more pseudogenes are found.
PPFINDER identifies processed (but not nonprocessed) pseudogenes by combining two homology-based approaches that are similar to previously described methods (Ohshima et al. 2003; Torrents et al. 2003). Similar to previous methods, it requires a database of potential parent genes from which the pseudogenes are derived. For the human genome, several large databases of known genes are available, but this is not the case for many other species. To render the PPFINDER procedure independent of external sequence databases, we used de novo gene predictions from N-SCAN (Gross and Brent 2006) as putative parent genes and found that we could reliably identify pseudogenes without a database of known genes. In fact, using N-SCAN predictions worked almost as well as did using databases of known genes. Thus, we have developed a bootstrapping method for removal of processed pseudogenes from gene predictions.
Results
Description of PPFINDER
PPFINDER uses two different methods of finding pseudogenes: the intron location method and the conserved synteny method. Both methods start with a gene model and try to find a parent gene from which it was derived by retroposition. If a parent gene is found, the homologous segment in the gene model is marked as a potential pseudogene fragment and put into a filtering procedure. This procedure aligns the parent to the potential pseudogene and discards those cases where the alignment contains an intron.
We developed and tested PPFINDER by using the human genome, NCBI build 35. Unless otherwise indicated, all mentions of genome and annotation sets refer to this build. Parameters were optimized by using the pseudogenes annotated on chromosome 7 (Hillier et al. 2003).
Intron location method
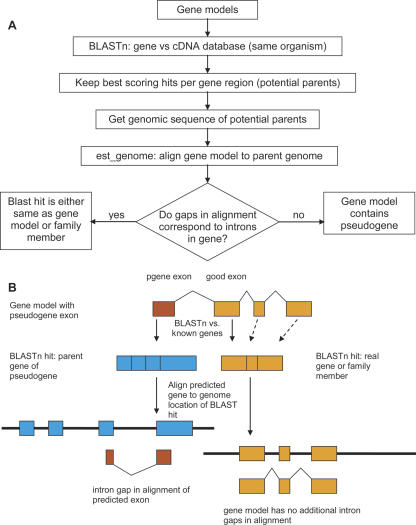
The intron location method identifies potential parent genes by using each gene model as a BLASTn query against a database of transcripts. It picks the highest scoring transcripts (all those with a score >75% of the best score) overlapping each nucleotide of the gene model (Fig. 1A). These transcripts are then aligned to the genomic locus of the gene model that was used as the query. If intron gaps in the alignment are found in different locations than the introns in the gene model, the model is marked as containing a potentially pseudogene-derived segment and put into the filtering procedure.
Figure 1.
The intron location method of pseudogene finding. (A) Flow diagram of the method. See text for details. (B) All predicted gene models are used for BLASTn against a database of known genes. When a pseudogene is incorporated in the gene model, it will hit its parent gene in the BLAST search (left side of diagram). Alignment with the genomic location of the parent gene will usually show intron gaps. Gene model segments that are not derived from pseudogenes may hit a family member elsewhere in the genome (right side of diagram). In this case, alignment of the prediction to the genomic region of the parent will typically include gaps where introns are predicted in the gene model.
Segments of the predicted gene that are not pseudogene-derived will have hits only with themselves and their family members. For pseudogene-derived gene segments, the best hit will be with their parent. To distinguish between family members and parent genes, the predicted gene is aligned to the genomic region of its best hits. Such alignments will introduce large gaps in the predicted gene, corresponding to introns. If the locations of these gaps do not correspond to the intron locations in the gene model, the part of the gene model that aligns to the parent is considered a potential pseudogene (Fig. 1B). When intron locations are not conserved among functional family members, this procedure yields false-positive pseudogenes, most of which are filtered out in the filtering step (see below).
A limitation of this method is that if the pseudogenic segment of a gene model aligns to a single exon of its parent gene, it will not be identified. In addition, this method will not identify pseudogenes with a single-exon parent gene, such as the olfactory receptors. (For a detailed discussion of how PPFINDER deals with the olfactory receptors, see the Supplemental data.)
Conserved synteny method
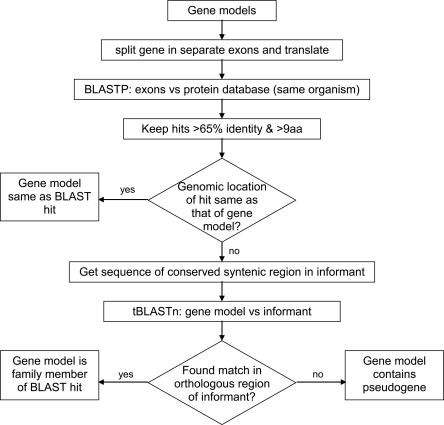
The conserved synteny method makes use of comparison between the genome being annotated (the target) and a second genome (the informant). It identifies potential parent genes by using translated exons as a BLASTp query against a database of proteins whose locus in the target genome is known (Fig. 2). Exons that match a protein from a different genomic location with 65% amino acid identity over at least nine amino acids are considered potential pseudogene fragments. This results in a large number of candidate pseudogenes that are further screened by determining whether they are of recent origin.
Figure 2.
Flow diagram for the conserved synteny method. See text for details.
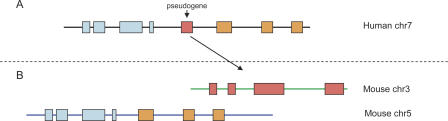
In general, processed pseudogenes are evolving neutrally (Ophir et al. 1999) and will disappear from genomes over time (Kimura 1968). This means that when the target genome is compared with an informant genome at an appropriate evolutionary distance, such as human to mouse, most ancestral pseudogenes are deteriorated or deleted. In a recent study, Zheng et al. (2005) found that ∼5% of processed pseudogenes on human chromosome 22 are preserved on the orthologous region in mouse, although they note that this may not be typical for the complete genome. Most functional genes, on the other hand, are ancestral. Therefore a mouse/human conserved synteny map can be used to identify pseudogenes in human gene models: If a putative pseudogene in the human genome interrupts a region of conserved synteny, it is likely to have arisen after the mouse–human split and hence to be a real pseudogene (Fig. 3). On the other hand, if it is found in the mouse at a position that corresponds to its position in the human genome, it is likely to be a functional gene.
Figure 3.
Use of conserved synteny in pseudogene finding. (A) A pseudogene (pink) on human chromosome 7 was inserted between two genes (blue and orange) (B). This part of human chromosome 7 is orthologous to a region on mouse chromosome 5. If the processed pseudogene in human was generated after the mouse–human split, it will not be present in the orthologous region in the mouse. Instead, the best match in the mouse genome is the location that is orthologous to the parent of the pseudogene.
To determine regions of conserved synteny in human, we made use of the Mouse Net Synteny map from UCSC (Karolchik et al. 2003). This map consists of the best mouse alignment for every part of the human genome and was generated by using BLASTZ (Schwartz et al. 2003). We set an empirical lower limit of 10 kb on regions of conserved synteny—shorter blocks are considered interruptions of conserved synteny. We do not attempt to determine whether blocks <10 kb were in fact derived by retroposition. The 10-kb cutoff was effective in removing even repeat-interrupted pseudogenes, without compromising the larger blocks of synteny.
PPFINDER will only look for conservation of a gene if it has a BLASTp hit in the procedure described at the beginning of this section. Although it would be possible to look for conservation of all gene models, doing so would remove all species-specific genes and exons from the annotation set as well as genes for which the conserved syntenic region in the informant is missing from the assembly.
A limitation of this method is that it does not identify ancestral pseudogenes, so its sensitivity depends on finding a sufficiently diverged informant genome (see Discussion).
Filtering
To verify potential pseudogenes, PPFINDER aligns the parent gene to the genomic region around the pseudogene and identifies all genomic bases to which the parent aligns. This part of the procedure allows PPFINDER to remove spurious pseudogenes.
Each of the methods described above finds false positives. In the intron location method, this occurs if gene family members differ in one or more intron locations. In the conserved synteny method, this happens if a predicted gene (1) is a member of a gene family, and (2) has one or more exons that do not fall in regions of conserved synteny, defined as blocks of at least 10 kb that map to a contiguous region in the informant genome. In most cases, alignment of the potential parent gene (which is in fact a family member) to the genomic region of the gene model will contain gaps, corresponding to introns. (The exceptions are single-exon gene models that are mislabeled as pseudogenes by the conserved synteny method.) Alignments of parent genes to pseudogene regions derived from them do not contain intron gaps. We used this to identify false positives.
To make this filtering step effective, it is necessary to distinguish true intron gaps from all other gaps. To allow for smaller gaps in the alignment, potential pseudogenes were considered real if the average length of interruptions (potential introns) was less than twice the average length of aligned segments (potential exons). We found that this cutoff works well for mammalian genomes. However, sometimes large gaps occur in parent-to-pseudogene alignments because repeats were inserted in pseudogenes after their formation. PPFINDER checks whether interruptions in the alignment contain mostly repeat sequence. If >75% of the interruption sequence is interspersed repeat, the pseudogene is considered verified.
The filtering step is very effective at removing false pseudogene candidates. However, it does allow a few false positives whose introns consist primarily of identifiable interspersed repeats. It also allows a few false positives whose putative parent has no introns. The intron location method cannot produce such false positives, but the conserved synteny method can. Finally, by using this filter we forgo the possibility of identifying nonprocessed pseudogenes. Because they often have a genelike structure with apparently normal introns, nonprocessed pseudogenes are difficult to distinguish reliably from functional genes.
During the filtering step, PPFINDER keeps track of which genomic nucleotides are covered by a parent-to-pseudogene alignment and outputs their coordinates. This output can be used to remove pseudogene-containing gene models or exons from the input annotation set. It can also be used to mask the pseudogene-derived nucleotides. Although this list can be used to annotate some of the pseudogene-derived nucleotides in a genome, PPFINDER is optimized for finding only those that affect the gene models in the input annotation.
Testing PPFINDER
To test PPFINDER, we ran it on the Human Conserved Coding Sequence (CCDS) gene set, a core set of human protein coding regions that are consistently annotated and of high quality (http://www.ncbi.nlm.nih.gov/CCDS/), and on the processed pseudogenes identified in the Vega project (see Methods; Ashurst et al. 2005). If PPFINDER works perfectly, no genes would be marked in the CCDS set and all pseudogenes would be marked in the Vega set.
For the intron location method, we used the human RefSeq mRNAs as a parent database (Pruitt et al. 2005). RefSeq’s annotated pseudogenes were omitted, and the rest were cleaned to remove likely errors (see Methods). We used the remaining 17,820 sequences as the nucleotide database. Our efforts were complicated by the presence of unannotated pseudogenes in RefSeq (see below). For the first stage of the conserved synteny method, we searched for potential parent genes among all human protein entries in the SWISS-PROT/TrEMBL/TrEMBLNew database for which a genome location was present in the UCSC Known Genes track (42,155 sequences).
Of 13,133 CCDS gene annotations, 37 were marked by PPFINDER as processed pseudogenes (0.3%). We manually inspected those hits and found that 23 were single-exon genes that are most likely to be functional retrogenes, because expressed sequence tags (ESTs) are found for each of them. The rest are genes from small gene families that have differences in their exon–intron boundaries in addition to large ratios of exon length to intron length. These genes are marked by the intron location method as putative pseudogenes and are not removed by the alignment filter because of their relatively small introns.
There are 2006 processed pseudogenes annotated in the Vega pseudogene track at UCSC. These pseudogenes were identified by the HAVANA group (http://www.sanger.ac.uk/HGP/havana/) because they are similar to known genes but contain frameshifts and/or stop codons and lack the exon–intron structures of their parent genes (Dunham et al. 1999). Tracks are currently available only for chromosomes 6, 9, 10, 13, 20, and X, and we used all of these. A total of 1567 genes were found by PPFINDER (78.1%). Manual inspection of the 42 annotated Vega pseudogenes missed on chromosome 13 showed that this is usually due to a low conservation between parent and pseudogene and/or fragmentation of the pseudogene, whereby only a small segment of the parent gene is found.
The intron location method identified 1283 pseudogenes in the Vega set, while the conserved synteny method found 1400 pseudogenes; 1116 pseudogenes were identified by both methods. This shows that the sensitivity of these two methods is similar at this evolutionary distance, but each finds pseudogenes that are missed by the other.
Effects of iterative pseudogene masking on gene prediction
Removing gene models that contain pseudogene fragments improves the accuracy of the annotation set by eliminating false positives. When the gene models are produced by a gene prediction program, however, masking out the pseudogenes and rerunning the program may produce a different annotation with more correct predictions than the original. The new gene models may also incorporate new pseudogene fragments that have not been masked. This can be addressed by alternating gene prediction and pseudogene masking until no more pseudogenes are found in the gene models.
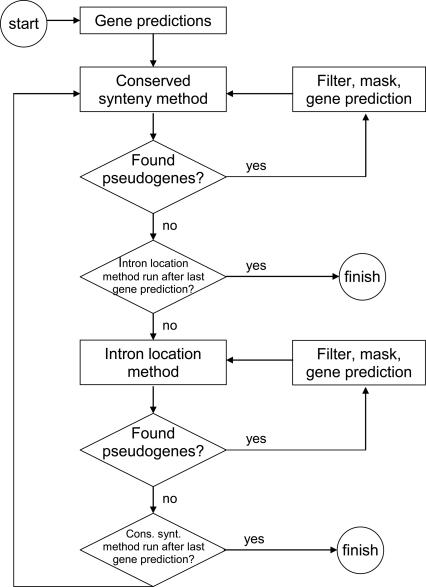
To test the effects of pseudogene masking on gene prediction, we used N-SCAN (Gross and Brent 2006), a de novo gene predictor that takes two or more genome sequences as its inputs. We ran N-SCAN with the human genome as the target and the mouse genome as the only informant and iteratively masked all predicted gene segments of pseudogenic origin (Fig. 4). After running four cycles of gene prediction and pseudogene masking, no more pseudogenes were found in the predictions (see Supplemental methods). PPFINDER masked out exons of 3947 N-SCAN genes with 1888 independent (i.e., nonoverlapping) parent genes. The result was a substantial decrease in the number of predicted genes, from 24,712 to 21,736 (Table 1, external databases column). Note that the reduction in gene number is not identical to the number of masked genes, because genes are repredicted after masking.
Figure 4.
Flow diagram for the bootstrap method that combines pseudogene finding with gene prediction. To iteratively mask pseudogenes and rerun gene prediction, PPFINDER is run with a masking step after each of the methods (conserved synteny and intron alignment). This nested looping is done to remove redundancy, because many pseudogenes will be found by both methods. First, the cycle of pseudogene finding and masking is run using the conserved synteny method, until no more pseudogenes are found. Then the same is done using the intron alignment method. PPFINDER will keep looping through both methods until neither finds any more pseudogenes. One masking/gene prediction loop is called one round.
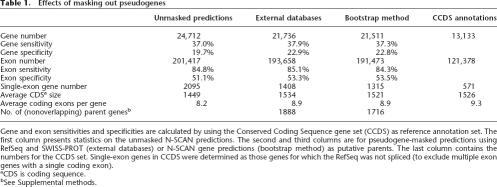
Table 1.
Effects of masking out pseudogenes
Gene and exon sensitivities and specificities are calculated by using the Conserved Coding Sequence gene set (CCDS) as reference annotation set. The first column presents statistics on the unmasked N-SCAN predictions. The second and third columns are for pseudogene-masked predictions using RefSeq and SWISS-PROT (external databases) or N-SCAN gene predictions (bootstrap method) as putative parents. The last column contains the numbers for the CCDS set. Single-exon genes in CCDS were determined as those genes for which the RefSeq was not spliced (to exclude multiple exon genes with a single coding exon).
aCDS is coding sequence.
bSee Supplemental methods.
We evaluated the gene predictions by using a gold standard set of annotated genes as described (Flicek et al. 2003) to determine what fractions of predicted open reading frames (ORFs) and coding exons were correct (gene and exon specificity, respectively) (Table 1). We also calculated the fractions of gold standard ORFs and coding exons that were predicted correctly (gene and exon sensitivity, respectively). We used the CCDS set described above as our gold standard. The results showed that this iterative masking procedure increased the sensitivity and specificity for both ORFs and coding exons, compared with N-SCAN alone. On average, both the number of exons per gene and the number of coding bases per gene increased, bringing them closer to the distributions for known genes (see Supplemental Fig. 1).
As expected, a sizeable number of single-exon gene predictions were masked out: 702 of the original gene predictions. Of these, 16 overlapped a single-exon gene in the CCDS set. This suggests that most of the masked single-exon gene models are incorrect. In addition, masking caused 85 nonmasked single-exon gene models to be incorporated into multi-exon genes. N-SCAN also predicted 122 new single-exon genes after iterative masking, of which 10 overlapped a single-exon CCDS gene. In total, the number of single-exon gene predictions decreased by 687.
We also compared the predictions before and after iterative pseudogene masking with the Vega pseudogene set. Before masking, the coding sequences of 749 N-SCAN predictions overlapped 783 annotated pseudogenes. Iterative masking and reprediction reduced these numbers to 101 and 106, respectively. Thus, the fraction of Vega pseudogenes incorporated into N-SCAN predictions was reduced from 39.0% to 5.3%.
As expected (Zhang et al. 2002), a substantial number of ribosomal pseudogenes were found: A total of 401 of the original gene predictions were masked out with a ribosomal parent gene.
Bootstrapping pseudogene detection from N-SCAN predictions
So far, we have used known genes as the database of potential parents for PPFINDER, but the primary application of gene predictors is to annotate genomes with few known genes. We therefore decided to repeat the experiment described above using N-SCAN’s initial predictions as the database of potential parents (for details, see Supplemental data). This bootstrap procedure masked out exons of 4280 genes in the original predicted set. Of these, 3355 were identical to those masked out by using external databases. As seen before, masking pseudogenes resulted in a substantial decrease in the number of gene models: from 24,712 to 21,511 (see bootstrap method in Table 1). Both the specificity and the sensitivity of gene prediction increased relative to the unmasked set. Surprisingly, the improvement in accuracy using N-SCAN predictions is nearly as good as that seen when using the external databases, even though the initial predictions contain many pseudogenic exons. For a detailed discussion of the differences between the bootstrap and external databases’ procedure, see Supplemental data.
To illustrate the application of this method to a genome with relatively few known genes, we ran it on the dog genome (Lindblad-Toh et al. 2005) with human as informant. This reduced the number of predicted genes and the number of single exon gene predictions and increased the average number of exons per gene. Overall, the effect on the statistical characteristics of the predicted dog gene set was similar to the effect seen in human (see Supplemental data). This suggests that the method can be used successfully on an unannotated mammalian genome. In order to test whether we could achieve the same results using known genes from dog, we ran PPFINDER by using the few available dog cDNA sequences as the parent database (Methods). This did not produce similar results (see Supplemental data).
All N-SCAN predictions and pseudogene-masked regions generated for this article are available in the Supplemental data at http://genes.cse.wustl.edu/vanbaren-06-pseudogene-data/. The UCSC Genome Browser is updated regularly with the most current predictions.
Applying PPFINDER to other sets of gene models
We used the pseudogene finding method to identify putative pseudogenes in other human genome annotation sets: GenScan (Burge and Karlin 1997), Geneid (Blanco et al. 2003), SGP-2 (Parra et al. 2003), and Ensembl (Curwen et al. 2004). We used the RefSeq and SWISS-PROT/TrEMBL/TrEMBLNew sequences as parent databases, as described in Testing PPFINDER above. In the NCBI build 35 GenScan set, we identified 4793 genes with pseudogene exons (11.3% of the predicted gene number); in Geneid, 4615 (14.5%); and in SGP-2, 5853 (17.5%). In Ensembl, PPFINDER identified 1378 transcripts of 1245 genes with putative pseudogene exons (5.3% of total gene number). Only 43 of those genes were marked as pseudogenes by Ensembl. Of the remaining genes, 357 mapped to the Vega annotated chromosomes and 173 of those overlapped a Vega pseudogene annotation. This suggests that at least half of the pseudogenes identified by PPFINDER are real.
These numbers comprise a substantial part of the gene annotations, and those methods may improve markedly if pseudogenes are removed. A list of these putative pseudogenes can be found at http://genes.cse.wustl.edu/vanbaren-06-pseudogene-data/.
We also ran PPFINDER on the RefSeq human mRNAs (Pruitt et al. 2005) as aligned to the genome on the UCSC Browser. PPFINDER found 305 putative pseudogenes that were not annotated as such in their GenBank record. Pseudogenes were found in all divisions of RefSeq including the reviewed set. Some of these may be expressed retrogenes, but manual inspection of a handful of sequences indicated that at least some of these are not functional genes, e.g., because no human ESTs or mRNAs overlap these RefSeqs (see http://genes.cse.wustl.edu/vanbaren-06-pseudogene-data/). It seems that a number of these genes were added to the set because they were published as a putative family member of a known gene (e.g., NM_001005192). Such studies sometimes rely on chromosomal mapping without verification of gene expression, which allows pseudogenes to enter the RefSeq gene set.
Discussion
PPFINDER is an accurate, standalone system for removing processed pseudogenes from any set of gene models. When applied to N-SCAN predictions, it reduces the number of pseudogenes incorporated into gene models by a factor of ∼8%. Its false-positive rate is only 0.3%, as estimated by comparison to the highly accurate CCDS collection of protein-coding gene annotations. This low false-positive rate may be due, in part, to the fact that PPFINDER is optimized for finding only those processed pseudogenes that overlap models of protein-coding genes. If we had designed PPFINDER to find all the processed pseudogenes in the input genome, we would have had to lower our threshold of evidence, thereby admitting more false positives. Additional pseudogenes can be found by alternately masking pseudogenes in gene models and rerunning a gene prediction program.
Using PPFINDER to remove pseudogenes from human genome predictions by N-SCAN, a state-of-the-art de novo gene prediction program, led to significant improvements in accuracy as evaluated by comparison to the CCDS gene models. Furthermore, PPFINDER made the statistical characteristics of the prediction set, including the fraction of genes that consist of a single exon and the average number of exons per gene, more like those of the CCDS gene models. Alternating gene prediction with pseudogene masking led N-SCAN to correctly predict exons it did not find before.
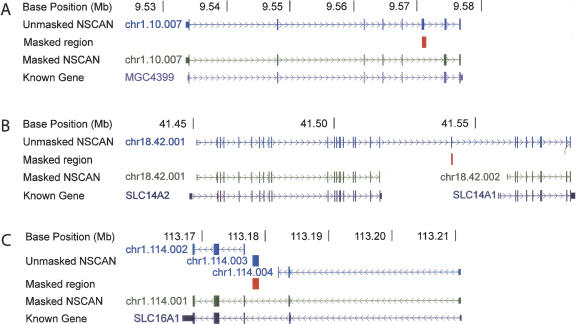
Masking pseudogenes and rerunning gene prediction improves gene prediction in two ways. First, it may result in a correct gene model that is similar to the original except for the absence of a pseudogene derived exon (Fig. 5A). Second, it may have a long distance effect on other parts of the gene model, such as causing it to be split into two correct models (Fig. 5B). Pseudogenic exons in gene models may also change the reading frame, causing real exons on either side of the pseudogenic exon to be omitted (not shown in Fig. 5). Finally, removing single-exon gene models that are based on pseudogenes in the introns of real genes allows N-SCAN to incorporate exons on both sides of the pseudogene into correct gene models (Fig. 5C). If a single-exon gene is predicted in an intron of a real gene, the real gene must be split in two because the current generation of de novo gene predictors does not predict overlapping transcripts.
Figure 5.
Improvement of gene prediction after pseudogene masking. (A) After masking out a pseudogene incorporated in the original gene model, the gene is predicted correctly. (B) A single gene model is split into two correct models after a pseudogene exon is masked. (C) Two gene models are merged into one correct model after masking the pseudogene in an intron of SLC16A1. UTRs are shown as thin blocks, coding exons as thicker blocks, and introns as lines. A gene is considered correctly predicted when the coding sequence is correct. This figure was modified from a screen shot of the UCSC Genome Browser at http://genome.ucsc.edu (Kent et al. 2002).
The number of pseudogenes PPFINDER found in other sets of de novo human gene predictions was similar to what it found in N-SCAN predictions. Interestingly, the gene set produced by the Ensembl annotation pipeline, which uses known transcripts to annotate the genome, also contained a substantial number of putative pseudogenes. Finally, we identified 305 previously unannotated, putative pseudogenes in the RefSeq gene set and found by manual curation that at least some of them are indeed pseudogenes.
One of our key findings is that PPFINDER can be effective even when there are no known genes to serve as potential parents. In that case, it can be run using gene predictions as the potential parents, including the same prediction set targeted for pseudogene removal. We found that using N-SCAN’s human genome predictions as the parent database was almost as effective for removing pseudogenes from those predictions as using known human genes. This bootstrapping capability is essential for removing pseudogenes from predictions in species with few known genes. An example is the dog genome, for which only a small number of mRNAs is present in GenBank. We show that the effects of pseudogene masking using the bootstrap method are similar to those seen in the human genome: It results in fewer predicted exons and gene models, and these gene models increase in length. When the dog mRNAs from GenBank were used instead, pseudogene finding appeared to be much less effective.
The effectiveness of the conserved synteny method depends on the divergence between the target and informant genomes. If the informant is too close, a large number of processed pseudogenes will be ancestral and hence undetectable by the conserved synteny method. The intron alignment method will therefore identify many more pseudogenes than the conserved synteny method, indicating that a more distant informant genome would yield greater sensitivity. On the other hand, if the informant is too distant, some functional genes may not fall in regions of identifiable conserved synteny. This will lead to a high number of false positives, most of which will be caught by the filter. The intron alignment method will not target most of these functional genes, since intron location is conserved over much longer evolutionary distances than is gene order. Therefore, if the intron alignment method identifies far fewer pseudogenes than the conserved synteny method, it may be better to use a more closely related informant. If no closer informant is available, PPFINDER can run the intron alignment method alone. Although the inability to use the conserved synteny method will reduce sensitivity, the data reported here suggest that the reduction will be modest.
In the future, we plan to enhance the filtering step to make PPFINDER even more broadly applicable. Currently, it relies on most introns being substantially longer than most exons. For species with relatively short introns and long exons, such as Caenorhabditis elegans, the filter cannot be used. In addition, some genes are masked out with paralogs despite a large intron-to-exon ratio because their introns consist largely of repeats. The next version of PPFINDER will rely more on splice site models and less on length for distinguishing true introns from interruptions caused by elements inserted into the pseudogene.
Another important task for the future is the development of an “NPPFINDER” for removing nonprocessed pseudogenes from gene models. Currently, no pseudogene finding method can reliably separate gene family members from nonprocessed pseudogenes, in part because the latter often do not have in-frame stop codons (Torrents et al. 2003). Removal of nonprocessed pseudogenes from gene predictions remains an interesting challenge that must be addressed by different methods than the ones described here.
Although there is always more work to be done, PPFINDER can now be used to significantly improve the accuracy of mammalian genome annotations, from well-studied genomes such as those of human and mouse to newly sequenced genomes such as those of dog and cow.
Methods
Sequences
All sequences were downloaded from UCSC (ftp://hgdownload. cse.ucsc.edu/goldenPath/). For details, see Supplemental data.
Synteny map and downloads
We downloaded the Mouse Alignment Net track from the UCSC Browser (NCBI build 35) (Karolchik et al. 2003) and removed all matches <10 kb. This size allows for repeat insertions in pseudogenes, which can extend a synteny block beyond the size of the pseudogene itself. The resulting tables were used to identify orthologous regions in the conserved synteny procedure.
The Known Genes track and sequences were also downloaded from UCSC in July 2004. A position table was created from the track, and the sequences were formatted for BLASTp. The RefSeq tracks (23,045 clones) were downloaded on March 13, 2005, and used for extracting the RefSeq sequences from the genome. Note that the annotated pseudogenes available in RefSeq (NG_ id numbers) are not part of this set. RefSeqs with obvious errors were removed (see Supplemental data).
Gene annotation sets were downloaded from ftp://hgdownload.cse.ucsc.edu/goldenPath/hg17/database on June 15, 2005.
The dog mRNA track was also downloaded from ftp://hgdownload.cse.ucsc.edu/goldenPath/canFam1/database/ in August 2005 and converted to GTF. No sequences were removed.
Validation sets
The CCDS and Vega Pseudogene tracks were downloaded from UCSC (ftp://hgdownload.cse.ucsc.edu/goldenPath/hg17/database) in March 2005 and converted to GTF. Overlapping CCDS genes were merged to a single gene with multiple transcripts. This ensured we would not count the same false positive twice. The Vega Pseudogene track contains many nongenic annotations and therefore was filtered so that only the processed pseudogenes remained (using the Table Browser at UCSC). Because many pseudogenes contain indels, a reading frame often cannot be properly assigned. Therefore we ran the conserved synteny method for translations of the pseudogenes in each of the three frames. The intron alignment method was run on the Vega set as is.
Selecting putative pseudogenes
For BLAST (http://blast.wustl.edu) (W. Gish, unpubl.) parameters, see Supplemental data. For both the intron location and the conserved synteny methods, only BLAST hits on the same strand as the prediction under review were considered. This avoids targeting genes that overlap pseudogenes located on the opposite strand. For the conserved synteny method, only HSPs longer than nine amino acids that had >65% identity were used. The putative pseudogenes found in this way were then used for tBLASTn against the orthologous region in mouse, as specified in the synteny map described above. If no BLAST hit was found, this means that the exon is not conserved between the species and is most likely a pseudogene. In both the intron location and the conserved synteny method, hits with transcripts (intron location) or proteins (conserved synteny) that mapped to the same genomic region as the gene model were skipped because they represent a correctly predicted gene.
The intron location method uses whole-gene models instead of single-exon translations. This means that when a pseudogene exon is incorporated in a gene model, this gene model can have BLAST hits with both the pseudogene parent and the actual gene. Therefore, for every hit, the range of overlap with the gene model was determined. Hits were kept if their score was at least 75% of the highest scoring hit, or if they overlapped a different segment of the gene model than all higher scoring hits and had a percentage identity of at least 75%. Every putative parent gene found in this way was used in the filtering step of PPFINDER.
N-SCAN evaluation
For details on how N-SCAN was run, see Supplemental data. We used the aligned human CCDS set downloaded from the UCSC Browser site as the basis on which to compare our predictions. The downloaded CCDS set contained 14,793 transcripts. After merging overlapping transcripts into genes and removing identical transcripts, 14,714 transcripts in 13,133 genes remained.
Only coding exons on whole chromosomes were used for evaluation of N-SCAN performance.
Segmental duplications
To find gene containing regions of high homology in the human genome, we took the RefSeqs that mapped to more than one location in the UCSC Genome Browser. These RefSeqs have a base identity level within 0.1% of the best alignment and at least 96% base identity with the genomic sequence. We took all N-SCAN predictions that overlapped any of these 535 loci, and for the 33 of these that were masked, we checked if the parent gene was derived from the paralog.
Acknowledgments
We thank LaDeana Hillier for providing the human chromosome 7 pseudogene set that started this work, and Mark Diekhans and Robert Baertsch for helpful discussions. This work was supported by grant HG02278 from the National Human Genome Research Institute to M.R.B.
Footnotes
[Supplemental material is available online at www.genome.org. N-SCAN and PPFINDER are open source software and may be obtained from http://genes.cse.wustl.edu/.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.4766206
References
- Alexandersson M., Cawley S., Pachter L., Cawley S., Pachter L., Pachter L. SLAM: Cross-species gene finding and alignment with a generalized pair hidden Markov model. Genome Res. 2003;13:496–502. doi: 10.1101/gr.424203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashurst J.L., Chen C.K., Gilbert J.G., Jekosch K., Keenan S., Meidl P., Searle S.M., Stalker J., Storey R., Trevanion S., Chen C.K., Gilbert J.G., Jekosch K., Keenan S., Meidl P., Searle S.M., Stalker J., Storey R., Trevanion S., Gilbert J.G., Jekosch K., Keenan S., Meidl P., Searle S.M., Stalker J., Storey R., Trevanion S., Jekosch K., Keenan S., Meidl P., Searle S.M., Stalker J., Storey R., Trevanion S., Keenan S., Meidl P., Searle S.M., Stalker J., Storey R., Trevanion S., Meidl P., Searle S.M., Stalker J., Storey R., Trevanion S., Searle S.M., Stalker J., Storey R., Trevanion S., Stalker J., Storey R., Trevanion S., Storey R., Trevanion S., Trevanion S., et al. The Vertebrate Genome Annotation (Vega) database. Nucleic Acids Res. 2005;33:D459–D465. doi: 10.1093/nar/gki135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco E., Parra G., Guigo R., Parra G., Guigo R., Guigo R.2003. Using geneid to identify genes. In Current protocols in bioinformatics (ed. D.B. Davison) pp. Unit 4.3. John Wiley & Sons Inc. New York [Google Scholar]
- Burge C., Karlin S., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Buzdin A.A. Retroelements and formation of chimeric retrogenes. Cell. Mol. Life Sci. 2004;61:2046–2059. doi: 10.1007/s00018-004-4041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curwen V., Eyras E., Andrews T.D., Clarke L., Mongin E., Searle S.M.J., Clamp M., Eyras E., Andrews T.D., Clarke L., Mongin E., Searle S.M.J., Clamp M., Andrews T.D., Clarke L., Mongin E., Searle S.M.J., Clamp M., Clarke L., Mongin E., Searle S.M.J., Clamp M., Mongin E., Searle S.M.J., Clamp M., Searle S.M.J., Clamp M., Clamp M. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I., Shimizu N., Roe B.A., Chissoe S., Hunt A.R., Collins J.E., Bruskiewich R., Beare D.M., Clamp M., Smink L.J., Shimizu N., Roe B.A., Chissoe S., Hunt A.R., Collins J.E., Bruskiewich R., Beare D.M., Clamp M., Smink L.J., Roe B.A., Chissoe S., Hunt A.R., Collins J.E., Bruskiewich R., Beare D.M., Clamp M., Smink L.J., Chissoe S., Hunt A.R., Collins J.E., Bruskiewich R., Beare D.M., Clamp M., Smink L.J., Hunt A.R., Collins J.E., Bruskiewich R., Beare D.M., Clamp M., Smink L.J., Collins J.E., Bruskiewich R., Beare D.M., Clamp M., Smink L.J., Bruskiewich R., Beare D.M., Clamp M., Smink L.J., Beare D.M., Clamp M., Smink L.J., Clamp M., Smink L.J., Smink L.J., et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Flicek P., Keibler E., Hu P., Korf I., Brent M.R., Keibler E., Hu P., Korf I., Brent M.R., Hu P., Korf I., Brent M.R., Korf I., Brent M.R., Brent M.R. Leveraging the mouse genome for gene prediction in human: From whole-genome shotgun reads to a global synteny map. Genome Res. 2003;13:46–54. doi: 10.1101/gr.830003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S.S., Brent M.R., Brent M.R. Using multiple alignments to improve gene prediction. J. Comput. Biol. 2006;13:379–393. doi: 10.1089/cmb.2006.13.379. [DOI] [PubMed] [Google Scholar]
- Hillier L.W., Fulton R.S., Fulton L.A., Graves T.A., Pepin K.H., Wagner-McPherson C., Layman D., Maas J., Jaeger S., Walker R., Fulton R.S., Fulton L.A., Graves T.A., Pepin K.H., Wagner-McPherson C., Layman D., Maas J., Jaeger S., Walker R., Fulton L.A., Graves T.A., Pepin K.H., Wagner-McPherson C., Layman D., Maas J., Jaeger S., Walker R., Graves T.A., Pepin K.H., Wagner-McPherson C., Layman D., Maas J., Jaeger S., Walker R., Pepin K.H., Wagner-McPherson C., Layman D., Maas J., Jaeger S., Walker R., Wagner-McPherson C., Layman D., Maas J., Jaeger S., Walker R., Layman D., Maas J., Jaeger S., Walker R., Maas J., Jaeger S., Walker R., Jaeger S., Walker R., Walker R., et al. The DNA sequence of human chromosome 7. Nature. 2003;424:157–164. doi: 10.1038/nature01782. [DOI] [PubMed] [Google Scholar]
- Hubbard T., Andrews D., Caccamo M., Cameron G., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., Andrews D., Caccamo M., Cameron G., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., Caccamo M., Cameron G., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., Cameron G., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., Clarke L., Coates G., Cox T., Cunningham F., Coates G., Cox T., Cunningham F., Cox T., Cunningham F., Cunningham F., et al. Ensembl 2005. Nucleic Acids Res. 2005;33:D447–D453. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Schwartz M., Sugnet C.W., Thomas D.J., Sugnet C.W., Thomas D.J., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N., Shin S., Lee S., Shin S., Lee S., Lee S. ASmodeler: Gene modeling of alternative splicing from genomic alignment of mRNA, EST and protein sequences. Nucleic Acids Res. 2004;32:W181–W186. doi: 10.1093/nar/gkh404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- Korf I., Flicek P., Duan D., Brent M.R., Flicek P., Duan D., Brent M.R., Duan D., Brent M.R., Brent M.R. Integrating genomic homology into gene structure prediction. Bioinformatics. 2001;17:S140–S148. doi: 10.1093/bioinformatics/17.suppl_1.s140. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas III E.J., Zody M.C., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas III E.J., Zody M.C., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas III E.J., Zody M.C., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas III E.J., Zody M.C., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas III E.J., Zody M.C., Kamal M., Clamp M., Chang J.L., Kulbokas III E.J., Zody M.C., Clamp M., Chang J.L., Kulbokas III E.J., Zody M.C., Chang J.L., Kulbokas III E.J., Zody M.C., Kulbokas III E.J., Zody M.C., Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Ohshima K., Hattori M., Yada T., Gojobori T., Sakaki Y., Okada N., Hattori M., Yada T., Gojobori T., Sakaki Y., Okada N., Yada T., Gojobori T., Sakaki Y., Okada N., Gojobori T., Sakaki Y., Okada N., Sakaki Y., Okada N., Okada N. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol. 2003;4:R74. doi: 10.1186/gb-2003-4-11-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ophir R., Itoh T., Graur D., Gojobori T., Itoh T., Graur D., Gojobori T., Graur D., Gojobori T., Gojobori T. A simple method for estimating the intensity of purifying selection in protein-coding genes. Mol. Biol. Evol. 1999;16:49–53. doi: 10.1093/oxfordjournals.molbev.a026037. [DOI] [PubMed] [Google Scholar]
- Parra G., Agarwal P., Abril J.F., Wiehe T., Fickett J.W., Guigo R., Agarwal P., Abril J.F., Wiehe T., Fickett J.W., Guigo R., Abril J.F., Wiehe T., Fickett J.W., Guigo R., Wiehe T., Fickett J.W., Guigo R., Fickett J.W., Guigo R., Guigo R. Comparative gene prediction in human and mouse. Genome Res. 2003;13:108–117. doi: 10.1101/gr.871403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K.D., Tatusova T., Maglott D.R., Tatusova T., Maglott D.R., Maglott D.R. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Baertsch R., Hardison R.C., Haussler D., Miller W., Hardison R.C., Haussler D., Miller W., Haussler D., Miller W., Miller W. Human–mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrents D., Suyama M., Zdobnov E., Bork P., Suyama M., Zdobnov E., Bork P., Zdobnov E., Bork P., Bork P. A genome-wide survey of human pseudogenes. Genome Res. 2003;13:2559–2567. doi: 10.1101/gr.1455503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanin E.F. Processed pseudogenes: Characteristics and evolution. Annu. Rev. Genet. 1985;19:253–272. doi: 10.1146/annurev.ge.19.120185.001345. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Gerstein M., Gerstein M. Large-scale analysis of pseudogenes in the human genome. Curr. Opin. Genet. Dev. 2004;14:328–335. doi: 10.1016/j.gde.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Harrison P., Gerstein M., Harrison P., Gerstein M., Gerstein M. Identification and analysis of over 2000 ribosomal protein pseudogenes in the human genome. Genome Res. 2002;12:1466–1482. doi: 10.1101/gr.331902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Harrison P.M., Liu Y., Gerstein M., Harrison P.M., Liu Y., Gerstein M., Liu Y., Gerstein M., Gerstein M. Millions of years of evolution preserved: A comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13:2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Zhang Z., Harrison P.M., Karro J., Carriero N., Gerstein M., Zhang Z., Harrison P.M., Karro J., Carriero N., Gerstein M., Harrison P.M., Karro J., Carriero N., Gerstein M., Karro J., Carriero N., Gerstein M., Carriero N., Gerstein M., Gerstein M. Integrated pseudogene annotation for human chromosome 22: Evidence for transcription. J. Mol. Biol. 2005;349:27–45. doi: 10.1016/j.jmb.2005.02.072. [DOI] [PubMed] [Google Scholar]