Abstract
Protein–protein interactions play key roles in protein function and the structural organization of a cell. A thorough description of these interactions should facilitate elucidation of cellular activities, targeted-drug design, and whole cell engineering. A large-scale comprehensive pull-down assay was performed using a His-tagged Escherichia coli ORF clone library. Of 4339 bait proteins tested, partners were found for 2667, including 779 of unknown function. Proteins copurifying with hexahistidine-tagged baits on a Ni2+-NTA column were identified by MALDI-TOF MS (matrix-assisted laser desorption ionization time of flight mass spectrometry). An extended analysis of these interacting networks by bioinformatics and experimentation should provide new insights and novel strategies for E. coli systems biology.
Escherichia coli is one of the best characterized organisms and has served as a model system to study many aspects of bacterial physiology and genetics of fundamental and applied interest. Among the 4339 predicted ORFs including previous prediction (Riley et al. 2006) in E. coli, nearly 50% are experimentally uncharacterized. In addition to functional analysis of individual ORFs, systematic analyses of relationships between constituent elements, such as gene regulatory networks, protein–protein interactions (PPIs), and metabolic networks, have only recently become feasible.
To date, comprehensive PPI studies have been based on the yeast two-hybrid system that detects binary interactions through activation of reporter gene expression (Fields and Song 1989; Uetz et al. 2000; Ito et al. 2001), and pull-down assays that detect large complexes by copurification of prey proteins through their interactions with bait proteins (Gavin et al. 2002; Ho et al. 2002) or protein chips (Zhu et al. 2001). In E. coli, a large-scale protein interaction network was recently carried out by pull-down assay using TAP-tagged bait proteins (Butland et al. 2005). We have already described a comprehensive E. coli ORF library (the ASKA library) as a new resource for E. coli biology (Kitagawa et al. 2005). Here, we report the use of this resource in a systematic analysis of PPIs using pull-down assays. With the advent of matrixassisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometry methods, it is feasible to identify PPIs on a proteome-wide scale.
We have carried out a large-scale identification of protein–protein interactions to gain further understanding of the E. coli model cell at the system level. Because E. coli is one of the best studied organisms, it should also be an excellent target for systems biology (Kitano 2002) and synthetic biology fields (Silver and Way 2004) approaches.
Results and Discussion
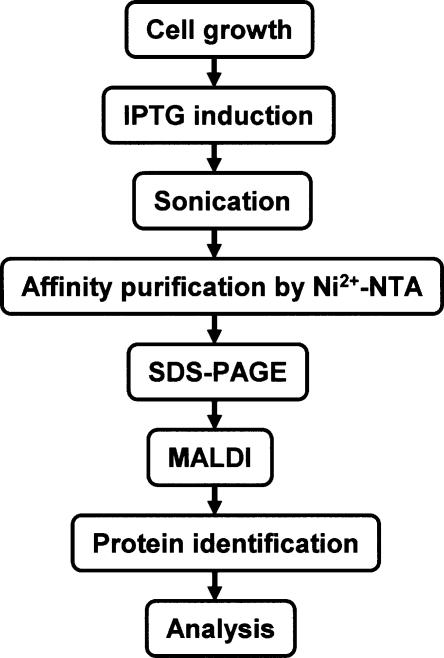
To purify protein–protein interaction partners or protein complexes of E. coli systematically, we developed the strategy depicted in Figure 1. Each His-tagged ORF clone (Kitagawa et al. 2005) was transformed into wild-type E. coli K-12 W3110. Transformants were grown to the early log phase (OD600 = 0.3), and bait protein expression was induced by adding 0.1 mM IPTG. After 2 h, proteins from mildly sonicated cells were purified on a Ni2+-NTA as described in the Methods section. Protein complexes were separated by SDS-PAGE gradient gel electrophoresis; individual protein bands were visualized by Coomassie Brilliant Blue, digested by trypsin in gel slice, analyzed by MALDI-TOF MS, and identified by the PS1 identification program.
Figure 1.
Schematic representation of the sequential steps used for the purification and identification of PPIs (see Methods for details).
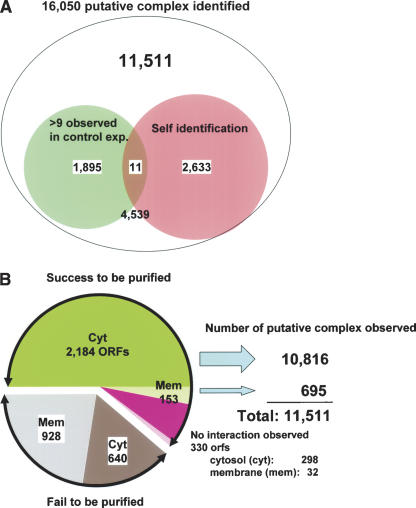
Of 4339 His-tagged bait proteins, 2667 proteins were successfully overproduced and purified. Of these, 2337 bait proteins copurified with an associated partner protein(s) and 330 bait proteins purified without any partners (Fig. 2B; Supplemental Table 2). The remaining 1568 bait proteins failed to be overproduced or purified (Fig. 2B; Supplemental Table 3). Of these, 928 were predicted to be membrane protein by SOSUI software (http://sosui.proteome.bio.tuat.ac.jp/sosuiframe0.html); the remaining 640 are cytosolic proteins. Some of the latter may show toxicity or aggregation when overproduced. In addition, 104 His-tagged bait proteins failed to be identified by mass spectrometry when overproduced as bait proteins and were removed from further analysis (Supplemental Table 9). In the end, 16,050 interacting partners were identified for the 2667 bait proteins examined in our experiments.
Figure 2.
Statistics of putative protein–protein interaction in E. coli. (A) The top number represents the total protein–protein interaction identified in this study. Of the total interactions, the small left circle (pale green) represents the number of interactions with proteins that may have affinity to Ni2+-NTA resin based on our control experiments. The right circle (pale red) shows the number of prey proteins identified as the same protein as histidine-tagged bait protein, which include 11,511 for further analyses. Other 4539 interactions were excluded because their protein assignment was ambiguous. (B) Left: The green region represents the number of cytosolic bait proteins successfully purified. The pink region represents the number of purified bait proteins that had no protein–protein interaction partner. The gray region shows the bait proteins that we failed to purify. Hatched regions represent the baits predicted or known as membrane proteins. Right: Numbers represent the putative PPIs observed.
To reduce identification of false positive interaction, we first carefully performed control experiments (n = 16) with exactly the same procedure described above, except by using the pCA24N vector to identify proteins with an intrinsic affinity for the Ni2+-NTA resin. Several proteins (TreC, GlmS, TufA, TufB, KatE, CstC, HisB, Crp, PykF, RplM) were frequently found (in at least 10 of 16 experiments) to be purified without any bait protein (Supplemental Table 5). In addition, 1906 (1895 plus 11 in Fig. 2A) interactions that identified these proteins as prey were discarded from further analysis.
We also identified 2644 (2633, self-identification, plus 11 in Fig. 2A) protein bands in SDS-PAGE gels that were identical to the His-tagged bait proteins. Whether these represented their endogenous gene products, as would be indicative of multimeric proteins or degradation products of His-tagged proteins, could not be distinguished. In total, we identified 11,511 protein partners in heterologous protein complexes in our analysis (Fig. 2; Supplemental Table 1).
To assess the quality of our results, we compared our data with the DIP database, catalogs of experimentally identified interaction between proteins (Xenarios et al. 2000). There are 447 PPIs in the DIP database with gene names for E. coli K-12. The bait proteins in our experiment matched one or both of 372-protein pairs in the DIP database. Of these, we succesfully detected 58 PPIs (15.6%) in our screening. In a different study, 101 of 447 PPIs (22.6%) were detected using matrix approach (Bader and Hogue 2002).
Recently, data for more than 5000 E. coli PPIs were reported by a large-scale pull-down analysis using TAP- or SPA-tagged bait protein expressed from the chromosome under control of the native promoter (Butland et al. 2005). Their strategy kept cellular stoichiometrics of target proteins; however, 2–4-L cultures were required as starting material to overcome sensitivity problems. We used an overproduction system with multicopy plasmid clones to focus on the identification of potential interactors. Our system, therefore, lost stoichiometry between bait and prey proteins but avoided sensitivity problems. Other differences are the position and size of tag fragments, larger TAP or SPA tags at C terminus versus smaller His tag at N terminus for our study. A total of 521 bait proteins were used in common; these detected 1168 prey proteins by the TAP/SPA system (Butland et al. 2005) and 995 prey proteins in our study, consisting of 5030 and 3088 interactions, respectively. Among these, 218 (4% or 7% interactions are in common. In spite of the different strategies, this overlap is similar to the comparison of the large-scale PPIs in yeast (Ito et al. 2002) and of value to apply for further biochemical and physiological analyses based on the result of both E. coli studies.
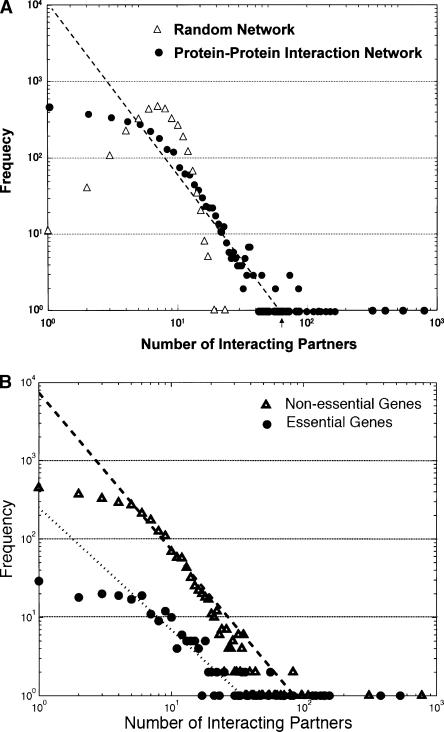
We also evaluated our results using overall topological properties. Although over one-half of the bait proteins had five or fewer interacting partners, some proteins, notably GroL and NadE, had hundreds. The frequency follows a power law, i.e., ∼k−2.23, where k is the number of interacting partners (Fig. 3A). The emergent property of this interaction network is that the average distance of protein pairs in the network will be short and will not be greatly influenced by random deletion of proteins, reflecting how the network evolved (Barabasi and Albert 1999). To evaluate statistically, we constructed a random network having the same number of edges as randomly choosing pairs of proteins from our result. The distribution was quite different from our observation (Fig. 3A); however, creating a random network by randomly shuffling bait proteins and subsequently prey proteins revealed similar topological properties (data not shown).
Figure 3.
Topological properties of protein–protein interaction network. (A) The frequency of all interacting partner proteins. x-axis represents the number of protein partners and y-axis represents the frequency. Experimental results and random generation of protein pairs having the same numbers of interactions are plotted. (B) The comparison of the frequency of interacting partner proteins between essential and nonessential gene products.
We recently identified 303 essential gene candidates in E. coli as genes that we were unable to delete from the chromosome (Baba et al. 2006). Using this information, we compared the number of interacting partners for essential and nonessential genes (Fig. 3B); we found that the connectivity is significantly higher for essential genes (17.8 in average) than for nonessential genes (6.7). This property is consistent with the observation in S. cerevisiae (Yu et al. 2004).
These results provide further support that the network of protein interactions identified in this study is sufficiently reliable and worthwhile to apply for further investigation of a global protein–protein interaction map in E. coli.
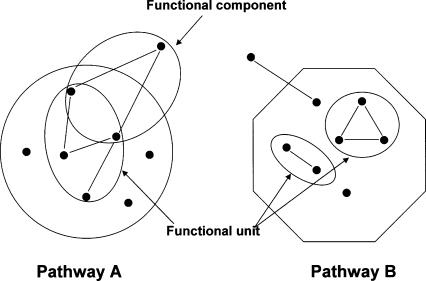
High-throughput protein–protein interaction screening increases the chance of identifying artificial partners that physically interact but that are not in close proximity in the cell, because they are in distinct subcellular locations or expressed at times. To reduce the number of false positives and to aid in extracting the substantial meaningful interactions, we classified interacting proteins by biological function according to the KEGG pathway database (http://www.genome.ad.jp/kegg/pathway.html; Kanehisa et al. 2002). Proteins were first classified into KEGG functional categories and interactions of proteins belonging to the same pathway were then analyzed by a single-linkage clustering algorithm as a way to find functional units (Fig. 4). In this way, we identified 107 functional units involved in metabolic pathways, transcriptional and translational machinery, recombination, ABC transport, or flagella assembly (Supplemental Table 6). To characterize the interaction of proteins not classified into KEGG pathway, we identified proteins that interact with at least two proteins belonging to the same pathway and defined these groups as functional components (Fig. 4). Sixty functional components, consisting of three or four proteins each, were identified. Of these, 42 components are involved in different metabolic pathways, 17 in RNA synthesis, and one in recombination (Supplemental Table 6). Therefore, protein–protein interactions in E. coli metabolic pathways were analyzed with respect to being in a functional unit.
Figure 4.
Schematic diagram for extracting protein–protein interaction pairs using KEGG pathway clusters. ORFs were clustered using KEGG database and encircled as Pathway A and B. Protein–protein interactions within the same cluster may be more plausible than those between different clusters and are defined as belonging to a functional unit. Proteins with an unknown function or proteins outside of a certain cluster that shows more than two independent interactions with proteins in the same cluster may also be plausible and were defined as functional components as encircled and marked by arrows. Node and arc represent protein and interaction, respectively.
Pyruvate dehydrogenase (PDH) is an enzyme complex for conversion of pyruvate to acetyl coenzyme A that funnels glycolytic carbon into the citric acid cycle. PDH is comprised of pyruvate dehydrogenase (E1P), lipoate acetyltransferase (E2P), and lipoamide dehydrogenase (E3) encoded by aceE, aceF, and lpdA, respectively (Quail et al. 1994). We identified a novel interaction of PDH with PdhR (Supplemental Table 6), which is a metabolite responsive repressor of the pdhR–aceE–aceF–lpdA operon. These data suggest that PdhR may function as a sensor of the enzymatic activity or cellular concentration of PDH.
The tricarboxylic acid (TCA) cycle is responsible for the oxidation of acetyl coenzyme A (CoA), which is derived mainly from pyruvate produced by glycolysis. We have identified the SucA–SucB complex that forms 2-ketoglutarate dehydrogenase and the succinate thiokinase complex (SucC–SucD; Buck et al. 1985). We detected an interaction between SucB and SucC in addition to the interactions of SucA–SucB and SucC–SucD, suggesting that a SucA–SucB–SucC–SucD complex may exist. The biological importance of such large complexes is unknown. In the TCA cycle, SucA–SucB and SucC–SucD catalyze separate reactions in a pathway; the interaction of SucA–SucB and SucC–SucD in a large complex may be important to prevent release of substrate from one reaction for the subsequent step.
We also identified a SucC–SdhA (succinate dehydrogenase) interaction, suggestive of formation of a SdhA–SucCD or SdhA–SucABCD complex, for mediating conversion of 2-ketoglutarate to fumarate in the TCA cycle. E. coli has three distinct fumarases encoded by fumA, fumB, and fumC (Bell et al. 1989; Guest 1992; Guest and Russell 1992). FumA and FumB are 90% identical and belong to a novel class of fumarases that are dimeric, oxygen-labile, iron-sulfur proteins; our analysis identified the FumA–FumB interaction (Supplemental Table 6).
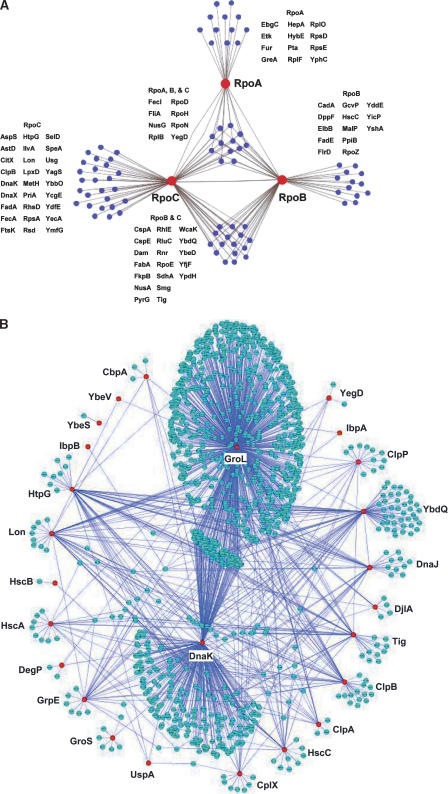
RNA polymerase (RNAP) is a multi-subunit complex for transcription, consisting of α, β and β′ core subunits and one of several σ factors for promoter selection. Our analysis identified many known interactions, including all σ factors except σ38, which regulates most of the genes in stationary phase, NusA, NusG, and Rho, enhancing of RNAP pausing (Lau et al. 1983; Sigmund and Morgan 1988) and Rho-dependent termination (Nehrke et al. 1993; Kainz and Gourse 1998). Network structure was drawn using Cytoscape 2.1 (http://www.cytoscape.org/) in Figure 5. Additionally, we identified several novel interactions including RNAP with RhlE, a putative ATP-dependent RNA helicase, and RNAP with VacB, a 3′- 5′ exoribonuclease (Fig. 5A). Earlier reports suggest that RhlE (Li et al. 1998) and VacB (Cheng et al. 1998) are important for RNA maturation. E. coli cold shock protein CspA, a homolog of eukaryotic Y-box DNA-binding proteins (Wolffe 1994; Sommerville and Ladomery 1996; Yamanaka et al. 1998) that binds strongly to the 110-bp segment of the hns promoter in the presence of RNAP (Brandi et al. 1994), was also identified in interaction with RpoB and RpoC (Fig. 5A), suggesting involvement in the formation of an open complex with RNAP. These data suggest that CspA may act as a transcriptional modulator.
Figure 5.
Subgraph of protein–protein interactions. (A) RNAP interacting protein network. Networks of proteins that interact directly (blue nodes) with each of the RNAP subunits, RpoA, RpoB, and RpoC (red nodes). (B) Molecular chaperone and their target network properties. Red nodes represent major chaperone proteins and blue nodes represent their targets. The largest cluster shows the GroL interaction network and the second largest one shows the network of DnaK.
Yet, we failed to identify interactions between RNAP and transcriptional factors, possibly due to their presence in low concentration of transient interactions with RNAP, or high affinity to target DNA. We did observe interactions between some chaperones (DnaK, ClpB, Tig) and RNAP (Supplemental Table 1). The DnaK (Hsp70) chaperone molecule has been reported to assist the proper folding and assembly of RNAP (Kimura and Ishihama 1996). Our data suggest that not only DnaK but also other chaperones (like ClpB and Tig) may assist the assembly of RNAP as well.
We constructed a global chaperone network based on our protein–protein interaction data showing the target specificities of the chaperones (Fig. 5B; Supplemental Table 7). Chaperones, such as DnaK, GroL, and Trigger Factor (Tig), are required for folding of newly synthesized polypeptides (Deuerling et al. 2003). We identified interactions of 776 ORFs with GroL. The molecular weights of most GroL (HSP60) substrates range from 20 to 80 kDa (data not shown), in agreement with earlier results (Houry et al. 1999). Furthermore, 310 DnaK (HSP70) substrates and 40 Tig substrates were identified. The functional relationship between these chaperone systems and the overlap of DnaK, GroL, and Tig is only partially understood; 92 proteins appear to be substrates of both DnaK and GroL. These proteins may require both systems for folding, degradation, or translocation. Interestingly, only six proteins were identified as overlapping targets of DnaK and Tig, in contrast to earlier observations (Deuerling et al. 2003). This inconsistency may be due to the use of different strains; nine proteins were identified as common substrates for Tig and GroL. These data suggest that Tig shares overlapping protein substrates or sequential process with other chaperones under normal growth conditions.
ClpB is unique among the Hsp100 family proteins of E. coli; ClpB cooperates with DnaK in the solubilization and refolding of unfolded proteins (Glover and Lindquist 1998; Goloubinoff et al. 1999; Mogk et al. 1999; Motohashi et al. 1999; Zolkiewski 1999). Of 43 candidate ClpB substrates, 11 proteins also interact with DnaK and 13 with GroL, suggesting that ClpB cooperates with DnaK/GroL (data not shown). YbdQ is an experimentally uncharacterized protein; however, it is a homolog of the universal stress protein UspA in E. coli and is induced following temperature shift (30°C–42°C; data not shown). We identified 83 target proteins of YbdQ, which mostly overlap with DnaK, DnaJ, and GroL. Our identification of multiple overlapping targets provides further insight into the process of protein folding by chaperones.
Prokaryotic genes are often organized into operons, clusters of genes that are transcribed together from the same mRNA. We estimated 653 transcriptional units based on correlation of expression profiles between genes (H. Maeno and S. Kanaya, unpubl.), which is consistent with the estimated total of 630–700 operons in E. coli (Salgado et al. 2000). The interaction between proteins encoded by the same operon was analyzed and interactions in 52 operons were identified (Supplemental Table 8).
A total of 798 proteins of unknown function were identified as prey. First of all, these data provide evidence that these proteins are synthesized in a cell (Supplemental Table 4). To further characterize interaction of these prey proteins, we carried out domain analysis of frequently identified proteins (data not shown). As mentioned above, we observed that YbdQ contains the conserved domain of universal stress protein (UspA). YciH and YcgE contain the conserved domain of translational initiation factor (COG0023, PF01253) and helix turn helix (COG0789, PF00376) domains, respectively (http://www.ncbi.nlm.nih.gov/COG/old/xognitor.html, http://www.sanger.ac.uk/Software/Pfam/search.shtml). Hence, our data offer insight into the functional prediction of uncharacterized proteins.
In summary, our data from pull-down assay using His-tagged bait proteins represent a global protein–protein interaction map in an E. coli cell and provide a framework for an understanding of cellular systems. The functions of most of these complexes are not known; however, much work over the next few years will expand our knowledge of PPIs and help to analyze the structure and function of protein complexes qualitatively and quantitatively.
Methods
Purification of the bait protein
E. coli K-12 W3110 cells producing histidine-tagged bait proteins were grown at 37°C in 5 mL LB containing Cm (50 μg/mL) medium to OD600 = 0.3. Samples were taken 2 h after the addition of 0.1 mM IPTG. The cells were collected by centrifugation (10,000 rpm, 3 min, 4°C) and resuspended in 400 μL of cold buffer I (50 mM Sodium phosphate [pH 7.0], 200 mM NaCl, Proteinase inhibitor [Roche]). All manipulations were done at 4°C. Crude cell extracts were obtained by sonication (5 × 5 sec, level 3, Astrason ultrasonic processor) and centrifugation (16,000 rpm, 15 min). Cell extracts were loaded onto a 30-μL Nickel (Ni2+)-column (prepared according to manufacturer’s instructions [Qiagen] and equilibrated with buffer I), washed three times with 1 mL of buffer 2 (20 mM imidazole and 0.05% n-octyl-β-glucoside [PIERCE]), and proteins were eluted with buffer 3 (50 mM Tris-HCl [pH 6.8], 2% SDS, 0.1% bromophenol blue, 10% glycerol, 100 mM DTT, 6 M Urea and 250 mM imidazole). Eluted proteins were analyzed by 7.5 ∼15% gradient SDS-PAGE (Bio Craft) followed by Coomassie Brilliant Blue (CBB-R250) staining. Electrophoresis was done at 20 mA current for 3 h by model BE-222 (Bio Craft).
Identification of prey proteins
Protein bands were excised as ∼1-mm2 pieces with a razor blade and transferred into buffer 3 (0.4 M ammonium carbonate and 50% acetonitrile). The gel was washed with Milli-Q water briefly and two times with 100 μL washing buffer (50% acetonitrile, 0.2 M ammonium hydrogen carbonate). After shrinking with 100% acetonitrile, the gel pieces were digested overnight at 37°C with the protease Trypsin (Sigma) (65 μL, 0.003 μg/μL, 10 mM HCl) in the reaction buffer (0.2 M ammonium bicarbonate). The resulting tryptic peptides were extracted from the gel with the elution buffer (50% acetonitrile, 0.3% trifluoroacetic acid) and desalted with ZipTipuC18 (Millipore). Less than 1μL of the sample was spotted to the sample plate of mass spectrometry (Voyager DE-PRO, Applied Biosystems) with matrix solution α-cyano-4-hydroxy cinnamic acid (α-CHCA) (<5 mg/mL, 50% acetonitrile, 0.3% trifluoroacetic acid). Bait and prey proteins were identified by Data Explorer and/or Proteomics solution 1 (PS1) (Applied Biosystems). ORF database used for protein identification by PS1 was constructed by ourselves (GenoBase, http://ecoli.aist-nara.ac.jp).
Comparison with published PPI data
To match our data with those from the use of the TAP/SPA system (Butland et al. 2005), protein names were compared in b-number, using values in Supplemental Table 1 and data provided by the authors. First, our 2337 and their 648 baits were compared to find 521 common baits; those interactions that do not result from the common baits were then excluded. Interactions excluded from our data are identified in Supplemental Table 1. The remaining 3088 and 5030 interactions from our and their experiments with the common baits were used to find 218 common interactions (Supplemental Table 1).
Acknowledgments
We thank Barry L. Wanner for critical reading and suggestions for improvement. This work was supported by a grant from CREST, JST (Japan Science and Technology) and in part from NEDO (New Energy and Industrial Technology Development Organization).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4527806
References
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Tomita M., Wanner B.L., Mori H., Wanner B.L., Mori H., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knock-out mutants—the Keio collection. Mol. Syst. Biol. 2006 doi: 10.1038/msb4100050. http://www.nature.com/msb/journal/v2/n1/full/msb4100050.html [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader G.D., Hogue C.W., Hogue C.W. Analyzing yeast protein–protein interaction data obtained from different sources. Nat. Biotechnol. 2002;20:991–997. doi: 10.1038/nbt1002-991. [DOI] [PubMed] [Google Scholar]
- Barabasi A.L., Albert R., Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Bell P.J., Andrews S.C., Sivak M.N., Guest J.R., Andrews S.C., Sivak M.N., Guest J.R., Sivak M.N., Guest J.R., Guest J.R. Nucleotide sequence of the FNR-regulated fumarase gene (fumB) of Escherichia coli K-12. J. Bacteriol. 1989;171:3494–3503. doi: 10.1128/jb.171.6.3494-3503.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandi A., Pon C.L., Gualerzi C.O., Pon C.L., Gualerzi C.O., Gualerzi C.O. Interaction of the main cold shock protein CS7.4 (CspA) of Escherichia coli with the promoter region of hns. Biochimie. 1994;76:1090–1098. doi: 10.1016/0300-9084(94)90035-3. [DOI] [PubMed] [Google Scholar]
- Buck D., Spencer M.E., Guest J.R., Spencer M.E., Guest J.R., Guest J.R. Primary structure of the succinyl-CoA synthetase of. Escherichia coli. Biochemistry. 1985;24:6245–6252. doi: 10.1021/bi00343a031. [DOI] [PubMed] [Google Scholar]
- Butland G., Peregrin-Alvarez J.M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Peregrin-Alvarez J.M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Starostine A., Richards D., Beattie B., Krogan N., Richards D., Beattie B., Krogan N., Beattie B., Krogan N., Krogan N., et al. Interaction network containing conserved and essential protein complexes in. Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- Cheng Z.F., Zuo Y., Li Z., Rudd K.E., Deutscher M.P., Zuo Y., Li Z., Rudd K.E., Deutscher M.P., Li Z., Rudd K.E., Deutscher M.P., Rudd K.E., Deutscher M.P., Deutscher M.P. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J. Biol. Chem. 1998;273:14077–14080. doi: 10.1074/jbc.273.23.14077. [DOI] [PubMed] [Google Scholar]
- Deuerling E., Patzelt H., Vorderwulbecke S., Rauch T., Kramer G., Schaffitzel E., Mogk A., Schulze-Specking A., Langen H., Bukau B., Patzelt H., Vorderwulbecke S., Rauch T., Kramer G., Schaffitzel E., Mogk A., Schulze-Specking A., Langen H., Bukau B., Vorderwulbecke S., Rauch T., Kramer G., Schaffitzel E., Mogk A., Schulze-Specking A., Langen H., Bukau B., Rauch T., Kramer G., Schaffitzel E., Mogk A., Schulze-Specking A., Langen H., Bukau B., Kramer G., Schaffitzel E., Mogk A., Schulze-Specking A., Langen H., Bukau B., Schaffitzel E., Mogk A., Schulze-Specking A., Langen H., Bukau B., Mogk A., Schulze-Specking A., Langen H., Bukau B., Schulze-Specking A., Langen H., Bukau B., Langen H., Bukau B., Bukau B. Trigger Factor and DnaK possess overlapping substrate pools and binding specificities. Mol. Microbiol. 2003;47:1317–1328. doi: 10.1046/j.1365-2958.2003.03370.x. [DOI] [PubMed] [Google Scholar]
- Fields S., Song O., Song O. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Rick J.M., Michon A.M., Cruciat C.M., Michon A.M., Cruciat C.M., Cruciat C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Glover J.R., Lindquist S., Lindquist S. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Goloubinoff P., Mogk A., Zvi A.P., Tomoyasu T., Bukau B., Mogk A., Zvi A.P., Tomoyasu T., Bukau B., Zvi A.P., Tomoyasu T., Bukau B., Tomoyasu T., Bukau B., Bukau B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. 1999;96:13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest J.R. Oxygen-regulated gene expression in Escherichia coli. The 1992 Marjory Stephenson Prize Lecture. J. Gen. Microbiol. 1992;138:2253–2263. doi: 10.1099/00221287-138-11-2253. [DOI] [PubMed] [Google Scholar]
- Guest J.R., Russell G.C., Russell G.C. Complexes and complexities of the citric acid cycle in. Escherichia coli. Curr. Top. Cell. Regul. 1992;33:231–247. doi: 10.1016/b978-0-12-152833-1.50018-6. [DOI] [PubMed] [Google Scholar]
- Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., Millar A., Taylor P., Bennett K., Boutilier K., Taylor P., Bennett K., Boutilier K., Bennett K., Boutilier K., Boutilier K., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Houry W.A., Frishman D., Eckerskorn C., Lottspeich F., Hartl F.U., Frishman D., Eckerskorn C., Lottspeich F., Hartl F.U., Eckerskorn C., Lottspeich F., Hartl F.U., Lottspeich F., Hartl F.U., Hartl F.U. Identification of in vivo substrates of the chaperonin GroEL. Nature. 1999;402:147–154. doi: 10.1038/45977. [DOI] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y., Ozawa R., Yoshida M., Hattori M., Sakaki Y., Yoshida M., Hattori M., Sakaki Y., Hattori M., Sakaki Y., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Ota K., Kubota H., Yamaguchi Y., Chiba T., Sakuraba K., Yoshida M., Ota K., Kubota H., Yamaguchi Y., Chiba T., Sakuraba K., Yoshida M., Kubota H., Yamaguchi Y., Chiba T., Sakuraba K., Yoshida M., Yamaguchi Y., Chiba T., Sakuraba K., Yoshida M., Chiba T., Sakuraba K., Yoshida M., Sakuraba K., Yoshida M., Yoshida M. Roles for the two-hybrid system in exploration of the yeast protein interactome. Mol. Cell. Proteomics. 2002;1:561–566. doi: 10.1074/mcp.r200005-mcp200. [DOI] [PubMed] [Google Scholar]
- Kainz M., Gourse R.L., Gourse R.L. The C-terminal domain of the αsubunit of Escherichia coli RNA polymerase is required for efficient ρ-dependent transcription termination. J. Mol. Biol. 1998;284:1379–1390. doi: 10.1006/jmbi.1998.2272. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Kawashima S., Nakaya A., Goto S., Kawashima S., Nakaya A., Kawashima S., Nakaya A., Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Ishihama A., Ishihama A. Subunit assembly in vivo of Escherichia coli RNA polymerase: Role of the amino-terminal assembly domain of α subunit. Genes Cells. 1996;1:517–528. doi: 10.1046/j.1365-2443.1996.d01-258.x. [DOI] [PubMed] [Google Scholar]
- Kitagawa M., Ara T., Arifuzzaman M., Ioka-Nakamichi T., Inamoto E., Toyonaga H., Mori H., Ara T., Arifuzzaman M., Ioka-Nakamichi T., Inamoto E., Toyonaga H., Mori H., Arifuzzaman M., Ioka-Nakamichi T., Inamoto E., Toyonaga H., Mori H., Ioka-Nakamichi T., Inamoto E., Toyonaga H., Mori H., Inamoto E., Toyonaga H., Mori H., Toyonaga H., Mori H., Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Kitano H. Systems biology: A brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- Lau L.F., Roberts J.W., Wu R., Roberts J.W., Wu R., Wu R. RNA polymerase pausing and transcript release at the λ tR1 terminator in vitro. J. Biol. Chem. 1983;258:9391–9397. [PubMed] [Google Scholar]
- Li Z., Pandit S., Deutscher M.P., Pandit S., Deutscher M.P., Deutscher M.P. Polyadenylation of stable RNA precursors in vivo. Proc. Natl. Acad. Sci. 1998;95:12158–12162. doi: 10.1073/pnas.95.21.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A., Tomoyasu T., Goloubinoff P., Rudiger S., Roder D., Langen H., Bukau B., Tomoyasu T., Goloubinoff P., Rudiger S., Roder D., Langen H., Bukau B., Goloubinoff P., Rudiger S., Roder D., Langen H., Bukau B., Rudiger S., Roder D., Langen H., Bukau B., Roder D., Langen H., Bukau B., Langen H., Bukau B., Bukau B. Identification of thermolabile Escherichia coli proteins: Prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18:6934–6949. doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K., Watanabe Y., Yohda M., Yoshida M., Watanabe Y., Yohda M., Yoshida M., Yohda M., Yoshida M., Yoshida M. Heat-inactivated proteins are rescued by the DnaK.J-GrpE set and ClpB chaperones. Proc. Natl. Acad. Sci. 1999;96:7184–7189. doi: 10.1073/pnas.96.13.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrke K.W., Zalatan F., Platt T., Zalatan F., Platt T., Platt T. NusG alters ρ-dependent termination of transcription in vitro independent of kinetic coupling. Gene Expr. 1993;3:119–133. [PMC free article] [PubMed] [Google Scholar]
- Quail M.A., Haydon D.J., Guest J.R., Haydon D.J., Guest J.R., Guest J.R. The pdhR-aceEF-lpd operon of Escherichia coli expresses the pyruvate dehydrogenase complex. Mol. Microbiol. 1994;12:95–104. doi: 10.1111/j.1365-2958.1994.tb00998.x. [DOI] [PubMed] [Google Scholar]
- Riley M., Abe T., Arnaud M.B., Berlyn M.K., Blattner F.R., Chaudhuri R.R., Glasner J.D., Mori G., Horiuchi T., Keseler I.M., Abe T., Arnaud M.B., Berlyn M.K., Blattner F.R., Chaudhuri R.R., Glasner J.D., Mori G., Horiuchi T., Keseler I.M., Arnaud M.B., Berlyn M.K., Blattner F.R., Chaudhuri R.R., Glasner J.D., Mori G., Horiuchi T., Keseler I.M., Berlyn M.K., Blattner F.R., Chaudhuri R.R., Glasner J.D., Mori G., Horiuchi T., Keseler I.M., Blattner F.R., Chaudhuri R.R., Glasner J.D., Mori G., Horiuchi T., Keseler I.M., Chaudhuri R.R., Glasner J.D., Mori G., Horiuchi T., Keseler I.M., Glasner J.D., Mori G., Horiuchi T., Keseler I.M., Mori G., Horiuchi T., Keseler I.M., Horiuchi T., Keseler I.M., Keseler I.M., et al. Escherichia coli K-12: A cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 2006;34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado H., Moreno-Hagelsieb G., Smith T.F., Collado-Vides J., Moreno-Hagelsieb G., Smith T.F., Collado-Vides J., Smith T.F., Collado-Vides J., Collado-Vides J. Operons in Escherichia coli: Genomic analyses and predictions. Proc. Natl. Acad. Sci. 2000;97:6652–6657. doi: 10.1073/pnas.110147297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigmund C.D., Morgan E.A., Morgan E.A. Nus A protein affects transcriptional pausing and termination in vitro by binding to different sites on the transcription complex. Biochemistry. 1988;27:5622–5627. doi: 10.1021/bi00415a034. [DOI] [PubMed] [Google Scholar]
- Silver P., Way J., Way J. Cells by design. Scientist. 2004;18:30–31. [Google Scholar]
- Sommerville J., Ladomery M., Ladomery M. Masking of mRNA by Y-box proteins. FASEB J. 1996;10:435–443. doi: 10.1096/fasebj.10.4.8647342. [DOI] [PubMed] [Google Scholar]
- Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Lockshon D., Narayan V., Srinivasan M., Pochart P., Narayan V., Srinivasan M., Pochart P., Srinivasan M., Pochart P., Pochart P., et al. A comprehensive analysis of protein–protein interactions in. Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. Bioessays. 1994;16:245–251. doi: 10.1002/bies.950160407. [DOI] [PubMed] [Google Scholar]
- Xenarios I., Rice D.W., Salwinski L., Baron M.K., Marcotte E.M., Eisenberg D., Rice D.W., Salwinski L., Baron M.K., Marcotte E.M., Eisenberg D., Salwinski L., Baron M.K., Marcotte E.M., Eisenberg D., Baron M.K., Marcotte E.M., Eisenberg D., Marcotte E.M., Eisenberg D., Eisenberg D. DIP: The database of interacting proteins. Nucleic Acids Res. 2000;28:289–291. doi: 10.1093/nar/28.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K., Fang L., Inouye M., Fang L., Inouye M., Inouye M. The CspA family in Escherichia coli: Multiple gene duplication for stress adaptation. Mol. Microbiol. 1998;27:247–255. doi: 10.1046/j.1365-2958.1998.00683.x. [DOI] [PubMed] [Google Scholar]
- Yu H., Greenbaum D., Xin Lu H., Zhu X., Gerstein M., Greenbaum D., Xin Lu H., Zhu X., Gerstein M., Xin Lu H., Zhu X., Gerstein M., Zhu X., Gerstein M., Gerstein M. Genomic analysis of essentiality within protein networks. Trends Genet. 2004;20:227–231. doi: 10.1016/j.tig.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Lan N., Jansen R., Bidlingmaier S., Houfek T., Jansen R., Bidlingmaier S., Houfek T., Bidlingmaier S., Houfek T., Houfek T., et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- Zolkiewski M. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from. Escherichia coli. J. Biol. Chem. 1999;274:28083–28086. doi: 10.1074/jbc.274.40.28083. [DOI] [PubMed] [Google Scholar]