Abstract
Oscillations of gene expression and physiological activity in suprachiasmatic nucleus (SCN) neurons result from auto-regulatory feedback loops of circadian clock gene transcription factors. In the present experiment, we have determined the pattern of PERIOD1, PERIOD2 and CLOCK expression within neuroendocrine dopaminergic neurons (NDNs) of ovariectomized (OVX) rats. We have also determined the effects of per1, per2 and clock mRNA knockdown in the SCN with antisense deoxyoligonucleotides (AS-ODN) on DA release from NDNs. Diurnal rhythms of PER1 and PER2 expression in tuberoinfundibular dopaminergic (TIDA) and periventricular hypophyseal dopaminergic (PHDA) neurons, peaked at CT18 and CT12, respectively. Rhythms of PER1 expression in tuberohypophyseal dopaminergic (THDA) neurons were undetectable. Rhythms of PER2 expression were found in all three populations of NDNs, with greater levels of PER2 expression between CT6-CT12. AS-ODN injections differentially affected DA turnover in the axon terminals of the median eminence (ME), neural lobe (NL) and intermediate lobe (IL) of the pituitary gland, resulting in a significant decrease in DA release in the early subjective night in the ME (TIDA), a significant increase in DA release at the beginning of the day in the IL (PHDA) and no effect in the NL (THDA). AS-ODN-treatment induced a rhythm of DA concentration in the anterior lobe, with greater DA levels in the middle of the day. These data suggest that clock gene expression, particularly PER1 and PER2, within NDNs may act to modulate diurnal rhythms of DA release from NDNs in the OVX rat.
Keywords: clock gene, prolactin, dopamine, suprachiasmatic nucleus, hypothalamus
INTRODUCTION
Dopamine (DA) of hypothalamic origin exerts tonic inhibitory control over prolactin (PRL) secretion (for review see (17)). DA is released directly into hypothalamo-hypophyseal portal blood from three populations of neuroendocrine dopaminergic neurons (NDNs, (5, 17)). These subpopulations include the tuberoinfundibular dopaminergic (TIDA) and tuberohypophyseal neuroendocrine dopaminergic (THDA,A12) neurons with cell bodies in the arcuate nucleus and periventricular hypophyseal dopaminergic (PHDA,A14) neurons with cell bodies in the periventricular region (5, 17). THDA and PHDA axons traverse the pituitary stalk and terminate on fenestrated short portal vessels within the neural (NL) and intermediate (IL) lobes of the pituitary gland (21). TIDA axons terminate on fenestrated capillary beds within the external zone of the median eminence that drain into long portal vessels (LPV), transporting DA to the anterior lobe of the pituitary gland (8, 10). TIDA neurons are well-established as the primary PRL inhibitory neurons, although growing importance has been assigned to both THDA and PHDA neurons in the regulation of PRL secretion (13, 21, 43, 52, 61).
We have observed diurnal rhythms of DA turnover in the nerve terminals of TIDA, THDA and PHDA neurons in the ovariectomized (OVX) rat (see (57, 58)). While both TIDA and PHDA neurons display circadian rhythms of DA release, THDA neurons failed to exhibit a free-running rhythm. In mammals, circadian rhythms are coordinated by the master biological clock located in the suprachiasmatic nucleus (SCN) of the anterior hypothalamus. The SCN contains several thousand individual autonomous oscillators shown to express circadian rhythms of gene expression and physiological activity. The molecular substrates for these sustained endogenous rhythms are a series of tightly regulated transcriptional/translational negative feedback loops of circadian clock gene transcription factor expression (15, 55). Several circadian clock gene transcription factors have been cloned in mammals, including the period gene family, clock, and bmal1 (15, 55). Recent data suggest that neural targets of the SCN, the primary circadian oscillator, express the putative clock genes with a circadian rhythm (1, 22, 23, 75, 76). These tissues appear to function as “damped” oscillators that are both entrained by photic cues transduced by the SCN and actively stimulated to maintain rhythmicity via SCN afferents (76). Therefore, in the absence of SCN input, these tissues cannot sustain a circadian rhythm of clock gene expression for more than 3-5 days, or alternatively, display desynchronized rhythms of gene expression masked by SCN-mediated coordination. These areas include the ARN, paraventricluar nucleus (PVN), pineal gland and pituitary gland (76). However, a notable exception to this pattern is the olfactory bulb, which appears to maintain free-running oscillations of PER1 expression in SCN lesioned rats (22). Moreover, recent experiments suggest that cultured NIH3T3 fibroblasts express the gene Rev-erb⟨ in a self-sustained cell-autonomous rhythm that continues in daughter cells after mitosis (48).
Recent reports suggest that NDNs within the rostral arcuate nucleus (RARN) express PER1 with a diurnal rhythm (33). As several studies indicate that clock genes play an active role in cellular physiology through regulation of protein-protein interactions and gene transcription, it seems plausible that clock genes may act within NDNs to drive expression of enzymes responsible for DA synthesis within the pre-synaptic cell and/or release of DA from the presynaptic terminal (18, 24, 69). A corresponding rhythm of clock gene expression within NDNs would suggest that NDNs may act as semi-autonomous or damped circadian oscillators. In fact, a rudimentary examination of the hypothalamic-specific promoter region of the tyrosine hydroxylase gene reveals a significant (>10) number of E-box (CACGTG)-like sequence motifs ((42) and Sellix and Freeman unpublished observation). CLOCK:BMAL1 heterodimers drive gene transcription through DNA recognition at the E-box motif, E-box sequences within the TH promoter may suggest a level of clock-controlled gene expression (CCG) in NDNs.
Given the novel and diverse rhythms of DA turnover we have observed from the TIDA, THDA and PHDA neurons under a standard 12:12 L:D cycle, we hypothesized that rhythms of clock gene expression in NDNs neurons correspond to the rhythmic synthesis and release of DA from nerve terminals. Using Western blot and immunocytochemistry, we have attempted to verify the expression of several of the clock gene products within tissues of the hypothalamo-pituitary-gonadal axis and to further characterize the rhythmic expression of these gene products within each of the three populations of hypothalamic DA neurons in the OVX rat. We have utilized antibodies against the DA synthesis rate-limiting enzyme tyrosine hydroxylase (TH) to identify DAergic neurons within the periventricular nucleus (PeVN, PHDA neurons), RARN (THDA neurons) and dorsomedial ARN (DMARN, TIDA neurons) and specific primary antibodies against PER1, PER2 and CLOCK to identify those DAergic neurons expressing clock genes in their nucleus under a 12:12 L:D cycle.
Several reports have shown that mutations affecting the core molecular clock network, including PER1/2, CLOCK and BMAL1, have dramatic and varied effects on circadian rhythms of locomotor activity (3, 25, 35, 59). Therefore, we hypothesize that acute transient knockdown of clock gene expression in the SCN may have a significant effect on the timing and/or amplitude of the diurnal rhythms of NDN activity. The inability of SCN-targeted antisense treatment to disrupt diurnal rhythms of DA turnover would suggest that NDNs maintain the capacity to release DA with an endogenous rhythm in the absence of timing cues from the SCN. In the current experiment we determined the effects of short-duration knockdown of per1, per2 and clock mRNA on the diurnal rhythms of NDN activity, as well as PRL and CORT secretion in the OVX rat. Given the potential relationship between diurnal rhythms of clock gene expression in SCN neurons and rhythms of DA turnover in NDNs, it is reasonable to assume that a molecular lesion of the SCN may lead to transient disruption of clock-controlled gene expression in SCN neurons. Disruption of clock-controlled genes, such as AVP and VIP, in SCN neurons would disrupt the ability of the SCN to communicate photoperiodic cues to target regions such as the mediobasal hypothalamus. We believe that our results will add to our general understanding of the functional role for clock genes in target oscillators within the brain that may or may not depend on the SCN for their synchronization.
Materials and Methods
Animals
Adult female Sprague-Dawley rats ( > 60 days of age) weighing 250-300g (Charles River Labs inc., Wilmington, MA) were housed under a standard 12:12 L:D cycle with lights on at 06:00h with constant temperature (25C) and humidity. Standard rat chow and water were available ad libitum. The room was illuminated with four 40 W fluorescent bulbs, producing a minimum illumination of 100 lux at cage level. During the 12-hour dark phase, animals were handled with the aid of infrared goggles (Unitec Inc, Night Vision Optics, Huntington Beach, CA) and sacrificed in dim red light ( < 1 lux). All experimental protocols were approved by the Florida State University Animal Care and Use Committee (ACUC).
Bilateral Ovariectomy and Analysis of Drinking Rhythms
Animals were anesthetized with halothane and OVX bilaterally. All animals were placed under a standard L:D cycle (lights on 06:00h-18:00h) for 2-3 days for habituation to the home cage. Feeding and drinking patterns are established methods for determining circadian time (CT), a subjective measure of time based on the activity of the animal, independent of the L:D cycle (14, 45, 68). Drinking was measured over the 24-hour day with an automated device (Dilog Instruments, Tallahassee, FL.) counting individual licks in 30-second bins over 24 hrs and the onset of the subjective activity period (CT12) was calculated. CT12 was used as a reference time for tissue collection under a standard 12:12 L:D cycle. The use of the CT scale allowed us to make direct comparisons between data collected in the current experiments and analyses of DA turnover performed under constant conditions in previous published reports (57, 58). Double plotted actograms of drinking activity (12-hour moving average of drinking activity around a central peak of activity) were produced with Circadia software (ver. 2.1.16, Behavioral Cybernetics, Inc., Tallahassee, FL.)
Tissue Preparation for Immunocytochemistry and Digital Microscopy
Animals were deeply anesthetized with Halothane and transcardially perfused through the descending aorta with 60 mls of pre-wash (0.1M PBS pH 7.35 containing 0.5% sodium nitrite and 10,000 I.U./L of heparin), followed by 200 ml of ice-cold 4% paraformaldehyde (Sigma-Aldrich, St. Louis, MO) in 0.1M PBS, pH 7.35. Following perfusion, brains were removed and placed in 4% paraformaldehyde to post-fix at 4C overnight. The following morning brains were blocked immediately anterior to the optic chiasm and just posterior of the mammillary bodies and then cryoprotected in a 20% sucrose solution for 36-48h. Brains were sectioned on a sliding microtome (Richard-Allan Scientific, Kalamazoo, MI) at 40-∝m thickness and collected in 4 adjacent series. Sections were collected in 12 well plates containing cryoprotectant solution (71) and stored at -20C.
Sections were processed for PER1, PER2, CLOCK and tyrosine hydroxylase (TH) immunoreactivity. Each series of sections was rinsed three times for 15 min in 0.1M PBS containing 0.1% Triton-X 100 and 0.1% sodium azide (PBX) to remove cryoprotectant (Triton-X and sodium azide, Sigma-Aldrich, St. Louis, MO). Non-specific binding was blocked in 10%normal goat serum (Chemicon, Temecula, CA) in PBX for 1h. Polyclonal primary antibodies against mouse PER1 (rabbit host, polyclonal used at 1:10,000, kind gift of Dr. David Weaver), mouse PER2 (rabbit host, polyclonal used at 1:1000, Alpha Diagnostics Inc, San Antonio, TX) or mouse CLOCK (goat host, polyclonal used at 1:10,000, Santa Cruz Biotechnology, Santa Cruz, CA) were incubated with monoclonal anti-mouse tyrosine hydroxylase (1:10,000, Chemicon, Temecula, CA) for 48h at 4C on a rotating bench top shaker. Mouse antigens were chosen due to the greater than 90% homology in amino acid sequence between mouse, rat, and human circadian clock proteins ((3, 16, 37, 55), NCBI Genebank). All primary and secondary antibodies were diluted in PBX. Sections were washed three times for 10 min with PBX between each step. Donkey anti-rabbit or anti-goat CY3 (Excitation=550 nm, Emission=570 nm) conjugated and anti-mouse CY2 (Excitation=492 nm, Emission=510 nm) conjugated secondary antibodies were added (1:600 in PBX, Jackson Immunochemicals, West Grove, PA) and sections were again incubated at 4C for 12-18h. Sections were then rinsed with PBX three times for 15 min, mounted and coverslips were applied with diluted aqua-polymount (Polysciences, Warrington, PA). After several hours the edges of the coverslips were sealed with nail polish. Controls included sections wherein primary antibody was excluded or PER1, PER2 and CLOCK primary antibodies were pre-absorbed with a substantial amount (10-100 fold higher concentration) of the peptide fragments they were raised against (data not shown).
Neurons in the rostral ARN (THDA), dorsomedial ARN (TIDA), and PeVN (PHDA) were identified as DAergic based on the presence of TH-IR as previously described (11, 38-40). PER1, PER2 or CLOCK and TH double-labeled neurons were identified and counted within the ARN (DMARN and RARN) and PeVN. Although clock proteins translocate back to the nucleus, like most proteins they are formed in the cytoplasm and require phosphorylation and dimerization to reach the nucleus (37). However, their transcriptional activation/repression function occurs within the nucleus. Therefore, we counted PER1, PER2 and CLOCK-IR nuclei within TH-IR neurons. Given the absence of nuclear staining for TH, we were able to clearly and efficiently identify clock protein-IR nuclei in TH-IR neurons (see Fig. 1). Images were taken with a Leica DMLB compound stereomicroscope fitted with short-pass filters (CY2, 488 nm, CY3 596 nm) and a SPOT-RT cooled CCD camera attached to a microcomputer. Image acquisition and analysis was conducted using Metamorph software (Universal Imaging, Downingtown, PA.). Grayscale images of circadian clock gene and TH-IR were overlaid and pseudocolored in Metamorph. For display purposes, representative videomicrographs were taken of each region for each clock protein with a Zeiss 510 confocal laser scanning microscope at 1∝m section thickness.
Figure 1.
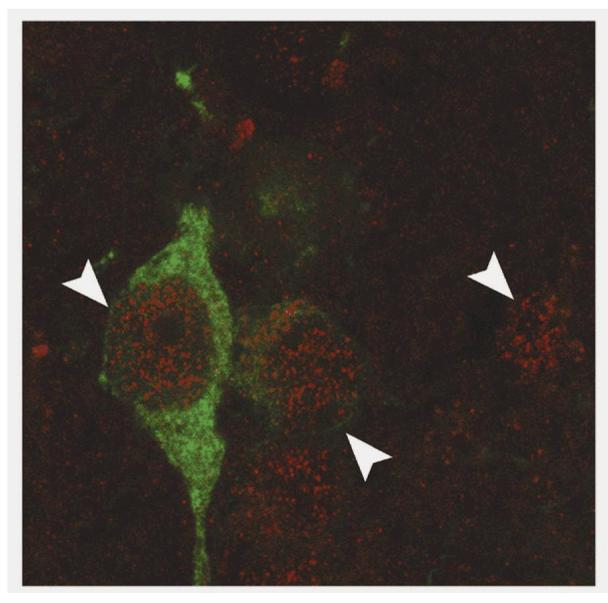
Clock gene expression within the nucleus of neuroendocrine dopaminergic neurons. CLOCK immunoreactivity (red stain; white arrowheads) within the nucleus of NDNs (TH-IR neuron; green stain). An absence of granular staining within the nucleolus is a clear indicator of nuclear staining. Several non-TH-IR neurons within the hypothalamus were also identified as CLOCK-IR.
Stereotaxic implantation of Bilateral SCN Cannulae and Intra-SCN Injection of Deoxyoligonucleotides (ODN)
A minimum of five days after OVX, rats were anesthetized (100∝l / 100g weight) with a mixture of ketamine (49mg/ml)/ xylazine (1.8mg/ml) and implanted stereotaxically with bilateral stainless steel guide tubes (1.5mm apart, 9.5mm in length, 27 gauge) whose tips were placed at the dorsal border of the SCN (0.8 mm posterior to bregma, 7.9 mm ventral to the dorsal surface of the dura mater). Bilateral 33 gauge solid steel mandrils were placed inside the guide tubes and dust caps were used to secure the apparatus. Animals were allowed to recover on a heated pad and returned to their home-cage for 2-3 days. After thorough recovery, animals were transferred, during the light portion of the L:D cycle between 1000h and 1400h, to the lickometer device and allowed to habituate for a minimum of 24h. Animals were placed in the Lickometer device in a 12:12 L:D cycle with lights on from 0600h-1800h for 5 days. On the fifth day under a 12:12 L:D cycle animals were injected intra-SCN with clock gene antisense cocktail. Briefly, animals were anesthetized with Halothane and 33 gauge bilateral mandrils were removed. Bilateral internal cannulae (33 gauge, 10.5 mm length, 1 mm extension) were inserted into the guide tubes and 800 nl of antisense deoxyoligonucleotides (AS-ODN) or random sequence oligos (RS-ODN) were injected at 200 nl/min with two 1∝l Hamilton syringes attached to an automated syringe pump (Kd Scientific, Fisher Scientific, Fair Lawn, NJ). AS-ODNs were generated against the 5’ transcription start site (5’INI) and 3’ cap site of per1, per2 and clock mRNA. Sequences for mper1 (Genebank accession number: NM 011065) and mper2 (Genebank accession number: NM 011066) AS-ODN were slightly modified from those used by Akiyama and colleagues (2, 70) in order to slightly increase the GC content of the oligonucleotide. Clock AS-ODNs were developed independently in our laboratory using the known sequence of clock mRNA (Genebank accession number: NM 007715). Per1, per2 and clock AS-ODN and RS-ODN sequences are listed in Table 1. According to the protocol developed by Akiyama et al (2, 70), we verified both time and dose dependent effects of AS-ODN on PER1, PER2 and CLOCK expression in the SCN with immunoblots for each clock protein (data not shown).
Table 1.
Antisense and random sequence deoxyoligonucleotide sequences
| Antisense Sequence | |
| Per1 - 5'INI | CCT*TCTAGGGGACCACT*CAT |
| Per1 – 3'CAP | GGT*GCTGTTTTCTTCTG*CAG |
| Per2 – 5'INI | TAT*CCATTCATGTCGGG*CTC |
| Per2 – 3'CAP | GAC*ACAAGCAGTCAAC*AAA |
| Clock – 5'INI | CAG*CTTTACGGTAAACAA*CAT |
| Clock – 3'CAP | AAG*GGTCAGTCAGGCT*GTC |
| Random Sequence | |
| Per1 – RS | GCT*CTGGTCTAGTACC*CTA |
| Per2 – RS | ATC*TGCTACTAGGTTC*GTC |
| Clock – RS | ACC*GTACTACTTCGGCT*GTC |
Indicates phosphorothioate linkage within the oligonucleotide sequence.
Preliminary experiments verified a significant knockdown of PER1, PER2 and CLOCK expression in the SCN by more than 60% within 6h of injection with 3 nmoles of AS-ODN (2.5 ∝g/∝l, 800 nl injection, data not shown). Per1, per2 and clock mRNA levels recovered to control values by 12h post-injection and remained at normal levels at 36 and 48h after infusion of AS-ODN (data not shown). Therefore, in order to verify the acute effects of clock gene mRNA knockdown all experimental animals were injected 6h before sacrifice. Control animals were injected with random-sequence ODNs with the same nucleotide content (%AGCT) as the AS-ODN but are not complementary to clock gene mRNA sequences (verified with Primer 3.1, MIT, AS-ODN and RS-ODN sequences listed in Table 1). Following sacrifice, small tissue blocks containing the bilateral SCN were collected and immunoblotting for PER1, PER2 and CLOCK was used to quantify the effects of AS-ODN and RS-ODN in the SCN. Optical density for each gene was normalized to ®-actin loading control for each sample and densitometry was carried out on a Bio-rad gel documentation system (Bio-rad, Hercules, CA). Optical density (OD units/mm2) of PER1, PER2, CLOCK and actin loading controls were determined and used to calculate relative protein abundance. Data represent the mean relative protein abundance of all AS-ODN and RS-ODN animals used for DA turnover analysis.
Reverse Transcription-Polymerase Chain Reaction (RT-PCR) analysis of clock gene expression
RT-PCR was used to determine the tissue distribution of several clock gene mRNAs in OVX female rat brain and pituitary gland. Animals kept under a standard L:D cycle were briefly anesthetized under hypercapnic (50% CO2) conditions and sacrificed by decapitation ∼ZT8, the approximate peak time of per1 and per2 mRNA expression in the SCN (55). Brains were rapidly removed and placed in a coronal brain matrix on ice (ASI instruments, Warren, MI). Individual 2 mm thick sections containing the SCN or the medial basal hypothalamus (including the ARN and median eminence) were placed on a diethylpyrocarbonate (DEPC) treated sterile glass slide. A single 4 mm2 cube including the entire SCN or ARN were excised from the coronal brain slices and placed in 1 ml of TRIZOL reagent (Invitrogen, Carlsbad, CA.), followed by homogenization on ice. After homogenization, 100 ∝l of chloroform was added and each tube was briefly vortexed and then placed in a centrifuge for 15 min at 10,000 rpm (12,000 x g). The aqueous phase was removed from each tube and pooled according to sample. An equal volume of isopropanol was added to each tube and all tubes were placed in a -80C freezer. After 6 hours at -80C, precipitated RNA was pelleted by centrifugation at 13,000 rpm for 20 minutes at 4C. The remaining isopropanol was removed and the pellets were washed twice with 70% ethanol, allowed to air-dry and then re-suspended in DEPC-treated water. Total RNA concentration was analyzed by UV/vis spectrophotometry and diluted to normalize the sample to 500 ng/∝l, of which 5 ∝g was used for reverse transcription (RT). Messenger RNA was reverse transcribed to cDNA using the SuperScript tm First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Following RT, clock genes were amplified using 2 ∝l of RT reaction with the Platinum tm PCR SuperMix (Invitrogen, Carlsbad, CA.) and gene specific primers for clock genes from mouse sequences including mper1,m per2, mclock and mbmal1 (10 pmoles/reaction, 1 ∝l, final vol. of 50 ∝l). PCR was performed in an MJ Research (Watertown, MA) PTC-200 thermocycler at 94C for 1 min. followed by 35 cycles of amplification (94C for 1 min, 58C for 1 min, 72C for 4 min) and a final extension at 74C for 15 min. Samples were stored at -20C until use. Reaction products were separated with 2% agarose (analytical grade, Invitrogen, Carlsbad, CA) gels in Tris:Acetate:EDTA (TAE, Fisher Scientific, Suwanee, GA) buffer. Gels were stained by immersion in ethidium bromide solution (0.1 ∝g/ml in TAE) for 10 min followed by two 20-30 min TAE rinses. Gels were photographed and analyzed using a KODAK Gel Logic 100 Imaging and Analysis System (Eastman Kodak Company, Rochester, NY). Remaining PCR products were purified with a Qiagen PCR purification Kit (Qiagen, Valencia, CA) and sequenced at the Florida State University core molecular analysis facility. NCBI blast sequence analysis confirmed amplification of rat clock gene products with mouse primers, and supported high homology ( > 90% in all cases) between mouse, rat, and human clock genes.
Western Blot and Immuno-detection of Clock Gene Products in Neuroendocrine Tissues
I. Extraction of clock proteins.
Proteins were extracted from neuroendocrine tissues according to the method of Lee and colleagues (37). Briefly, animals maintained under a standard 12:12 L:D cycle were sacrificed at CT12 under hypercapnic conditions and decapitated (CT12 represents the approximate peak of PER1 and PER2-IR in the SCN, (3, 16, 55)). The brain and pituitary gland were rapidly removed and the brain was placed in a chilled brain-sectioning matrix (ASI Instruments Inc., Warren MI) on ice while the pituitary gland was rapidly frozen in liquid nitrogen. Two mm thick frontal sections of the brain including the SCN and the ARN were made with a sterile razor blade and placed on a chilled glass slide. Using a scalpel, a 3mm x 3mm cube including the SCN and a 3mm x 4mm angled dissection of the ARN were removed, as well as the cerebellum and a majority of the piriform cortex from both hemispheres. Following dissection, tissues were rapidly frozen in liquid nitrogen and stored at -80C until protein extraction. Tissues were thawed on ice in chilled homogenization buffer containing: 20 mM HEPES, 100 mM NaCl, 0.05% triton-X 100, 1mM DTT, 5 mM sodium-® glycerophosphate, 1 mM Na orthovanadate, 1 mM EDTA, 0.5 mM PMSF, and a cocktail of protease inhibitors including aprotonin (10∝g/ml), leupeptin (5∝g/ml) and pepstatin A (2∝g/ml, (37)). Tissue samples were sonicated on ice with a mini-homogenizer and disposable pestle (Kimble-Kontes, Vineland, NJ) using 10-15 strokes (3-4 sec/stroke). After centrifugation at 12,000 x g for 15 min, supernatant was transferred to a fresh tube and re-centrifuged. Supernatant was removed and extracted protein concentration was determined by the micro-modified bichonchoninic acid protein detection system (Pierce, Rockford IL).
II. SDS-PAGE and Electroblotting.
Extracted proteins were resolved by SDS-PAGE electrophoresis according to the established protocol of Lee and colleagues (37). As clock proteins vary significantly in MW (i.e., CLOCK MW ∼85-100 kD, PER1 and PER2 ∼180 kD) we utilized separate gel concentrations for each to maximize resolution (CLOCK 8%, PER1/2 7%). Samples were mixed with equal volume of 2X sample buffer containing: 100 mM Tris-HCl pH 6.8, 4% SDS, 20% glycerol, 5% ®-mercaptoethanol (®-ME), 2 mM EDTA and 0.1 mg/ml bromophenol blue and heated at 95C for 5 min (37). Samples were briefly centrifuged at 12,000g and cooled to room temperature. Samples and MW markers were loaded into 10 well pre-set polyacrylamide gels using the mini-protean II system (Bio-Rad, Hercules CA) and electrophoresis was carried out using 150 V of constant voltage for 45 min-1h in Tris-glycine-SDS buffer (8mM Tris-base, 40 mM glycine, 0.1% SDS). Electroblotting was carried out with standard buffers. Briefly, gels were placed in transfer buffer (Tris-glycine-SDS-methanol, 8mM Tris-base, 40mM glycine, 0.1% SDS and 20% methanol) for 1h to overnight before electroblotting with constant 100V for 90 min. Transfer was verified by visual inspection of Pagemarker Pre-stained molecular weight marker (VWR Inc.) and following brief staining with fast-green protein stain.
III. Immuno-detection
Following verification of successful transfer, blots were placed in 5% non-fat milk for 30 min at RT, followed by primary antibody (anti-mouse CLOCK guinea pig 1:1000, anti-mouse PER1/2 guinea pig 1:1000 (kind gift of Dr. Choogon Lee, The Florida State University College of Medicine, Tallahassee, FL.) for 12-16h at 4C. As described above, mouse antigens were chosen due to the greater than 90% homology in amino acid sequence between mouse, rat and human circadian clock proteins ((3, 16, 37, 55), NCBI Genebank). Blots were rinsed three times for 10 min with Tris-buffered saline with 0.1% Tween-20 (TTBS) and placed in secondary antibody (anti-guinea pig IgG conjugated to horseradish peroxidase at 1:5,000, Jackson Immunolabs, West Grove, PA) for 1h at RT. Following 6-8 10 min washes in TTBS, CLOCK or PER1/2 protein levels were visualized using ECL chemiluminescence (Amersham Inc., Piscataway NJ) according to the manufacturer’s specifications. Verification of clock gene staining within tissue extracts was verified by comparison with protein extracts from liver of wild-type (WT) and PER1/2 double knock-out mice or kidney proteins from WT and CLOCK KO mice (kindly provided by Dr. Choogon Lee, data not shown). Blots were stripped of primary/secondary antibody complex according to the protocol provided by Pierce using their stripping and re-probing buffer and re-probed with anti-actin mouse monoclonal primary antibody (1:1000, Sigma-Aldrich, St. Louis, MO).
Tissue Preparation and Measurement of Dopamine (DA) and Dihydroxyphenylacetate (DOPAC) by High Performance Liquid Chromatography with Electrochemical Detection (HPLC-EC)
Six hours after AS- or RS- ODN injection animals were briefly sedated by inducing hypercapnia (50% CO2: O2) and then rapidly decapitated. Trunk blood was collected and serum samples were frozen at -20C until assayed for PRL and CORT concentrations by RIA. The brain and pituitary gland were quickly removed and placed on a chilled glass slide. The median eminence, neural, intermediate and anterior lobes of the pituitary gland were carefully dissected, placed in homogenization buffer (0.2 N perchlorate with 50 ∝M EGTA) and rapidly (∼30 sec.) frozen in an ArticIce tube transport block (USA Scientific Inc., Ocala FL.). Tissue samples were stored at -80C until assayed for DA and DOPAC. On the day of analysis for catecholamines, tissue samples were thawed and processed for HPLC-EC analysis as previously described (58). The HPLC-EC technique has been well established in our laboratory (12). The concentrations of DA and DOPAC, a primary metabolite of DA, were measured in tissue extracts from the pituitary gland and mediobasal hypothalamus as previously described (58). The amount of catecholamine in each sample was estimated by direct comparison to the area under each peak for known amounts of catecholamine. The amount of 3,4-dihydroxybenzylamine (DHBA, RT = 6.5 min) recovered was compared to the amount of DHBA added as internal standard and corrected for sample loss (usually < 5%). Assay sensitivities are 30 pg for DA and 15 pg for DOPAC. DA turnover is defined as the exocytotic release of DA from neuroendocrine DAergic nerve terminals, DA re-uptake, and the degradation of DA to DOPAC by monoamine oxidase (MAO) in the pre-synaptic terminal (41).
Protein Assay
The amount of protein in samples for HPLC-EC and Western blot analysis were measured using a micro-modified form of the Pierce Bichonchoninic Acid (BCA) Protein Assay Kit (Pierce, Rockford, IL) as previously described (58). Assay sensitivity was 1 ∝g protein and the intra-assay coefficient of variation was 5-10%.
Radioimmunoassay
The concentration of PRL in serum was determined by RIA using NIDDK materials supplied through the National Pituitary Hormone Distribution Program (A.F. Parlow) as previously described (11). Serum concentrations of PRL are expressed as ng/ml in terms of the rat PRL RP-3 standard. Serum corticosterone (CORT) concentration was determined using the commercially available Coat-a-Count® rat corticosterone RIA kit (Diagnostic Products Corp., Los Angeles, CA) according to the manufacturer’s specifications. For both PRL and CORT, assay sensitivity was 1 ng/ml and the inter-assay and intra-assay coefficients of variation were 10% and 5%, respectively.
Experimental Design
In order to establish the relationship between diurnal rhythms of DA turnover in NDN neurons and rhythmic clock gene expression we have identified the pattern of PER1, PER2 and CLOCK-immunoreactivity (IR) in DAergic neurons of OVX animals under a standard 12:12 L:D cycle. We utilized RT-PCR for clock gene products in order to verify their expression within tissues of the HPG axis, including the ARN and pituitary gland. Tissues were obtained from OVX animals perfused at CT0, 6, 12 or 18; the peak times for PER1 and PER2 mRNA expression in the SCN (∼ZT8, 14:00h). Brain sections were stained for PER1, PER2, CLOCK and TH-IR. The percentage of PER1, PER2 or CLOCK/TH double-labeled cells was determined in the PeVN, rostral ARN and dorsomedial ARN. For antisense knockdown experiments, four adult female OVX Sprague-Dawley were housed individually in cages attached to the automated drinking device under L:D conditions for 5 days. On the fifth day under a 12:12 L:D cycle animals were injected with antisense ODN (AS-ODN) or random sequence ODN (RS-ODN). Four animals were each sacrificed six hours after ODN-injection at CT 0,6,9,12,15 or 18 and tissue was collected for HPLC-EC and Western blot analysis. Therefore, animals injected with AS- or RS- ODN at CT18 were sacrificed at CT0 and animals injected at CT0 were sacrificed at CT6. Serum was collected to determine serum PRL and corticosterone (CORT) by RIA.
Data Analysis
The percentage of PER1, PER2 or CLOCK/TH double-labeled cells and the number of single-labeled cells represent the mean + SEM of 5 images/region/animal and a total of 3 animals per time point (15 total images/region/timepoint), as a function of circadian time. Although they exhibit a distinct rhythm, all of our data do not conform to a sine/cosine wave function, which prohibits a non-linear regression analysis to present the data as a function of time. Moreover, as samples were obtained from each animal following decapitation or perfusion with fixative at individual time points over a 24 h period, it is difficult to extrapolate accurate phase and period measures. While it is preferable when performing circadian studies to collect serial samples of individual animals, analyses of recovered tissue preclude such an approach in our experiments. To facilitate direct comparisons between current and previous experiments, all data points regardless of treatment condition were aligned by circadian time. The percentage of PER1, PER2 and CLOCK/TH double-labeled neurons were analyzed with ANOVA for time of day effects, followed by Bonferroni post-hoc statistical tests. Serum PRL, serum CORT, DA turnover in the ME, NL, IL and DA concentration in the AL are expressed as mean (ng/ml, ng/ml and DOPAC:DA ratio, respectively) + SEM of 4 animals and were analyzed with two-way ANOVA for (1) time of day effects, (2) ODN effects and the interaction between time and ODN treatment, followed by Bonferroni paired post-hoc statistical tests. Relative protein abundance from AS-ODN and RS-ODN animals were analyzed with ANOVA for ODN effects, followed by Bonferroni post-hoc tests. In all experiments, significant differences were considered at P values < 0.05. ANOVA were performed and graphs were created with Graph-pad Prism software (Graphpad Software Inc., San Diego, CA.)
RESULTS
Diurnal rhythms of PER1-IR in TIDA and PHDA neurons, but not THDA neurons
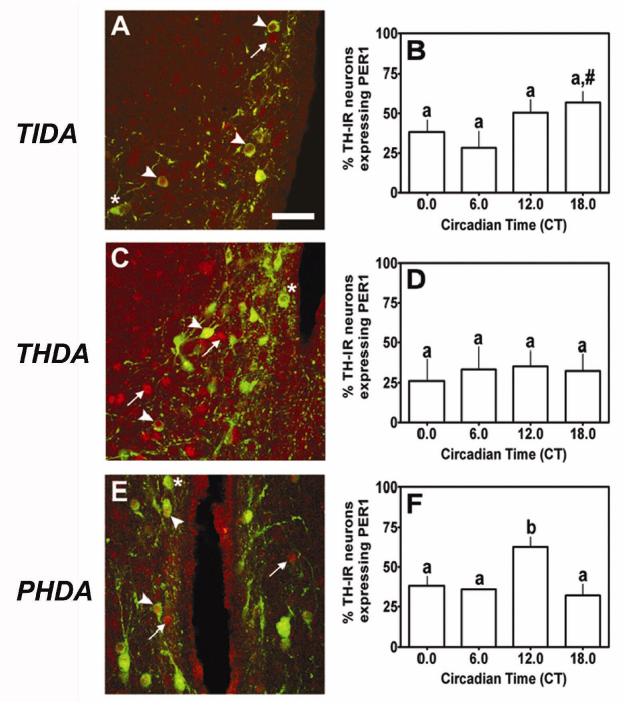
Circadian clock protein immunoreactivity within NDNs is characterized by dense, granular nuclear staining and an absence of staining within the nucleolus (see Fig. 1). PER1-IR nuclei were clearly labeled within TIDA, THDA and PHDA neurons located within the DMARC, RARN and PeVN, respectively (arrowhead, Fig.2A,C,E). More than 50% of NDNs within the DMARN, RARN and PeVN expressed nuclear PER1-IR during the light period. We observed several PER1-IR nuclei that were not double-labeled with TH (arrow, Fig. 2A,C,E), as well as several TH-IR neurons that did not express PER1 (asterisk, Fig. 2A,C,E). Analysis of PER1-IR within the DMARN of the hypothalamus as a function of circadian time (CT) revealed a main effect of time (F=4.85, p < 0.05, Fig. 2B). Pairwise comparisons as a function of circadian time revealed a significant diurnal rhythm of nuclear PER1 expression in TIDA neurons with a peak at CT18 (p < 0.05) compared with a nadir at CT6 (Fig. 2B). Therefore, we observed a significant diurnal rhythm of PER1-IR within TIDA neurons. We have previously determined that TIDA neurons display a diurnal rhythm of DA release with a significant acrophase between CT0 and CT6 (57, 58). Analysis of PER1-IR within the RARN of the hypothalamus did not reveal a significant effect of circadian time (F=0.52, p > 0.05, Fig. 2D), suggesting that THDA neurons do not display diurnal rhythms of nuclear PER1 expression (Fig. 2D). Analysis of PER1-IR within the PeVN of the hypothalamus revealed a significant main effect of time-of-day (F=4.91, p < 0.05, Fig. 2F). Individual comparisons revealed a significant diurnal rhythm of PER1-IR within PHDA neurons characterized by an acrophase at CT12, compared with basal levels at CT0 (p < 0.05), CT6 (p < 0.01) and CT18 (p < 0.01, Fig. 2F). Together, these data suggest that diurnal rhythms of DA synthesis and release from TIDA and PHDA neurons may be regulated by both SCN afferents and local regulation by nuclear PER expression.
Figure 2.
PER1 expression in NDNs under a standard 12:12 L:D cycle. We observed strong nuclear PER1 expression (red; arrowhead) within (A) TIDA neurons (green), (C) THDA neurons and (E) PHDA neurons. Several neurons within these regions expressed TH-IR but failed to show strong PER1 staining (asterisk) or exhibited strong PER1 staining without detectable TH-IR (arrow). PER1 expression peaked in (B) TIDA neurons at CT18 under a standard L:D cycle. (D) We failed to detect a significant rhythm of PER1 expression within THDA neurons. (F) PER1 expression peaked at CT12 under a standard 12:12 L:D cycle in PHDA neurons. In B, D and F, differing letters indicate significance across circadian time and # indicates a significant peak above basal level regardless of adjacent differences (P < 0.05; see results). Scale bar in A = 50 ∝m
Diurnal rhythms of PER2-IR in TIDA, THDA and PHDA neurons
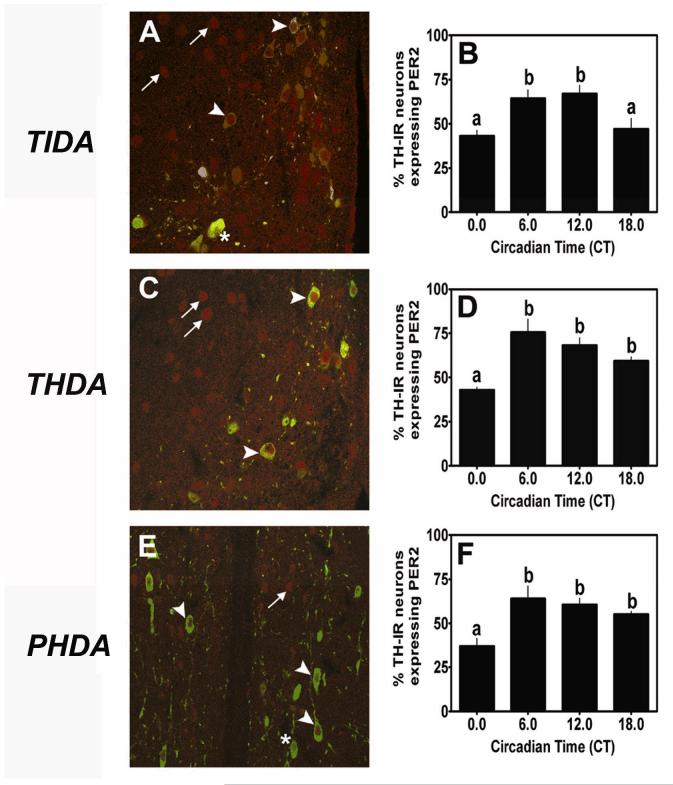
PER2-IR nuclei were clearly labeled within TIDA, THDA and PHDA neurons located within the DMARN, RARN and PeVN, respectively (arrowhead, Fig. 3A,C,E). More than 50% of NDNs within the DMARN, RARN and PeVN expressed nuclear PER2-IR during the subjective day. We observed several PER2-IR nuclei that were not double-labeled with TH (arrow), as well as several TH-IR neurons that did not express PER2 (*, Fig. 3A,C,E). Analysis of PER2-IR within TIDA revealed a main effect of time (F=5.10, p < 0.05, Fig. 3B). Pairwise comparisons exposed a significant diurnal rhythm of PER2-IR within TIDA neurons characterized by peak levels of PER2 expression at CT6 (p < 0.05) and CT12 (p < 0.05), compared with a nadir at CT0 (Fig. 3B). PER2-IR levels returned to basal levels at CT18 (p < 0.05, compared with peak levels at CT12). In contrast with PER1-IR, which peaked at CT18 under a standard L:D cycle in TIDA neurons,PER2-IR peaked between CT6 and CT12 in animals housed under a standard L:D cycle (Fig. 2B vs. Fig. 3B). Analysis of PER2-IR within the RARN of the hypothalamus revealed a main effect of time (F=3.70, p < 0.05, Fig. 3D). Comparisons within animals under a L:D cycle as a function of time delineate a significant diurnal rhythm of PER2 expression within THDA neurons defined by a significant increase from CT0 to CT6 (p < 0.05), followed by a sustained level throughout the remainder of the subjective day (Fig. 3D). Analysis of PER2-IR within the PeVN of the hypothalamus revealed a significant main effect of time (F=6.93, p < 0.01, Fig. 3F). Pairwise comparisons within animals housed under a standard 12:12 L:D cycle as a function of time revealed a significant diurnal rhythm of PER2 expression with a significant peak at CT6 (p < 0.05, compared with a nadir at CT0), followed by a sustained level of PER2 expression throughout the remainder of the subjective day (Fig. 3F). These data suggest a potential role for PER2 within all three populations of NDNs in the regulation of rhythmic DA release and metabolism.
Figure 3.
PER2 expression in NDNs under a standard 12:12 L:D cycle. We observed strong nuclear PER2 expression (red; arrowhead) within (A) TIDA neurons (green), (C) THDA neurons and (E) PHDA neurons. Several neurons within these regions expressed TH-IR but failed to show strong PER2 staining (asterisk) or exhibited strong PER2 staining without detectable TH-IR (arrow). PER2 expression peaked in (B) TIDA neurons at CT6 and CT12 under L:D conditions. (D) PER2 expression peaked at CT6 within THDA neurons under L:D conditions. (F) PER2 expression peaked at CT6 under L:D in PHDA neurons. In B, D and F, differing letters indicate significance across circadian time and # indicates a significant peak above basal level regardless of adjacent differences (P < 0.05; see results). Scale bar in A = 50 ∝m
Constitutive expression of CLOCK protein in TIDA, THDA and PHDA neurons
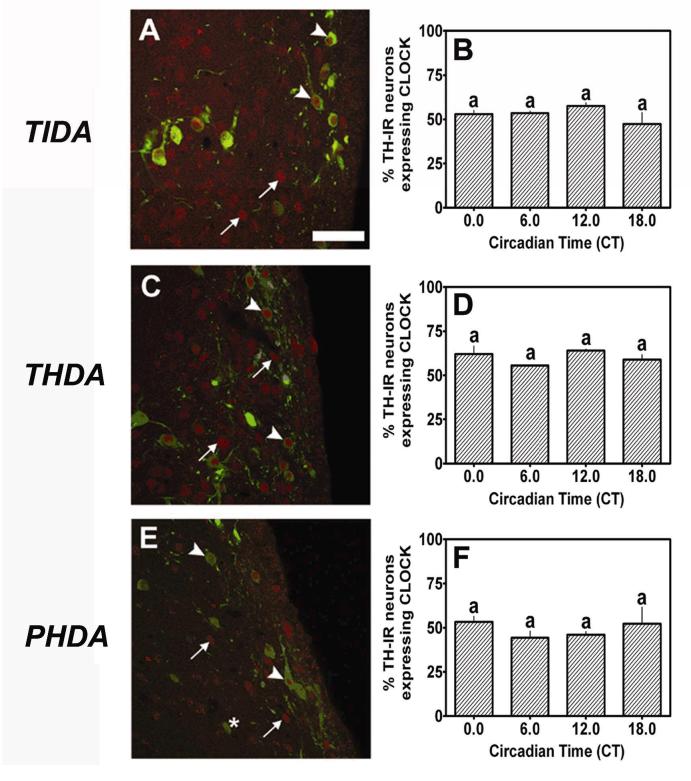
CLOCK-IR nuclei were intensely labeled within TIDA, THDA and PHDA neurons located within the DMARN, RARN and PeVN, respectively (arrowhead, Fig.4A,C,E). More than 50% of NDNs within the DMARN, RARN and PeVN expressed nuclear CLOCK-IR during the subjective day. We observed several CLOCK-IR nuclei that were not double-labeled with TH (arrow), as well as several TH-IR neurons that did not express CLOCK (*, Figs. 4A,C,E). One-factor analysis of CLOCK-IR within the DMARN (F= 2.03, p > 0.05), RARN (F= 0.05, p > 0.05) and PeVN (F=3.07, p > 0.05) failed to reveal a significant main effect of time in any regions. Therefore, we did not observe a significant rhythm of CLOCK expression within TIDA neurons (Fig. 4B), THDA neurons (Fig. 4D), or PHDA neurons (Fig. 4F) under a standard 12:12 L:D cycle. Thus, data from these experiments suggest that CLOCK protein is constitutively expressed within all three populations of NDNs (Figs. 4B,D,F). Given the established interactions between the PER family of proteins and the CLOCK protein in SCN neurons, our data suggest a potential role in the regulation of circadian rhythms of gene expression and physiological activity within NDNs. However, without functional evidence, we cannot assume that the circadian clock genes drive circadian rhythms in NDNs in the absence of signals from the SCN.
Figure 4.
CLOCK expression in NDNs under a standard 12:12 L:D cycle. We observed strong nuclear CLOCK expression (red; arrowhead) within (A) TIDA neurons (green), (C) THDA neurons and (E) PHDA neurons. Several neurons within these regions expressed TH-IR but failed to show strong CLOCK staining (asterisk) or exhibited strong CLOCK staining without detectable TH-IR (arrow). Constitutive CLOCK expression was observed in (B) TIDA, (D) THDA and (F) PHDA neurons under a standard 12:12 L:D cycle. Scale bar in A = 50 ∝m
Acute knockdown of per1, per2 and clock mRNA expression disrupts diurnal rhythms of drinking behavior
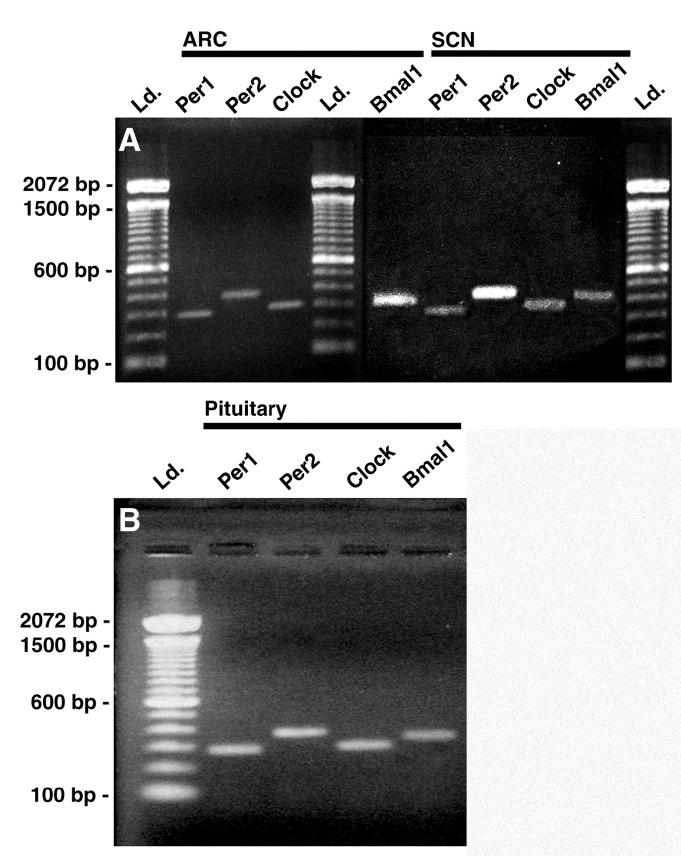
Prior to antisense knockdown of circadian clock gene expression in NDN, we attempted to verify the expression of circadian clock gene mRNA within the ARN and pituitary gland of OVX rats. As shown in Figure 5, per1, per2, bmal1 and clock gene mRNA are expressed within the SCN, ARN and pituitary of OVX rats. Gene products were detected with RT-PCR as described in methods and were verified by DNA-sequencing at our core facility. To verify a functional circadian clock and to determine a reference point for AS-ODN injection and tissue collection, animals were placed in our lickometer device following stereotaxic surgery. The beginning of the 12-hour activity period, identified as CT12, was determined on the two days prior to tissue collection for each animal and averaged to predict the onset of activity on the following day. We predicted CT12 under entrained conditions with an assumed error of 10-15 minutes, given a variance in activity onset among our rats (generally 10-15 minutes from cycle to cycle), which we consider acceptable with a sampling frequency of 2-4 hours. In order to determine the effects of AS-ODN on the rhythm of drinking activity, a pair of animals were placed in a standard 12:12 L:D for a single day, followed by injection of AS-ODN or RS-ODN on the second day under either condition (grey arrowhead, Fig. 6). Animals were allowed to remain in the lickometer device for several days following AS-ODN or RS-ODN injection. In both AS-ODN and RS-ODN injected animals under a standard 12:12 L:D cycle, CT12 was approximately 1730+0.25h (Fig. 6). AS-ODN-treatment eliminated the diurnal rhythm of drinking activity when compared with a RS-ODN controls which recovered after approximately 72h (Fig. 6). Drinking behavior dropped to a minimum during this period, marked by an average number of water bottle licks below 10 licks/10 minute bin. These data suggest that AS-ODN cocktail transiently disrupted the function of the central molecular oscillator within the SCN.
Figure 5.
Reverse-Transcriptase Polymerase Chain Reaction (RT-PCR) amplification of per1, per2, clock and bmal1 mRNA from SCN, ARN and pituitary from OVX rats. Animals were sacrificed at approximately ZT8 under hypercapnic conditions. Individual 2 mm thick tissue punches containing the suprachiasmatic nucleus (SCN) or the medial basal hypothalamus (including the arcuate nucleus (ARN) and median eminence) and the entire anterior lobe of the pituitary were dissected and used for total RNA extraction. RT using oligo-DT primers was carried out to amplify cDNA. Gene specific primers for per1, per2, clock and bmal were used to amplify cDNA. PCR products were separated with agarose gel electrophoresis and stained with ethidium bromide.
Figure 6.
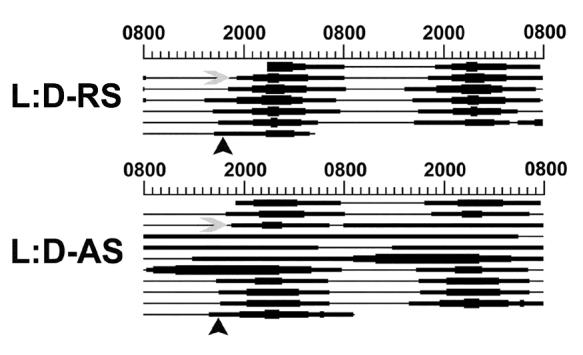
Injection of clock gene AS-ODN disrupts rhythms of drinking behavior. Injection of RS-ODN into the SCN failed to disrupt diurnal rhythms of drinking activity. Following AS-ODN injection into the SCN, diurnal rhythms of drinking activity were abolished for up to 72 hours, marked by a constitutively low amount of drinking. In all figures, black arrowheads indicate approximate CT12. Data shown are individual animals representative of each treatment group.
AS-ODN cocktail against per1, per2 and clock mRNA reduces PER1, PER2 and CLOCK protein expression in the SCN
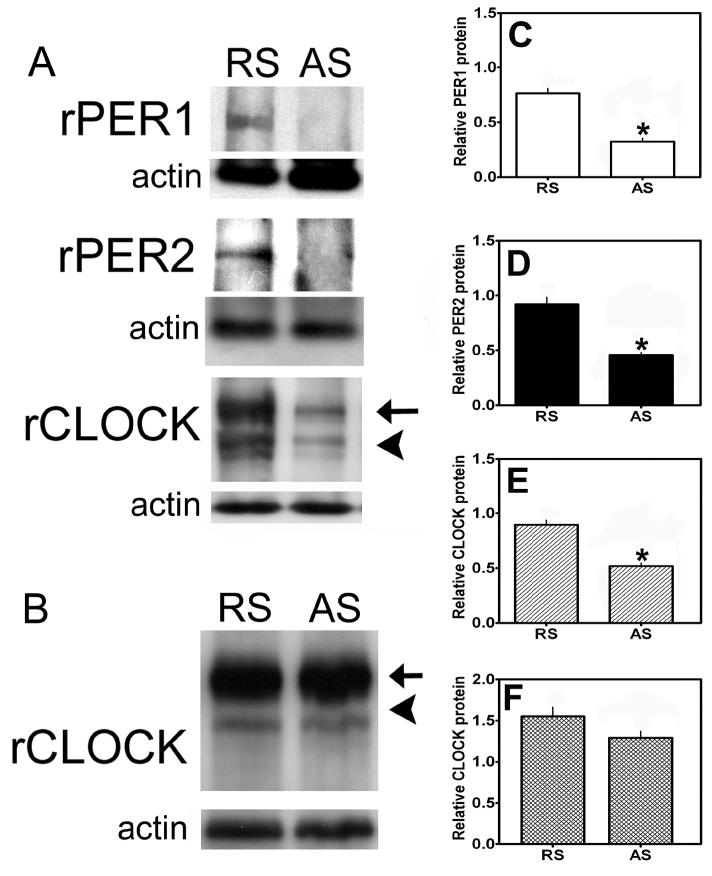
Following injection with AS- or RS- ODN, animals were sacrificed as described above and SCN tissue blocks were collected and clock protein levels were analyzed with Western blot. Given the necessity for acute sacrifice of animals for DA turnover analysis, we could not perfuse the animals and therefore could not verify the physical placement of cannulae in the SCN. However, we verified the effects of AS-ODN on the level of clock proteins in the SCN with Western blot. Thus, animals that received AS-ODN but did not show a significant ( > 50%) knockdown of circadian clock proteins were not included in the experiment. Moreover, we did perfuse animals that were treated with AS- or RS- ODN and analyzed for drinking behavior (as described above) and stained them for PER1, PER2 and CLOCK to verify successful cannula placement in these representative animals (data not shown). One-factor analysis of PER1, PER2 and CLOCK expression within SCN tissue extracts revealed a significant main effect of ODN treatment (F=57.19, p < 0.01). Densitometry of Western blots containing SCN samples from animals injected with AS-ODN or RS-ODN revealed significant decreases in PER1, PER2 and CLOCK protein levels within the SCN following AS-ODN injection (see Fig. 7A). As shown in figure 7C-E, SCN tissue samples from AS-ODN injected rats contained significantly less PER1 (p < 0.01, Fig. 7C), PER2 (p < 0.01, Fig. 7D) and CLOCK (p < 0.01, Fig. 7E) protein than RS-ODN injected controls. Tissue samples from the piriform cortex (PC), a brain region known to express PER1, PER2 and CLOCK but located well outside of the hypothalamus, were collected and analyzed for CLOCK protein expression as a positive control (Fig. 7B). Analysis of CLOCK expression as a function of ODN treatment failed to reveal a significant effect of AS-ODN injection in the SCN on CLOCK expression in the PC (Fig. 7F, F=6.037, p > 0.05). Similar observations were found for PER1 and PER2 expression within the piriform cortex (data not shown). These data support a localized and specific reduction of PER1, PER2 and CLOCK expression within the SCN following precise AS-ODN injection.
Figure 7.
Injection of clock gene AS-ODN reduces PER1, PER2 and CLOCK expression within the SCN. Injection of AS-ODN significantly reduced (A,C) PER1, (A,D) PER2 and (A,E) CLOCK expression within the SCN compared to RS-ODN-treated controls. (B,F) SCN injection of AS-ODN failed to reduce CLOCK gene expression within the piriform cortex (PC). Similar results were observed for PER1 and PER2 (data not shown). In A and B, blots are single tissues from representative animals of each group. In C-F, bars represent pooled values or relative protein abundance units for all animals used in each group (AS and RS -ODN treated, n=24). In A and B, arrows indicate non-specific binding of CLOCK primary antibody, while arrowheads indicate CLOCK specific staining. In C-F, * indicate significant effects of AS-ODN vs. RS-ODN-treatment (P < 0.05).
Acute knockdown of PER1, PER2 and CLOCK protein expression in the SCN differentially affects diurnal rhythms of PRL and CORT secretion
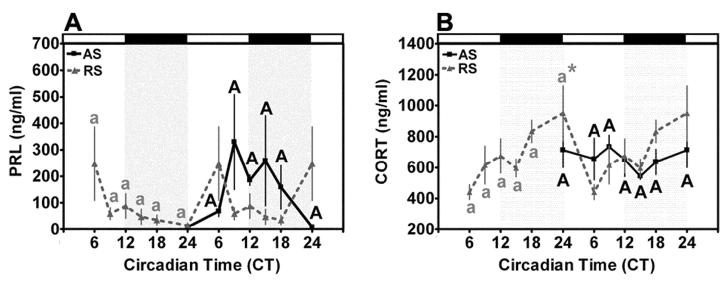
Two-factor analysis of serum PRL levels in AS-ODN and RS-ODN-treated animals maintained under a standard L:D cycle as a function of treatment and circadian time failed to reveal a significant main effect of time (F= 0.84, p > 0.05), treatment (F=1.949, p > 0.05) or an interaction between time x treatment (F=0.71, p > 0.05). In agreement with previous reports from our laboratory and others (12, 38), we failed to detect a significant diurnal rhythm of serum PRL in control RS-ODN-treated OVX rats. Moreover, treatment with AS-ODN failed to affect the timing or magnitude of PRL secretion in OVX animals under a standard L:D cycle (Fig. 8A). Thus, we cannot conclude from these data that AS-ODN-induced disruption of the molecular oscillator exerted a significant effect on PRL secretion in the OVX rat. Twin-factor analysis of serum CORT levels in AS-ODN and RS-ODN-treated animals maintained under a standard L:D cycle as a function of time and treatment revealed a significant effect of time (F=3.04, p < 0.05), treatment (F=7.48, p < 0.01) but not an interaction between time x treatment (F=1.12, p > 0.10). Individual comparisons within RS-ODN-treated animals under a 12:12 L:D cycle established a significant rhythm of CORT secretion with a rise to peak level above baseline at CT24 (p < 0.05 when compared with basal levels at CT6 and CT9, Fig. 8B). Treatment with AS-ODN eliminated this diurnal rhythm of CORT secretion (p > 0.05 across time within AS-ODN-treated animals under a standard 12:12 L:D cycle). Thus, AS-ODN-treatment disrupted the transcriptional feedback loop in SCN neurons, leading to disruption of the diurnal rhythm of CORT secretion in the OVX rat.
Figure 8.
Clock gene knockdown disrupts diurnal rhythms of CORT secretion, but not PRL secretion. (A) Serum PRL did not display a significant diurnal rhythm in RS-ODN animals maintained under a 12:12 L:D cycle. Treatment with AS-ODN failed to affect PRL secretion in these animals as we did not observe a significant difference at any time between AS or RS-injected animals. (B) Serum CORT exhibited a significant diurnal rhythm in RS-ODN-treated rats with a rise to peak at CT0 that was abolished by AS-ODN treatment. Data from RS (dashed line)-treated animals are double-plotted, while data from AS animals (solid line) are single plotted on the right. Data are mean values + SEM for n=4 rats. Differing letters within treatment (lowercase, RS-ODN treated; uppercase, AS-ODN treated) indicate significant effects of time (P < 0.05). * indicates significant peaks above basal level regardless of adjacent differences (P < 0.05). # indicates significant effects of ODN-treatment (P < 0.05).
Acute knockdown of PER1, PER2 and CLOCK protein expression in the SCN differentially affects diurnal rhythms of DA turnover in NDNs and DA concentration within the AL
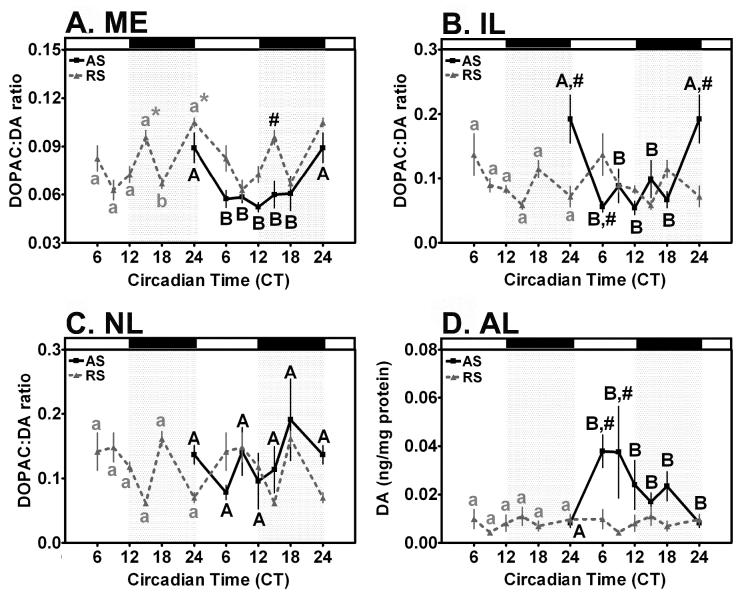
Two-way ANOVA of DA turnover within the ME of animals maintained under a 12:12 L:D cycle as a function of time and ODN treatment revealed a significant effect of time (F=12.97, p < 0.001) and treatment (F=20.25, p < 0.001), but not the interaction between time x treatment (F=0.88, p > 0.05). Individual comparisons within RS-ODN-treated rats under a standard L:D cycle revealed a significant biphasic diurnal rhythm of DA turnover defined by peaks at CT0 (p < 0.05) and CT15 (p < 0.01) above basal levels at CT9 and CT18 (Fig. 9A). Treatment with AS-ODN abolished the peak at CT15 (p < 0.05 when compared with RS-ODN rats), resulting in a U-shaped rhythm with a single peak above baseline at CT0 (p < 0.05, Fig. 9A). Thus, AS-ODN modified the magnitude of the diurnal rhythm of DA turnover in the ME in the early night, when compared with RS-ODN controls.
Figure 9.
Clock gene knockdown differentially affects diurnal rhythms of DA turnover in the ME, NL and IL, and DA concentration in the AL. (A) DA turnover within the ME displayed a significant diurnal rhythm in RS-ODN-injected controls that was influenced by AS-ODN injection into the SCN. (B) DA turnover within the IL displayed a significant diurnal rhythm in both RS-ODN and AS-ODN injected rats. (C) DA turnover within the NL failed to display a rhythm in either RS-ODN-treated controls or AS-ODN-injected rats. (D) DA concentration in the AL of RS-ODN treated animals failed to exhibit a significant diurnal rhythm. Treatment with AS-ODN induced a diurnal rhythm of DA in the anterior lobe under a 12:12 L:D cycle. In A-D, data from RS-ODN-treated animals are double-plotted (dashed line), while data from AS-ODN-treated animals are single plotted on the right (solid line). For A-C, DA turnover is expressed as DOPAC:DA ratio within DA synaptic terminals while (D) shows total DA in the anterior lobe of the pituitary, which contains few, if any, DAergic synapses. Data are mean values + SEM for n=4 rats. Differing letters within treatment (lowercase, RS-ODN treated; uppercase, AS-ODN treated) indicate significant effects of time (P < 0.05). * indicate significant peaks above baseline within treatment regardless of adjacent differences (P < 0.05). # indicate significant effects of ODN-treatment (P < 0.05).
Two-way ANOVA of DA turnover within the IL of animals maintained under a 12:12 L:D cycle as a function of time and ODN treatment revealed a significant effect of treatment(F=3.90, p < 0.05) and an interaction between time and treatment (F=4.10, p < 0.001), but no main effect of time (F=2.15, p=0.07). Pairwise comparisons within RS-ODN-treated controls under a standard 12:12 L:D revealed a significant diurnal rhythm of DA turnover in the IL (Fig. 9B). DA turnover within the IL of RS-ODN controls under a 12:12 L:D cycle exhibited an acrophase at CT0 (p < 0.05), compared with a nadir at CT15. AS-ODN-treatment significantly adjusted the shape of this rhythm by increasing DA turnover within the IL at CT0 (p < 0.001, AS-ODN vs. RS-ODN) and decreasing DA turnover at CT6 (p < 0.05, AS-ODN vs. RS-ODN, Fig. 9B). Thus, AS-ODN affected the magnitude and timing of the diurnal rhythm of DA turnover within the IL of OVX rats by advancing the peak of DA turnover from CT6 to CT0. From these data, we can conclude that AS-ODN treatment modulates rhythms of DA turnover within the IL. These data agree with the results for DA turnover from the ME presented above. As these two populations displayed similar rhythms of DA turnover in previous experiments (58), it is not surprising that they exhibit similar responses with respect to AS-ODN treatment (compare Figs. 9A and 9B)
Two-factor analysis of DA turnover within the NL of animals maintained under a standard L:D cycle as a function of time and ODN treatment did not show a significant effect of time (F=1.76, p > 0.05) and treatment (F=1.86, p > 0.05), or a significant interaction between time x treatment (F=1.11, p > 0.05). In contrast with our prior results, pairwise comparisons within RS-ODN-treated rats maintained in a standard 12:12 L:D cycle failed to reveal significant rhythms of DA turnover in the NL (Fig. 9C). Moreover, animals treated with AS-ODN also failed to display a significant diurnal rhythm of DA turnover in the NL (Fig. 9C). Thus, although AS-ODN injection into the SCN failed to significantly affect the timing or magnitude of DA turnover in the NL, our inability to detect a rhythm of DA turnover in the current experiment obviates any conclusions regarding the effects of AS-ODN on the THDA neurons.
Two-factor analysis of DA concentration within the AL of animals maintained under a standard 12:12 L:D cycle as a function of time and ODN treatment revealed a significant effect of time (F=2.80, p < 0.05) and treatment (F=8.84, p < 0.001), but not an interaction between time x treatment (F=1.18, p > 0.05). Comparisons as a function of time within RS-ODN-treated rats under a standard L:D cycle failed to reveal a significant diurnal rhythm of DA concentration within the AL (Fig. 9D). AS-ODN injection into the SCN induced a significant diurnal rhythm of DA concentration in the AL with a significant acrophase between CT6 and CT9 (p < 0.05),when compared with a nadir at CT0 (Fig. 9D). The diurnal rhythm of DA concentration in the AL induced by AS-ODN-treatment represents a significant increase above RS-ODN controls at both CT6 (p < 0.05) and CT9 (p < 0.01).
DISCUSSION
The purpose of these experiments was to determine the pattern of PER1, PER2 and CLOCK expression within NDNs and ascertain the functional relationship between rhythms of clock gene expression within NDNs and previously determined rhythms of DA turnover within TIDA, THDA and PHDA neurons. Previously, we reported significant circadian rhythms of DA turnover in the OVX rat (58). We hypothesized that these circadian rhythms of DA release, which dictate the timing of the ovarian steroid induced PRL surge on the afternoon of proestrus, are facilitated by autonomous rhythms of clock gene expression within NDNs. We attempted to validate this hypothesis using antisense knockdown to disrupt the transcriptional feedback loops regulating circadian rhythms of activity in SCN neurons. Given the established relationship between the SCN and DAergic target neurons in the ARN, we hypothesized that disruption of the circadian clock in the SCN would abolish diurnal rhythms of DA release from NDN synaptic terminals.
PER1-, PER2- and CLOCK-IR nuclei are clearly labeled within TH-IR neurons in the DMARN, RARN and PeVN. Generally, circadian clock protein-IR was limited to the nucleus and is readily distinguishable from TH-IR cytoplasmic staining. Only those neurons expressing a clear clock protein-IR nucleus were considered clock protein-IR cells. While only TH-IR and clock protein/TH-double labeled cells were counted in the current experiment, we observed numerous PER1, PER2 and CLOCK-IR nuclei within the analyzed regions that were not TH-IR. Under a standard L:D cycle, PER1-IR within TIDA neurons displayed a diurnal rhythm with a peak late in the subjective day. In addition, PER2 expression displayed a diurnal rhythm of expression with significant peaks at CT6 and CT12. We have previously reported a diurnal rhythm of DA turnover within TIDA neurons with a biphasic pattern, defined by peaks at CT6 and CT12 (57, 58). Thus, PER2 expression, but not PER1 expression, peaked in parallel with the time of peak DA turnover within these neurons. Like the SCN, TIDA neurons express CLOCK-IR in a constitutive manner under a standard 12:12 L:D cycle. Based on these cumulative data, we can assume that TIDA neurons may act as dampened oscillators, with light-entrained or light-activated rhythms of clock gene expression. Recent findings agree with this conclusion, suggesting that neurons within the ARN are unable to express free-running rhythms of PER expression in isolated cell culture for more than 2-3 cycles (75, 76).
Unlike TIDA neurons, THDA neurons failed to exhibit a diurnal rhythm of PER1 expression, but did display a diurnal rhythm of PER2 expression with a significant peak at CT6. Like TIDA neurons, THDA exhibit a diurnal rhythm of DA turnover with a biphasic pattern defined by peaks at CT6 and CT12. Therefore, the rhythm of PER2 expression, but not PER1 expression, within THDA neurons corresponds to the rhythm of DA turnover observed for these neurons under a standard L:D cycle. These data are surprising, given our previous report that THDA neurons exhibit clear diurnal rhythms with significant peaks near CT6 and CT12 (57, 58). It remains to be seen whether PER1 and PER2 have redundant roles in the regulation of molecular feedback loops of circadian clock genes and/or DA synthetic enzyme expression in NDNs. Additional evidence regarding the molecular interactions of PER proteins in NDNs is required to either confirm or refute this hypothesis. Like TIDA neurons, THDA neurons express CLOCK-IR in a constitutive manner under L:D conditions. In PHDA neurons, both PER1 and PER2 expression displayed diurnal rhythms with significant peaks at CT12 and CT6, respectively. We have shown that DA turnover within the IL peaks between CT10 and CT14, followed by a trough at CT18 (57, 58). In agreement with data from both TIDA and THDA neurons, PER expression patterns within PHDA neurons correspond to the timing of DA turnover within the IL, both of which occur primarily during the light-phase. Like both TIDA and THDA neurons, PHDA neurons express CLOCK in a constitutive manner. Several experiments have concluded that CLOCK protein is also constitutively expressed within the SCN (7, 26, 28, 44, 54, 60). Therefore, it would appear that constitutive CLOCK expression is a defining feature of the molecular clock found within both the central circadian oscillator in the SCN and its primary central targets, including the NDNs of the hypothalamus. Thus, our results suggest a potential role for the core molecular feedback loops driving the core circadian clock in the timing of molecular and physiological events in NDNs.
Recently, Kriegsfeld and colleagues (33) reported a diurnal rhythm of PER1 expression in neuroendocrine cells of the ARN in the female mouse, using the same primary antiserum, with a significantly greater number of PER1::GFP/TH double labeled cells at CT10 than CT22. These data suggest that PER1 expression peaks in the latter portion of the subjective day and reaches a nadir at or near the middle of the subjective night (CT18-22, (33)). Further, the results of Bae and colleagues revealed that PER1-IR peaked within the SCN at or near CT12 (3, 16, 25). These data would suggest that the peak in PER1-IR in NDNs parallels the peak of PER1 expression within the SCN. However, experiments indicate that PER1 expression in peripheral tissues peaks approximately 6-12 hours after PER expression within the SCN. Our data show that PER1-IR peaks at CT18, approximately 6-12 hours after the peak of PER1 expression we and others have observed within the SCN (3, 16, 25).
To our best knowledge, we are the first to examine the rhythmic expression of these proteins in the NDNs of steroid-depleted OVX rats. Several experiments suggest that ovarian steroid receptors are expressed in SCN neurons and that steroids exert direct effects on gene expression and physiological activity within the SCN (34, 46, 47, 49, 63, 64). Therefore, it is difficult to predict what effect removal of endogenous ovarian steroids would have on clock gene expression within the SCN and NDNs. We can assume that light-entrained rhythms of PER expression, which are highly dependent on neural input from the retina, are likely less dependent on the influence of ovarian steroids. Of course, we cannot rule out the dramatic effects of ovarian steroid hormone withdrawal on steroid-sensitive neuronal oscillators within the SCN and/or NDNs. Future experiments using steroid hormone replacement models will address this fundamental issue.
Several hypotheses could be offered to explain the function of the diurnal rhythms of clock protein expression we have observed. We have determined in previous experiments that NDNs express VPAC2 receptors that are affected by the level of circulating ovarian steroid hormones (19). Further, we have shown that disruption of VIP peptide expression within the SCN affects the activity of NDNs under a standard 12:12 L:D cycle (20). Numerous studies have shown that VIP peptide displays a diurnal rhythm of expression within the SCN characterized by a significant increase in VIP expression in the late subjective night between CT18 and CT22 (4, 27, 51, 53, 62, 65-67, 77). We can conclude from these studies that VIP release from SCN neurons entrains the activity of NDNs in the late subjective night. Moreover, additional evidence suggests VIP induces PER1 and PER2 expression in the SCN during the late subjective night (50). Therefore, we can assume that VIP, released from SCN afferents within the DMARN, RARN and PeVN, binds to VIP type-2 receptors and differentially activates PER1 and PER2 expression through increased intracellular cAMP and CREB mediated signaling (36). However, this hypothesis cannot explain the phase relationships between PER1 and PER2 expression in TIDA neurons, or the absence of PER1 expression in THDA neurons. Additional experiments are necessary to verify the functional redundancy of PER1 and PER2 within NDNs. Although we lack the evidence, we cannot rule out a role for arginine vasopressin of SCN origin in our model. Further experiments are necessary to delineate the precise role of both VIP and AVP in the activation and maintenance of clock gene expression within NDNs.
In the present study we have determined the effects of transient per1, per2 and clock mRNA knockdown on the light-entrained rhythms of PRL secretion, CORT secretion, DA concentration within the anterior lobe of the pituitary and DA turnover within NDN synaptic terminals. We have attempted to ascertain the influence of clock gene-controlled physiological activity and gene expression within the SCN on the rhythm of DA release from NDNs. Using a cocktail of per1, per2 and clock AS-ODN, we have generated a temporary “molecular” lesion of the central circadian oscillator. Based on data from our previous experiments (57, 58), we hypothesized that NDNs may continue to oscillate with a diurnal rhythm in the absence of photoperiodic cues from the SCN. Given that we have already determined that both PER1 and PER2 display diurnal rhythms of expression within TIDA, THDA and PHDA neurons, we hypothesized that clock gene expression within NDNs may act to facilitate diurnal rhythms of DA release from NDNs in the absence of afferent input from the SCN. A negative effect of clock gene AS-ODN injection in the SCN on diurnal rhythms of DA turnover in NDNs would indicate the capacity of the NDN to behave as a semi-autonomous or damped circadian oscillator.
Injection of AS-ODN into the SCN successfully disrupted the diurnal rhythm of drinking activity in the OVX rat. Interestingly, treatment with AS-ODN appeared to induce a long lasting arrhythmia in drinking behavior for up to 72 hours. Although we observed a long duration effect on drinking behavior, we failed to see a similar decrease in PER1, PER2 and CLOCK expression in SCN tissue extracts beyond 12-18 hours (data not shown). Therefore, we can conclude that our transient molecular knockdown exerted long-lasting effects (up to 72 hours) on downstream targets of the SCN, although the overall level of clock protein expression returned to normal within 24 hours. As we did not measure the rhythm of PER1, PER2 or CLOCK expression in the SCN after AS or RS -ODN injection, we cannot rule out significant long-lasting effects of our ODNs on the phase and/or period of the oscillations.
As in previous experiments, we failed to detect a rhythm of PRL secretion in the OVX rat (12, 38). Our inability to detect a significant PRL secretory rhythm precludes determination of AS-ODN treatment affects on PRL secretion. Additional experiments, using both steroid-primed and cycling rats, could provide additional insight into the role of clock genes within the SCN in the control of PRL secretion. Unlike PRL, serum CORT exhibited a significant diurnal rhythm in RS-ODN-treated controls that was disrupted by AS-ODN treatment. Data from numerous experiments suggest that AVPergic afferents of SCN origin terminate on CRH neurons within the medial parvicellular paraventricular nucleus (6, 30-32, 56, 72, 73). Further evidence suggest that AVP and corticotrophin releasing factor (CRF) mRNA are synthesized in PVN neurons with a distinct circadian rhythms, possibly under the direct control of SCN afferents (74). Thus, our ability to disrupt the diurnal rhythm of CORT secretion in OVX rats, like the effects we observed on drinking activity, support successful molecular lesion of the SCN with our AS-ODN cocktail.
In agreement with our previous experiments (57, 58), TIDA and PHDA neurons displayed diurnal rhythms of DA turnover with significant peaks in the early subjective day (CT0 in TIDA and PHDA neurons) and early subjective night (CT15 in TIDA neurons). Treatment with AS-ODN against per1/2 and clock mRNA significantly adjusted the magnitude of DA turnover, leading to a significant change in the shape of the rhythm. Treatment with AS-ODN eliminated the second peak of DA turnover within TIDA neurons that occurred at CT15 (approximately 2030h) but failed to affect the peak of DA turnover at CT0. AS-ODN treatment advanced the acrophase of DA turnover in the IL from CT6 to CT0. Moreover, AS-ODN treatment increased DA turnover at CT15, such that it no longer represents the absolute nadir of DA turnover, as it does in RS-ODN controls. These data suggest that AS-ODN adjusted the magnitude of DA release but failed to completely eliminate the diurnal rhythm. Further, these results suggest an overall increase in the level of DA release, indicating disruption of a potential DA-release inhibiting factor of SCN or hypothalamic origin. Nonetheless, this effect correlates with our previous experiments, suggesting that treatment with AS-ODN against VIP affected the pattern of immediate early gene expression within NDNs (20). Several studies suggest that AVP expression within neurons of the SCN shell or dorsomedial SCN displays a free-running endogenous rhythm under the direct control of CLOCK:BMAL1 enhancers, while VIP expression exhibits a light-entrained, but not free-running, circadian rhythm (29, 53). However, recent studies suggest that VIP mRNA synthesis is under the direct control of the molecular oscillator in the mouse (9). Although we cannot rule out the potential influence of AVP from the SCN shell, we can assume that, if VIP is the primary neurotransmitter of the SCN-NDN tract, that light-induced or endogenous rhythms of VIP release in the mediobasal hypothalamus are responsible for similar rhythms of DA release from NDNs. Within the AL, we failed to detect a significant rhythm of DA concentration within RS-ODN control animals. However, AS-ODN injection induced a significant diurnal rhythm with peaks between CT6 and CT9. As mentioned above, this response is not expected, given the generally variable, basal level of DA release from all three populations in the OVX rat observed in previous experiments (12). Variation in the rhythmic release of DA from each individual population, as a result of ovarian steroid hormone withdrawal and the absence of a significant PRL rhythm may result in the distinct, albeit dissociated rhythm we have detected here. Overall, these data suggest that AS-ODN treatment may reveal a significant diurnal rhythm of DA turnover and total DA concentration by removing rhythmic inhibition of DA release from a presently unknown inhibitory pathway.
We can conclude, therefore, from these studies and others that NDNs may function as damped oscillators, expressing clock genes in a L:D cycle under the direct influence of input from the SCN. Additional experiments are needed to gain a more thorough understanding of the role for each clock gene product in NDNs with regards to the synthesis and release of DA. Further, future experiments will attempt to garner a better understanding of the effects of ovarian steroids on the molecular clock in both the SCN and NDNs. Our current report indicated the presence of circadian clock gene products in numerous non-DAergic neurons within the arcuate and periventricular nuclei. Future experiments will provide us with a better understanding of the form and function of these neurons, as well as their role as potential target oscillators in the hypothalamus. For example, we cannot make a definitive claim regarding the function of NDNs as damped oscillators without evidence that they display free-running and cell-autonomous rhythms of gene expression and electrical activity. Our data show that AS-ODN treatment significantly reduced PER1, PER2 and CLOCK expression within NDNs and affected the diurnal rhythms of drinking behavior, CORT secretion and NDN activity in the OVX rat. Therefore, our current data and previous evidence suggest that the diurnal rhythms of DA release in TIDA, THDA and PHDA neurons are semi-dependent on rhythmic output from SCN neurons. Evidence suggests that disruption of clock-controlled genes, such as VIP, either directly via VIP AS-OSN as in previous experiments (20) or indirectly through disruption of per1, per2 and clock transcription (current report), may modulate, but not abolish, rhythms of DA release. This hypothesis relies on information regarding the relationship between clock gene expression in SCN neurons and VIP mRNA synthesis (20). Further experiments are needed to strengthen the potential relationship between clock controlled gene expression in NDNs, VIP synthesis and release from SCN afferents and DA release from NDN synaptic terminals.
Footnotes
This work was supported by grant NIH DK43200 and NIH DA19356
Reference List
- 1.Abe M, Herzog ED, Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. Circadian rhythms in isolated brain regions. J Neurosci. 2002;22:350–356. doi: 10.1523/JNEUROSCI.22-01-00350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akiyama M, Kouzu Y, Takahashi S, Wakamatsu H, Moriya T, Maetani M, Watanabe S, Tei H, Sakaki Y, Shibata S. Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J Neurosci. 1999;19:1115–1121. doi: 10.1523/JNEUROSCI.19-03-01115.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae K, Jin XW, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 4.Ban Y, Shigeyoshi Y, Okamura H. Development of vasoactive intestinal peptide mRNA rhythm in the rat suprachiasmatic nucleus. J Neurosci. 1997;17:3920–3931. doi: 10.1523/JNEUROSCI.17-10-03920.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben Jonathan N, Hnasko R. Dopamine as a prolactin (PRL) inhibitor. Endocr Rev. 2001;22:724–763. doi: 10.1210/edrv.22.6.0451. [DOI] [PubMed] [Google Scholar]
- 6.Buijs RM, Wortel J, Van Heerikhuize JJ, Feenstra MG, Ter Horst GJ, Romijn HJ, Kalsbeek A. Anatomical and functional demonstration of a multisynaptic suprachiasmatic nucleus adrenal (cortex) pathway. Eur J Neurosci. 1999;11:1535–1544. doi: 10.1046/j.1460-9568.1999.00575.x. [DOI] [PubMed] [Google Scholar]
- 7.Chang DC, Reppert SM. The circadian clocks of mice and men. Neuron. 2001;29:555–558. doi: 10.1016/s0896-6273(01)00230-6. [DOI] [PubMed] [Google Scholar]
- 8.Dahlstrom A, Fuxe K. Evidence for the existance of monoaminergic containing neurons in the central nervous system. I. Demonstration of monoamines in cell bodies of brain stem neurons. Acta Physiol Scand. 1964;62(Suppl 232):1–55. [PubMed] [Google Scholar]
- 9.Dardente H, Menet JS, Challet E, Tournier BB, Pevet P, Masson-Pevet M. Daily and circadian expression of neuropeptides in the suprachiasmatic nuclei of nocturnal and diurnal rodents. Brain Res Mol Brain Res. 2004;124:143–151. doi: 10.1016/j.molbrainres.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Demarest KT, Moore KE. Lack of a high affinity transport system for dopamine in the median eminence and posterior pituitary. Brain Res. 1979;171:545–551. doi: 10.1016/0006-8993(79)91060-6. [DOI] [PubMed] [Google Scholar]
- 11.DeMaria JE, Lerant AA, Freeman ME. Prolactin activates all three populations of hypothalamic neuroendocrine dopaminergic neurons in ovariectomized rats. Brain Res. 1999;837:236–241. doi: 10.1016/s0006-8993(99)01667-4. [DOI] [PubMed] [Google Scholar]
- 12.DeMaria JE, Livingstone JD, Freeman ME. Ovarian steroids influence the activity of neuroendocrine dopaminergic neurons. Brain Res. 2000;879:139–147. doi: 10.1016/s0006-8993(00)02763-3. [DOI] [PubMed] [Google Scholar]
- 13.DeMaria JE, Zelena D, Vecsernyes M, Nagy GM, Freeman ME. The effect of neurointermediate lobe denervation on hypothalamic neuroendocrine dopaminergic neurons. Brain Res. 1998;806:89–94. doi: 10.1016/s0006-8993(98)00740-9. [DOI] [PubMed] [Google Scholar]
- 14.Eichler VB, Moore RY. Pineal hydroxyindole-O-methyltransferase and gonadal responses to blinding or continuous darkness blocked by pineal denervation in the male hamster. Neuroendocrinology. 1971;8:81–85. doi: 10.1159/000121995. [DOI] [PubMed] [Google Scholar]
- 15.Emery P, Reppert SM. A rhythmic Ror. Neuron. 2004;43:443–446. doi: 10.1016/j.neuron.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Field MD, Maywood ES, O’Brien JA, Weaver DR, Reppert SM, Hastings MH. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25:437–447. doi: 10.1016/s0896-6273(00)80906-x. [DOI] [PubMed] [Google Scholar]
- 17.Freeman ME, Kanyicska B, Lerant A, Nagy G. Prolactin: structure, function, and regulation of secretion. Physiol Rev. 2000;80:1523–1631. doi: 10.1152/physrev.2000.80.4.1523. [DOI] [PubMed] [Google Scholar]
- 18.Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U. The mammalian circadian timing system: from gene expression to physiology. Chromosoma. 2004;113:103–112. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- 19.Gerhold LM, Horvath TL, Freeman ME. Vasoactive intestinal peptide fibers innervate neuroendocrine dopaminergic neurons. Brain Res. 2001;919:48–56. doi: 10.1016/s0006-8993(01)02993-6. [DOI] [PubMed] [Google Scholar]
- 20.Gerhold LM, Sellix MT, Freeman ME. Antagonism of vasoactive intestinal peptide mRNA in the suprachiasmatic nucleus disrupts the rhythm of FRAs expression in neuroendocrine dopaminergic neurons. J Comp Neurol. 2002;450:135–143. doi: 10.1002/cne.10307. [DOI] [PubMed] [Google Scholar]
- 21.Goudreau JL, Falls WM, Lookingland KJ, Moore KE. Periventricular-hypophysial dopaminergic neurons innervate the intermediate but not the neural lobe of the rat pituitary gland. Neuroendocrinology. 1995;62:147–154. doi: 10.1159/000126999. [DOI] [PubMed] [Google Scholar]
- 22.Granados-Fuentes D, Prolo LM, Abraham U, Herzog ED. The suprachiasmatic nucleus entrains, but does not sustain, circadian rhythmicity in the olfactory bulb. J Neurosci. 2004;24:615–619. doi: 10.1523/JNEUROSCI.4002-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Granados-Fuentes D, Saxena MT, Prolo LM, Aton SJ, Herzog ED. Olfactory bulb neurons express functional, entrainable circadian rhythms. Eur J Neurosci. 2004;19:898–906. doi: 10.1111/j.0953-816x.2004.03117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guillaumond F, Sage D, Deprez P, Bosler O, Becquet D, Francois-Bellan AM. Circadian binding activity of AP-1, a regulator of the arylalkylamine N-acetyltransferase gene in the rat pineal gland, depends on circadian Fra-2, c-Jun, and Jun-D expression and is regulated by the clock’s zeitgebers. J Neurochem. 2000;75:1398–1407. doi: 10.1046/j.1471-4159.2000.0751398.x. [DOI] [PubMed] [Google Scholar]
- 25.Hastings MH, Field MD, Maywood ES, Weaver DR, Reppert SM. Differential regulation of mPER1 and mTIM proteins in the mouse suprachiasmatic nuclei: new insights into a core clock mechanism. J Neurosci. 1999;19:RC11. doi: 10.1523/JNEUROSCI.19-12-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honma S, Ikeda M, Abe H, Tanahashi Y, Namihira M, Honma K, Nomura M. Circadian oscillation of BMAL1, a partner of a mammalian clock gene Clock, in rat suprachiasmatic nucleus. Biochem Biophys Res Commun. 1998;250:83–87. doi: 10.1006/bbrc.1998.9275. [DOI] [PubMed] [Google Scholar]
- 27.Isobe Y, Muramatsu K. Day-night differences in the contents of vasoactive intestinal peptide, gastrin-releasing peptide and Arg-vasopressin in the suprachiasmatic nucleus of rat pups during postnatal development. Neurosci Lett. 1995;188:45–48. doi: 10.1016/0304-3940(95)11391-9. [DOI] [PubMed] [Google Scholar]
- 28.Isojima Y, Okumura N, Nagai K. Molecular mechanism of mammalian circadian clock. J Biochem (Tokyo) 2003;134:777–784. doi: 10.1093/jb/mvg219. [DOI] [PubMed] [Google Scholar]
- 29.Jin X, Shearman LP, Weaver DR, Zylka MJ, De Vries GJ. Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 30.Kalsbeek A, Drijfhout WJ, Westerink BH, Van Heerikhuize JJ, Van der Woude TP, van d V. Buijs RM. GABA receptors in the region of the dorsomedial hypothalamus of rats are implicated in the control of melatonin and corticosterone release. Neuroendocrinology. 1996;63:69–78. doi: 10.1159/000126937. [DOI] [PubMed] [Google Scholar]
- 31.Kalsbeek A, Van Heerikhuize JJ, Wortel J, Buijs RM. A diurnal rhythm of stimulatory input to the hypothalamo-pituitary-adrenal system as revealed by timed intrahypothalamic administration of the vasopressin V1 antagonist. J Neurosci. 1996;16:5555–5565. doi: 10.1523/JNEUROSCI.16-17-05555.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalsbeek A, van d V, Buijs RM. Decrease of endogenous vasopressin release necessary for expression of the circadian rise in plasma corticosterone: a reverse microdialysis study. J Neuroendocrinol. 1996;8:299–307. doi: 10.1046/j.1365-2826.1996.04597.x. [DOI] [PubMed] [Google Scholar]
- 33.Kriegsfeld LJ, Korets R, Silver R. Expression of the circadian clock gene Period 1 in neuroendocrine cells: an investigation using mice with a Per1::GFP transgene. Eur J Neurosci. 2003;17:212–220. doi: 10.1046/j.1460-9568.2003.02431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kruijver FP, Swaab DF. Sex hormone receptors are present in the human suprachiasmatic nucleus. Neuroendocrinology. 2002;75:296–305. doi: 10.1159/000057339. [DOI] [PubMed] [Google Scholar]
- 35.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 36.Laburthe M, Couvineau A. Molecular pharmacology and structure of VPAC receptors for VIP and PACAP. Reg Pept. 2002;108:165–173. doi: 10.1016/s0167-0115(02)00099-x. [DOI] [PubMed] [Google Scholar]
- 37.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 38.Lerant A, Freeman ME. Dopaminergic neurons in periventricular and arcuate nuclei of proestrous and ovariectomized rats: Endogenous diurnal rhythm of Fos-related antigens expression. Neuroendocrinology. 1997;65:436–445. doi: 10.1159/000127207. [DOI] [PubMed] [Google Scholar]
- 39.Lerant A, Freeman ME. Ovarian steroids differentially regulate the expression of PRL-R in neuroendocrine dopaminergic neuron populations: a double label confocal microscopic study. Brain Res. 1998;802:141–154. doi: 10.1016/s0006-8993(98)00583-6. [DOI] [PubMed] [Google Scholar]
- 40.Lerant AA, DeMaria JE, Freeman ME. Decreased expression of fos-related antigens (FRAs) in the hypothalamic dopaminergic neurons after immunoneutralization of endogenous prolactin. Endocrine. 2001;16:181–187. doi: 10.1385/ENDO:16:3:181. [DOI] [PubMed] [Google Scholar]
- 41.Lindley SE, Gunnet JW, Lookingland KJ, Moore KE. 3,4-Dihydroxyphenylacetic acid concentrations in the intermediate lobe and neural lobe of the posterior pituitary gland as an index of tuberohypophysial dopaminergic neuronal activity. Brain Res. 1990;506:133–138. doi: 10.1016/0006-8993(90)91209-y. [DOI] [PubMed] [Google Scholar]
- 42.Liu J, Merlie JP, Todd RD, O’Malley KL. Identification of cell type-specific promoter elements associated with the rat tyrosine hydroxylase gene using transgenic founder analysis. Brain Res Mol Brain Res. 1997;50:33–42. doi: 10.1016/s0169-328x(97)00163-0. [DOI] [PubMed] [Google Scholar]
- 43.Lookingland KJ, Farah JM, Lovell KL, Moore KE. Differential regulation of tuberohypophysial dopaminergic neurons terminating in the intermediate lobe and in the neural lobe of the rat pituitary gland. Neuroendocrinology. 1985;40:145–151. doi: 10.1159/000124066. [DOI] [PubMed] [Google Scholar]
- 44.Maywood ES, O’Brien JA, Hastings MH. Expression of mCLOCK and other circadian clock-relevant proteins in the mouse suprachiasmatic nuclei. J Neuroendocrinol. 2003;15:329–334. doi: 10.1046/j.1365-2826.2003.00971.x. [DOI] [PubMed] [Google Scholar]
- 45.Moore RY, Eichler VB. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 1972;42:201–206. doi: 10.1016/0006-8993(72)90054-6. [DOI] [PubMed] [Google Scholar]
- 46.Morin LP. Effect of ovarian hormones on synchrony of hamster circadian rhythms. Physiol Behav. 1980;24:741–749. doi: 10.1016/0031-9384(80)90406-0. [DOI] [PubMed] [Google Scholar]
- 47.Morin LP, Fitzgerald KM, Zucker I. Estradiol shortens the period of hamster circadian rhythms. Science. 1977;196:305–307. doi: 10.1126/science.557840. [DOI] [PubMed] [Google Scholar]
- 48.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts, cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 49.Nakamura TJ, Shinohara K, Funabashi T, Kimura F. Effect of estrogen on the expression of Cry1 and Cry2 mRNAs in the suprachiasmatic nucleus of female rats. Neurosci Res. 2001;41:251–255. doi: 10.1016/s0168-0102(01)00285-1. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen HS, Hannibal J, Fahrenkrug J. Vasoactive intestinal polypeptide induces per1 and per2 gene expression in the rat suprachiasmatic nucleus late at night. Eur J Neurosci. 2002;15:570–574. doi: 10.1046/j.0953-816x.2001.01882.x. [DOI] [PubMed] [Google Scholar]
- 51.Okamura H, Kawakami F, Tamada Y, Geffard M, Nishiwaki T, Ibata Y, Inouye ST. Circadian change of VIP mRNA in the rat suprachiasmatic nucleus following p-chlorophenylalanine (PCPA) treatment in constant darkness. Brain Res Mol Brain Res. 1995;29:358–364. doi: 10.1016/0169-328x(94)00278-m. [DOI] [PubMed] [Google Scholar]
- 52.Pan J-T, Gala RR. Central nervous system regions involved in the estrogen-induced afternoon prolactin surge. I. Lesion studies. Endocrinology. 1985;117:382–387. doi: 10.1210/endo-117-1-382. [DOI] [PubMed] [Google Scholar]
- 53.Piggins HD, Cutler DJ. The roles of vasoactive intestinal polypeptide in the mammalian circadian clock. J Endocrinol. 2003;177:7–15. doi: 10.1677/joe.0.1770007. [DOI] [PubMed] [Google Scholar]
- 54.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 55.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 56.Sakamoto H, Yasukawa H, Masuhara M, Tanimura S, Sasaki A, Yuge K, Ohtsubo M, Ohtsuka A, Fujita T, Ohta T, Furukawa Y, Iwase S, Yamada H, Yoshimura A. A Janus kinase inhibitor, JAB, is an interferon-gamma-inducible gene and confers resistance to interferons. Blood. 1998;92:1668–1676. [PubMed] [Google Scholar]
- 57.Sellix MT, Egli M, Henderson RP, Freeman ME. Ovarian steroid hormones modulate circadian rhythms of neuroendocrine dopaminergic neuronal activity. Brain Res. 2004;1005:164–181. doi: 10.1016/j.brainres.2004.01.049. [DOI] [PubMed] [Google Scholar]
- 58.Sellix MT, Freeman ME. Circadian rhythms of neuroendocrine dopaminergic neuronal activity in ovariectomized rats. Neuroendocrinology. 2003;77:59–70. doi: 10.1159/000068334. [DOI] [PubMed] [Google Scholar]
- 59.Shearman LP, Jin X, Lee C, Reppert SM, Weaver DR. Targeted disruption of the mPer3 gene: subtle effects on circadian clock function. Mol Cell Biol. 2000;20:6269–6275. doi: 10.1128/mcb.20.17.6269-6275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 61.Shieh KR, Pan JT. Sexual differences in the diurnal changes of tuberoinfundibular dopaminergic neuron activity in the rat: role of cholinergic control. Biol Reprod. 1996;54:987–992. doi: 10.1095/biolreprod54.5.987. [DOI] [PubMed] [Google Scholar]
- 62.Shinohara K, Funabashi T, Kimura F. Temporal profiles of vasoactive intestinal polypeptide precursor mRNA and its receptor mRNA in the rat suprachiasmatic nucleus. Brain Res Mol Brain Res. 1999;63:262–267. doi: 10.1016/s0169-328x(98)00289-7. [DOI] [PubMed] [Google Scholar]
- 63.Shinohara K, Funabashi T, Mitushima D, Kimura F. Effects of estrogen on the expression of connexin32 and connexin43 mRNAs in the suprachiasmatic nucleus of female rats. Neurosci Lett. 2000;286:107–110. doi: 10.1016/s0304-3940(00)01102-2. [DOI] [PubMed] [Google Scholar]
- 64.Shinohara K, Funabashi T, Nakamura TJ, Kimura F. Effects of estrogen and progesterone on the expression of connexin-36 mRNA in the suprachiasmatic nucleus of female rats. Neurosci Lett. 2001;309:37–40. doi: 10.1016/s0304-3940(01)02022-5. [DOI] [PubMed] [Google Scholar]
- 65.Shinohara K, Honma S, Katsuno Y, Abe H, Honma K. Circadian rhythms in the release of vasoactive intestinal polypeptide and arginine-vasopressin in organotypic slice culture of rat suprachiasmatic nucleus. Neurosci Lett. 1994;170:183–186. doi: 10.1016/0304-3940(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 66.Shinohara K, Tominaga K, Inouye SIT. Phase dependent response of vasoactive intestinal polypeptide to light and darkness in the suprachiasmatic nucleus. Neurosci Res. 1999;33:105–110. doi: 10.1016/s0168-0102(98)00122-9. [DOI] [PubMed] [Google Scholar]
- 67.Shinohara K, Tominaga K, Isobe Y, Inouye S-IT. Photic regulation of peptides located in the ventrolateral subdivision of the suprachiasmatic nucleus of the rat: Daily variations of vasoactive intestinal polypeptide, gastrin-releasing peptide, and neuropeptide Y. J Neurosci. 1993;13:793–800. doi: 10.1523/JNEUROSCI.13-02-00793.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci U S A. 1972;69:1583–1586. doi: 10.1073/pnas.69.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U S A. 2002 doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wakamatsu H, Takahashi S, Moriya T, Inouye ST, Okamura H, Akiyama M, Shibata S. Additive effect of mPer1 and mPer2 antisense oligonucleotides on light-induced phase shift. NeuroReport. 2001;12:127–131. doi: 10.1097/00001756-200101220-00033. [DOI] [PubMed] [Google Scholar]
- 71.Watson RE, Wiegand SJ, Clough RW, Hoffman GE. Use of cryoprotectant to maintain longterm peptide immunoreactivity and tissue morphology. Peptides. 1986;7:155–163. doi: 10.1016/0196-9781(86)90076-8. [DOI] [PubMed] [Google Scholar]
- 72.Watts AG, Swanson LW. Efferent projections of the suprachiasmatic nucleus: II, studies using retrograde transport of fluorescent dyes and simultaneous peptide immunohistochemistry in the rat. J Comp Neurol. 1987;258:230–252. doi: 10.1002/cne.902580205. [DOI] [PubMed] [Google Scholar]
- 73.Watts AG, Swanson LW, Sanchez-Watts G. Efferent projections of the suprachiasmatic nucleus: I studies using anterograde transport of Phaseolus vulgaris leucoagglutinin in the rat. J Comp Neurol. 1987;258:204–229. doi: 10.1002/cne.902580204. [DOI] [PubMed] [Google Scholar]
- 74.Watts AG, Tanimura S, Sanchez-Watts G. Corticotropin-releasing hormone and arginine vasopressin gene transcription in the hypothalamic paraventricular nucleus of unstressed rats: daily rhythms and their interactions with corticosterone. Endocrinology. 2004;145:529–540. doi: 10.1210/en.2003-0394. [DOI] [PubMed] [Google Scholar]
- 75.Wilsbacher LD, Yamazaki S, Herzog ED, Song EJ, Radcliffe LA, Abe M, Block G, Spitznagel E, Menaker M, Takahashi JS. Photic and circadian expression of luciferase in mPeriod1-luc transgenic mice invivo. Proc Natl Acad Sci U S A. 2002;99:489–494. doi: 10.1073/pnas.012248599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 77.Yang J, Cagampang FRA, Nakayama Y, Inouye S-IT. Vasoactive intestinal polypeptide precursor mRNA exhibits diurnal variation in the rat suprachiasmatic nuclei. Mol Brain Res. 1993;20:259–262. doi: 10.1016/0169-328x(93)90049-u. [DOI] [PubMed] [Google Scholar]