Abstract
Interferons (IFNs) are antiviral cytokines that selectively regulate gene expression through several signaling pathways including nuclear factor κB (NFκB). To investigate the specific role of NFκB in IFN signaling, we performed gene expression profiling after IFN treatment of embryonic fibroblasts derived from normal mice or mice with targeted deletion in NFκB p50 and p65 genes. Interestingly, several antiviral and immunomodulatory genes were induced higher by IFN in NFκB knockout cells. Chromatin immunoprecipitation (ChIP) experiments demonstrated that NFκB was basally bound to the promoters of these genes, while IFN treatment resulted in the recruitment of STAT1 and STAT2 to these promoters. However, in NFκB knockout cells IFN induced STAT binding as well as the binding of the IFN regulatory factor-1 (IRF1) to the ISG promoters. IRF1 binding closely correlated with enhanced gene induction. Moreover, NFκB suppressed both antiviral and immunomodulatory actions of IFN against influenza virus. Our results identify a novel negative regulatory role of NFκB in IFN-induced gene expression and biological activities, and suggest that modulating NFκB activity may provide a new avenue for enhancing the IFN's therapeutic effectiveness.
Type I IFNs (IFNα, IFNβ, IFNω, and IFNτ) are multifunctional cytokines that are critical in the host defense to infectious agents by modulating innate and adaptive immune responses. IFNs induce their biological effects by regulating the expression of a family of early response genes, called IFN-stimulated genes (ISGs), through JAK-mediated tyrosine-phosphorylation of the STAT factors, STAT1 and STAT2. The phosphorylated STAT proteins dimerize, translocate into nucleus, and bind to the conserved IFN-stimulus response element (ISRE) within the promoters of ISGs (1). In addition, transcription factors of the IFN regulatory factor (IRF) family have been shown to regulate ISG expression (2). Accumulating evidence indicates that IFNs also activate the nuclear factor κB (NFκB) transcription factor in a serine/threonine kinase-dependent signaling pathway (3,4).
The mammalian NFκB proteins, p50, p52, RelA (p65), RelB, and c-Rel form homodimers and heterodimers to regulate the expression of genes involved in the immune response, inflammation and cell survival (5). In most cell types, the predominant form of NFκB, the p50:p65 heterodimer, is bound to Iκ B inhibitory proteins in the cytoplasm of unstimulated cells. Similar to various other stimuli, IFNα/β activates a phosphatidylinositol-3 kinase/Akt pathway, which results in the dissociation of the inactive cytosolic NFκB/IκB complexes followed by NFκ B nuclear translocation and DNA binding (3,4). We previously demonstrated that mouse embryonic fibroblasts (MEFs) derived from mice with germ-line deletions of the p50 and p65 genes were resistant to IFN-induced NFκB activation and were sensitized to the antiviral action of IFNβ against vesicular stomatitis virus (6).
The present study was undertaken to further define the role of NFκB in IFN-induced gene expression and the biological actions of IFN. Gene expression profiling identified 35 ISGs whose induction was highly regulated by NFκB. A subset of genes that were induced higher by IFN in NFκB-KO cells encoded GTP-binding and antigen presentation proteins, which play critical roles in IFN's antiviral and immunomodulatory activities, respectively. Quantitative RT-PCR demonstrated that these ISGs were induced more rapidly and at significantly lower IFN concentrations in NFκB-KO cells relative to wild-type (WT) MEFs. Chromatin immunoprecipitation (ChIP) assays demonstrated that NFκB dimers containing p50 and p65 were basally bound to the promoters of these ISGs. IFN induced the binding of STAT1 and STAT2 to these promoters. However, the kinetics of ISG induction in NFκB-KO MEFs by IFN correlated with the promoter binding of IRF1. These results suggest that IFN induction of these genes is negatively regulated by NFκB. We also found that NFκB suppressed not only the direct antiviral action of IFN against influenza virus, but also IFN-induced influenza-specific MHC class I antigen presentation. Together, our results suggest that NFκB not only regulates the induction of a subset of IFN-induced genes, but also IFN's antiviral and immunomodulatory activities.
EXPERIMENTAL PROCEDURES
Biological Reagents and Cell Culture. Recombinant rat IFNβ was obtained from Biogen-Idec, Inc. (7). Antibodies directed against the following proteins were used: p65, p50, IRF1, STAT1, STAT2, Tap1 and β-actin from Santa Cruz Biotechnology (Santa Cruz, CA); Mx1 (Dr. Otto Haller); phospho-Stat2 from Upstate Biotechnology (Charlottesville, VA); and TFIIB from ActiveMotif (Carlsbad, CA). WT and NFκB-KO MEFs (8) were plated at 1 x 104 cells/cm2 every 3 days in Dulbecco's modified Eagle's medium supplemented with 10% DCS (Hyclone Laboratories, Logan, UT), and 100 μg/ml penicillin G, and 100 μg/ml streptomycin.
RNA Preparation and Microarray Analysis. Total cellular RNA from control and IFNβ-treated (2,500 U/ml for 5 hrs) WT or NFκB-KO MEFs was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Approximately 10 μg of RNA was submitted to Genome Explorations Inc. (Memphis, TN) for labeling and hybridization to murine U74Av2 GeneChips (Affymetrix Inc.) according to the manufacturer's protocols. Expression values were determined using Affymetrix Microarray Suite (MAS) 5.0 software. All data analysis was performed using GeneSpring software 7.0 (Silicon Genetics, Inc.). The MAS 5.0 gene expression values for each gene were normalized as described previously (6). Fold induction by IFN was calculated in matched pairs of WT and NFκB-KO MEFs, respectively. The average of fold induction from three independent sets of GeneChip data for both WT and NFκB-KO MEFs was subjected to non-parametric t-test. The difference of fold induction for each gene by IFN was calculated by subtracting fold induction in WT MEFs from fold induction in NFκB-KO MEFs.
Quantitative Real-Time PCR. Total RNA was isolated from untreated and IFNβ-treated MEFs using TRIzol reagent (Invitrogen, Carlsbad, CA). Quantitative real-time PCR (RT-PCR) was performed on a SmartCycler (Cepheid, Sunnyvale, CA) using the AccessQuick™ RT-PCR system (Promega, Madison, WI), and SYBR green I (Molecular Probes, Eugene, Oregon) according to the manufacturer's instructions. The following forward and reverse primers were used for each gene respectively: Ifi47, 5′-CTCGGACGGTTCTTCTTATC-3′, 5′-AGCACCCTCCTCTCTTCATG-3′;
Tap1, 5′-TTGCCTGAACAAGAACAGTG-3′, 5′-AAGTTCCCCCTTGATGTCTG-3′;
Mx1, 5′-GACTACCACTGAGATGACCC-3′, 5′-CTCTATTTCCTCCCCAAATG-3′;
β-actin,5′-AAGGAGATTACTGCTCTGGC-3′,5′-ACATCTGCTGGAAGGTGGAC-3′.
Reverse transcription was performed at 48°C for 45 min and RT-PCR cycling parameters were as follows: denaturation at 95°C for 2 min, amplification at 94°C for 30 sec, and 62°C for 30 sec for 35 cycles. The product size was initially monitored by agarose gel electrophoresis and melting curves were analyzed to control for specificity of PCR reactions. The data on IFN-induced genes was normalized to the expression of the housekeeping gene β-actin. The relative units were calculated from a standard curve, plotting 3 different concentrations against the PCR cycle number at the cycle threshold (with a 10-fold increment equivalent to ∼3.1 cycles).
Immunoblotting. At various times after IFNβ treatment (1,000 U/ml), MEFs were lysed directly in RIPA buffer (whole cell lysate), or nuclear extracts were prepared with the NE-PER Kit (Pierce, Rockford, IL). Equivalent amounts of protein were subjected to SDS-PAGE. Proteins were transferred to nitrocellulose membranes, immunoblotted for Tap1, Mx1, β-actin, or TFIIB and visualized by enhanced chemiluminescence (Pierce, Rockford, IL).
Chromatin Immunoprecipitation. ChIP experiments were performed using the ChIP-IT™ Chromatin Immunoprecipitation Kit (Active Motif, Carlsbad, CA) according to the manufacturer's instructions. DNA was sheared to an average size of ∼600 bp. The following forward and reverse primers were used for each gene respectively:
Ifi47, 5′-CATCTCTTTCATCCTTGTCC-3′, 5′-AGAAGCCTGGAAGATTCAAG-3′;
Tap1, 5′-CACTTCTAGTCAGCTCCACC-3′, 5′-AGAGTCTGGTCCTAGCCTGG-3′;
Mx1, 5′-CCAGAGGAGAATTGAAACCG-3′, 5′-TCCCAACCTCAGTACCAAGC-3′.
Antiviral Assay. To determine the cellular sensitivity to IFN's ability to reduce Influenza A virus titer, cell cultures were pre-incubated overnight with IFN, followed by infection with influenza A/Puerto Rico/8/34 (PR8) virus for 1 hr at 1 plaque-forming unit (pfu) per cell. At 24 hrs post-infection, the virus yield in the medium was assayed by plaque formation on Madin-Darby canine kidney cells as described previously (9).
Influenza-specific immune assays. To assay for MHC class I functional expression, 5 × 103 WT or NFκB MEF cells were co-cultured with 1 × 105 4-39 hybridoma cells in individual wells of 96-well plates for 24 hrs in a 1:1 mixture of DMEM and Complete Tumor Media (CTM) containing 0.1 μg/ml DbNP-366 peptide and IFNβ. The 4-39 hybridoma line reacts positively by IL-2 production with the immunodominant DbNP-366 peptide, but negatively with DbPA-224, KbPB1-703, KbNS2-114 and DbPB1-F2-62 peptides (10,11). Alternatively, to assay for MHC Class I antigen processing and presentation following infection with influenza virus, 5 × 103 MEF cells were cultured in individual wells of a 96-well plate in DMEM overnight in the presence or absence of IFNβ. Then MEF cells were infected with PR8 influenza A virus at 1 pfu/cell in serum-free DMEM for 1 hr, followed by co-culture with 4-39 hybridoma cells (1 × 105) without or with IFNβ in DMEM plus CTM (1:1) for 24 hrs. IL-2 production, which in this assay correlates directly to the MHC class I expression on the MEF cell surface, was measured by ELISA using purified anti-IL-2 and biotin-anti-IL-2 (BD Pharmingen) antibodies following the manufacturer's directions.
RESULTS
Identification of ISGs that are NFκB-dependent. We previously reported that NFκB plays a role in regulating ISG expression (6). Subsequently, a functional genomics approach was used to monitor the role of NFκB in gene expression changes induced by IFN. For these experiments, we used fibroblasts derived from mice with germ-line deletions of the p50 and p65 genes. These MEFs were shown to be resistant to IFN-induced NFκB activation (6), and hence can be considered as NFκB knockout (NFκB-KO). In contrast, MEFs expressing p50 and p65 genes were IFN-responsive and considered wild-type (WT). In three independent experiments, WT and NFκB-KO MEFs were treated in the presence or absence of IFNβ (2,500 U/ml for 5 hrs), and total RNA was extracted, labeled and hybridized to murine U74Av2 GeneChips (Affymetrix). Control cultures received no IFN but were treated in parallel. Expression values were determined using Affymetrix Microarray Suite (MAS) 5.0 software, and the data were filtered and analyzed using GeneSpring software 7.0 (Silicon Genetics, Inc.) as described previously (6).
After data processing, a total of 7412 probe sets were used to compare gene expression between MEFs differing with respect to genotype (NFκB-KO and WT) and treatment (control and IFN-treatment). Parametric two-way ANOVA (p<0.05, n=3 for each group) identified 1375 probe sets whose expression was different between any two of four groups (control WT MEFs, IFN-treated WT MEFs, control NFκB-KO MEFs, IFN-treated NFκB-KO MEFs). The induction of 35 genes by IFN was significantly (p<0.05, non-parametric t-test, n=3 for each group) different between WT and NFκB-KO cells. Functional classification of NFκB-regulated ISGs using EASE (Expression Analysis Systematic Explorer) analysis (12) revealed that genes encoding GTP-binding and antigen presentation proteins were significantly overrepresented (variant one-tailed Fisher exact probability test, EASE scores < 0.01). The NFκB-regulated GTP-binding ISGs included the 65-67 kDa guanylate-binding proteins (Gbp 1 and Gbp2), the Mx proteins (Mx1 and Mx2), and the 47-kDa GTPase Ifi47 (also called Irg47) (13). GBPs play important roles in resistance to viruses (14-16), as well as intracellular protozoa and bacteria (13). On the other hand, the NFκB-regulated antigen presentation ISGs (Tap1, Tap2, Psmb9/Lmp2, and Psmb8/Lmp7) are involved in degrading intracellular proteins into antigenic peptides, and contribute to the transport of these peptides to endoplasmic reticulum where they bind to the assembled MHC class I molecules (17). As shown in Table 1, Gbp1, Ifi47, Mx1, Mx2, Tap1, Psmb9/Lmp2 and Psmb8/Lmp7 were induced by IFN to a greater extent in NFκB-KO MEFs than in WT MEFs, while Gbp2 and Tap2 were induced more in WT MEFs than in NFκB-KO MEFs. These findings suggested that NFκB regulates the expression of a subset of ISGs that play important roles in antiviral and immune responses.
Table 1.
ISGs regulated by NFκB in WT and NFκB-KO MEFs.
| Entrez Gene ID | Gene Symbol | WT Fold Inductionc | NFκB-KO Fold Inductionc | Fold Induction Differenced |
|---|---|---|---|---|
| 15953 | Ifi47a | 111.8 | 295.5 | 183.6 |
| 17857 | Mx1a | 114.3 | 286.6 | 172.2 |
| 17858 | Mx2a | 35.0 | 56.9 | 21.9 |
| 14468 | Gbp1a | 38.5 | 779.5 | 741.0 |
| 14469 | Gbp2a | 411.2 | 175.6 | -235.6 |
| 21354 | Tap1b | 11.87 | 34.35 | 22.5 |
| 21355 | Tap2b | 10.58 | 2.92 | -7.7 |
| 16913 | Pmsb8b | 38.24 | 58.96 | 20.7 |
| 16912 | Pmsb9b | 10.29 | 18.43 | 8.1 |
| 15018 | H2-Q7b | 19.83 | 52.78 | 33.0 |
NFκB-regulated GTP-binding ISGs.
NFκB-regulated antigen presentation ISGs.
Fold induction of genes by IFN was the average of the ratio of gene induction in IFN-treated/control MEFs from three independent sets of GeneChip data.
The difference of fold induction of genes by IFN was calculated by subtracting fold induction in WT MEFs from fold induction in NFκB-KO MEFs.
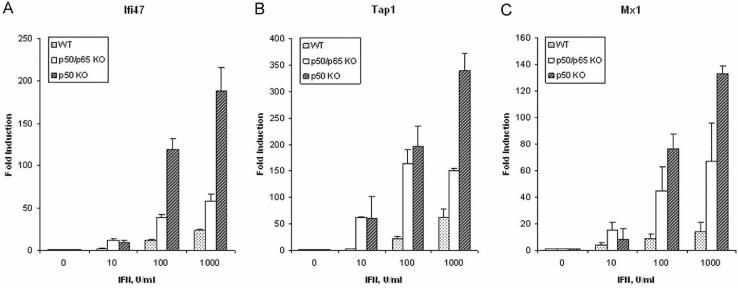
Dose-dependence and kinetics of NFκB-dependent ISGs. To further investigate the induction of ISGs that were negatively regulated by NFκB, quantitative real-time PCR assays were performed for Ifi47, Tap1 and Mx1 using RNA from WT and NFκB-KO MEFs treated with IFNβ. Consistent with the microarray results, Mx1, Ifi47 and Tap1 (Fig. 1) were induced by IFNβ in a dose-dependent manner to higher expression levels in NFκB-KO cells than in WT cells. Similarly, we showed previously that Mx1 and Nmi were also induced more in NFκB-KO cells than in WT cells by IFN in a dose-dependent manner (6).
Figure 1.
Dose-dependent induction by IFN of Ifi47 (A), Tap1 (B) and Mx1 (C) expression. Real-time PCR was performed on cDNAs prepared from MEFs (WT, NFκB-KO and p50-KO) treated with IFNβ at varying concentrations for 5 hrs. Gene expression was normalized to actin expression in each sample. Data are shown as fold-induction relative to untreated fibroblasts, and are mean values ± the SEM (n=3).
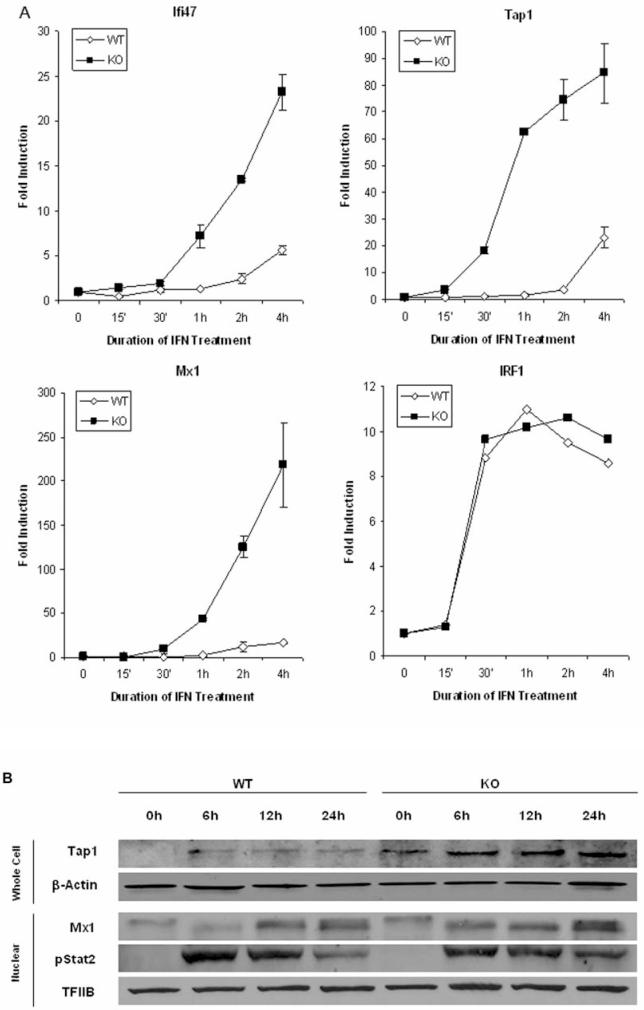
We also examined the time course of IFN induction of these genes. Ifi47, Tap1 and Mx1 (Fig. 2A) were induced more rapidly in NFκB-KO cells compared to WT cells. For example in WT MEFs, Mx1 was induced by 2 hrs of IFN treatment and reached a maximal induction level of ∼15 fold after 4 hrs. In sharp contrast, Mx1 was induced ∼10 fold after 30 min and ∼220 fold after 4 hrs of IFN treatment in NFκB-KO cells. Moreover, the changes in the mRNA levels were accompanied by changes in the protein levels for Tap1 and Mx1 proteins (Fig. 2B). The effect of NFκB on regulation of these ISGs is specific since the expression profile of another ISG, IRF1, was not regulated by NFκB (Fig. 2A). In addition, the effects of NFκB on IFN-induced gene expression did not reflect altered JAK-STAT signaling, since STAT2 activation was similar in WT and NFκB-KO MEFs (Fig. 2B). These results suggested that NFκB selectively decreased and delayed the transcription of a subset of ISGs.
Figure 2.
Time course of IFN induced gene expression. (A) Real-time PCR was performed for Ifi47, Tap1, Mx1 and IRF1 using cDNAs prepared from MEFs treated with IFNβ at 1000 U/ml for varying times. Gene expression was normalized to actin expression in each sample. Data are shown as fold-induction relative to untreated fibroblasts, and are mean values +/- the SEM (n=3). (B) Immunoblotting was performed with indicated antibodies on whole cell or nuclear lysates prepared from MEFs treated with IFNβ (1000 U/ml) for the indicated times. Protein loading was evaluated by immunoblotting with anti-actin (cell) or -TFIIB (nuclear) antibodies. Similar results were obtained in at least two independent experiments.
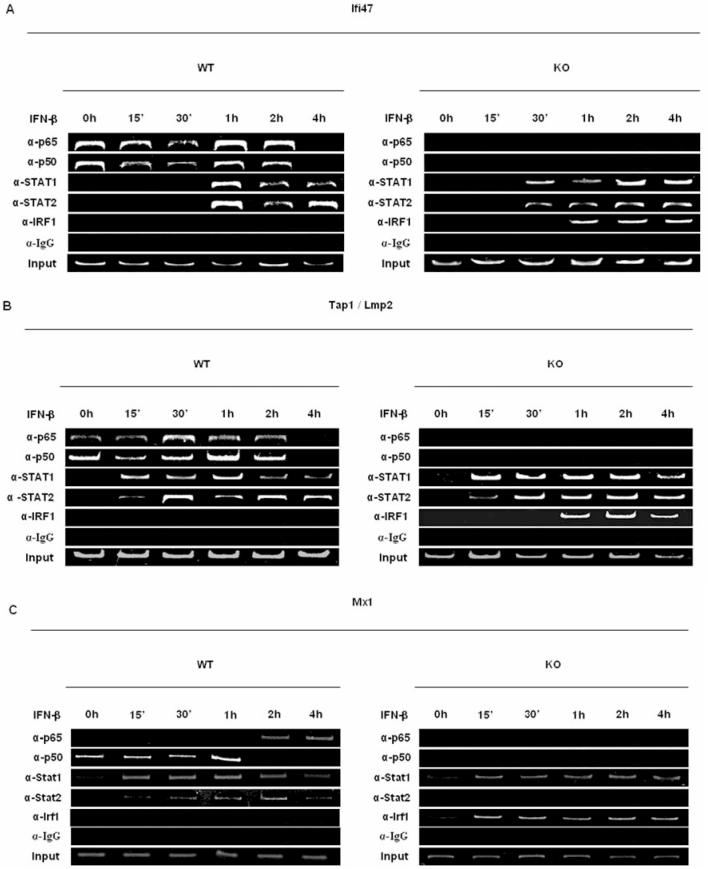
The binding of p50 and p65 to the promoters of NFκB-dependent ISGs. To investigate which NFκB proteins regulated the transcription of Ifi47, Tap1 and Mx1, we performed ChIP assays. Although STAT proteins were not basally bound to the promoters of Ifi47, Tap1 and Mx1, IFN treatment induced the binding of Stat1 and Stat2 to these promoters in WT and NFκB-KO MEFs between 15 to 60 min of IFN treatment (Fig. 3A-C). This result was expected since the binding of these STAT proteins is necessary for IFN-induced activation of ISG transcription. In contrast to STAT proteins, the p50 NFκB protein was basally bound to the promoter of these ISGs in WT MEFs, while p65 was basally bound to Ifi47 and Tap1 promoters. Since the genes for p50 and p65 were both deleted in NFκB-KO mice, neither protein was found bound to the promoter of these ISGs in NFκB-KO MEFs. Interestingly, in NFκB-KO MEFs IFN treatment resulted in the recruitment of IRF1 to the promoters of all three ISGs, and IRF1 recruitment closely correlated with the rapid and enhanced induction of the genes upon IFN treatment. Although IRF1 itself is an ISG, we found that IRF1 expression was not NFκB-regulated; IFN induction of IRF1 was similar in WT and NFκB-KO MEFs as determined by microarray analysis (∼6-fold) or quantitative real-time PCR (Fig. 2A). Taken together these results demonstrate that NFκB directly binds to the promoters of a subset of ISGs and thereby inhibits their induction by IFN, perhaps by inhibiting recruitment of IRF1 to their promoters.
Figure 3.
Transcription factors binding profiles to NFκB-regulated ISG promoters in WT and NFκB-KO fibroblasts. ChIP assays were performed on extracts from control and IFN-treated MEFs using the indicated antibodies for precipitation and various primers that targeted specific regions in the Ifi47 (A), Tap1 (B), and Mx1 (C) promoters (see methods). Similar results were obtained in at least three independent experiments.
p50 was bound to the promoters of all three ISG promoters and correlated with diminished ISG expression. Therefore, we examined the effect of the p50 protein alone on the induction of NFκB-regulated ISGs in MEFs derived from p50 knockout mice. As shown in Figure 1, Ifi47, Tap1 and Mx1 were induced by IFNβ to higher expression levels in p50-KO than in either NFκB-KO cells or in WT cells. These results suggest that p50 is the predominant negative regulator of these ISGs.
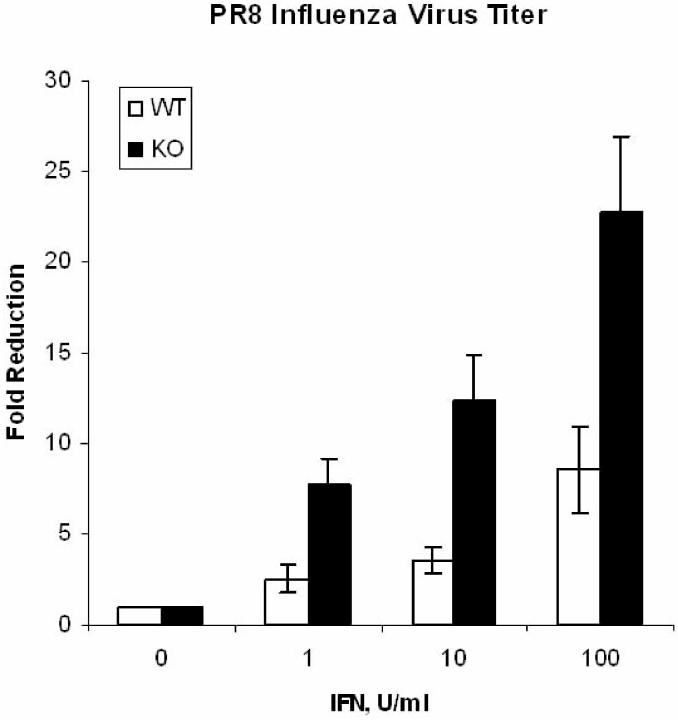
NF κ B dampens the antiviral and immunomodulatory activity of IFN against the influenza virus. The NFκB-regulated ISG Mx1 has selective antiviral activity in mice against the influenza virus (16). Therefore, we investigated whether NFκB modulated the anti-influenza activity of IFN. WT and NFκB-KO MEFs were infected with the PR8 strain of influenza A in the absence or presence of different concentrations of IFN, and viral replication was measured after 24 hrs. As shown in Fig. 4, IFN (100 U/ml) results in only a 10-fold reduction in virus titer in WT MEFs. However, a 100-fold lower IFN concentration (1 U/ml) was sufficient to induce equivalent antiviral effects in NFκB-KO as compared to WT MEFs.
Figure 4.
IFN-induced antiviral response to influenza infection in WT and NFκB-KO fibroblasts. To determine the ability of IFNβ to reduce virus titers in influenza-infected WT and NFκB-KO MEFs, fibroblasts were pre-incubated overnight with IFNβ, infected with influenza-PR8 virus, and at 24 hrs the virus yield was assayed by plaque formation (9). Viral titers in untreated WT and NFκB-KO MEFs were 9.5 ± 4.5 X 104 and 7.6 ± 2.2 X 104 pfu/ml, respectively.
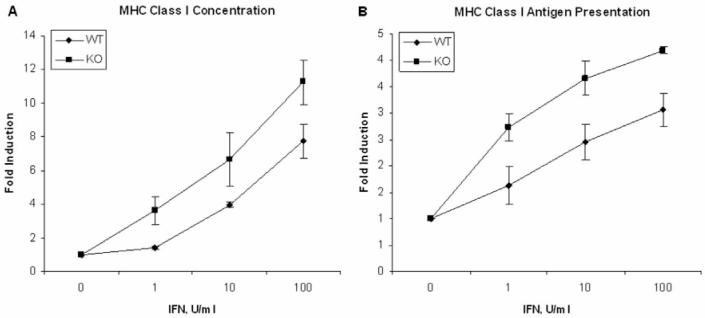
The NFκ B-regulated ISG Tap1 is required for antigen presentation of an immunodominant influenza viral epitope (18). Since IFN also modulates this facet of the host immune response, we next determined the effect of IFN on influenza-specific MHC class I antigen presentation in WT and NFκ B-KO MEFs. Using a T cell hybridoma specific for the immunodominant DbNP-366 influenza peptide as responder cells, we determined that antigen presentation levels were significantly enhanced by IFN treatment in NFκ B-KO MEFs as compared to WT cells (Fig. 5). Taken together, these results indicate that NFκB dampens several functionally important IFN activities.
Figure 5.
IFN-induced immune response to influenza infection in WT and NFκB-KO fibroblasts. To determine the effect of IFN on the immune response to influenza virus, (A) the MHC class I levels (10), or (B) MHC class I antigen presentation (11) on control and IFN-treated MEFs was determined. The data represent the average of duplicate determinations from three independent experiments.
DISCUSSION
The purpose of the present study was to characterize the role of NFκB in the biological actions of IFN using a functional genomic approach. We show that NFκB negatively regulates the induction of a subset of ISGs by IFN, and that NFκB suppresses IFN's antiviral and immunomodulatory activities in the context of influenza virus infection. Both IFN and NFκB play critical roles in the host defense to various pathogens. However, the role of NFκB in IFN's biological functions is relatively undefined. We previously found that the deficiency of p65 and p50 NFκB subunits sensitized MEFs to antiviral action of type I IFNs against vesicular stomatitis virus (VSV) (6). Indeed, NFκB increases the susceptibility of human cells to influenza virus infection (19). Our findings provide a possible molecular mechanism for this observation whereby p50-containing NFκB dimers basally inhibit a subset of IFN antiviral genes via binding directly to their promoters.
The role of the JAK-STAT pathway in gene induction and in the generation of many of the biological effects of IFN is firmly established. However, it is now apparent that the activation of the JAK-STAT pathway alone is insufficient to account for all of the biological actions of IFNs. Some of these pathways operate independently of the JAK-STAT pathway, while other pathways may cooperate with STATs to regulate the transcription of target genes. For example, although the role of IRF9 together with the STAT1/STAT2 dimer in the transcriptional regulation of ISGs is well described, other IRF proteins can also act as transcriptional activators and/or repressors of IFN-regulated genes (2). In addition, the p300/CBP co-activators can interact with STAT proteins to regulate ISG transcription (20,21).
We show that, although NFκB is basally bound to ISG promoters, the IFN-induced binding of STAT1 and STAT2 to these promoters was unimpeded. Therefore, NFκB does not negatively regulate ISG induction through affecting STAT binding to ISG promoters. However, NFκB proteins appear to modulate IRF1 promoter binding. IRF1 is also an IFN-induced transcriptional activator of ISG expression (22). We show that in NFκB-KO MEFs, but not in WT MEFs, IRF1 is recruited to ISG promoters upon IFN treatment, and this recruitment closely correlates with the more rapid and enhanced ISG expression in NFκB-KO cells. Moreover, other co-activators of ISG expression (i.e., p300/CBP) are also bound to ISG promoters in the absence of NFκB proteins (unpublished observations LW, RH and LMP). Therefore, NFκB may negatively regulate ISG expression by directly or indirectly inhibiting the promoter binding of co-activators of ISG expression.
The notion that NFκ B suppresses transcription is contrary to its classical role of stimulating the transcription of target genes. However, several recent studies indicate that NFκB may downregulate cellular responsiveness to specific cytokines. For example, tumor necrosis factor-mediated JNK signaling is enhanced in NFκB-KO mice (23,24). Moreover, we previously showed that the IFN-induced expression of some ISGs (Mx1 and Nmi) is negatively regulated by NFκB, while the expression of other ISGs, such as Ifit1 and Isg15, is enhanced by NFκ B (6). In the present study, we show that NFκB has opposing effects on different ISGs that map to a single genetic locus. For example, the induction by IFN of Gbp1 and Gbp2, which are located adjacent to one another on mouse chromosome 6, are negatively and positively regulated by NFκB, respectively (Table 1). Similarly, the induction of Tap1 and Tap2, which are adjacent to one another on mouse chromosome 17, is also affected in opposite directions by NFκB (Table 1). These results suggest that NFκB may act as a transcriptional switch to fine-tune the expression of neighboring ISG family members in a locus. It will be important to determine the cellular consequences of this regulatory pathway with regard to the specific functions of Gbp and Tap family proteins.
The ability to integrate multiple signaling pathways to achieve unique responses is a critical requirement for development and homeostasis in all metazoans (25). In this study, we have characterized the relationship of the classical JAK-STAT pathway to NFκB pathway in the transcriptional regulation of Ifi47, Tap1 and Mx1. Moreover, we demonstrate that NFκB regulates IFN's antiviral activity and the promotion of T cell activation. Type I IFNs are broadly used in clinical treatment of viral infections, multiple sclerosis and cancer. We show that IFN's efficacy as an antiviral agent against influenza virus is significantly enhanced in NFκ B-KO MEFs. Interestingly, NFκB inhibitors are currently used to treat inflammatory diseases and autoimmune disease. Thus, we propose that the combination of IFN and NFκB inhibitors may be used to maximize therapeutic benefit of IFN in the treatment of human disease, while minimizing the dosage and possibly its undesirable side effects.
ACKNOWLEDGEMENTS
We thank Dr. Otto Haller (University of Freiburg, Freiburg, Germany), Dr. Darren Baker (Biogen-Idec, Cambridge, MA), and Drs. David Baltimore (California Institute of Technology, Pasadena, CA) and Alexander Hoffmann (University of California-San Diego, La Jolla,CA) for kindly providing anti-Mx1 antibody, IFNβ, and MEF cultures, respectively. We also thank Dennis Carrigan and Dr. Jong-Gwam Kim for advice and technical support. Supported by NIH grant CA73753 (L.M.P.) and by funds from the Muirhead Chair Endowment at the University of Tennessee Health Science Center.
REFERENCES
- 1.Darnell JEJ, Kerr IM, Stark GR. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 2.Barnes B, Lubyova B, Pitha PM. J Interferon Cytokine Res. 2002;22:59–71. doi: 10.1089/107999002753452665. [DOI] [PubMed] [Google Scholar]
- 3.Yang CH, Murti A, Basu L, Kim JG, Pfeffer LM. Proc Natl Acad Sci USA. 2000;97:13631–13636. doi: 10.1073/pnas.250477397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang CH, Murti A, Pfeffer SR, Kim JG, Donner DB, Pfeffer LM. J. Biol. Chem. 2001;276:13756–13761. doi: 10.1074/jbc.M011006200. [DOI] [PubMed] [Google Scholar]
- 5.Hayden MS, Ghosh S. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 6.Pfeffer LM, Kim JG, Pfeffer SR, Carrigan DJ, Baker DP, Wei L, Homayouni R. J. Biol. Chem. 2004;279:31304–31311. doi: 10.1074/jbc.M308975200. [DOI] [PubMed] [Google Scholar]
- 7.Arduini RM, Li Z, Rapoza A, Gronke R, Hess DM, Wen D, Miatkowski K, Coots C, Kaffashan A, Viseux N, Delaney J, Domon B, Young CN, Boynton R, Chen LL, Chen L, Betzenhauser M, Miller S, Gill A, Pepinsky RB, Hochman PS, Baker DP. Protein Expr Purif. 2004;34:229–242. doi: 10.1016/j.pep.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Beg AA, Baltimore D. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 9.Govorkova EA, Fang HB, Tan M, Webster RG. Antimicrob Agents Chemother. 2004;48:4855–4863. doi: 10.1128/AAC.48.12.4855-4863.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belz GT, Xie W, Altman JD, Doherty PC. J Virol. 2000;74:3486–3493. doi: 10.1128/jvi.74.8.3486-3493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O'Neill R, Schickli J, Palese P, Henklein P, Bennink JR, Yewdell JW. Nat Med. 2001;7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 12.Hosack DA, Dennis G, Jr., Sherman BT, Lane HC, Lempicki RA. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor GA, Feng CG, Sher A. Nat Rev Immunol. 2004;4:100–109. doi: 10.1038/nri1270. [DOI] [PubMed] [Google Scholar]
- 14.Anderson SL, Carton JM, Lou J, Xing L, Rubin BY. Virology. 1999;256:8–14. doi: 10.1006/viro.1999.9614. [DOI] [PubMed] [Google Scholar]
- 15.Lee SH, Vidal SM. Genome Res. 2002;12:527–530. doi: 10.1101/gr.20102. [DOI] [PubMed] [Google Scholar]
- 16.Haller O, Frese M, Kochs G. Rev Sci Tech. 1998;17:220–230. doi: 10.20506/rst.17.1.1084. [DOI] [PubMed] [Google Scholar]
- 17.Ploegh HL.Nutr Rev 200058S25–30.discussion S63-73 [DOI] [PubMed] [Google Scholar]
- 18.Uger RA, Barber BH. J Immunol. 1997;158:685–692. [PubMed] [Google Scholar]
- 19.Nimmerjahn F, Dudziak D, Dirmeier U, Hobom G, Riedel A, Schlee M, Staudt LM, Rosenwald A, Behrends U, Bornkamm GW, Mautner J. J Gen Virol. 2004;85:2347–2356. doi: 10.1099/vir.0.79958-0. [DOI] [PubMed] [Google Scholar]
- 20.Horvath CM, Darnell JE. Curr Opin Cell Biol. 1997;9:233–239. doi: 10.1016/s0955-0674(97)80067-1. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Takami K, Lo MS, Huang G, Yu Q, Roswit WT, Holtzman MJ. J Biol Chem. 2005;280:34306–34315. doi: 10.1074/jbc.M503263200. [DOI] [PubMed] [Google Scholar]
- 22.Pine R. J.Virol. 1992;66:4470–4478. doi: 10.1128/jvi.66.7.4470-4478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Smaele E, Zazzeroni F, Papa S, Nguyen DU, Jin R, Jones J, Cong R, Franzoso G. Nature. 2001;414:308–313. doi: 10.1038/35104560. [DOI] [PubMed] [Google Scholar]
- 24.Tang G, Minemoto Y, Dibling B, Purcell NH, Li Z, Karin M, Lin A. Nature. 2001;414:313–317. doi: 10.1038/35104568. [DOI] [PubMed] [Google Scholar]
- 25.Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]