Abstract
The role of nonlinear DNA in replication, recombination, and transcription has become evident in recent years. Although several studies have predicted and characterized regulatory elements at the sequence level, very few have investigated DNA structure as regulatory motifs. Here, using G-quadruplex or G4 DNA motifs as a model, we have researched the role of DNA structure in transcription on a genome-wide scale. Analyses of >61,000 open reading frames (ORFs) across 18 prokaryotes show enrichment of G4 motifs in regulatory regions and indicate its predominance within promoters of genes pertaining to transcription, secondary metabolite biosynthesis, and signal transduction. Based on this, we predict that G4 DNA may present regulatory signals. This is supported by conserved G4 motifs in promoters of orthologous genes across phylogenetically distant organisms. We hypothesized a regulatory role of G4 DNA during supercoiling stress, when duplex destabilization may result in G4 formation. This is in line with our observations from target site analysis for 55 DNA-binding proteins in Escherichia coli, which reveals significant (P < 0.001) association of G4 motifs with target sites of global regulators FIS and Lrp and the sigma factor RpoD (σ70). These factors together control >1000 genes in the early growth phase and are believed to be induced by supercoiled DNA. We also predict G4 motif-induced supercoiling sensitivity for >30 operons in E. coli, and our findings implicate G4 DNA in DNA-topology-mediated global gene regulation in E. coli.
DNA adopts several secondary structure motifs, although the Watson-Crick duplex is its predominant natural state in genomes. The role of non-B DNA motifs in recombination, replication, and particularly, regulation of gene expression has been implicated and generally appreciated in recent years, although still relatively less understood (Sinden 1994; Perez-Martin and de Lorenzo 1997; Pedersen et al. 2000; Bacolla and Wells 2004). It is now evident that DNA sequence also encodes for spatial structures, much like protein sequence, apart from protein-coding and cis-acting regulatory elements. Cells use these structural motifs in such a way that DNA sequence information per se has a minimal role other than facilitating formation of the structural motifs. Several reports have implicated the role of non-B DNA structures in the context of gene regulation, both in prokaryotes (for review, see Hatfield and Benham 2002) and eukaryotes (for review, see Rich and Zhang 2003; Bacolla and Wells 2004). Many studies have predicted and determined regulatory elements at the sequence level (Wasserman et al. 2000; Beer and Tavazoie 2004; Xie et al. 2005); however, very few have investigated DNA structure in this context (Florquin et al. 2005). We have focused on searching and investigating the role of a particular type of non-B DNA motif—the G-quadruplex or G4 DNA as a structural regulatory signal.
Guanine-rich sequences attain unique four-stranded conformations known as G4 DNA (Gellert et al. 1962; Sen and Gilbert 1988; Balagurumoorthy and Brahmachari 1994). G4 DNA stabilized by charge coordination with monovalent cations (especially K+) within a planar array of four-hydrogen-bonded guanines (G-quartets or tetrads) may result from intramolecular or intermolecular association of four DNA strands in parallel or antiparallel orientation (Fig. 1; for review, see Gilbert and Feigon 1999). Chromosomal regions containing guanine-rich sequence capable of forming G4 DNA include immunoglobin heavy-chain switch regions (Dunnick et al. 1993), G-rich minisatellites (Weitzmann et al. 1997), rDNA (Hanakahi et al. 1999), and telomeres (Parkinson et al. 2002). G4 DNA has been implicated in regulation of the human oncogene c-myc (Siddiqui-Jain et al. 2002; Seenisamy et al. 2004) and as an “at-risk motif” involved in genome rearrangements in the nematode Caenorhabditis elegans (Cheung et al. 2002).
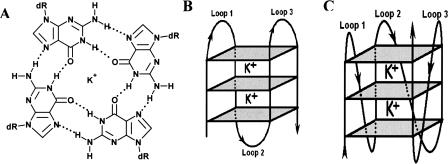
Figure 1.
Schematic representation of G4 motif. (A) Hydrogen-bonded G-tetrad with K+ (Na+ also stabilizes a G-tetrad); each guanine in this planar array is contributed from different G-runs, which are separated by intervening loops in an intramolecular motif. (B,C) Intramolecular folding pattern showing stem and loop organization in an antiparallel (B) and parallel (C) conformation of a G4 motif, where the planes represent each tetrad unit and are stacked to form the stem of the motif.
In vivo structure formation by DNA may have deleterious consequences as established by human neurodegenerative diseases caused by triplet repeat expansions (McMurray 1999; Sinden 1999; Cummings and Zoghbi 2000). Furthermore, non-B DNA structures are targeted by the cellular mismatch repair factors, wherein any lacking factors cause repeat instability in Saccharomyces cerevisiae (Strand et al. 1993) and tumors in humans (Kolodner 1995; Modrich and Lahue 1996). DNA secondary structures, particularly G4 DNA, also play a central role in telomere extension and are the focus of targeted anticancer drug development (Zahler et al. 1991; Neidle and Read 2000; Incles et al. 2004). It is known that the Escherichia coli RecQ can unwind G4 DNA and that the family of RecQ helicases is conserved and is essential for genomic stability in organisms from E. coli to humans (Shen and Loeb 2000; Wu and Maizels 2001; Bachrati and Hickson 2003). However, no systematic investigation of G4 DNA in prokaryotes exists, except one recent study showing in vivo existence of G4 DNA in E. coli (Duquette et al. 2004). On the other hand, non-B DNA forms have been implicated as regulatory signals in E. coli under supercoiling stress. Specific roles have been illustrated in a few cases like the ilvGMEDA, leuV, and ilvYC operons (Sheridan et al. 1999; Opel and Hatfield 2001; for review, see Hatfield and Benham 2002). In this context, it is interesting to consider that G4 DNA might be important in gene regulation and genetic stability in prokaryotes.
Using a nucleic acid pattern recognition program, we searched 18 representative prokaryote genomes for G4 DNA sequences and analyzed their genomic distribution and association with genes. Our analysis indicated enrichment of G4 DNA within the near upstream region of genes relative to other non-coding regions across all organisms. A comparative functional analysis (using 23 classes from COGS) of >61,000 open reading frames (ORFs) indicated that transcription, amino acid biosynthesis, and signal transduction genes could be predominantly controlled by G4 DNA. We also observed that the motifs were conserved within promoters of orthologous genes across phylogenetically distant organisms. Additionally, randomly selected potential G4 forming sequences from E. coli were observed to adopt quadruplex structure in solution under physiological conditions. Transcription-factor-binding site analysis of 55 DNA-binding proteins in the region flanking G4 DNA sequences in E. coli indicated significant association with global regulators, which are known to be supercoiling sensitive. Taken together, our findings indicate a putative role of G4 DNA in prokaryotic gene regulation. Based on our observations in E. coli, we predict that G4 DNA may be one of the factors involved in DNA-topology-mediated gene expression.
Results
Definition of G4 motifs, classification, and genome-wide search strategy
Intramolecular G4 DNA motifs comprise four runs of guanines (constituting the stem of G4 motif) interspersed with nucleotide bases, which form three intervening loops (Fig. 1; Balagurumoorthy and Brahmachari 1994; Gilbert and Feigon 1999). We developed a pattern search algorithm to identify potential G4 DNA sequences wherein four consecutive G-runs were identified, after allowing for three intervening loops (see Methods). In order to avoid overestimation of G4 DNA motifs, overlapping patterns (with more than four G-runs) were stitched together and the sequence was designated as a tract, which can adopt multiple G4 motifs but is most likely to present only one exclusive motif. In the following text, we refer to such tracts as PG4 (potential G4) motifs. Applying our search strategy in a genome-wide screen, we collated two basic forms of information for mapping and comparative analyses: (1) the frequency of the bases comprising the tracts and (2) association of the tracts with the regulatory regions of genes.
Results of genome searches
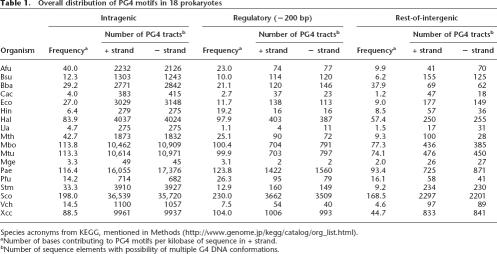
We applied our search strategy to 18 complete prokaryote genomes representing different phylogenetic origins. All PG4 motifs identified within the respective genomic regions—intragenic, putative regulatory (up to 200 bp upstream of genes), or “rest-of-intergenic” (see Methods)—for 18 organisms are listed, organized according to the above criteria, on our Web site (http://www.igib.res.in/prokaryote/PG4.htm). Table 1 shows a summary of the distribution in both + and − strands.
Table 1.
Overall distribution of PG4 motifs in 18 prokaryotes
Species acronyms from KEGG, mentioned in Methods (http://www.genome.jp/kegg/catalog/org_list.html).
aNumber of bases contributing to PG4 motifs per kilobase of sequence in + strand.
bNumber of sequence elements with possibility of multiple G4 DNA conformations.
The overall number of motifs was similarly distributed in both the strands and appeared to be higher in organisms with a high GC%, which was expected as the motifs were G-rich (Table 1; Supplemental Table S1). Interestingly, the frequency of PG4 motifs (number of bases involved in motif formation per kilobase) varied considerably among the three regions. It was interesting to observe that >98% of the motifs in both the + and − strands had two tetrad units in the stem, and a tract size below 40 bases was prevalent (>95%) across all organisms in both strands (Supplemental Tables S3 and S4). The variation in size of the three loops was also analyzed and is represented in mosaic plots (Supplemental Fig. S2; Friendly 1994) for each genomic region across all organisms. The overall distribution indicates no preference in any particular loop size combination in the intergenic regions (plots A and B), while the intragenic region (plot C) showed a preference for a loop size of four in all combinations. All genomes were enriched in PG4 motifs vis-à-vis the probability of random occurrence. By using BLAST for each identified PG4 motif sequence against the respective organism, we observed that the probability of random occurrence was very low for most sequences except ones that were <14 bases and occurred multiple times (Supplemental Table S2) (an extensive set from five different organisms is available at http://www.igib.res.in/prokaryote/PG4.htm). Several other independent observations emphasize this (see Supplemental material).
Distinct genomic distribution was observed for PG4 motifs
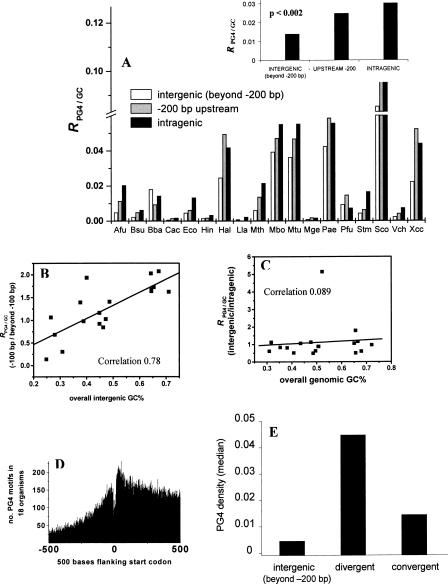
The frequency of motifs (both strands together) in the different genomic regions is listed in Table 1. Streptomyces coelicolor A3(2) (Sco), with the highest GC% (72.11%) (Supplemental Table S1) had the highest density of PG4 motifs, and the near upstream region harbored the major proportion of them. Similarly, Pseudomonas aeruginosa PAO1, Halobacterium sp. NRC-1, and Xanthomonas campestris pv. Campestris str. ATCC 33,913 also showed a high PG4 motif frequency in the −200-bp region. On the other hand, the low-GC-content (<40%) genomes of Clostridium acetobutylicum ATCC 824, Lactococcus lactis subsp. lactis Il1403, Haemophilus influenzae Rd KW20, and Mycoplasma genitalium G-37 had a low frequency of motifs. Figure 2A shows the distribution of PG4 motifs in intragenic, intergenic (beyond −200 bp), and putative regulatory regions for the + strand in 18 bacterial genomes. Owing to the observed overall correlation between motif density and GC% in the respective region, as expected for G-rich elements, we analyzed the variation in motif density across the three genomic regions after normalizing for GC%. Here, we excluded the downstream intergenic regions between convergent genes to avoid PG4 motifs, which could be putative terminators—as indicated by G4 DNA-induced polymerase “falloff” in several studies (Simonsson et al. 1998; Siddiqui-Jain et al. 2002). These regions have been independently analyzed relative to regulatory regions (see below). PG4 motifs in each region were expressed as a ratio of the frequency of GC bases in the respective region (RPG4/GC) (Fig. 2A). A higher R was observed in the intragenic regions relative to the intergenic region (beyond −200 bp) in the + strand (P < 0.001; medianintragenic = 0.030, medianintergenic (beyond −200 bp) = 0.014) (Fig. 2A, inset). More interestingly, the frequency of PG4 motifs in the putative regulatory region (up to −200 bp) was observed to be higher in comparison to the intergenic region (P < 0.0025; median−200 bp = 0.025) (Fig. 2A, inset; Supplemental Table S5). The observed difference was true for both the + and the − strands independently (Supplemental Fig. S3 shows − strand distribution); however, the difference in distribution of PG4 motifs between the strands was not statistically significant in any region (Supplemental Table S5). Sequence with multiple G-runs that would not adopt a G4 DNA motif (control pattern) did not show enrichment in the putative regulatory regions (Supplemental Fig. S9; Supplemental Table S12). We also checked the distribution of PG4 motifs using the variable stem size parsing method (Huppert and Balasubramanian 2005; Todd et al. 2005) and found it to be consistent with our observations (Supplemental Fig. S8).
Figure 2.
Putative regulatory regions in prokaryotes are enriched in PG4 motifs. (A) Genome-wide distribution of PG4 motifs within the + strand in 18 prokaryotes showing frequency of the bases forming PG4 motifs in each region expressed as a ratio of the GC frequency of the respective region (RPG4/GC) for each organism. (Inset) Median ratio (RPG4/GC) for each region calculated from the distribution in the respective regions across all organisms. (Supplemental Table S5 shows the mean and standard deviation, and Supplemental Fig. S3 shows a similar distribution for the − strand.) The intergenic (beyond −200 bp) region includes all intergenic regions except the downstream region between two convergently oriented genes. (B) GC-rich organisms have selected for PG4 motifs in their immediate upstream regions. Ratio of the frequency of PG4 motifs (after controlling for GC% in the respective regions) in the −100-bp region versus beyond −100 bp within the intergenic region shows a high correlation with the GC% of the intergenic region for respective organisms. (C) The motif frequency of intergenic versus intragenic regions does not depend on the GC% of the genome. The ratio-plot for intergenic versus intragenic regions against overall (genome-wide) GC% of the organism shows very low correlation. M. genitalium shows a high ratio (>5.0) because of a very low intergenic basepair length (correlation on excluding M. genitalium was 0.24). (D) The number of PG4 motifs decreases sharply on moving upstream of genes relative to the intragenic regions. Data were plotted from all 61,355 ORFs in 18 organisms within the flanking 500 bases of the start codon of all ORFs. The center of each motif sequence was used for mapping with respect to the start codon (i.e., for a sequence of length n, the n/2-th base was used as its coordinate). (E) Promoter-rich regions have a higher density of PG4 motifs. Intergenic regions separating divergently (promoter-rich) and convergently (possibly promoter-less) oriented gene pairs were mapped in all 18 organisms for comparison. The median of PG4 density (number of bases involved in motif pattern normalized for sequence length of the respective region) is shown along with the density in the intergenic regions (beyond −200 bp, as in A). The difference between the divergent and convergent (P < 0.007) and the divergent and intergenic (P < 0.025) regions was significant, while the difference between the convergent and intergenic regions was not significant (P = 0.199). All statistical comparisons were done in a pairwise mode for the different genomic regions, and significance was estimated using the two-tailed nonparametric Signed Wilcoxon Test. The organism acronyms are as obtained from KEGG and are mentioned in Methods.
Motif frequency decreases beyond 50–100 bases upstream of start codon
We tested the implications of the above observation with respect to the near upstream region by plotting the number of bases making PG4 motifs in the + strand within blocks of 50 bases up to 500 bases upstream of all genes, excluding coding regions (Supplemental Fig. S4). The motif frequency decreased sharply on moving upstream from the start codon in nearly all organisms, indicating a prevalence of PG4 motifs in near upstream regions. We checked whether the GC% of the entire intergenic region affected the motif frequency in the near upstream (−100 bp) region. The ratio of RPG4/GC in the −100-bp region and RPG4/GC in the entire non-coding region excluding the first −100 bases from a gene (RPG4/GC(−100 bp)/RPG4/GC(beyond −100 bp)) was plotted against the overall GC% of the entire intergenic region of all 18 organisms (Fig. 2B). Interestingly, a positive correlation was observed in this case (Spearman’s ρ [nonparametric correlation coefficient] = 0.72, P < 0.001). As a control, we analyzed the frequency ratios in the entire intergenic versus intragenic region (RPG4/GC(intergenic)/RPG4/GC(intragenic)) against the overall genomic GC%. In this case, the correlation was not significant (Spearman’s ρ = 0.09, P = 0.499) (Fig. 2C). This indicated that GC-rich genomes positively selected for PG4 motifs in the near upstream region relative to respective far intergenic regions. A particular case is the GC-rich S. coelicolor genome, where we clearly observed a higher occurrence of PG4 motifs in the putative regulatory region relative to the other genomic regions (Fig. 2A) and also relative to other genomes (Supplemental Fig. S4). Overall distribution of motifs flanking 0.5 kb of the start codon of genes (in 61,355 ORFs) across 18 organisms (Fig. 2D) highlighted the decrease in PG4 frequency (on moving away from start codon) in the near upstream region vis-à-vis the coding region and also indicated that sequence overlapping start codons was relatively less dense in motifs. Interestingly, intragenic regions close to the start codon also showed high PG4 motif density, which could be potential signals for repression of transcription. This is in line with observations of G4-motif-induced arrest in DNA synthesis (Woodford et al. 1994; Simonsson et al. 1998; Siddiqui-Jain et al. 2002). Additionally, the possibility of attenuation or antitermination signals by G4 motif formation in the transcribed mRNA cannot be ruled out (Christiansen et al. 1994; Horsburgh et al. 1996).
Intergenic regions separating divergently oriented genes are enriched in PG4 motifs
We analyzed intergenic regions separating divergently and convergently oriented genes in 18 organisms for PG4 motif density. It was observed that the motif density was significantly higher in the divergent intergenic regions (mediandivergent: 0.0469 vs. medianconvergent: 0.0158; P < 0.007) (Fig. 2E). We also observed that PG4 density in divergent intergenic regions was higher than the density observed in the intergenic region (medianintergenic(beyond −200 bp): 0.0055; P < 0.025). Although a higher PG4 density was observed in divergent intergenic relative to putative regulatory regions, this was not statistically significant (P = 0.199) (Supplemental Table S10). Similarly, the difference with intragenic regions was also not significant. PG4 density in convergent intergenic regions, however, did not show significant difference when compared to intergenic, intragenic, or regulatory regions (Supplemental Table S10). Thus, although a functional role of PG4 motifs as terminators cannot be ruled out, enrichment in divergent intergenic regions relative to convergent ones suggests a functional role of PG4 motifs with regulatory consequences.
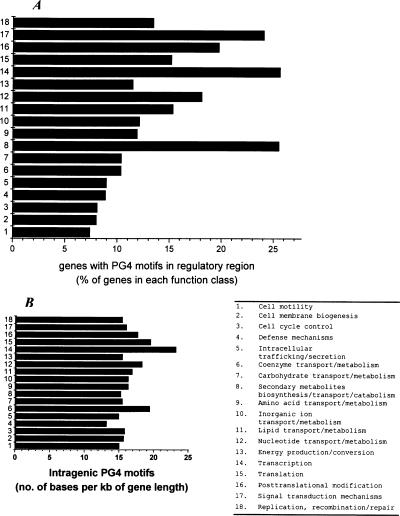
ORFs with PG4 motifs in regulatory region show distinct functional distribution
We analyzed 37,974 ORFs across 18 species in 23 different functional classes from the COGS database (Tatusov et al. 1997). These ORFs were considered after excluding genes belonging to undefined functional classes (i.e., function unknown and general function prediction only). Of these, 5574 (14.7%) ORFs had at least one motif in the + or the − strand within the −200 bp of the start codon. We observed that the functional classes secondary metabolite biosynthesis, transport and catabolism (25.52%), transcription (25.64%), and signal transduction (24.08%) had more genes, which harbored one or more PG4 motifs in their regulatory regions relative to others (P < 0.004; the average genes in other classes is 11.97%, SD = 3.75%) (Fig. 3A). Interestingly, >17% (968 genes) of all ORFs having motifs in the regulatory region pertained to transcription (P < 10−8; <10% in any other class) (Supplemental Fig. S5). We did not observe much variation in the intragenic PG4 motif frequency between the function classes (∼15–23 bases per kilobase of gene) on analyzing the coding region of all 37,974 ORFs (Fig. 3B). Transcription factor genes showed a somewhat higher motif frequency than the average; however, this was not significant (P = 0.108). It must be mentioned that regulatory control, considering operon organization within bacterial genomes, may not be necessarily exerted from the immediate upstream region of the ORFs, and thus our analysis gives a global view of the PG4 motifs vis-à-vis their putative functional role. Analysis in the context of operons in E. coli is presented below.
Figure 3.
Genes harboring PG4 motifs in their regulatory region show distinct functional distribution in a comparative analysis comprising 37,974 ORFs from 18 organisms. (A) The distribution of genes with at least one PG4 motif within the −200-bp region is shown as the percentage of total genes in the respective function class—secondary metabolite biosynthesis, transcription, and translation related genes show significant difference (P < 0.004). (B) The intragenic PG4 motif density indicates that the distribution is not significantly different across the functional classes (P = 0.108). The PG4 motif density was calculated as the number of bases involved in motif formation per kilobase of gene length. Two classes, chromatin structure and dynamics and RNA processing and modification, which constitute only 0.054% and 0.09% of the distribution, were not included in the plots. Extracellular structure, nuclear structure, and cytoskeleton genes do not have any motifs in their regulatory regions. Undefined classes like function unknown and general function prediction have been excluded from analysis along with genes not found in the COGS database. All function information was obtained from the COGS database. A plot showing distribution across the functional classes with respect to the total ORFs (5574) with PG4 motifs in −200-bp regions is shown in Supplemental Figure S5.
Orthologs in distantly related species conserve PG4 motifs within regulatory region
Conservation of motifs across species, especially if the species belong to evolutionarily distant groups, indicates biological significance as functional signals. We hypothesized that if PG4 elements serve as regulatory motifs, they are liable to be conserved in the regulatory region of orthologous genes. We analyzed orthologous groups from COGS for E. coli genes harboring PG4 motifs in the –200 region and checked whether the corresponding orthologs (in the 17 other organisms) also had one or more PG4 motifs in their upstream region. We found 40 genes where PG4 motifs were conserved (Table 2). In 36 of these, at least one species was from an evolutionarily distant group (von Mering et al. 2003), and 20 genes showed conservation in orthologs across at least two distant groups. It was interesting to observe that a majority of the genes with conserved motifs pertained to metabolism (amino acid, carbohydrate, and vitamins/cofactors), membrane transport (ABC and ion), transcription, and translation.
Table 2.
Representative table showing Escherichia coli genes and orthologs with conserved PG4 motifs (within regulatory region)
The full list is given in Supplemental Table S6.
aSpecies acronyms as in KEGG (http://www.genome.jp/kegg/catalog/org_list.html). Groups of phylogenetically related organisms were obtained from the STRING server (see Methods).
bAnnotation information from COGS, the GO server, and NCBI.
Analysis of PG4 motifs in E. coli
Mapping of PG4 motifs to the regulatory network in E. coli
Based on our findings, which collectively indicated that PG4 motifs may be biologically significant as functional regulators, we focused on E. coli as a reference organism for mapping the motifs to known regulatory networks. E. coli was chosen for this analysis as by far it is the most studied organism in this respect. We mapped all the PG4 motifs found within the upstream region (200 bases) on the + strand of genes in E. coli in the context of characterized/predicted promoters and operons (Salgado et al. 2004) (see Supplemental material). Operons with PG4 motifs in the regulatory region are listed in Table 3.
Table 3.
Representative table showing Escherichia coli operons with PG4 motifs in the putative regulatory region (up to −200 bp upstream of operon) for the + strand
The full list is given in Supplemental Table S7.
aPredicted or characterized operon from RegulonDB.
Target sites for global supercoiling-sensitive regulators are predominantly associated with PG4 motifs in E. coli
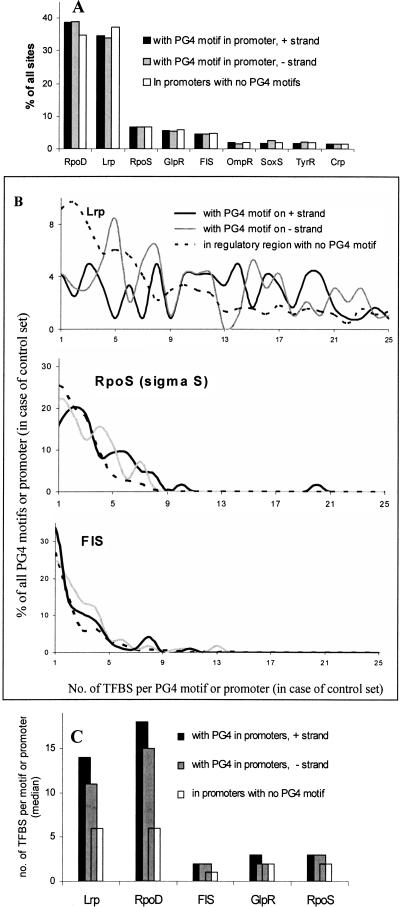
We mapped transcription-factor-binding sites (TFBS) to sequence flanking (100 bases) PG4 motifs, which were present within −200 bases of the start codon. The target sites of nine DNA-binding proteins (Crp, FIS, GlpR, Lrp, OmpR, RopD, RpoS, SoxS, and TyrR) were prominent (with >50 sites associated with 118 PG4 motifs in the + strand, for each factor). RpoD (38.8%) and Lrp (34.7%) constituted the majority of 6493 sites observed for 28 factors on the + strand (Fig. 4A). Similarly, out of 4250 sites for 26 factors associated with 96 PG4 motifs present on the − strand, RpoD and Lrp comprised 38.9% and 33.9% of the sites. FIS, GlpR, and RpoS constituted 4%–6%, and Crp, OmpR, TyrR, and SoxS had ∼1%–2% sites each associated with PG4 motifs present on the + or − strand. As a control set, 445 putative promoter regions (up to 200 bases upstream of start codons) devoid of PG4 motifs were also considered for TFBS. Out of a total of 14,292 TFBS in this case, RpoD and Lrp constituted 34.8% and 37.1% of the sites, respectively. The frequency of each target site in the three sets—that is, sequence flanking PG4 motifs present on the + strand, flanking regions of motifs present on the − strand, and control promoter regions with no motifs—was compared. Interestingly, a significant (P < 0.001; nonparametric comparison using the Mann-Whitney Test) difference in the frequency of individual sites was observed in the flanking region of PG4 motifs (both, when present on the + or − strand) with respect to the control set, for five of the nine factors considered: Lrp, FIS, GlpR, RpoS, and RpoD. The frequency distribution of target sites was not different when associated with PG4 motifs present on the + or − strand (P > 0.05), except in the case of GlpR. The P-values for all comparisons are given in Supplemental Table S8. Individual frequency distribution plots for target sites of the nine factors in the respective regions and average sites (median) per motif (or promoter, in case of the control set) for the five factors with significant difference are shown in Figure 4, B and C (also Supplemental Fig. S6). We used predicted factor-binding sites as observed before without any further change. A large number of binding sites for factors like RpoD and Lrp result from the presence of numerous contiguous sites (overlapping within 2–3 bases at times); however, this is not expected to affect our conclusions since it holds for the control set also.
Figure 4.
Global regulators Lrp, FIS, and GlpR and sigma factors σ70 and σS are predominantly associated with PG4 motifs in Escherichia coli. We computationally mapped target sites for 55 DNA-binding proteins in the region flanking (100 bp) PG4 motifs present within −200 bp of start codons in the + strand (118 motifs) and − strand (96 motifs). Sites were also mapped to 445 promoter regions (within −200 bp of start codon) devoid of PG4 motifs as a control set. (A) Overall representation of sites (for nine factors with >1% sites) as a percentage of total sites for 55 DNA-binding proteins is shown for the respective regions. (B) Frequency distribution of TFBS. Motifs or promoters (%) were plotted against the number of sites found either flanking the motifs or within the promoter (in case of control set); representative plots for three factors are shown (for others, see Supplemental Fig. S6). Distributions were observed to be significantly (P < 0.001) different for Lrp, RpoD, FIS, RpoS, and GlpR when compared between the + or − strand and the control set, while SoxS, TyrR, Crp, and OmpR did not show a statistically significant difference (P > 0.05). (C) Target sites (median) per motif (+/− strand) or promoter (control set) are shown for five factors with significantly different distribution. Nonparametric comparisons were done using the Mann-Whitney U-test; the P-values for respective comparisons are shown in Supplemental Table S8.
E. coli PG4 sequences adopt G-quadruplex motifs in solution
All PG4 motifs identified by us were based on previous information about sequence patterns, which could adopt quadruplex motifs. We selected 11 sequences randomly from the upstream region of different genes in E. coli and checked their potential to adopt a quadruplex motif in solution under physiological conditions using CD. CD profiles for both strand orientations of the quadruplex motifs, parallel and antiparallel, have been well established (Balagurumoorthy and Brahmachari 1994). Nine out of 11 sequences readily formed structure in the presence of monovalent cation (both Na+ and K+). The CD spectra for all 11 sequences are included in Supplemental Figure S7.
Discussion
Our analysis shows overrepresentation of G-quadruplex or G4 DNA motifs in putative regulatory regions in the non-coding genome of prokaryotes (Fig. 2A–E). Interestingly, a detailed analysis in regulatory regions of E. coli indicated that the target sites of transcription factors Lrp, FIS, GlpR, RpoS, and RpoD were predominantly associated with G4 motifs. This is the first genome-wide comparative study of G4 DNA in prokaryotes, and collectively our observations suggest that PG4 motifs may be biologically relevant as regulatory signals in prokaryotes. This is further supported by the fact that genomes with high PG4 motif frequency in their regulatory region (Fig. 2A) also show strong emphasis on regulation [12.3% and 8.4% of the coded proteins in S. coelicolor A3(2) and P. aeruginosa PAO1, respectively, are predicted to be involved in regulation relative to 3.0% in Mycobacterium tuberculosis, 5.8% in E. coli, and 5.3% in Bacillus subtilis] (Stover et al. 2000; Bentley et al. 2002).
Recent studies show evidence of in vivo presence of G4 DNA in E. coli (Duquette et al. 2004) and prevalenceof G4 DNA in the human genome, which indicated a possible functional role of these motifs (Huppert and Balasubramanian 2005; Todd et al. 2005). In comparison to the motifs reported from the human genome, the loop size distribution in prokaryotes appears quite contrasting. Our analysis indicated loop sizes of >3 nt to be predominant (Supplemental Fig. S2) compared to an overrepresentation of single nucleotide loops in the human genome (Huppert and Balasubramanian 2005). Based on analysis of the CD spectra of several G-rich oligonucleotides (Dapic et al. 2003; Hazel et al. 2004), this indicates the likelihood of G4 motifs with parallel strand orientation (Fig. 1) being preferred in the human genome, while the bacterial genomes appear to predominantly harbor motifs that could adopt both parallel and antiparallel structures (Supplemental Fig. S7).
Growth phase response of sigma factor RpoD (σ70) and global regulators FIS and Lrp may be mediated through G4 motif formation in their target sites during supercoiling stress
Transcription-factor-binding site analysis in E. coli indicated five regulators, Lrp, FIS, GlpR, including the sigma factors σ70 and σS (product of rpoD and rpoS genes, respectively), to be predominantly associated with PG4 motifs. Except σS, which is essential for transcription of stationary-phase genes, σ70, Lrp, FIS, and GlpR are “switched on” in the exponential growth phase following nutritional upshift and together control >1000 genes (Ishihama 1999 and references therein; Martinez-Antonio and Collado-Vides 2003). It is interesting to consider the relevance of association with PG4 motifs for these crucial regulatory factors.
An increase in the ATP/ADP ratio (energy charge) due to nutritional upshift enhances gyrase activity (because of the enhanced phosphorylation potential of the cell) resulting in higher negative superhelicity (Balke and Gralla 1987; Travers et al. 2001; Hatfield and Benham 2002). The stress-induced duplex destabilization (SIDD) model of Benham et al. indicates that high superhelical density results in the formation of localized non-B DNA motifs to counter superhelical stress, which exert regulatory control in E. coli (Singh et al. 1995; Hatfield and Benham 2002). In line with the SIDD model, we envisage that occurrence of local sites of partially destabilized duplex states may induce strand- and sequence-specific formation of G4 motifs on strand separation (Wang et al. 2004). Based on our finding that regulatory regions harboring G4 sites are strongly associated with target sites of FIS, Lrp, GlpR, σ70, and σS (Fig. 4), we predict that the regulatory response of these factors is mediated by G4 DNA formation in the supercoiled state. The actual mechanism, however, may involve presentation of specific recognition elements along with a combinatorial effect of diverse factors ranging from DNA topology to cellular interacting partners. The association of the target sites over a broad region (100 bp flanking G4 motifs) is consistent with the observation that transcription factor binding induces transmission of superhelical destabilization to distal promoter sites (Sheridan et al. 1999; Opel et al. 2004).
Our prediction is also supported by a high incidence of in vivo G4 motifs in supercoiled DNA relative to other topoisomers (58% vs. 31% and 24% in relaxed and linear templates, respectively) in E. coli (Duquette et al. 2004). The association of FIS-binding sites with G4 motifs is consistent with several reports indicating that FIS operates by stabilizing local DNA architecture and that supercoiling-responsive promoters harbor FIS-binding sites (Schneider et al. 2000). Additionally, it has been demonstrated that topA induces G4 motif formation (Arimondo et al. 2000). This is in line with our prediction, as FIS has been observed to induce topA expression during oxidative stress (Weinstein-Fischer et al. 2000).
Target sites for σS, which regulates >100 stationary-phase-specific genes (Ishihama 1999; Martinez-Antonio and Collado-Vides 2003), were also associated with PG4 motifs. Decreased DNA superhelicity in stationary-phase E. coli cells induces σS activity while repressing that of σ70 (Kusano et al. 1996; Bordes et al. 2003). On the other hand, σS response can also be induced by high osmolarity and heat shock during growth phase (Hengge-Aronis 1993; Jishage et al. 1996). Thus, although σS response is complex, it is tempting to speculate that G4 motif formation in growth phase may assist repression of σS target sites and thereby contribute in switching of the σ70/σS response during the transition from growth to stationary phase (Ishihama 1999).
A recent study demonstrates that the osmotic stress response (OSR) in E. coli is induced by supercoiling. Interestingly, they observed that the upstream region of genes in OSR are significantly enriched in binding sites for FIS, Lrp, GlpR, and RpoS (Table 5 of Cheung et al. 2003), which is consistent with our prediction. We also observed that several genes with statistically significant expression in OSR, like apt, poxB, nhaB, ydcF, ygfF, and yibK, had one or more PG4 motifs in their regulatory regions. In another recent genome-wide expression study of genes responsive to DNA relaxation in E. coli, we observed that several significantly induced (cls, ycgL, insB_2, and insA_2) and repressed (eaeH, yeeS, mazG, and gidA) genes may be potentially regulated by PG4 motifs, present either in the respective promoters or upstream of respective operons (Peter et al. 2004).
It must be noted that our conclusions are based on mostly predicted transcription-factor-binding sites and unlike the human c-myc and other cases (where G4 is implicated to be regulatory) (Howell et al. 1996; Siddiqui-Jain et al. 2002; Etzioni et al. 2005), no experimental proof exists of G4 DNA-mediated regulation in bacteria. Genome-wide ChIP analysis for binding sites in conjunction with molecules or factors that specifically bind to G4 motifs and in vitro specificity assays will be required to confirm our findings (Schaffitzel et al. 2001; Rezler et al. 2005).
Our findings provide several bioinformatics insights. Significant among them are tentative results showing that transcription, secondary metabolite biosynthesis, and signal transduction classes have more genes (relative to other classes) that could be under G4 motif control (Fig. 3A). This is largely consistent with the fact that supercoiling-induced DNA topology controls transcription of several regulons and stimulons during the exponential growth phase (Schneider et al. 1999; Hatfield and Benham 2002; Peter et al. 2004). Cheung et al. (2003) have made similar observations with respect to clusters of genes belonging to macromolecule, amino acid, and building block biosynthesis, which are significantly overexpressed as a result of superhelical stress during the osmotic stress response (OSR). Another interesting observation indicated that GC-rich genomes selected positively for G4 motifs in the near upstream regions (Fig. 2B). Based on the SIDD model of Hatfield and Benham, it is tempting to speculate that in GC-rich organisms, where duplex destabilization is energetically more demanding, favorable G4 motif formation may relax superhelical density (Hatfield and Benham 2002). Our previous reports suggesting favorable kinetics of G4 motif formation within the nuclease-hypersensitive element of the human c-myc promoter support this hypothesis (Halder and Chowdhury 2005; Halder et al. 2005). However, such a possibility has to be clearly demonstrated.
Taken together, these findings suggest that G4 DNA may be biologically relevant as regulatory signals in prokaryotes. The motifs may be particularly important in translating environmental stimuli (nutritional upshift) into a functional message by presenting target sites for orchestrating the activity of global transcriptional regulators in E. coli.
Methods
Organisms
18 bacterial organisms were used for our analysis after downloading their genomes from the NCBI database (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi). (The abbreviations used are from the KEGG database; http://www.genome.jp/kegg/catalog/org_list.html; Kanehisa and Goto 2000; they are also mentioned in the Supplemental material). Organisms were chosen such that apart from two groups of closely related organisms—Vch, Stm, Eco and Mtu, Mbo, Sco—all others belonged to evolutionarily distant clades. The STRING server (http://string.embl.de/; von Mering et al. 2003), which is based on various phylogenetic distance measures, was used for this purpose.
PG4 motif searching, genomic mapping, and analysis
Potential G4 motifs (i.e., G-quadruplex-forming sequences) in the 18 genomes were searched with a customized program written using Perl. We adopted a general pattern: Gn-NL1-Gn-NL2-Gn-NL3-Gn, where G is guanine and N is any nucleotide including G. The number of guanines constituting the stem of G4 DNA (Fig. 1) is given by n. n was varied from 2 to 5 but restricted to be constant within a particular motif. This does not exclude identification of G4 sequence with variable G-runs as Gs were included in loops, and enables ready detection of the number of tetrads possible in a given sequence (details of the parsing method, including comparison with previously published methods, is discussed in the Supplemental material). The number of nucleotides in the three loops L1, L2, and L3 was allowed to vary from 1 to 5, such that the size of loops may vary within a given G4 motif. The program was rerun with cytosine instead of guanine to identify motifs on the − strand and appropriately corrected for orientation before mapping their position in the context of genes. We restricted our program to the above values of stem and loop length after considering the following points. First, single G-tetrads have been observed only in very high (millimolar) guanine solutions and may not be physiologically relevant (Gellert et al. 1962), and a tetrad length exceeding five guanines was not found by us except in only one or two cases. Similar observations have been made before (Todd et al. 2005). We included a stem size of two guanines as various previous reports indicated G4 DNA and RNA with two tetrads as biologically relevant (see Supplemental material) (Wells et al. 1988; Darnell et al. 2001). The loop length was arbitrarily restrained to a maximum of 5 nt for practical reasons. An unrestrained loop length would make searching difficult; moreover, we found that G4 motifs exist as short nucleic acids (a length between 10 and 39 bases was predominant) (Supplemental Table S4), which is also supported by earlier evidence (Hazel et al. 2004). Considering the possible variability in the loops within a motif, we analyzed the loop distribution using a mosaic plot (Friendly 1994) wherein the predominant loop distributions can be readily identified. A single putative quadruplex sequence may present multiple quadruplex topologies with variation in both loop and tetrad compositions. Furthermore, overlapping patterns may be present with more than four G-runs where only one motif is possible at a time (Supplemental Fig. S1). This complicates the analysis for exactly determining the number of possible motifs in a given sequence. We have addressed this by stitching overlapping patterns to present tracts. This tract information has been used for all genomic comparative analysis. However, for analysis of tetrad size and loop combinations, all possible PG4 motifs were considered. This was particularly done to check the prevalence of any structural type or tetrad/loop combination of G4 DNA on a genome-wide scale.
We divided each genome into three regions for mapping of the PG4 motifs: (1) intragenic; (2) putative regulatory (up to 200 bases) upstream of the gene’s start codon; and (3) “rest-of-intergenic,” comprising all other non-coding intergenic regions (including the downstream intergenic region separating convergently oriented genes). Region 2 comprises the actual intergenic distance when two genes are separated by <200 bases. This partitioning was used for all analysis except where mentioned. The relative abundance of PG4 motifs in different genomic regions was statistically compared, and the significance levels were estimated using the nonparametric Signed Wilcoxon Ranks Test (Wilcoxon 1945). As a control for the significance of PG4 motifs vis-à-vis their distinct genomic distribution, we searched for sequence patterns with multiple G-runs, which were restricted such that they would be unable to adopt a G4 motif (see Supplemental material). The programs written for genome-wide searching, mapping, and analyzing PG4 motifs are available upon request.
In prokaryotes, it is difficult to predict regulatory regions. For a gene within an operon, its promoter may be several genes upstream. In certain cases, a gene within an operon may have its own promoter also. It is difficult to predict operons, and moreover, the first gene in an operon in less well-studied organisms (McGuire et al. 2000). On the other hand, we noticed that a majority of operons in E. coli consist of only two to three genes (Salgado et al. 2004). For comparative function analyses across all organisms, we considered the near upstream region of all ORFs as the putative regulatory region. The upstream distance was taken to be −200 bp as the majority of binding sites for DNA-binding proteins in bacteria are found within the first 200 bases upstream of the start codon. Even in cases when there is a site further upstream, one finds another site close to the promoter (Gralla and Collado-Vides 1996). Similarly, very few regulatory sites are present downstream of the start codon; hence, we analyzed only the non-coding regions proximal to the start codon for PG4 motifs, as motifs in this region are expected to be most relevant in gene regulation.
Function classification of genes with PG4 motifs in regulatory region in 18 organisms
We considered 61,974 ORFs in 18 organisms classified in 23 functional classes as defined by the COGS database (Tatusov et al. 1997) for this analysis. All ORFs with one or more PG4 motif(s) within 200 bp of the start codon in each function class were identified. These data were analyzed in two ways after excluding genes that were either not present or not well defined (i.e., general function prediction and function unknown) in COGS. First, we found the percentage of positive hits (genes with PG4 motif in −200 bp) in each class. Second, we found out the function classes with a higher number of positive hits. As a control, we also analyzed the intragenic PG4 motif density of all the 61,974 ORFs. First, the intragenic PG4 motifs were identified, and then the number of bases constituting a motif was counted in each case. This was expressed as “per kb of gene sequence” (motif frequency). The value of motif frequency across all genes in each function class was evaluated for comparative analysis.
Identification of orthologs with conserved PG4 motifs in regulatory region
For finding orthologous genes with conserved PG4 motifs in regulatory regions within the 18 bacterial organisms studied by us, we used the COGS database. The E. coli genes with PG4 motifs within the −200-bp region were considered as the reference in this case and used to search for orthologs in the 17 other organisms. Identification of one or more PG4 motif(s) within the −200 region of the ortholog was considered a positive hit, and these genes were classified into the phylogenetic groups defined by STRING server (von Mering et al. 2003). Identification of orthologs with conserved PG4 motif(s) in the regulatory region in distantly related organisms was considered to be significant.
Mapping of PG4 motifs to the transcription regulatory network in E. coli
We mapped the regulatory regions with PG4 motifs in E. coli in the context of its regulatory network, that is, characterized and predicted operons and promoters. Characterized promoters for 16 genes were obtained from the PromEC database (http://margalit.huji.ac.il/promec/) (Hershberg et al. 2001), and 178 predicted operons and 151 predicted promoters from RegulonDB (http://www.cifn.unam.mx/Computational_Genomics/regulondb/) were used (Salgado et al. 2004).
Mapping of transcription-factor-binding sites to PG4 motifs in the −200-bp region of genes in E. coli
We searched for transcription-factor-binding sites (TFBS) associated with all PG4 motifs found in the regulatory region of E. coli genes. TFBS (DNA sequence matrices) for 55 E. coli DNA-binding proteins from http://arep.med.harvard.edu/ecoli_matrices/ (Robison et al. 1998) with high confidence scores, which comprise both functionally characterized as well as predicted sites, were used. Sites from the library of matrices were validated against the gene list file for E. coli from NCBI (http://www.ncbi.nlm.nih.gov/genomes/) before mapping to flanking regions (100 bases) of PG4 motifs, which are present within 200 bp upstream of the start codon. A window size of 100 bases was used as the flanking region based on previous studies indicating that formation of non-B DNA motifs may have a regulatory effect at a distant promoter site (Hatfield and Benham 2002); however, this distance can be >100 bases. Similarly, we mapped TFBS on 445 putative regulatory regions (comprising 200 bp upstream of start codons) that did not harbor any PG4 motifs, as a control set. All matrices were mapped as reported in the database, without introducing any change for overlapping target sites.
Circular dichroism
Circular dichroism (CD) measurements were performed on a Jasco Spectropolarimeter (model J 715) as described previously (Mathur et al. 2004). See also Supplemental material.
Acknowledgments
We are grateful to Samir K. Brahmachari for support and encouragement. S.C. thanks Partha P. Majumder of the Indian Statistical Institute, Kolkata for helpful discussion; Munia Ganguli, IGIB, for critical reading of the manuscript; and all members of the Chowdhury Lab for useful discussions. K.H. acknowledges a research fellowship (JRF) from CSIR. This work was supported by grants from the CSIR task force project CMM 0017. We also thank the referees for assisting us in enriching the manuscript content.
Footnotes
[Supplemental material is available online at www.genome.org and http://www.igib.res.in/prokaryote/PG4.htm.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.4508806
References
- Arimondo P.B., Riou J.F., Mergny J.L., Tazi J., Sun J.S., Garestier T., Helene C., Riou J.F., Mergny J.L., Tazi J., Sun J.S., Garestier T., Helene C., Mergny J.L., Tazi J., Sun J.S., Garestier T., Helene C., Tazi J., Sun J.S., Garestier T., Helene C., Sun J.S., Garestier T., Helene C., Garestier T., Helene C., Helene C. Interaction of human DNA topoisomerase I with G-quartet structures. Nucleic Acids Res. 2000;28:4832–4838. doi: 10.1093/nar/28.24.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati C.Z., Hickson I.D., Hickson I.D. RecQ helicases: Suppressors of tumorigenesis and premature aging. Biochem. J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacolla A., Wells R.D., Wells R.D. Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 2004;279:47411–47414. doi: 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- Balagurumoorthy P., Brahmachari S.K., Brahmachari S.K. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994;269:21858–21869. [PubMed] [Google Scholar]
- Balke V.L., Gralla J.D., Gralla J.D. Changes in the linking number of supercoiled DNA accompany growth transitions in. Escherichia coli. J. Bacteriol. 1987;169:4499–4506. doi: 10.1128/jb.169.10.4499-4506.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer M.A., Tavazoie S., Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- Bentley S.D., Chater K.F., Cerdeno-Tarraga A.M., Challis G.L., Thomson N.R., James K.D., Harris D.E., Quail M.A., Kieser H., Harper D., Chater K.F., Cerdeno-Tarraga A.M., Challis G.L., Thomson N.R., James K.D., Harris D.E., Quail M.A., Kieser H., Harper D., Cerdeno-Tarraga A.M., Challis G.L., Thomson N.R., James K.D., Harris D.E., Quail M.A., Kieser H., Harper D., Challis G.L., Thomson N.R., James K.D., Harris D.E., Quail M.A., Kieser H., Harper D., Thomson N.R., James K.D., Harris D.E., Quail M.A., Kieser H., Harper D., James K.D., Harris D.E., Quail M.A., Kieser H., Harper D., Harris D.E., Quail M.A., Kieser H., Harper D., Quail M.A., Kieser H., Harper D., Kieser H., Harper D., Harper D., et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- Bordes P., Conter A., Morales V., Bouvier J., Kolb A., Gutierrez C., Conter A., Morales V., Bouvier J., Kolb A., Gutierrez C., Morales V., Bouvier J., Kolb A., Gutierrez C., Bouvier J., Kolb A., Gutierrez C., Kolb A., Gutierrez C., Gutierrez C. DNA supercoiling contributes to disconnect σS accumulation from σS-dependent transcription in. Escherichia coli. Mol. Microbiol. 2003;48:561–571. doi: 10.1046/j.1365-2958.2003.03461.x. [DOI] [PubMed] [Google Scholar]
- Cheung I., Schertzer M., Rose A., Lansdorp P.M., Schertzer M., Rose A., Lansdorp P.M., Rose A., Lansdorp P.M., Lansdorp P.M. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 2002;31:405–409. doi: 10.1038/ng928. [DOI] [PubMed] [Google Scholar]
- Cheung K.J., Badarinarayana V., Selinger D.W., Janse D., Church G.M., Badarinarayana V., Selinger D.W., Janse D., Church G.M., Selinger D.W., Janse D., Church G.M., Janse D., Church G.M., Church G.M. A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of. Escherichia coli. Genome Res. 2003;13:206–215. doi: 10.1101/gr.401003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen J., Kofod M., Nielsen F.C., Kofod M., Nielsen F.C., Nielsen F.C. A guanosine quadruplex and two stable hairpins flank a major cleavage site in insulin-like growth factor II mRNA. Nucleic Acids Res. 1994;22:5709–5716. doi: 10.1093/nar/22.25.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings C.J., Zoghbi H.Y., Zoghbi H.Y. Trinucleotide repeats: Mechanisms and pathophysiology. Annu. Rev. Genomics Hum. Genet. 2000;1:281–328. doi: 10.1146/annurev.genom.1.1.281. [DOI] [PubMed] [Google Scholar]
- Dapic V., Abdomerovic V., Marrington R., Peberdy J., Rodger A., Trent J.O., Bates P.J., Abdomerovic V., Marrington R., Peberdy J., Rodger A., Trent J.O., Bates P.J., Marrington R., Peberdy J., Rodger A., Trent J.O., Bates P.J., Peberdy J., Rodger A., Trent J.O., Bates P.J., Rodger A., Trent J.O., Bates P.J., Trent J.O., Bates P.J., Bates P.J. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J.C., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B., Jin P., Brown V., Warren S.T., Darnell R.B., Brown V., Warren S.T., Darnell R.B., Warren S.T., Darnell R.B., Darnell R.B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Dunnick W., Hertz G.Z., Scappino L., Gritzmacher C., Hertz G.Z., Scappino L., Gritzmacher C., Scappino L., Gritzmacher C., Gritzmacher C. DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Res. 1993;21:365–372. doi: 10.1093/nar/21.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquette M.L., Handa P., Vincent J.A., Taylor A.F., Maizels N., Handa P., Vincent J.A., Taylor A.F., Maizels N., Vincent J.A., Taylor A.F., Maizels N., Taylor A.F., Maizels N., Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes & Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzioni S., Yafe A., Khateb S., Weisman-Shomer P., Bengal E., Fry M., Yafe A., Khateb S., Weisman-Shomer P., Bengal E., Fry M., Khateb S., Weisman-Shomer P., Bengal E., Fry M., Weisman-Shomer P., Bengal E., Fry M., Bengal E., Fry M., Fry M. Homodimeric MyoD preferentially binds tetraplex structures of regulatory sequences of muscle-specific genes. J. Biol. Chem. 2005;280:26805–26812. doi: 10.1074/jbc.M500820200. [DOI] [PubMed] [Google Scholar]
- Florquin K., Saeys Y., Degroeve S., Rouze P., Vande P.Y., Saeys Y., Degroeve S., Rouze P., Vande P.Y., Degroeve S., Rouze P., Vande P.Y., Rouze P., Vande P.Y., Vande P.Y. Large-scale structural analysis of the core promoter in mammalian and plant genomes. Nucleic Acids Res. 2005;33:4255–4264. doi: 10.1093/nar/gki737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friendly M. Mosaic displays for multi-way contingency tables. J. Am. Stat. Assoc. 1994:190–200. [Google Scholar]
- Gellert M., Lipsett M.N., Davies D.R., Lipsett M.N., Davies D.R., Davies D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert D.E., Feigon J., Feigon J. Multistranded DNA structures. Curr. Opin. Struct. Biol. 1999;9:305–314. doi: 10.1016/S0959-440X(99)80041-4. [DOI] [PubMed] [Google Scholar]
- Gralla J.D., Collado-Vides J., Collado-Vides J.1996. Organization and function of transcription regulatory elements. In Escherichia coli andSalmonella: Molecular and cellular biology ed. F.C. Neidhardt), pp. 1232–1245. ASM Press; Washington, DC [Google Scholar]
- Halder K., Chowdhury S., Chowdhury S. Kinetic resolution of bimolecular hybridization versus intramolecular folding in nucleic acids by surface plasmon resonance: Application to G-quadruplex/duplex competition in human c-myc promoter. Nucleic Acids Res. 2005;33:4466–4474. doi: 10.1093/nar/gki750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder K., Mathur V., Chugh D., Verma A., Chowdhury S., Mathur V., Chugh D., Verma A., Chowdhury S., Chugh D., Verma A., Chowdhury S., Verma A., Chowdhury S., Chowdhury S. Quadruplex–duplex competition in the nuclease hypersensitive element of human c-myc promoter: C to T mutation in C-rich strand enhances duplex association. Biochem. Biophys. Res. Commun. 2005;327:49–56. doi: 10.1016/j.bbrc.2004.11.137. [DOI] [PubMed] [Google Scholar]
- Hanakahi L.A., Sun H., Maizels N., Sun H., Maizels N., Maizels N. High affinity interactions of nucleolin with G-G-paired rDNA. J. Biol. Chem. 1999;274:15908–15912. doi: 10.1074/jbc.274.22.15908. [DOI] [PubMed] [Google Scholar]
- Hatfield G.W., Benham C.J., Benham C.J. DNA topology-mediated control of global gene expression in. Escherichia coli. Annu. Rev. Genet. 2002;36:175–203. doi: 10.1146/annurev.genet.36.032902.111815. [DOI] [PubMed] [Google Scholar]
- Hazel P., Huppert J., Balasubramanian S., Neidle S., Huppert J., Balasubramanian S., Neidle S., Balasubramanian S., Neidle S., Neidle S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- Hengge-Aronis R. Survival of hunger and stress: The role of rpoS in early stationary phase gene regulation in. E. coli. Cell. 1993;72:165–168. doi: 10.1016/0092-8674(93)90655-a. [DOI] [PubMed] [Google Scholar]
- Hershberg R., Bejerano G., Santos-Zavaleta A., Margalit H., Bejerano G., Santos-Zavaleta A., Margalit H., Santos-Zavaleta A., Margalit H., Margalit H. PromEC: An updated database of Escherichia coli mRNA promoters with experimentally identified transcriptional start sites. Nucleic Acids Res. 2001;29:277. doi: 10.1093/nar/29.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh B.C., Kollmus H., Hauser H., Coen D.M., Kollmus H., Hauser H., Coen D.M., Hauser H., Coen D.M., Coen D.M. Translational recoding induced by G-rich mRNA sequences that form unusual structures. Cell. 1996;86:949–959. doi: 10.1016/S0092-8674(00)80170-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell R.M., Woodford K.J., Weitzmann M.N., Usdin K., Woodford K.J., Weitzmann M.N., Usdin K., Weitzmann M.N., Usdin K., Usdin K. The chicken β-globin gene promoter forms a novel “cinched” tetrahelical structure. J. Biol. Chem. 1996;271:5208–5214. doi: 10.1074/jbc.271.9.5208. [DOI] [PubMed] [Google Scholar]
- Huppert J.L., Balasubramanian S., Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incles C.M., Schultes C.M., Kempski H., Koehler H., Kelland L.R., Neidle S., Schultes C.M., Kempski H., Koehler H., Kelland L.R., Neidle S., Kempski H., Koehler H., Kelland L.R., Neidle S., Koehler H., Kelland L.R., Neidle S., Kelland L.R., Neidle S., Neidle S. A G-quadruplex telomere targeting agent produces p16-associated senescence and chromosomal fusions in human prostate cancer cells. Mol. Cancer Ther. 2004;3:1201–1206. [PubMed] [Google Scholar]
- Ishihama A. Modulation of the nucleoid, the transcription apparatus, and the translation machinery in bacteria for stationary phase survival. Genes Cells. 1999;4:135–143. doi: 10.1046/j.1365-2443.1999.00247.x. [DOI] [PubMed] [Google Scholar]
- Jishage M., Iwata A., Ueda S., Ishihama A., Iwata A., Ueda S., Ishihama A., Ueda S., Ishihama A., Ishihama A. Regulation of RNA polymerase σ subunit synthesis in Escherichia coli: Intracellular levels of four species of σ subunit under various growth conditions. J. Bacteriol. 1996;178:5447–5451. doi: 10.1128/jb.178.18.5447-5451.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R.D. Mismatch repair: Mechanisms and relationship to cancer susceptibility. Trends Biochem. Sci. 1995;20:397–401. doi: 10.1016/s0968-0004(00)89087-8. [DOI] [PubMed] [Google Scholar]
- Kusano S., Ding Q., Fujita N., Ishihama A., Ding Q., Fujita N., Ishihama A., Fujita N., Ishihama A., Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase E σ70 and E σ38 holoenzymes. Effect of DNA supercoiling. J. Biol. Chem. 1996;271:1998–2004. doi: 10.1074/jbc.271.4.1998. [DOI] [PubMed] [Google Scholar]
- Martinez-Antonio A., Collado-Vides J., Collado-Vides J. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 2003;6:482–489. doi: 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Mathur V., Verma A., Maiti S., Chowdhury S., Verma A., Maiti S., Chowdhury S., Maiti S., Chowdhury S., Chowdhury S. Thermodynamics of i-tetraplex formation in the nuclease hypersensitive element of human c-myc promoter. Biochem. Biophys. Res. Commun. 2004;320:1220–1227. doi: 10.1016/j.bbrc.2004.06.074. [DOI] [PubMed] [Google Scholar]
- McGuire A.M., Hughes J.D., Church G.M., Hughes J.D., Church G.M., Church G.M. Conservation of DNA regulatory motifs and discovery of new motifs in microbial genomes. Genome Res. 2000;10:744–757. doi: 10.1101/gr.10.6.744. [DOI] [PubMed] [Google Scholar]
- McMurray C.T. DNA secondary structure: A common and causative factor for expansion in human disease. Proc. Natl. Acad. Sci. 1999;96:1823–1825. doi: 10.1073/pnas.96.5.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P., Lahue R., Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- Neidle S., Read M.A., Read M.A. G-quadruplexes as therapeutic targets. Biopolymers. 2000;56:195–208. doi: 10.1002/1097-0282(2000)56:3<195::AID-BIP10009>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Opel M.L., Hatfield G.W., Hatfield G.W. DNA supercoiling-dependent transcriptional coupling between the divergently transcribed promoters of the ilvYC operon of Escherichia coli is proportional to promoter strengths and transcript lengths. Mol. Microbiol. 2001;39:191–198. doi: 10.1046/j.1365-2958.2001.02249.x. [DOI] [PubMed] [Google Scholar]
- Opel M.L., Aeling K.A., Holmes W.M., Johnson R.C., Benham C.J., Hatfield G.W., Aeling K.A., Holmes W.M., Johnson R.C., Benham C.J., Hatfield G.W., Holmes W.M., Johnson R.C., Benham C.J., Hatfield G.W., Johnson R.C., Benham C.J., Hatfield G.W., Benham C.J., Hatfield G.W., Hatfield G.W. Activation of transcription initiation from a stable RNA promoter by a Fis protein-mediated DNA structural transmission mechanism. Mol. Microbiol. 2004;53:665–674. doi: 10.1111/j.1365-2958.2004.04147.x. [DOI] [PubMed] [Google Scholar]
- Parkinson G.N., Lee M.P., Neidle S., Lee M.P., Neidle S., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- Pedersen A.G., Jensen L.J., Brunak S., Staerfeldt H.H., Ussery D.W., Jensen L.J., Brunak S., Staerfeldt H.H., Ussery D.W., Brunak S., Staerfeldt H.H., Ussery D.W., Staerfeldt H.H., Ussery D.W., Ussery D.W. A DNA structural atlas for Escherichia coli. J. Mol. Biol. 2000;299:907–930. doi: 10.1006/jmbi.2000.3787. [DOI] [PubMed] [Google Scholar]
- Perez-Martin J., de Lorenzo V., de Lorenzo V. Clues and consequences of DNA bending in transcription. Annu. Rev. Microbiol. 1997;51:593–628. doi: 10.1146/annurev.micro.51.1.593. [DOI] [PubMed] [Google Scholar]
- Peter B.J., Arsuaga J., Breier A.M., Khodursky A.B., Brown P.O., Cozzarelli N.R., Arsuaga J., Breier A.M., Khodursky A.B., Brown P.O., Cozzarelli N.R., Breier A.M., Khodursky A.B., Brown P.O., Cozzarelli N.R., Khodursky A.B., Brown P.O., Cozzarelli N.R., Brown P.O., Cozzarelli N.R., Cozzarelli N.R. Genomic transcriptional response to loss of chromosomal supercoiling in. Escherichia coli. Genome Biol. 2004;5:R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezler E.M., Seenisamy J., Bashyam S., Kim M.Y., White E., Wilson W.D., Hurley L.H., Seenisamy J., Bashyam S., Kim M.Y., White E., Wilson W.D., Hurley L.H., Bashyam S., Kim M.Y., White E., Wilson W.D., Hurley L.H., Kim M.Y., White E., Wilson W.D., Hurley L.H., White E., Wilson W.D., Hurley L.H., Wilson W.D., Hurley L.H., Hurley L.H. Telomestatin and diseleno sapphyrin bind selectively to two different forms of the human telomeric G-quadruplex structure. J. Am. Chem. Soc. 2005;127:9439–9447. doi: 10.1021/ja0505088. [DOI] [PubMed] [Google Scholar]
- Rich A., Zhang S., Zhang S. Timeline: Z-DNA: The long road to biological function. Nat. Rev. Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- Robison K., McGuire A.M., Church G.M., McGuire A.M., Church G.M., Church G.M. A comprehensive library of DNA-binding site matrices for 55 proteins applied to the complete Escherichia coliK-12 genome. J. Mol. Biol. 1998;284:241–254. doi: 10.1006/jmbi.1998.2160. [DOI] [PubMed] [Google Scholar]
- Salgado H., Gama-Castro S., Martinez-Antonio A., Diaz-Peredo E., Sanchez-Solano F., Peralta-Gil M., Garcia-Alonso D., Jimenez-Jacinto V., Santos-Zavaleta A., Bonavides-Martinez C., Gama-Castro S., Martinez-Antonio A., Diaz-Peredo E., Sanchez-Solano F., Peralta-Gil M., Garcia-Alonso D., Jimenez-Jacinto V., Santos-Zavaleta A., Bonavides-Martinez C., Martinez-Antonio A., Diaz-Peredo E., Sanchez-Solano F., Peralta-Gil M., Garcia-Alonso D., Jimenez-Jacinto V., Santos-Zavaleta A., Bonavides-Martinez C., Diaz-Peredo E., Sanchez-Solano F., Peralta-Gil M., Garcia-Alonso D., Jimenez-Jacinto V., Santos-Zavaleta A., Bonavides-Martinez C., Sanchez-Solano F., Peralta-Gil M., Garcia-Alonso D., Jimenez-Jacinto V., Santos-Zavaleta A., Bonavides-Martinez C., Peralta-Gil M., Garcia-Alonso D., Jimenez-Jacinto V., Santos-Zavaleta A., Bonavides-Martinez C., Garcia-Alonso D., Jimenez-Jacinto V., Santos-Zavaleta A., Bonavides-Martinez C., Jimenez-Jacinto V., Santos-Zavaleta A., Bonavides-Martinez C., Santos-Zavaleta A., Bonavides-Martinez C., Bonavides-Martinez C., et al. RegulonDB (version 4.0): Transcriptional regulation, operon organization and growth conditions in Escherichia coli K-12. Nucleic Acids Res. 2004;32:D303–D306. doi: 10.1093/nar/gkh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffitzel C., Berger I., Postberg J., Hanes J., Lipps H.J., Pluckthun A., Berger I., Postberg J., Hanes J., Lipps H.J., Pluckthun A., Postberg J., Hanes J., Lipps H.J., Pluckthun A., Hanes J., Lipps H.J., Pluckthun A., Lipps H.J., Pluckthun A., Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R., Travers A., Kutateladze T., Muskhelishvili G., Travers A., Kutateladze T., Muskhelishvili G., Kutateladze T., Muskhelishvili G., Muskhelishvili G. A DNA architectural protein couples cellular physiology and DNA topology in Escherichia coli. Mol. Microbiol. 1999;34:953–964. doi: 10.1046/j.1365-2958.1999.01656.x. [DOI] [PubMed] [Google Scholar]
- Schneider R., Travers A., Muskhelishvili G., Travers A., Muskhelishvili G., Muskhelishvili G. The expression of the Escherichia coli fis gene is strongly dependent on the superhelical density of DNA. Mol. Microbiol. 2000;38:167–175. doi: 10.1046/j.1365-2958.2000.02129.x. [DOI] [PubMed] [Google Scholar]
- Seenisamy J., Rezler E.M., Powell T.J., Tye D., Gokhale V., Joshi C.S., Siddiqui-Jain A., Hurley L.H., Rezler E.M., Powell T.J., Tye D., Gokhale V., Joshi C.S., Siddiqui-Jain A., Hurley L.H., Powell T.J., Tye D., Gokhale V., Joshi C.S., Siddiqui-Jain A., Hurley L.H., Tye D., Gokhale V., Joshi C.S., Siddiqui-Jain A., Hurley L.H., Gokhale V., Joshi C.S., Siddiqui-Jain A., Hurley L.H., Joshi C.S., Siddiqui-Jain A., Hurley L.H., Siddiqui-Jain A., Hurley L.H., Hurley L.H. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J. Am. Chem. Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- Sen D., Gilbert W., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- Shen J.C., Loeb L.A., Loeb L.A. The Werner syndrome gene: The molecular basis of RecQ helicase-deficiency diseases. Trends Genet. 2000;16:213–220. doi: 10.1016/s0168-9525(99)01970-8. [DOI] [PubMed] [Google Scholar]
- Sheridan S.D., Benham C.J., Hatfield G.W., Benham C.J., Hatfield G.W., Hatfield G.W. Inhibition of DNA supercoiling-dependent transcriptional activation by a distant B-DNA to Z-DNA transition. J. Biol. Chem. 1999;274:8169–8174. doi: 10.1074/jbc.274.12.8169. [DOI] [PubMed] [Google Scholar]
- Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H., Grand C.L., Bearss D.J., Hurley L.H., Bearss D.J., Hurley L.H., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsson T., Pecinka P., Kubista M., Pecinka P., Kubista M., Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R.R. DNA structure and function. Academic Press; San Diego.: 1994. [Google Scholar]
- Sinden R.R. Biological implications of the DNA structures associated with disease-causing triplet repeats. Am. J. Hum. Genet. 1999;64:346–353. doi: 10.1086/302271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J., Mukerji M., Mahadevan S., Mukerji M., Mahadevan S., Mahadevan S. Transcriptional activation of the Escherichia coli bgl operon: Negative regulation by DNA structural elements near the promoter. Mol. Microbiol. 1995;17:1085–1092. doi: 10.1111/j.1365-2958.1995.mmi_17061085.x. [DOI] [PubMed] [Google Scholar]
- Stover C.K., Pham X.Q., Erwin A.L., Mizoguchi S.D., Warrener P., Hickey M.J., Brinkman F.S., Hufnagle W.O., Kowalik D.J., Lagrou M., Pham X.Q., Erwin A.L., Mizoguchi S.D., Warrener P., Hickey M.J., Brinkman F.S., Hufnagle W.O., Kowalik D.J., Lagrou M., Erwin A.L., Mizoguchi S.D., Warrener P., Hickey M.J., Brinkman F.S., Hufnagle W.O., Kowalik D.J., Lagrou M., Mizoguchi S.D., Warrener P., Hickey M.J., Brinkman F.S., Hufnagle W.O., Kowalik D.J., Lagrou M., Warrener P., Hickey M.J., Brinkman F.S., Hufnagle W.O., Kowalik D.J., Lagrou M., Hickey M.J., Brinkman F.S., Hufnagle W.O., Kowalik D.J., Lagrou M., Brinkman F.S., Hufnagle W.O., Kowalik D.J., Lagrou M., Hufnagle W.O., Kowalik D.J., Lagrou M., Kowalik D.J., Lagrou M., Lagrou M., et al. Complete genome sequence of Pseudomonas aeruginosaPA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Strand M., Prolla T.A., Liskay R.M., Petes T.D., Prolla T.A., Liskay R.M., Petes T.D., Liskay R.M., Petes T.D., Petes T.D. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- Tatusov R.L., Koonin E.V., Lipman D.J., Koonin E.V., Lipman D.J., Lipman D.J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Todd A.K., Johnston M., Neidle S., Johnston M., Neidle S., Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A., Schneider R., Muskhelishvili G., Schneider R., Muskhelishvili G., Muskhelishvili G. DNA supercoiling and transcription in Escherichia coli: The FIS connection. Biochimie. 2001;83:213–217. doi: 10.1016/s0300-9084(00)01217-7. [DOI] [PubMed] [Google Scholar]
- von Mering C., Huynen M., Jaeggi D., Schmidt S., Bork P., Snel B., Huynen M., Jaeggi D., Schmidt S., Bork P., Snel B., Jaeggi D., Schmidt S., Bork P., Snel B., Schmidt S., Bork P., Snel B., Bork P., Snel B., Snel B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Noordewier M., Benham C.J., Noordewier M., Benham C.J., Benham C.J. Stress-induced DNA duplex destabilization (SIDD) in the E. coli genome: SIDD sites are closely associated with promoters. Genome Res. 2004;14:1575–1584. doi: 10.1101/gr.2080004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman W.W., Palumbo M., Thompson W., Fickett J.W., Lawrence C.E., Palumbo M., Thompson W., Fickett J.W., Lawrence C.E., Thompson W., Fickett J.W., Lawrence C.E., Fickett J.W., Lawrence C.E., Lawrence C.E. Human–mouse genome comparisons to locate regulatory sites. Nat. Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- Weinstein-Fischer D., Elgrably-Weiss M., Altuvia S., Elgrably-Weiss M., Altuvia S., Altuvia S. Escherichia coli response to hydrogen peroxide: A role for DNA supercoiling, topoisomerase I and Fis. Mol. Microbiol. 2000;35:1413–1420. doi: 10.1046/j.1365-2958.2000.01805.x. [DOI] [PubMed] [Google Scholar]
- Weitzmann M.N., Woodford K.J., Usdin K., Woodford K.J., Usdin K., Usdin K. DNA secondary structures and the evolution of hypervariable tandem arrays. J. Biol. Chem. 1997;272:9517–9523. doi: 10.1074/jbc.272.14.9517. [DOI] [PubMed] [Google Scholar]
- Wells R.D., Collier D.A., Hanvey J.C., Shimizu M., Wohlrab F., Collier D.A., Hanvey J.C., Shimizu M., Wohlrab F., Hanvey J.C., Shimizu M., Wohlrab F., Shimizu M., Wohlrab F., Wohlrab F. The chemistry and biology of unusual DNA structures adopted by oligopurine.oligopyrimidine sequences. FASEB J. 1988;2:2939–2949. [PubMed] [Google Scholar]
- Wilcoxon F. Individual comparisons by ranking methods. Biometrics. 1945:80–83. [Google Scholar]
- Woodford K.J., Howell R.M., Usdin K., Howell R.M., Usdin K., Usdin K. A novel K+-dependent DNA synthesis arrest site in a commonly occurring sequence motif in eukaryotes. J. Biol. Chem. 1994;269:27029–27035. [PubMed] [Google Scholar]
- Wu X., Maizels N., Maizels N. Substrate-specific inhibition of RecQ helicase. Nucleic Acids Res. 2001;29:1765–1771. doi: 10.1093/nar/29.8.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lindblad-Toh K., Lander E.S., Kellis M., Lander E.S., Kellis M., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler A.M., Williamson J.R., Cech T.R., Prescott D.M., Williamson J.R., Cech T.R., Prescott D.M., Cech T.R., Prescott D.M., Prescott D.M. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]