Abstract
By constructing nearly isogenic lines (NILs) that differ only at a single quantitative trait locus (QTL), we fine-mapped the yield-improving QTL qGY2–1 to a 102.9-kb region on rice chromosome 2. Comparison analysis of the genomic sequences in the mapped QTL region between the donor (Dongxiang wild rice, Oryza rufipogon Griff.) and recurrent (Guichao2, Oryza sativa ssp. indica) parents used for the development of NILs identified the haplotypes of a leucine-rich repeat receptor kinase gene cluster, which showed extensive allelic variation. The sequences between genes in the cluster had a very high rate of divergence. More importantly, the genes themselves also differed between two haplotypes: Only 92% identity was observed for one allele, and another allele was found to have completely lost its allelic counterpart in Guichao2. The other six shared genes all showed >98% identity, and four of these exhibited obvious regulatory variation. The same haplotype segments also differed in length (43.9-kb in Guichao2 vs. 52.6-kb in Dongxiang wild rice). Such extensive sequence variation was also observed between orthologous regions of indica (cv. 93–11) and japonica (cv. Nipponbare) subspecies of Oryza sativa. Different rates of sequence divergence within the cluster have resulted in haplotype variability in 13 rice accessions. We also detected allelic expression variation in this gene cluster, in which some genes gave unequal expression of alleles in hybrids. These allelic variations in structure and expression suggest that the leucine-rich repeat receptor kinase gene cluster identified in our study should be a particularly good candidate for the source of the yield QTL.
To practice efficient breeding through genetic engineering techniques, it is important to identify the genetic component of important agronomic traits, which often take the form of quantitative phenotypes. The genetic variation underlying quantitative phenotypes results from the segregation of numerous quantitative trait loci (QTLs), each responsible for a portion of the total variation (Mackay 2001). Although a large number of QTLs have been mapped in plants, because of their complex genetic basis, only a few have been cloned. Molecular dissection of QTL is an important area of research and has so far received little investigation in plants. With the development of nearly isogenic lines (NILs) in which a single genomic segment contains the QTL in an otherwise uniform genetic background, the molecular basis underlying allelic variation at QTL is similar to the identified variation for simple Mendelian loci, namely, alterations in gene expression or protein function (Paran and Zamir 2003). The genetic variation underlying most cloned QTL in plants has been attributed to altered protein function or loss of function resulting from sequence variation in coding regions; however, the allelic sequence divergence of the two cloned QTLs, tb1, which largely controls differences in plant architecture between maize and its wild relative, and fw2.2, which accounts for as much as 30% of the difference in fruit size between wild and cultivated tomato, is confined to regulatory regions of these genes (Doebley et al. 1997; Frary et al. 2000).
Although comparative genetic mapping has revealed extensive conservation of gene order and gene content among closely related grass species (Paterson et al. 2000), a much higher degree of sequence diversity at the microstructural level has been detected in even largely collinear genomic regions (Bennetzen 2000). Sequence comparisons in the same species are more informative; investigation of the bz genomic region in two different maize inbreds revealed that such microstructural diversity extends even to allelic regions, thus demonstrating that genetic microcolinearity can be violated within species (Fu and Dooner 2002). With the rapidly increasing amounts of available genomic sequence data, the mechanisms and biological significance of this intraspecies sequence variation can be investigated by comparative sequence analysis of orthologous genomic regions from the same species of crops. Recently, the nature, mechanisms, and specificities of evolutionarily recent changes in sequence that have led to rapid and dramatic divergence of the indica and japonica rice subspecies genomes have been identified by using the African rice Oryza glaberrima as a reference (Ma and Bennetzen 2004). In another study, Song and Messing not only surveyed the genomic organization but also analyzed the expression patterns of z1C loci, and they found that when inbreds with haplotype diversity were reciprocally crossed, hybrids exhibited an unexpected shift of expression patterns so that “overdominance” instead of “dominance complementation” of allelic and nonallelic gene expression occurred (Song and Messing 2003). Furthermore, to compare expression of alleles in the same genetic context, Guo et al. examined parental transcript accumulation in a maize hybrid using allele-specific RT-PCR analysis and found that allelic variation in expression occurred in hybrids, ranging from unequal expression of the two alleles to expression of a single allele regardless of its parent-of-origin (Guo et al. 2004).
Numerous studies have provided evidence that such allelic variation of gene expression occurs frequently in animals. For example, among over 600 heterozygous genes in the human genome, 54% exhibited preferential expression of one allele (at least a twofold difference) in at least one individual; moreover, 28% were found to have a greater than fourfold difference between the two alleles (Lo et al. 2003). This result is consistent with another article reporting that six out of 13 genes in humans had significant differences between expressions of the two alleles (Yan et al. 2002). Furthermore, Cowles et al. screened murine genes for allele-specific effects by studying F1 intercrosses from four inbred mouse strains and concluded that the differences in transcript levels were due to cis-acting sequence variation but were not the result of parental imprinting; in the F1 mice the alleles were exposed to common trans-acting factors and environmental influences (Cowles et al. 2002). These results show that naturally occurring regulatory variants can be detected at a significant frequency in a genomic survey and suggest that the quantitative variation of gene expression may have important consequences for organismal phenotype and evolution.
The rice species Oryza sativa is one of the most important crops in the world and is considered to be a model plant for genome research because of its small genome size and synteny with other cereal crops (Gale and Devos 1998; Sasaki and Burr 2000). The use of its wild relatives provides a more powerful way to exploit a wide range of allelic variation because they have adapted to specific environmental conditions (Yano 2001). Moreover, despite their overall inferior agronomic performance, wild species are likely to contain genetic factors that can improve the yield of modern varieties (Tanksley et al. 1996), and although many other deleterious genes often mask their effects, some favorable genes (e.g., yld1.1 and yld2.1) and QTLs (e.g., gw3.1) from wild rice have been identified (Xiao et al. 1996; Li et al. 2004). The publication of the genome sequence for the japonica (cv. Nipponbare) and indica (cv. 93–11) subspecies of rice (Goff et al. 2002; Yu et al. 2002) provides an unprecedented opportunity for analyzing genomic sequence variation within the same species and studying gene function relevant to important agronomic traits in this important crop plant. Together with the availability of thousands of molecular markers for genotyping, this availability will greatly facilitate map-based cloning of QTLs with allelic variation from wild rice.
In this study, we detected the haplotype of a leucine-rich repeat receptor kinase gene cluster from NILs of a single QTL. To accomplish this, we compared the collinear genomic sequences in the whole-mapped yield-improving QTL region between two parents used in developing NILs. Sequence analysis indicated that this gene cluster is a particularly good candidate for the source of the yield-related QTL. We further analyzed part of the genomic sequences in this gene cluster from 13 rice accessions to further investigate haplotype diversity at this locus. We also used RT-PCR to evaluate the expression level of genes from rice accessions with different haplotypes at this locus and their combinations in hybrid crosses. Furthermore, to make comparisons between alleles within the same genotype and eliminate complications caused by the parental genetic background, we analyzed allele-specific expression variation in one of the genes from the cluster in rice hybrids. Results from this study may help us understand the molecular basis of QTL and genetic mechanism of heterosis in rice, and may provide a clue to identifying agriculturally important genes with functional regulatory variants.
Results
Fine mapping of the QTL qGY2–1 for improved yield in rice
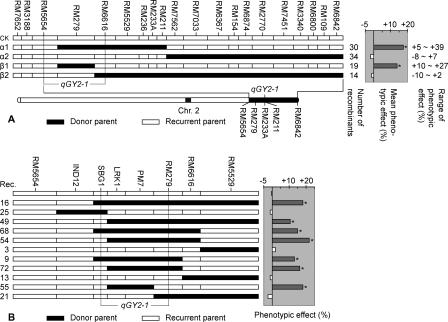
The QTL qGY2–1, associated with grain yield per plant, was identified in Dongxiang wild rice (Oryza rufipogon Griff.) using advanced backcross QTL (AB-QTL) analysis. qGY2–1 was quickly mapped to the short arm of chromosome 2 near SSR marker RM233A and was considered a major gene that could increase the grain yield of the indica cultivar Guichao2 by 25.9% (Li et al. 2002). To fine-map this QTL, we used an introgression line, BIL19, to develop a set of NILs of the target QTL. This line from BC4F4 was selected because it contained an Oryza rufipogon introgressive fragment in qGY2–1 region and relatively few nontarget background introgressions. An NIL heterozygous for target QTL was selfed to generate a segregating population (NIL-F2). A total of 1500 NIL-F2 progenies were subjected to SSR analysis with 20 markers between RM7652 and RM6842, revealing 97 recombinants. For each of the 97 recombinant families, 40 selfed progenies were genotyped with the appropriate segregating markers and analyzed for grain yield per plant. To simplify data representation, we subdivided the 97 recombinant families into four recombination groups (Fig. 1A). Group α1 includes 30 recombinants between RM6842 and RM211 (30 families); the recombinants between RM7562 and RM211 (three families, which contain the shortest introgressed segment in the α1 group) were used to set the limits for the combined graphical representation of the group. All of the recombinants in group α1 show a significant increase in grain yield per plant. The reciprocal recombination group α2 contained 34 families with an Oryza rufipogon segment proximal to RM7562. None of the recombinants showed a significant effect on grain yield per plant. The α groups placed the QTL proximal to RM7562.
Figure 1.
Fine mapping and physical positioning of qGY2–1. (A) Phenotypic analysis of the recombination groups in the short arm of chromosome 2 near SSR marker RM233A. Each group is composed of families whose genotype is represented by a bar divided into black (Dongxiang wild rice, Oryza rufipogon) and empty (Guichao2, Oryza sativa ssp. indica) segments. The genomic composition of the α1, β1 group and α2, β2 group are, respectively, defined by the shortest and the longest O. rufipogon segments detected in the included recombinant families. The borders between bars are arbitrarily drawn midway between positive and negative markers for the introgressed O. rufipogon segment. To the right are the number of recombinants in the group, the mean phenotypic effect of the group, and the minimum and maximum effects in the included families. An asterisk indicates that the phenotypic effect of each of the included families was significant (P < 0.05). The genetically mapped interval of qGY2–1 is indicated by broken lines. (B) Genotypic and phenotypic analysis of 11 recombinant families. Asterisks in the right panel denote a significant phenotypic effect (P < 0.05). The physical location of qGY2–1 is indicated by a broken line between SBG1 and RM279.
Using the same procedure, group β located qGY2–1 between RM6616 and RM5654. To further narrow the position of qGY2–1, four new markers IND12, SBG1, LRK1, and PM7 (Supplemental Table 1) were designed and genetically mapped by using 97 recombinants. Two of these (PM7 and LRK1) showed a complete cosegregation with the QTL. Based on the five markers, IND12, SBG1, LRK1, PM7, and RM279, we identified 11 families as recombinants in the interval between RM 6616 and RM5654. The phenotypic effects for each of the 11 families were used to determine the location of qGY2–1 in a manner consistent with the mapping of the rest of the recombinant families. Recombinants 25, 49, 54, 72, and 55 delimited the QTL to the region upstream of SBG1, and recombinants 55 and 21 delimited the QTL to the region downstream of RM279. Thus, the position of the QTL was further narrowed down to the region between markers RM279 and SBG1 (Fig. 1B).
Sequence analysis of the genomic region associated with the mapped QTL
According to the fine mapping of qGY2–1, the significant yield effect in the NILs resulted from the introgression of the genomic region between marker RM279 and SBG1 of Dongxiang wild rice in the genetic background Guichao2. This finding suggested that sequence variation should present in the same genomic regions between Guichao2 and Dongxiang wild rice and that the variation in this region should be associated with the QTL for yield. To further dissect the molecular basis of this QTL, we first retrieved the genomic sequences between markers SBG1 and RM279 of the rice subspecies japonica (cv. Nipponbare) and indica (cv. 93–11) from RGP (http://rgp.dna.affrc.go.jp/) and NCBI (http://www.ncbi.nlm.nih.gov/), and then performed sequence comparisons between the two orthologous regions. The identical sequences were used to design PCR primers whose products overlap each other by at least 200 bp, based on the sequences in Nipponbare. A total of 35 primer pairs were designed covering the whole genomic regions between markers SBG1 and RM279 (Supplemental Table 1). PCRs were then performed with total genomic DNA from Guichao2 and Dongxiang wild rice, respectively, using these primers. All amplified fragments were sequenced and then assembled in reference to the orthologous sequences from Nipponbare and 93–11. As a result, a 94.1-kb sequence of Guichao2 and 102.9-kb sequence of Dongxiang wild rice in the genomic regions between markers SBG1 and RM279 were submitted to GenBank with accession numbers of DQ195081 and AY756174, respectively. Sequence alignment was then performed between these two orthologous segments to reveal variations associated with the yield QTL.
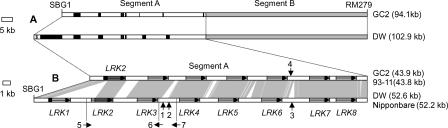
Based on the extent of sequence divergence, the orthologous genomic regions between markers SBG1 and RM279 could be divided into two segments (Fig. 2A). Segment B showed >99% sequence identity between Guichao2 and Dongxiang wild rice, and the variations in this segment were evenly dispersed small insertion/deletions (indels) and base substitutions of a total size of no more than 80 bp. Further analysis indicated that these variations did not alter the amino acid sequence encoded by the predicted genes in segment B. In contrast, extensive sequence variation was detected from segment A between Guichao2 (43.9-kb genomic region) and Dongxiang wild rice (52.6-kb genomic region); at least a 13-kb sequence could not be aligned between these two rice accessions (Fig. 2A). Thus, this analysis indicates that the sequence variation in segment A is responsible for the mapped QTL.
Figure 2.
Identification of the LRK gene cluster associated with the mapped QTL. (A) Schematic diagram of the sequence comparison in the QTL region flanked by the markers SBG1 and RM279 between Guichao2 (GC2) and Dongxiang wild rice (DW). Based on the extent of sequence divergence, the orthologous segment was divided into two sections. Segment B (represented by the lightly shaded rectangles) showed 99% sequence identity. The sequence of segment A exhibited extensive variation (represented by black bars or lines). (B) Sequence alignment of the LRK gene cluster in segment A between Guichao2 (or 93–11) and Dongxiang wild rice (or Nipponbare). Conserved sequences are connected by vertical areas. Horizontal arrows in shaded rectangles indicate polarity and position of LRK gene copies. Upward vertical arrows 1, 2, and 3 indicate position 337, 36, and 66-bp indels between Dongxiang wild rice and Nipponbare, respectively; downward vertical arrow 4 indicates the position of a 38-bp indel between Guichao2 and 93–11; horizontal arrows 5, 6, and 7 indicate polarity and the position of three gene fragments within the LRK cluster.
On the basis of the available sequence annotation for the japonica cultivar Nipponbare, there are 11 predicted genes in the 52.2-kb orthologous region of segment A in Nipponbare. Of these genes, three had no function annotation and seemed from their small sizes to be gene fragments (207, 538, and 223 bp, respectively), whereas the other eight were all annotated as putative phytosulfokine receptor, a leucine-rich repeat receptor kinase, each with a gene size of more than 3 kb, thus forming a dense gene cluster throughout the whole region (Supplemental Table 2) (Fig. 2B). Sequence comparison analysis indicated that this orthologous region of segment A between Dongxiang wild rice and Nipponbare showed a high degree of identity; besides a number of small indels and substitution mutations of 1 or 2 bp, there was only variation in three indels with a total size of 439 bp in the intergenic region. These analyses of sequence comparisons indicated that the genomic sequence of segment A in Dongxiang wild rice is also composed of three gene fragments and the leucine-rich repeat receptor kinase gene cluster. The three gene fragments did not express any detectable transcripts in any rice parents or their hybrids used in our research (data not shown); thus, we concluded that the sequence variation of this leucine-rich repeat receptor kinase gene cluster in the mapped QTL region between Guichao2 and Dongxiang wild rice (Fig. 2B) was most likely to associate with the QTL for yield. This finding also suggested that the orthologous region in Nipponbare might contain this yield-improving QTL and that rice cultivar Nipponbare is from a relatively recent introduction of a lineage related to the Dongxiang wild rice.
Haplotype divergence of the leucine-rich repeat receptor kinase gene cluster
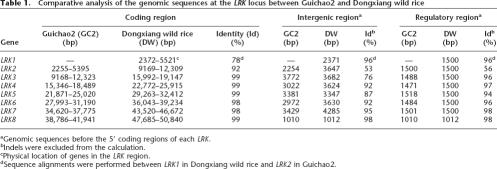
Sequence analysis with FGENESH revealed that this leucine-rich repeat receptor kinase (designated LRK) gene cluster consists of eight intronless LRK gene copies (designated LRK1 to LRK8) arranged in a tandem fashion throughout the 52.6-kb genomic region in Dongxiang wild rice; there are only seven LRK gene copies tandemly arrayed throughout the 43.9-kb orthologous genomic region in Guichao2 (Fig. 2B). Coding sequences of the first LRK gene (which is proximal to marker SBG1) at the LRK locus in Guichao2 showed the highest identity (92%) to the second gene at this locus in the Dongxiang wild rice, whereas the other six shared genes all showed more than 98% identity in the coding region between the two rice accessions (Table 1). These findings indicate that in Guichao2, the allelic counterpart of LRK1 (3150 bp in the coding region) has been lost completely, and the gene cluster contains only the other seven LRK members (i.e., LRK2 to LRK8).
Table 1.
Comparative analysis of the genomic sequences at the LRK locus between Guichao2 and Dongxiang wild rice
aGenomic sequences before the 5′ coding regions of each LRK.
bIndels were excluded from the calculation.
cPhysical location of genes in the LRK region.
dSequence alignments were performed between LRK1 in Dongxiang wild rice and LRK2 in Guichao2.
In contrast to the high sequence conservation in the coding regions, the intergenic sequences in the 5′ coding regions of the LRK genes (except for LRK8) showed extensive variations between Guichao2 and Dongxiang wild rice. Numerous indels offer an obvious source of this sequence variation in the intergenic region. A total of 252 indels were identified in the orthologous intergenic regions based on sequence alignments. Of these indels, 120 appear to be deletions from Dongxiang wild rice relative to Guichao2, whereas 132 appear to be deletions from Guichao2 relative to Dongxiang wild rice, accounting for a total of 675 bp and 6286 bp of the DNA sequence, respectively (Supplemental Table 3). Hence, indel variations result in about 3.4% and 24.6% of the sequence difference in the 19.8-kb intergenic regions of Guichao2 and 25.6-kb intergenic regions of Dongxiang wild rice, respectively; the average of 13.9% was similar to the general frequency revealed in the comparison of large orthologous regions between japonica and indica subspecies of rice (Ma and Bennetzen 2004). A large indel of 3647 bp before the 5′ coding regions of LRK2 and two other smaller indels with sizes of 780 bp and 809 bp before the 5′ coding regions of LRK6 and LRK7, respectively, are primarily responsible for the different size of the intergenic region at the LRK locus between the two rice accessions. Small indels with sizes of 1 or 2 bp far outnumber the larger ones, contributing little to genome size variation in these regions (Supplemental Table 3).
Pairwise comparison of the sequences in the homologous regions extending 1.5 kb upstream of the start codons of LRK revealed that the sequence variation occurs in the putative 5′ regulatory region of some LRK genes (Table 1). Interestingly, although the intergenic sequence of the 5′ coding region of LRK7 is highly divergent between Guichao2 and Dongxiang wild rice, no obvious sequence variation was observed in the regulatory region. More important, the regulatory region of LRK2 between the two rice accessions exhibited a low sequence identity (56%), whereas the regulatory sequence of LRK2 in Guichao2 showed 96% identity to that of LRK1 in Dongxiang wild rice after indels were excluded from the calculation of sequence identity (Table 1). A different extent of sequence variations was also observed in the regulatory regions of LRK3 to LRK6 between the two rice accessions. Such large haplotype divergence in both a coding and regulatory region at a single gene cluster has rarely been reported in rice and suggests that the LRK gene cluster should be a particularly good candidate for the source of the yield QTL detected in our study.
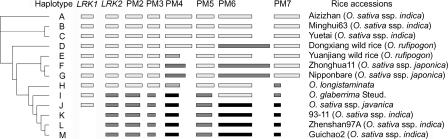
We also found that besides numerous small indels and base substitutions of 1 or 2 bp, sequence variation at the LRK locus between Guichao2 and 93–11 was confined to only one indel with a size of 38 bp in the intergenic region near the 3′ coding region of LRK6 (Fig. 2B). Because sequence comparison in the LRK regions between Dongxiang wild rice and Nipponbare also showed a high degree of identity, we concluded that the same pattern of haplotype divergence at the LRK locus existed between the japonica (cv. Nipponbare) and indica (cv. 93–11) subspecies of rice; pairwise sequence alignment at the LKR locus between Nipponbare and 93–11 (data not shown) confirmed this conclusion, thus demonstrating that genetic colinearity in this region could be violated within the same rice species. To further investigate haplotype diversity at the LRK locus in rice, we analyzed part of the genomic sequences in this gene cluster from other cultivar and wild rice, using primers from LRK1 and LRK2 and primers designed to detect sequence polymorphisms before the 5′ coding region of LRK2 to LRK7 between Guichao2 (or 93–11) and Dongxiang wild rice (or Nipponbare) (Supplemental Table 1). All PCR products were sequenced to identify small sequence variations. Base on the presence/absence of LRK1, the divergence of LRK2, and the sequence variation before the 5′ coding region of LRK2 to LRK7, 13 haplotypes (A to M) were detected among 13 rice accessions (Fig. 3; Supplemental Table 4). The phylogenetic analysis based on the divergent sequences is shown in Figure 3.
Figure 3.
Haplotype diversity of the LRK gene cluster in 13 rice accessions. Based on the presence/absence of LRK1, the divergence of LRK2, and the sequence variation before the 5′ coding region of LRK2 to LRK7 (designated by PM2 to PM7), 13 haplotypes (A–M) were detected among 13 rice accessions. Different shaded rectangles indicate the size and sequence variation in the polymorphous regions. The small sequence variation within a size of 2 bp in the segments represented by the same rectangles are not shown in this figure. To the left is the phylogenic tree generated by ClustalX using the neighbor-joining method based on the divergent sequences in the LRK cluster.
Allelic variation in gene expression at the LRK locus
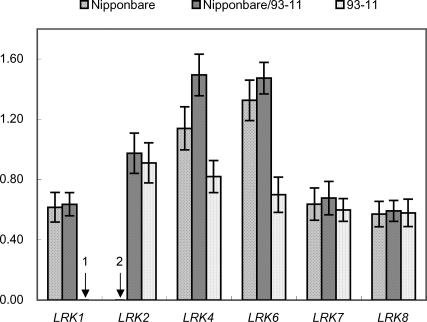
Because the genomic sequences at the LRK locus between Nipponbare and 93–11 exhibit the same pattern of haplotype variation as that described between Guichao2 and Dongxiang wild rice, and for the convenience of acquiring experimental materials, we measured the expression level of each individual LRK gene in Nipponbare, 93–11, and their hybrid cross Nipponbare/93–11 by using RT-PCR to investigate whether sequence variation at this locus affects the expression of LRK genes. Seedlings at the three-leaf stage were selected for the expression analysis. We found that although the gene copies at the LRK locus differed between the Nipponbare and 93–11 haplotypes, the number of genes expressed at a detectable level was the same. Alleles of LRK4, LRK6, LRK7, and LRK8 were expressed in both haplotypes, whereas LRK1 was expressed in Nipponbare and LRK2 was only expressed in 93–11 (Fig. 4). Within the same haplotype, the active genes were not uniformly expressed at the same level. In Nipponbare, the highest expressed gene was LRK6, with expression level about 2.3× higher than that of LRK8 (the lowest expressed gene) (P < 0.01). In 93–11, the highest-expressed gene was LRK2, with expression level about 1.9× higher than that of LRK8 (the lowest-expressed gene) (P < 0.01).
Figure 4.
Allelic expression variation of LRK genes in Nipponbare, 93–11, and their hybrid cross Nipponbare/93–11. Rice seedlings at the three-leaf stage were processed for the measurement of the transcript level using RT-PCR. Expression levels of LRK genes were evaluated relative to that of actin1. Error bars, standard deviation (SD). Vertical arrow 1 indicates loss of LRK1 in 93–11, and vertical arrow 2 indicates that transcript was not detected in Nipponbare.
It is interesting that in the hybrid, LRK1 seems to be expressed at the same level as it is in the homozygous parent, even though there is only one genomic copy of the allele in the hybrid cross Nipponbare/93–11 versus two in Nipponbare. Furthermore, although LRK2 was present in both haplotypes, only the allele in 93–11 was expressed, perhaps because the deletion of LRK1 brings functional regulatory sequences in close proximity to LRK2 in 93–11. Further expression analysis using allelic-specific primers indicated that only the 93–11 allele of LRK2 is expressed in the hybrid, with an expression level similar to that of the homozygous parent 93–11. We also found that both LRK7 and LRK8 exhibited similar levels of allelic expression between hybrids and the two parents, probably attributable to the high degree of sequence conservation in their regulatory regions. However, both LRK4 and LRK6 showed much higher expression levels in Nipponbare than in 93–11 (P < 0.01), probably the result of the sequence variation in their regulatory regions. More important, in hybrid cross Nipponbare/93–11, these two genes exhibited an unexpected shift in expression pattern, in which there was a significantly higher level of expression for LRK4 in the hybrid compared to either parent (P < 0.01), typical of transgressive variation, and slightly higher levels of expression for LRK6 in the hybrid than in Nipponbare. Furthermore, we also found that LRK genes at the same locus in Guichao2, BIL19 (which contained a homozygous introgression chromosomal segment of Dongxiang wild rice across the whole QTL region in the Guichao2 genetic background) and in their hybrid cross Guichao2/BIL19 show the same pattern of allelic expression variation as described above (Supplemental Table 5). However, no significant differences in allele expression were detected between parents with the similar LRK haplotypes and their hybrid crosses (e.g., Guichao2/93–11) (data not shown).
Allele-specific transcript accumulation of LRK6 in hybrids
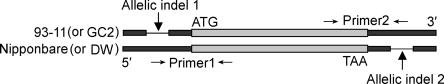
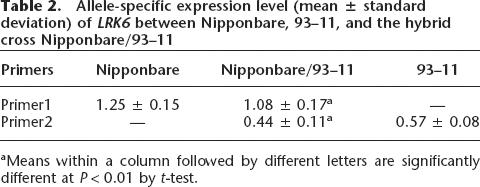
Although LRK4 and LRK6 exhibited an elevated expression level in the hybrid compared with the parents, the proportion of transcript contributed by each parental allele expressed in the hybrid was unknown. Thus, it was necessary to examine parental transcript accumulation in the hybrid using allele-specific expression analysis. Because allelic sequence polymorphisms are often located in the untranslated region, we compared the 5′ and 3′ untranslated sequences of LRK4 and LRK6 between Nipponbare (or Dongxiang wild rice) and 93–11 (or Guichao2) in detail. We found that one 23-bp indel, which appeared to be a deletion in 93–11 (or Guichao2) relative to Nipponbare (or Dongxiang wild rice) at the 5′ untranslated region of LRK6, and another 17 bp indel, which appeared to be a deletion in Nipponbare (or Dongxiang wild rice) relative to 93–11 (or Guichao2) in the 3′ untranslated region of LRK6. Accordingly, the allele-specific primers for LRK6 based on these polymorphisms were designed to perform allele-specific RT-PCR analysis in Nipponbare, 93–11, and their hybrid cross Nipponbare/93–11 (Fig. 5). Our results revealed that, although both parental alleles of LRK6 were expressed, they did not contribute equally to total transcript accumulation in the hybrid (Table 2). In the hybrid cross Nipponbare/93–11, the expression level of the Nipponbare allele (1.08 ± 0.17) was about 2.45× higher than that of 93–11 allele (0.44 ± 0.11) (P < 0.01), which is consistent with the results in Figure 4. Because the allelic ratio of the genomic PCR product in the hybrid was not significantly different from the expected allelic ratio of 1.0 (data not shown), the allele-specific expression variation of LRK6 generated from the cDNA PCR was not likely the result of any amplification bias. However, the allele-specific transcript levels of LRK6 in the hybrid (i.e., 1.08 ± 0.17 for the Nipponbare allele and 0.44 ± 0.11 for the 93–11 allele) were significantly lower (P < 0.05) than transcript levels in their originating parents (i.e., 1.25 ± 0.15 for the Nipponbare allele and 0.57 ± 0.08 for the 93–11 allele) (Table 2).
Figure 5.
Schematic diagram of designing primers used for allelic-specific expression analysis of LRK6. Two small allelic indels in the 5′ and 3′ noncoding regions of LRK6 were used for designing the allelic-specific primers. Lightly shaded rectangles, coding regions; darkly shaded rectangles, non-coding regions; black lines, allelic indels. Primer1, primer specific for Nipponbare (or Dongxiang wild rice, DW); Primer2, primer specific for 93–11 (or Guichao2, GC2). RT-PCR was performed using one allelic-specific primer and another primer derived from the coding region.
Table 2.
Allele-specific expression level (mean ± standard deviation) of LRK6 between Nipponbare, 93–11, and the hybrid cross Nipponbare/93–11
aMeans within a column followed by different letters are significantly different at P < 0.01 by t-test.
A similar pattern of allele-specific expression variation of LRK6 was also detected in the hybrid cross Guichao2/BIL19, in which the expression level of Dongxiang wild rice allele in BIL19 was about 2.23× higher than that of the Guichao2 allele (P < 0.01) (data not shown). The allele-specific expression variation pattern of LRK6 in hybrid crosses Nipponbare/93–11 and Guichao2/BIL19, where the first accession denotes the female parent, appeared to suggest that there was no parent-of-origin effect for the expression of LRK6 in hybrids.
Discussion
Haplotype variability results from different rates of sequence divergence
Our studies uncovered exceptional haplotype variability at the LRK locus in the rice genome. The LRK gene is located in an unusually gene-dense region, in which seven or eight LRK genes were found within a 40–50 kb genomic interval uninterrupted by transposon elements. Colinearity was restricted to the coding region of six genes and one intergenic region, and a single gene (LRK1) was absent in three indica haplotypes (Fig. 3). Such non-colinear haplotypes were also found in a comparison of the same genomic intervals at the z1C and bz locus in maize inbred lines and at the Rph7 locus in two barley cultivars (Fu and Dooner 2002; Song and Messing 2003; Lai et al. 2005; Scherrer et al. 2005). The apparent intraspecific violation of genetic colinearity at these loci was partly attributed to genes missing and moving by transposable elements, including transposons and retrotransposons. To investigate whether a similar situation occurred at the LRK locus, we surveyed the genomic sequences surrounding the LRK gene cluster in Nipponbare and 93–11 and found that, on the basis of the available sequence annotation (http://rgp.dna. affrc.go.jp/), an 8.5-kb transposon lies immediately proximal to the 5′ end of Nipponbare LRK cluster, while it is absent in the orthologous regions of 93–11. Because transposons are known often to induce chromosome breakage and hence chromosomal rearrangements (Dooner and Belachew 1991), the fact that the gene missing from 93–11 is next to a transposon block in Nipponbare implicated the transposon in the deletion/insertion process of LRK1 and its following intergenic sequence. Interestingly, a sequence comparison in the coding and regulatory region indicated that the LRK2 in 93–11 (or Guichao2) seemed to be a chimera containing segments from LRK1and LRK2 in Nipponbare (or Dongxiang wild rice) (Fig. 2B), thus implying that the unequal recombination may also be the source of gene number differences in LRK gene clusters. This characteristic is similar to that revealed in the studies of the Rp1 locus, which was associated with disease resistance in maize (Richter et al. 1995; Ramakrishna et al. 2002). Furthermore, the ratio of nonsynonymous to synonymous substitutions (Ka/Ks) for the coding region of LRK2 between cultivar rice (Guichao2) and wild rice (Dongxiang wild rice, Oryza rufipogon) was only 0.48, thus implying that the orthologous gene pair of LRK2 is likely to be under purifying selection.
Because the absence of LRK1 was only found in three indica cultivar groups and all eight LRK gene members could be detected in all other sampled accessions of rice, we speculated that haplotypes K, L, and M resulted from the deletion of LRK1, and that duplications of LRK occurred before the separation of these haplotypes. This model of ancient large-scale duplication followed by selective gene loss has been proposed to be the major factor in the evolution of dicot family genomes (Ku et al. 2000). Based on the sequence divergence of LRK2 and polymorphs of PM2, PM3, and PM5, haplotypes in the sampled accessions could be divided into two groups (haplotype A to H for group I, and haplotype I to M for group II). Sequence variation before the 5′ coding region of LRK4, LRK6, and LRK7 results in the haplotype diversity in group I, and variation before the 5′ coding region of LRK6 and LRK7 results in the haplotype diversity in group II. It is interesting that haplotypes H and I in two different groups, respectively, share the same sequence variation before the 5′ coding region of LRK6 and LRK7, implying that divergence in these regions already occurred before separation of haplotypes between two groups. This haplotype variability resulting from different rates of sequence variation at the homologous LRK loci could provide a basis for the molecular mechanism of the generation of diversity in the rice genome. Moreover, the result of phylogenetic analysis suggested the independent domestication of several indica and japonica subspecies of Oryza sativa. Further complete sequencing of the LRK genomic interval in more accessions of rice could reveal more haplotype divergence at the LRK locus, thus providing an unprecedented opportunity for analyzing genomic evolution and for investigating the origin and differentiation of cultivated rice.
Naturally occurring allelic variation underlying complex quantitative traits
Genetic variation in nature often takes the form of a quantitative phenotypic range with an approximately normal distribution, rather than of qualitative phenotypes that fall into discrete categories (Paran and Zamir 2003). Although it is difficult to identify and characterize the definite genomic regions associated with the quantitative differences, several recent studies in plants have revealed that naturally occurring allelic variation underlies the inheritance of complex quantitative traits (Paran and Zamir 2003). Naturally occurring allelic variation can be classified as coding variation, which alters the amino acid sequence of the encoded protein, or regulatory variation, which affects the level or pattern of gene expression and often results in quantitative differences. In the present study, sequence comparison of the LRK gene cluster in rice indicated that both coding regions (LRK1 and LRK2) and regulatory regions of the expressed genes (LRK4 and LRK6) exhibited variation at a single QTL. Although the allelic variation such as presence/absence of LRK1 and the sequence divergence in the coding region of LRK2 were important in dissecting the QTL, a recent study has provided evidence that regulatory variation in allelic expression is also important in determining phenotypic diversity (Cowles et al. 2002).
Because the allele-specific expression variation of LRK6 in hybrids controls for trans-acting effects and environmental influences, our study suggested that the effects of cis-acting elements in the LRK cluster on the allelic expression variation may also contribute to the explanation of the molecular basis of the QTL identified in our research. This mechanism was revealed in a study of tb1 in maize (Doebley et al. 1997) and fw2.2 in tomato (Nesbitt and Tanksley 2002). Furthermore, sequence comparison between alleles from wild relatives and cultivated varieties at these two loci has shown that the effects of selection are also limited to the regulatory region (Wang et al. 1999; Frary et al. 2000). Because of this, naturally occurring allelic variation in gene expression at the LRK locus between Guichao2 and BIL19 (which contained an introgression fragment from wild rice in Guichao2 genetic background) suggested that the regulatory variation of LRK genes may be also involved in natural or artificial selection, contributing to our understanding of the domestication of the complex quantitative trait locus in rice species.
On the other hand, because genes at the LRK locus are highly correlated, it is difficult or impossible to determine which one of them is the genetic component of the QTL in our study. Moreover, LRK genes in the cluster may also act in an additive or interactive manner to cause phenotype variation; this behavior has been illustrated with the identification of gene clusters corresponding to a single QTL affecting a physiological trait, in which several tandem repeated genes contribute to the phenotypic effect and may interact genetically (Kroymann et al. 2003). Thus, from this perspective, the LRK gene cluster with naturally occurring allelic variation probably acts as a single Mendelian locus in determining phenotype effect.
According to sequence annotation (http://rgp.dna.affrc.go.jp), all gene members in the LRK cluster identified in our research encoded the putative kinase receptor of phytosulfokine, a peptide plant hormone. Because the leucine-rich repeat receptor kinase regulates a wide variety of developmental and defense-related processes in plants (Torii 2004), our research suggested that expression variation resulting from haplotype divergence in the structure of the LRK gene cluster may contribute to the complex quantitative trait in rice by regulating plant development; further transgenic research and functional complementation with these candidate LRK genes are required to demonstrate a causative relationship.
Allelic variation of gene expression in rice hybrids
Recent studies in maize (Song and Messing 2003; Guo et al. 2004) and mouse (Cowles et al. 2002) have provided evidence for significant allelic expression difference in hybrids. Because two alleles were compared in the same genetic context, their results implicated allelic diversity as the underlying mechanism of heterosis; whether or not such allelic variation indeed causes any phenotypic variation remains to be tested. With the available genomic sequences and their annotations in the databases, it is more important to detect allelic variation in potentially functional genomic regions and to evaluate its effect on the expression and function of genes in parents and their hybrids. In our study, the allelic variation of gene expression in hybrids at the LRK locus that associated with the yield QTL ranged from unequal expression of the two alleles simultaneously to expression of a single allele regardless of the parent-of-origin. In addition, both dominance complementation and some overdominance of allelic expression within a single gene cluster occurred in hybrids. The concept of “dominance complementation” fits the expression pattern of LRK1 and LRK2, in which only one allele from the parents is expressed in hybrids and no obvious dosage effect was detected. On the other hand, the concept of “overdominance” fits the expression pattern of LRK4, in which significantly higher expression levels of LRK4 in hybrid crosses were detected than in either parent, typical of the heterosis phenomenon. Although only slightly higher levels of expression for LRK6 in the hybrids were detected compared to either parent, further allele-specific RT-PCR analysis revealed that this variation of gene expression in hybrids result from additive effect of allelic expression in parents. The alleles do not contribute equally to transcript accumulation in hybrids. Because in the hybrid the two alleles are exposed to the common trans-acting regulatory factors, the allelic sequence variation of the cis-regulatory elements may cause a target gene to interact or bind differentially with the transcriptional factors, thus resulting in differential transcription between alleles (Guo et al. 2004). However, more direct evidence is needed to determine the regulatory mechanism of the allelic expression differences. Moreover, we also found that allele-specific expression of LRK6 in hybrids was significantly lower (P < 0.05) than expression in their originating parents and that the hybrids exhibited a lower expression level of LRK6 than the sum of the two parental expression levels (Table 2). This outcome may be the result of the transcription interference between two alleles in the hybrid, which warrants further investigation. Regardless, since the increase in hybrid vigor might have occurred by selecting alleles at the right set of loci that produce the best combinations in hybrids to bring about heterosis (Birchler et al. 2003), the allelic expression variation revealed in this study may help us understand the molecular basis of heterosis in rice and provide us with a clue about how to exploit heterosis by breeding hybrids with superior haplotype combinations.
Methods
Plant materials and QTL analysis
A BC4F2 population was constructed for QTL mapping, and a derivative BC4F4 population was used to develop a set of introgression lines using an Oryza sativa ssp. indica cultivar, Guichao2, as the recurrent parent and an accession of Dongxiang wild rice (Oryza rufipogon Griff.) as the donor parent (Li et al. 2002). One of the introgression lines, BIL19, which contained an Oryza rufipogon introgression in the yield-improving QTL qGY2–1 that had been mapped previously, and relatively few nontarget background introgressions, was selected as the starting material for the development of NILs and fine mapping of the QTL. NILs were constructed by backcrossing BIL19 to the indica rice Guichao2 for two generations followed by selfing to eliminate nontarget genomic regions. An NIL heterozygous for target QTL was selfed to generate a segregating population (NIL-F2). A total of 1500 NIL-F2 progeny were screened with 20 SSR markers in the interval between RM7652 and RM6842, and recombinant plants were selected. For each recombinant, 40 selfed progenies (F2:3 family) were genotyped with the appropriate segregating markers and analyzed for grain yield per plant. The required density of molecular markers in the qGY2–1 region was achieved by using published SSRs (McCouch et al. 2002). To further narrow the position of qGY2–1, four new markers were designed from publicly available rice genome sequence and the likelihood of detecting polymorphism between Guichao2 and Doxiang wild rice was predicted by comparing sequences from the japonica cultivar, cv. Nipponbare (http://rgp.dna.affrc.go.jp/), and the indica cultivar, cv. 93–11 (http://rise.genomics.org.cn/rice/index2.jsp). The F2:3 progeny for the open-field trial (48 plants per family) were planted in Sanya (18°N, 109°E), Hainan Province, in a completely randomized design. Agricultural practices, phenotypic measurements, data processing, and QTL analysis were as described previously (Li et al. 2002).
DNA isolation, PCR, and sequencing
DNA was extracted using a CTAB (cetyltrimethyl ammonium bromide) method (Murray and Thompson 1980). The PCR primers used for the amplification were designed by using Primer 3 (http://www.broad.mit.edu/cgi-bin/primer/primer3_www.cgi) and PCRs were performed using Taq plus DNA polymerase (Shenergy Biocolor) in the PTC-200 thermal cycling system (MJ Research). Single-band PCR products were cloned into the pGEM-T vector (Promega) and all selected clones were sequenced with T7 and SP6 primers using a dye terminator cycle sequencing kit (Perkin–Elmer) and an automated capillary DNA sequencer (Genetic Analyzer ABI 310, Perkin–Elmer).
Genomic sequence analysis and comparison
The genomic sequences of rice subspecies japonica cv. Nipponbare (GenBank accession no. AP007224) in the QTL region between markers SBG1 and RM279 were retrieved from NCBI (http://www.ncbi.nlm.nih.gov/) or RGP (http://rgp.dna. affrc.go.jp/). These sequences were used in BLASTN (http://www. ncbi.nlm.nih.gov/BLAST/) searches against the rice nonredundant genomic sequence database from NCBI (http://www. ncbi.nlm.nih.gov/) and BGI-RIS (http://rise.genomics.org.cn/rice/index2.jsp) to determine their orthologous regions in indica subspecies cv. 93–11. Sequence comparisons in the orthologous regions were performed by using the BLAST2 (Tatusova and Madden 1999) (http://www.ncbi.nlm.nih.gov/BLAST/) and ClustalX (Thompson et al. 1997) programs. Sequence analysis and gene prediction were performed by using the gene-finding programs FGENESH (http://www.softberry.com/berry.phtml). K-Estimator was used to estimate the ratios of nonsynonymous to synonymous substitutions (Ka/Ks) for the coding regions (Comeron 1999).
Gene expression analysis
Rice plants were cultivated under controlled environmental conditions. Fresh leaves were harvested separately from seedlings at the three-leaf stage, immediately frozen in liquid nitrogen, and stored at –80°C. Total RNA was extracted using TRIzol reagent (Gibco BRL) according to the manufacturer’s recommendations. Contaminating genomic DNA was removed from RNA samples by treatment with RNase-free DNase I (Sigma) at 37°C for 1 h. The total RNA was purified by phenol/chloroform extraction followed by precipitation in ethanol. mRNA was extracted by using a Poly(A) Tract kit (Promega) and then was transcribed with SuperScript II RNase H- reverse transcriptase (Invitrogen) according to the manufacturer’s recommendations. Approximately equal amounts of first strand cDNAs were used as templates for PCR amplification using gene-specific primers for 28–36 cycles. The rice actin1 gene (GenBank accession no. X16280.1) was amplified simultaneously using primers a1 (5′-CTGTCTTCCCCAGCATT GTC-3′) and a2 (5′-GGTCTTGGCAGTCTCCATTTC-3′) to serve as a positive control for quantification of the relative amounts of cDNA. Optimal conditions for RT-PCR were determined with reference to the published protocol of semiquantitative RT-PCR analysis (Marone et al. 2001). Amplified DNA products were analyzed with SmartView 5 (Furi). The level of allelic expression was evaluated relative to that of actin1. Three different sets of experiments were carried out for parents and hybrids, in which amplifications were repeated three times for each experiment, and the reported data were the means of three independent experiments. For analysis of allele-specific expression variation, genomic DNA PCR in hybrid crosses was conducted simultaneously to test any PCR amplification bias for either allele, which may confound the allele expression data.
Acknowledgments
We thank Xianyou Sun for maintaining the rice materials and Huifang Qin for helping with the experiments. We also thank anonymous referees for review of the manuscript. We thank Dr. Paul Kretchmer for his assistance in editing this manuscript. This research was supported by the China “973” Foundation (Grant No. 2001CB108805), and by a grant from the Program of Conservation and Utilization of Agro-Wild Plants of the Ministry of Agriculture of China, and the National Natural Science Foundation of China (No. 30270803).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4814006
References
- Bennetzen J.L. Comparative sequence analysis of plant nuclear genomes: microcolinearity and its many exceptions. Plant Cell. 2000;12:1021–1029. doi: 10.1105/tpc.12.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Auger D.L., Riddle N.C., Auger D.L., Riddle N.C., Riddle N.C. In search of the molecular basis of heterosis. Plant Cell. 2003;15:2236–2239. doi: 10.1105/tpc.151030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J.M. K-Estimator: Calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics. 1999;15:763–764. doi: 10.1093/bioinformatics/15.9.763. [DOI] [PubMed] [Google Scholar]
- Cowles C.R., Hirschhorn J.N., Altshuler D., Lander E.S., Hirschhorn J.N., Altshuler D., Lander E.S., Altshuler D., Lander E.S., Lander E.S. Detection of regulatory variation in mouse genes. Nat. Genet. 2002;32:432–437. doi: 10.1038/ng992. [DOI] [PubMed] [Google Scholar]
- Doebley J., Stec A., Hubbard L., Stec A., Hubbard L., Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- Dooner H.K., Belachew A., Belachew A. Chromosome breakage by pairs of closely linked transposable elements of the AC-DS family in maize. Genetics. 1991;129:855–862. doi: 10.1093/genetics/129.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary A., Nesbitt T.C., Frary A., Grandillo S., Knaap E., Cong B., Liu J., Meller J., Elber R., Alpert K.B., Nesbitt T.C., Frary A., Grandillo S., Knaap E., Cong B., Liu J., Meller J., Elber R., Alpert K.B., Frary A., Grandillo S., Knaap E., Cong B., Liu J., Meller J., Elber R., Alpert K.B., Grandillo S., Knaap E., Cong B., Liu J., Meller J., Elber R., Alpert K.B., Knaap E., Cong B., Liu J., Meller J., Elber R., Alpert K.B., Cong B., Liu J., Meller J., Elber R., Alpert K.B., Liu J., Meller J., Elber R., Alpert K.B., Meller J., Elber R., Alpert K.B., Elber R., Alpert K.B., Alpert K.B., et al. fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- Fu H., Dooner H.K., Dooner H.K. Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. 2002;99:9573–9578. doi: 10.1073/pnas.132259199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M.D., Devos K.M., Devos K.M. Comparative genetics in the grasses. Proc. Natl. Acad. Sci. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.A., Ricke D., Lan T.H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Ricke D., Lan T.H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Lan T.H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Glazebrook J., Sessions A., Oeller P., Varma H., Sessions A., Oeller P., Varma H., Oeller P., Varma H., Varma H., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Guo M., Rupe M.A., Zinselmeier C., Habben J., Bowen B.A., Smith O.S., Rupe M.A., Zinselmeier C., Habben J., Bowen B.A., Smith O.S., Zinselmeier C., Habben J., Bowen B.A., Smith O.S., Habben J., Bowen B.A., Smith O.S., Bowen B.A., Smith O.S., Smith O.S. Allelic variation of gene expression in maize hybrids. Plant Cell. 2004;16:1707–1716. doi: 10.1105/tpc.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann J., Donnerhacke S., Schnabelrauch D., Mitchell-Olds T., Donnerhacke S., Schnabelrauch D., Mitchell-Olds T., Schnabelrauch D., Mitchell-Olds T., Mitchell-Olds T. Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc. Natl. Acad. Sci. 2003;100:14587–14592. doi: 10.1073/pnas.1734046100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku H.M., Vision T., Liu J., Tanksley S.D., Vision T., Liu J., Tanksley S.D., Liu J., Tanksley S.D., Tanksley S.D. Comparing sequenced segments of the tomato and Arabidopsis genomes: Large-scale duplication followed by selective gene loss creates a network of synteny. Proc. Natl. Acad. Sci. 2000;97:9121–9126. doi: 10.1073/pnas.160271297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J.S., Li Y.B., Messing J., Dooner H.K., Li Y.B., Messing J., Dooner H.K., Messing J., Dooner H.K., Dooner H.K. Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc. Natl. Acad. Sci. 2005;102:9068–9073. doi: 10.1073/pnas.0502923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.J., Sun C.Q., Fu Y.C., Li C., Zhu Z.F., Chen L., Cai H.W., Wang X.K., Sun C.Q., Fu Y.C., Li C., Zhu Z.F., Chen L., Cai H.W., Wang X.K., Fu Y.C., Li C., Zhu Z.F., Chen L., Cai H.W., Wang X.K., Li C., Zhu Z.F., Chen L., Cai H.W., Wang X.K., Zhu Z.F., Chen L., Cai H.W., Wang X.K., Chen L., Cai H.W., Wang X.K., Cai H.W., Wang X.K., Wang X.K. Identification and mapping of genes for improving yield from Chinese common wild rice (O. rufipogon Griff.) using advanced backcross QTL analysis. Chin. Sci. Bull. 2002;47:1533–1537. [Google Scholar]
- Li J., Thomson M., McCouch S.R., Thomson M., McCouch S.R., McCouch S.R. Fine mapping of a grain-weight quantitative trait locus in the pericentromeric region of rice chromosome 3. Genetics. 2004;168:2187–2195. doi: 10.1534/genetics.104.034165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H.S., Wang Z., Hu Y., Yang H.H., Gere S., Buetow K.H., Lee M.P., Wang Z., Hu Y., Yang H.H., Gere S., Buetow K.H., Lee M.P., Hu Y., Yang H.H., Gere S., Buetow K.H., Lee M.P., Yang H.H., Gere S., Buetow K.H., Lee M.P., Gere S., Buetow K.H., Lee M.P., Buetow K.H., Lee M.P., Lee M.P. Allelic variation in gene expression is common in the human genome. Genome Res. 2003;13:1855–1862. doi: 10.1101/gr.1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Bennetzen J.L., Bennetzen J.L. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. 2004;101:12404–12410. doi: 10.1073/pnas.0403715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T.F. The genetic architecture of quantitative traits. Annu. Rev. Genet. 2001;35:303–339. doi: 10.1146/annurev.genet.35.102401.090633. [DOI] [PubMed] [Google Scholar]
- Marone M., Mozzetti S., Ritis D.D., Pierelli L., Scambia G., Mozzetti S., Ritis D.D., Pierelli L., Scambia G., Ritis D.D., Pierelli L., Scambia G., Pierelli L., Scambia G., Scambia G. Semiquantitative RT-PCR analysis to assess the expression levels of multiple transcripts from the same sample. Biol. Proced. Online. 2001;3:19–25. doi: 10.1251/bpo20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCouch S.R., Teytelman L., Xu Y.B., Lobos K.B., Clare K., Walton M., Fu B.Y., Maghirang R., Li Z.K., Xing Y.Z., Teytelman L., Xu Y.B., Lobos K.B., Clare K., Walton M., Fu B.Y., Maghirang R., Li Z.K., Xing Y.Z., Xu Y.B., Lobos K.B., Clare K., Walton M., Fu B.Y., Maghirang R., Li Z.K., Xing Y.Z., Lobos K.B., Clare K., Walton M., Fu B.Y., Maghirang R., Li Z.K., Xing Y.Z., Clare K., Walton M., Fu B.Y., Maghirang R., Li Z.K., Xing Y.Z., Walton M., Fu B.Y., Maghirang R., Li Z.K., Xing Y.Z., Fu B.Y., Maghirang R., Li Z.K., Xing Y.Z., Maghirang R., Li Z.K., Xing Y.Z., Li Z.K., Xing Y.Z., Xing Y.Z., et al. Development and mapping of 2240 new ssr markers for rice (Oryza sativa L.). DNA Res. 2002;9:199–207. doi: 10.1093/dnares/9.6.199. [DOI] [PubMed] [Google Scholar]
- Murray M.G., Thompson W.F., Thompson W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbitt T.C., Tanksley S.D., Tanksley S.D. Comparative sequencing in the genus Lycopersicon: Implications for the evolution of fruit size in the domestication of cultivated tomatoes. Genetics. 2002;162:365–379. doi: 10.1093/genetics/162.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paran I., Zamir D., Zamir D. Quantitative traits in plants: Beyond the QTL. Trends Genet. 2003;19:303–306. doi: 10.1016/S0168-9525(03)00117-3. [DOI] [PubMed] [Google Scholar]
- Paterson A.H., Bowers J.E., Burow M.D., Draye X., Elsik C.G., Jiang C.X., Katsar C.S., Lan T.H., Lin Y.R., Ming R., Bowers J.E., Burow M.D., Draye X., Elsik C.G., Jiang C.X., Katsar C.S., Lan T.H., Lin Y.R., Ming R., Burow M.D., Draye X., Elsik C.G., Jiang C.X., Katsar C.S., Lan T.H., Lin Y.R., Ming R., Draye X., Elsik C.G., Jiang C.X., Katsar C.S., Lan T.H., Lin Y.R., Ming R., Elsik C.G., Jiang C.X., Katsar C.S., Lan T.H., Lin Y.R., Ming R., Jiang C.X., Katsar C.S., Lan T.H., Lin Y.R., Ming R., Katsar C.S., Lan T.H., Lin Y.R., Ming R., Lan T.H., Lin Y.R., Ming R., Lin Y.R., Ming R., Ming R., et al. Comparative genomics of plant chromosomes. Plant Cell. 2000;12:1523–1539. doi: 10.1105/tpc.12.9.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna W., Emberton J., Ogden M., SanMiguel P., Bennetzen J.L., Emberton J., Ogden M., SanMiguel P., Bennetzen J.L., Ogden M., SanMiguel P., Bennetzen J.L., SanMiguel P., Bennetzen J.L., Bennetzen J.L. Structural analysis of the maize Rp1 complex reveals numerous sites and unexpected mechanisms of local rearrangement. Plant Cell. 2002;14:3213–3223. doi: 10.1105/tpc.006338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter T.E., Pryor T.J., Bennetzen J.L., Hulbert S.H., Pryor T.J., Bennetzen J.L., Hulbert S.H., Bennetzen J.L., Hulbert S.H., Hulbert S.H. New rust resistance specificities associated with recombination in the Rp1 complex in maize. Genetics. 1995;141:373–381. doi: 10.1093/genetics/141.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Burr B., Burr B. International rice genome sequencing project: The effort to completely sequence the rice genome. Curr. Opin. Plant Biol. 2000;3:138–141. doi: 10.1016/s1369-5266(99)00047-3. [DOI] [PubMed] [Google Scholar]
- Scherrer B., Isidore E., Klein P., Kim J.S., Bellec A., Chalhoub B., Keller B., Feuillet C., Isidore E., Klein P., Kim J.S., Bellec A., Chalhoub B., Keller B., Feuillet C., Klein P., Kim J.S., Bellec A., Chalhoub B., Keller B., Feuillet C., Kim J.S., Bellec A., Chalhoub B., Keller B., Feuillet C., Bellec A., Chalhoub B., Keller B., Feuillet C., Chalhoub B., Keller B., Feuillet C., Keller B., Feuillet C., Feuillet C. Large intraspecific haplotype variability at the Rph7 locus results from rapid and recent divergence in the barley genome. Plant Cell. 2005;17:361–374. doi: 10.1105/tpc.104.028225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R., Messing J., Messing J. Gene expression of a gene family in maize based on noncollinear haplotypes. Proc. Natl. Acad. Sci. 2003;100:9055–9060. doi: 10.1073/pnas.1032999100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley S.D., Grandillo S., Fulton T.M., Zamir D., Eshed T., Petiard V., Lopez J., Beck-Bunn T., Grandillo S., Fulton T.M., Zamir D., Eshed T., Petiard V., Lopez J., Beck-Bunn T., Fulton T.M., Zamir D., Eshed T., Petiard V., Lopez J., Beck-Bunn T., Zamir D., Eshed T., Petiard V., Lopez J., Beck-Bunn T., Eshed T., Petiard V., Lopez J., Beck-Bunn T., Petiard V., Lopez J., Beck-Bunn T., Lopez J., Beck-Bunn T., Beck-Bunn T. Advanced backcross QTL analysis in a cross between an elite processing line of tomato and its wild relative L. pimpinellifolium. Theor. Appl. Genet. 1996;92:213–224. doi: 10.1007/BF00223378. [DOI] [PubMed] [Google Scholar]
- Tatusova T.A., Madden T.L., Madden T.L. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G., Plewniak F., Jeanmougin F., Higgins D.G., Jeanmougin F., Higgins D.G., Higgins D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii K.U. Leucine-rich repeat receptor kinases in plants: Structure, function, and signal transduction pathway. Int. Rev. Cytol. 2004;234:1–46. doi: 10.1016/S0074-7696(04)34001-5. [DOI] [PubMed] [Google Scholar]
- Wang R.L., Stec A., Hey J., Lukens L., Doebley J., Stec A., Hey J., Lukens L., Doebley J., Hey J., Lukens L., Doebley J., Lukens L., Doebley J., Doebley J. The limits of selection during maize domestication. Nature. 1999;398:236–239. doi: 10.1038/18435. [DOI] [PubMed] [Google Scholar]
- Xiao J.H., Grandillo S., Ahn S.N., McCouch S.R., Tanksley S.D., Li J.M., Yuan L.P., Grandillo S., Ahn S.N., McCouch S.R., Tanksley S.D., Li J.M., Yuan L.P., Ahn S.N., McCouch S.R., Tanksley S.D., Li J.M., Yuan L.P., McCouch S.R., Tanksley S.D., Li J.M., Yuan L.P., Tanksley S.D., Li J.M., Yuan L.P., Li J.M., Yuan L.P., Yuan L.P. Genes from wild rice improve yield. Nature. 1996;384:223–224. [Google Scholar]
- Yan H., Yuan W.S., Velculescu V.E., Vogelstein B., Kinzler K.W., Yuan W.S., Velculescu V.E., Vogelstein B., Kinzler K.W., Velculescu V.E., Vogelstein B., Kinzler K.W., Vogelstein B., Kinzler K.W., Kinzler K.W. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- Yano M. Genetic and molecular dissection of naturally occurring variation. Curr. Opin. Plant Biol. 2001;4:130–135. doi: 10.1016/s1369-5266(00)00148-5. [DOI] [PubMed] [Google Scholar]
- Yu J., Hu S., Wang J., Wong G.K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Hu S., Wang J., Wong G.K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Wang J., Wong G.K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Wong G.K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Deng Y., Dai L., Zhou Y., Zhang X., Dai L., Zhou Y., Zhang X., Zhou Y., Zhang X., Zhang X., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]