Abstract
C1-tetrahydrofolate (THF) synthase is a trifunctional enzyme found in eukaryotes which contains the activities 10-formyl-THF synthetase, 5,10-methenyl-THF cyclohydrolase, and 5,10-methylene-THF dehydrogenase. The cytoplasmic isozyme of C1-THF synthase is well characterized from a number of mammals, including humans, but a mitochondrial isozyme has been previously identified only in the yeast Saccharomyces. Here we report the identification and characterization of the human gene encoding a functional mitochondrial C1-THF synthase. The gene spans 236 kbp on chromosome 6 and consists of 28 exons plus one alternative exon. The gene encodes a protein of 978 amino acids, including an N-terminal mitochondrial targeting sequence. The mitochondrial isozyme is 61% identical to the human cytoplasmic isozyme. Expression of the gene was detected in most human tissues, but transcripts were highest in placenta, thymus, and brain. Two mRNAs were detected, a 3.6 kb transcript and a 1.1 kb transcript, and both transcripts were observed in varying ratios in each tissue. The shorter transcript results from an alternative splicing event, where exon 7 is spliced to exon 8a instead of exon 8. Exon 8a is derived from an exonized Alu sequence, sharing no homology with exon 8 of the long transcript, and encodes just 15 amino acids followed by a stop codon and a polyadenylation signal. This short transcript potentially encodes a bifunctional enzyme lacking 10-formyl-THF synthetase activity. Both transcripts initiate at the same 5′ site, 107 nucleotides upstream of the ATG start codon. The full-length (2934 bp) cDNA fused to a C-terminal V5 epitope tag was expressed in CHO cells. Immunoblots of subfractionated cells revealed a 107 kD protein only in the mitochondrial fractions of these cells, confirming the mitochondrial localization of the protein. Yeast cells expressing the full-length human cDNA exhibited elevated 10-formyl-THF synthetase activity, confirming its identification as the human mitochondrial C1-THF synthase.
Keywords: mitochondria, folate, one-carbon metabolism
Introduction
C1-tetrahydrofolate (THF1) synthase is a trifunctional enzyme found in eukaryotes which contains the activities 10-formyl-THF synthetase (EC 6.3.4.3), 5,10-methenyl-THF cyclohydrolase (EC 3.5.4.9), and 5,10-methylene-THF dehydrogenase (EC 1.5.1.5) (Fig. 1, reactions 1-3). These activities, along with serine hydroxymethyltransferase (Fig. 1, reaction 4), are central to the interconversion of the one-carbon units carried by the biologically active form of folic acid, THF. The activated one-carbon units are used in a variety of cellular processes, including de novo purine and thymidylate synthesis, serine and glycine interconversion, methionine biosynthesis, and protein synthesis in mitochondria and chloroplasts.
Figure 1.
Compartmentation of folate-mediated one-carbon metabolism in eukaryotes. Reactions 1, 2, and 3, 10-formyl-THF synthetase (EC 6.3.4.3), 5,10-methenyl-THF cyclohydrolase (EC 3.5.4.9) and 5,10-methylene-THF dehydrogenase (EC 1.5.1.5), respectively, are catalyzed by C1-tetrahydrofolate synthase. The other reactions are catalyzed by 4, serine hydroxymethyltransferase (EC 2.1.2.1); 5, glycine cleavage system (EC 2.1.2.10); 6, methylenetetrahydrofolate reductase (EC 1.5.1.20); 7, methionine synthase (EC 2.1.1.13); 8, methionyl-tRNA formyltransferase (EC 2.1.2.9); 9, methenyltetrahydrofolate synthetase (EC 6.3.3.2). Not all coenzymes are shown for all reactions.
In eukaryotic cells, the mitochondrial and cytosolic compartments each contain a parallel set of one-carbon-unit interconverting enzymes (1). For example, in the yeast Saccharomyces cerevisiae, mitochondrial and cytoplasmic isozymes of C1-THF synthase (encoded by the nuclear genes MIS1 and ADE3, respectively) have been purified and characterized (2,3). Both isozymes exist as homodimers of 100 kDa subunits. Each subunit consists of a C-terminal 10-formyl-THF synthetase domain of about 70 kDa and an N-terminal bifunctional dehydrogenase/cyclohydrolase domain of about 30 kDa linked via a proteolytically sensitive connector region. This subunit size and domain structure is shared by cytoplasmic isozymes from mammalian and avian sources (4-9).
All three activities of C1-THF synthase are found in mammalian mitochondria as well (10,11). Our studies with intact rat liver mitochondria and mitochondrial extracts demonstrated the ability of these organelles to oxidize carbon 3 of serine to formate by a folate-dependent pathway (Fig. 1, reactions 1-4) (11). However, the existence, structure, and function of the folate-interconverting activities of C1-THF synthase in mammalian mitochondria has been controversial. MacKenzie and coworkers characterized a bifunctional NAD-dependent 5,10-methylene-THF dehydrogenase/5,10-methenyl-THF cyclohydrolase, originally isolated from ascites tumor cells (12,13). This bifunctional enzyme lacks the large C-terminal domain catalyzing the 10-formyl-THF synthetase activity, and thus is unable to produce formate. This enzyme was shown to be a nuclear-encoded mitochondrial protein (14,15), detectable only in transformed mammalian cells and embryonic or nondifferentiated tissues (12). Among adult differentiated tissues, NAD-dependent 5,10-methylene-THF dehydrogenase activity is detectable only in rat adrenal tissue (16), although the mRNA encoding this enzyme is present at low levels in all tissues examined (17). MacKenzie has argued that mammalian mitochondria lack a C1-THF synthase, and that the bifunctional NAD-dependent dehydrogenase/cyclohydrolase is the mammalian homolog of the mitochondrial trifunctional enzyme (18,19).
Here we report the identification and characterization of the human gene encoding a functional mitochondrial C1-THF synthase. We show that it is expressed widely in adult human tissues, and the full-length cDNA encodes a protein that localizes to mitochondria when expressed in CHO cells. These data confirm the existence of C1-THF synthase in mammalian mitochondria, completing the folate-interconverting pathway shown in Fig. 1.
Experimental Procedures
Materials— All chemicals were of the highest available commercial quality. DIFCO media components were obtained from VWR (West Chester, PA). Restriction enzymes, shrimp alkaline phosphatase, calf intestinal alkaline phosphatase and T4 DNA ligase were purchased from Gibco-BRL (Rockville, MD). Primers for PCR and sequencing were made by IDT (Coralville, IA). [α-32P]dATP (3000Ci/mmol) was purchased from NEN Life Science Products (Boston, MA).
Construction of full-length cDNA—A partial cDNA clone (DKFZp586G1517), constructed by the German Genome Project (RZPD)(20), was identified in GenBank (AL117452) by a BLAST search using the cDNA sequence of the human cytoplasmic C1-THF synthase (21). This cDNA contains 390 nt of 3′ noncoding sequence and a poly(A) tail, but lacks a start codon, indicating that it is truncated at the 5′ end. The truncated cDNA clone was obtained from RZPD (Germany) and its sequence confirmed by the DNA Analysis Facility of The University of Texas at Austin. The Human Genome Database contains the entire gene corresponding to this cDNA, and predicts an additional 5′ exon that encodes 60 additional N-terminal amino acids. The missing 5′ exon (exon 1, Fig. 4) was PCR amplified from a genomic PAC clone (dJ44A20) obtained from the Sanger Centre (UK). The PCR amplified product was gel purified using Qiagen gel extraction kit, subcloned into pGEM-T Easy vector (Promega, Madison WI) and its sequence verified. It was necessary to use MasterAmp Tfl DNA polymerase (Epicentre, Madison, WI) in the PCR reaction due to the high G+C content of exon 1 (see Results). The partial cDNA clone and the exon 1 clone were then used as templates in a Splice Overlap Extension PCR (SOE PCR) (22) to produce the full-length cDNA. The exon 1 fragment (230 bp) was amplified using Tfl polymerase and primers TOPO5′ (5′CACCATGGGCACGCGTCTGCCGCTC3′; ATG start codon underlined) and humitoSOE3′ (5′CTTCTCTGACGATGGAGTCCCG3′). The 2719 bp cDNA fragment was PCR amplified using Pfu polymerase and primers GS5′SOE (5′GGGACTCCATCGTCAGAGAAG3′) and TOPO3′ (5′GAACAAGCCTTTAACTTGTTCTGTTTC3′). TOPO3′ is complementary to the last 9 codons of the ORF before the stop codon. Both products were gel purified using a Qiagen gel extraction kit. The 230 bp and 2719 bp PCR products served as templates in the SOE PCR reaction using primers TOPO5′ and TOPO3′ and Tfl polymerase. The full-length cDNA product (2934 bp) was gel purified, and cloned into the mammalian expression vector pcDNA3.1D/V5-His-TOPO (Invitrogen) using directional TOPO cloning according to the manufacturer's instructions. The TOPO cloning reaction was transformed into One-Shot Chemically Competent E. coli (Invitrogen) by chemical transformation, and positive colonies were selected on YT (0.8% tryptone, 0.5% yeast extract, 0.5% NaCl) plates containing 50 μg/ml ampicillin. The colonies were screened by PCR with a vector primer and a gene-specific primer, and positive plasmids were prepared using a Qiagen mini plasmid preparation kit. Sequence analysis revealed a base substitution in the full-length clone, compared to the original cDNA and genomic sequences, presumably incorporated during the PCR reactions. (Tfl polymerase, which was chosen due to the high GC content of exon 1, lacks a 3′→5′ proofreading activity). This substitution was repaired using the Quick Change Site Directed Mutagenesis kit from Stratagene. The repaired full-length cDNA clone, pcDNA3.1-humito, was sequenced completely and the correct sequence confirmed (GenBank Accession # AY374130).
Figure 4.
Intron/exon structure of human mitochondrial C1-THF synthase gene. Exons are shown as numbered black bars; introns as thin horizontal lines. Exon and intron sizes and positions are drawn roughly to scale, with the exception of intron 26, which is 55,350 bp. The entire gene spans 236 kbp. The actual sizes of each exon and intron are listed in Table II.
CHO Cell Transfection-- 1.5 x 105 CHO cells were plated on 35-mm diameter dishes and cultured in α-MEM supplemented with 10% (v/v) fetal bovine serum. Duplicate plates were then transfected with 2 μg of pcDNA/humito per plate by the Lipofectamine 2000 Reagent (Invitrogen) method. After transfection, cells were cultured for an additional 48 hours in regular medium before a G418-containing selective medium (0.8 mg/ml) was applied. The selective medium was applied for about one week until antibiotic-resistant colonies developed. Resistant colonies were picked, re-plated, cultured and collected.
Preparation of Cell Homogenates and Subcellular Fractions-- Transfected cells were cultured in two 150 cm2 T-flasks to yield 1-2 X 108 cells. The mono-layer was rinsed with phosphate-buffered saline (PBS) (4 X 5 ml, 4°C), and then incubated with PBS containing 10 mM EDTA (10 ml) at room temperature until the cells detached (5-10 min). The flasks were tapped gently to dislodge the cells, and the cells were transferred to a 50-ml plastic conical tube. Cells were pelleted by centrifugation at 300 X g for 5 min at room temperature, and the cell pellet was washed with 15 ml homogenization solution (HMS, 250 mM sucrose, 1 mM EDTA, pH 6.9) at 4°C. The cell pellet was resuspended in HMS (2 ml, 4°C) and transferred to a nitrogen cavitation device (Kontes) and exposed to a pressure of 36 psi for 30 min at 4°C. The suspension of disrupted cells was collected into a 3-ml conical ground glass Duall tissue grinder and further disrupted with four strokes of the homogenizer (23).
Nuclei and unbroken cells were sedimented by centrifugation (900 X g, 6 min). The supernatant was removed carefully and transferred to another centrifuge tube and stored on ice. The pellet was resuspended in HMS (1 ml) and further dispersed by four strokes in the grinder. After centrifugation (900 X g, 6 min), the supernatant was combined with the first supernatant and stored on ice. The pellet was washed (3 X 1 ml HMS) and the final viscous pellet (nuclear fraction) was resuspended in HMS (1 ml). The combined supernatants were centrifuged (900 x g, 5 min), and any pellet was discarded. The volume of the supernatant [total post-nuclear supernatant (PNS) fraction] was increased to 5 ml by the addition of HMS.
The PNS was centrifuged (10,000 X g, 15 min), and the pellet was stored on ice. The supernatant was recentrifuged (10,000 X g, 15 min) to give a final supernatant (cytosolic fraction). The second pellet was combined with the first, washed with HMS (2 ml), and resuspended in HMS (1 ml) to give the mitochondrial fraction. Glutamate dehydrogenase activity (24) was used as a mitochondrial marker, and lactate dehydrogenase activity (25) was used as a cytoplasmic marker.
Immunoblotting--The protein concentration of the cytosolic and mitochondrial fractions was determined using the Bradford assay (26) with bovine serum albumin as a standard. Eighty μg of cytosolic and mitochondrial protein from transfected and untransfected CHO cells were fractionated on a 7.5% SDS-polyacrylamide gel for 50 minutes at 180 V. One half of the gel was stained and the proteins on the other half were transferred onto a nitrocellulose membrane (Midwest Scientific, Valley Park, MO) by electroblotting for 90 minutes at 250 mAmps. The membrane was then washed 3X with distilled water, 5 minutes each, and blocked in 2% dry milk in Tris-buffered saline (TBS) (10 mM Tris base, 0.15 M NaCl, pH 8.0) for 1 h at room temperature. The blocked membrane was then incubated with primary antibody (Mouse anti-V5 Ab; 1:1000 dilution; Invitrogen) diluted in TBS with 1% dry milk for 1 h at room temperature. The membrane was then washed with TBST (10 mM Tris base, 0.15 M NaCl, 0.0025% Tween-20, pH 8.0) 3 X 5 minutes each, and incubated with secondary antibody (Goat-anti-mouse Ab; 1:2000 dilution; Zymed, San Francisco, CA) for 1 hr at room temperature. The membrane was finally washed with TBST and TBS 2X, 5 minutes each, and rinsed with water before visualizing the bands. Reacting bands were visualized using enhanced chemiluminescence (ECL; Amersham Pharmacia Biotech) detection.
Expression in Yeast and Enzyme Assays— The full length human cDNA was subcloned from pcDNA3.1-humito into the BamH1 and XhoI site of the yeast expression vector, pVT103U (27). In this construct, pVT-humito, the entire human mitochondrial C1-THF synthase ORF, including the mitochondrial presequence, is expressed from the ADH promoter of the vector. Yeast strain DAY3 (ser1 ura3-52 trp1 leu2 ade3-130) (28) was transformed with pVT-humito or empty pVT103U vector using a lithium acetate method (29) modified as described at http://www.cm.utexas.edu/appling/YEASTtrafo.html. Cells were grown in synthetic minimal medium and extracts prepared and assayed for NAD+- and NADP+-dependent methylenetetrahydrofolate dehydrogenase activity as described (30). 10-Formyl-THF synthetase activity was determined according to Kirksey and Appling (31).
Northern analysis— A FirstChoice Northern Human Blot I kit was obtained from Ambion (Austin, TX), with poly(A)+ mRNA from the following adult human tissues: brain, placenta, skeletal muscle, heart, kidney, pancreas, liver, lung, spleen, and thymus. Probes were synthesized by asymmetric PCR using reagents supplied in the kit and [α-32P]dATP according to the kit manufacturer's instructions. The two probes represented the 5′ and 3′ ends of the putative mitochondrial C1-THF synthase cDNA. The 5′ end probe was synthesized using the primers GS5′SOE on the sense strand and 5′CCGCTCGAGCAAGGCATTGAGGACTTTGTTGCT3′ on the antisense strand (GSI). This 304 bp probe covered nt +215 to +518 (The A of the ATG start codon is designated +1). The 3′ end probe was synthesized using the primers 5′GATGCAGTCCCCTGCTATCA3′ for the sense strand (DRA3) and 5′GAACAAGCCTTTAACTTGTTCTGTTTC3′ for the antisense strand (TOPO3′). This 465 bp probe covered nt +2469 to +2933, ending just before the stop codon.
A probe was also synthesized for detection of the human cytoplasmic C1-THF synthase. The plasmid pUC13/HS230 (obtained from R. E. MacKenzie, McGill Univ.), which contains a 230 bp fragment near the 3′ end of the human cytoplasmic C1-THF synthase cDNA (21), was linearized by digestion with SacI. A linear PCR amplification method, following the kit manufacturer's instructions, was used to synthesize the probe. The antisense primer used was 5′GTAAAACGACGGCCAGT3′, which is complementary to the vector sequences flanking the insert.
The membrane was subjected to a 1 h prehybridization at 42°C with Ultrahyb Ultrasensitive Hybridization Buffer (Ambion). Probe was added at 106 cpm/ml of hybridization buffer and allowed to hybridize at 42°C overnight in a roller bottle. The membrane was then washed twice with NorthernMax Low Stringency wash solution (Ambion; equivalent to 2XSSC) for 10 minutes and twice with NorthernMax High Stringency wash solution (equivalent to 0.1XSSC) for 30 minutes, all at 42°C. The membrane was then exposed to a Storage Phosphor Screen (Molecular Dynamics) for 48 hours and imaged using a Molecular Dynamics 445 SI PhosphoImager. The same blot was stripped and reconstituted for hybridization with each probe according to the kit manufacturer's instructions.
Transcript Mapping-- The 5′ and 3′ ends of the transcripts were mapped by RNA Ligase Mediated Rapid Amplification of cDNA Ends (RLM-RACE) using the First Choice RLM-RACE kit from Ambion. Total human placental RNA (Ambion) was used to map the 5′ end of the transcript. Nested antisense primers specific to the cDNA were designed for use with the two nested 5′ RACE primers provided in the kit. The cDNA-specific inner primer (GSI2: 5′CGCCTCGAGACGGCTGGTTCTCAGGGGACAC3′; XhoI site underlined) was complementary to nt -9 to -30 in the 5′UTR. The cDNA-specific outer primer (GSO2: 5′AGCGCGACAGGGCACACGGAG3′) was complementary to nt +93 to +73. The 5′ RACE inner primer and the cDNA-specific inner primer had BamH1 and Xho1 sites, respectively at their 5′ ends to facilitate cloning.
For mapping the 3′ end of the 1.1 kb transcript, first strand cDNA was synthesized from total human placental RNA, using the supplied 3′ RACE adapter. Nested sense primers specific to the cDNA were designed for use with the two nested 3′ RACE primers provided in the kit. The cDNA-specific inner primer (3′RACE GSI: 5′CGCCTCGAGGAACTTGTTTAGCAACAAAGTCCT3′; XhoI site underlined) was equivalent to +485 to +508. The cDNA-specific outer primer (3′RACE GSO: 5′CGCCTCGAGCTCCCTCCAGATAGCAGTGAA3′) was equivalent to +390 to +410. The 3′ RACE inner primer and the cDNA-specific inner primer had BamH1 and Xho1 sites, respectively at their 5′ ends to facilitate cloning.
PCR fragments generated in the ‘inner’ PCR reactions of both 5′ and 3′ RACE were gel-purified, digested with BamHI and XhoI, and ligated separately into BamH1/Xho1-digested pBluescript II KS (+) vector (Stratagene, La Jolla, CA). The ligation reactions were transformed into chemically competent XL1-Blue cells (Stratagene) and positive colonies were selected on YT + ampicillin plates. Colonies were screened by PCR using T7 reverse (5′GTAATACGACTCACTATAGGGC3′) and T3 forward (5′AATTAACCCTCACTAAAGGG 3′) vector primers and plasmids were prepared for sequence analysis. This 1.1 kb cDNA has GenBank Accession # AY374131.
Results
cDNA Identification and Cloning – A cDNA encoding an ORF with high similarity to human cytoplasmic C1-THF synthase was cloned from human uterine RNA by the German Genome Project (RZPD) (GenBank AL117452). The homology extended the length of the proteins, suggesting that this encoded another trifunctional C1-THF synthase (Fig. 2). This cDNA encoded 917 amino acids plus 390 nt of 3′ noncoding sequence and a poly(A) tail, but lacked a start codon, suggesting that it was truncated at the 5′ end. Blasting this sequence against the Human Genome Database (NCBI) revealed the corresponding gene on chromosome 6, at 6q25.2. This gene, spanning 236 kbp, encodes the entire cDNA sequence in 27 exons, plus an additional 5′ exon that encodes 60 additional N-terminal amino acids. The predicted initiator codon sits within a near-perfect expanded Kozak consensus sequence (32). The first half of this N-terminal extension has the characteristics of a mitochondrial leader sequence, including the potential to form a postively charged amphipathic α helix. Truncation of the original cDNA clone was due to the presence of a Not1 site near the 3′ end of the first exon; Not1 was used in the cDNA cloning procedure (20). Subsequently, the RIKEN Mouse Gene Encyclopedia Project (33) identified a full-length mouse cDNA (ID:22289) that predicts a protein with 88% identity to the human protein, including the N-terminal extension (Fig. 2). The mouse cDNA lacks the Not1 site that caused truncation of the human cDNA. These data suggested that the gene on human chromosome 6 encodes a mitochondrial C1-THF synthase.
Figure 2.
Alignment of human and mouse mitochondrial C1-THF synthase with human cytoplasmic C1-THF synthase. A black box denotes identity, and a white box denotes conservative substitutions or identities in two out of three proteins. The alignment was produced by the INRA server at the Laboratoire de Génétique cellulaire (http://prodes.toulouse.inra.fr/multalin/multalin.html) using the MultAlin algorithm (56) and the output was generated by the ESPript program at the same site. hmito; human mitochondrial C1-THF synthase; mmito, mouse mitochondrial C1-THF synthase; hcyto, human cytoplasmic C1-THF synthase.
Attempts to construct a full-length cDNA by RACE using human uterine RNA were unsuccessful, probably due to the extremely high G+C content (>80%) of the first exon. Instead, a genomic PAC clone (dJ44A20; Sanger Centre, UK) was used to PCR amplify the 5′ exon. This was then spliced to the remaining cDNA by SOE PCR to construct a full-length cDNA encoding the human protein (GenBank Accession # AY374130).
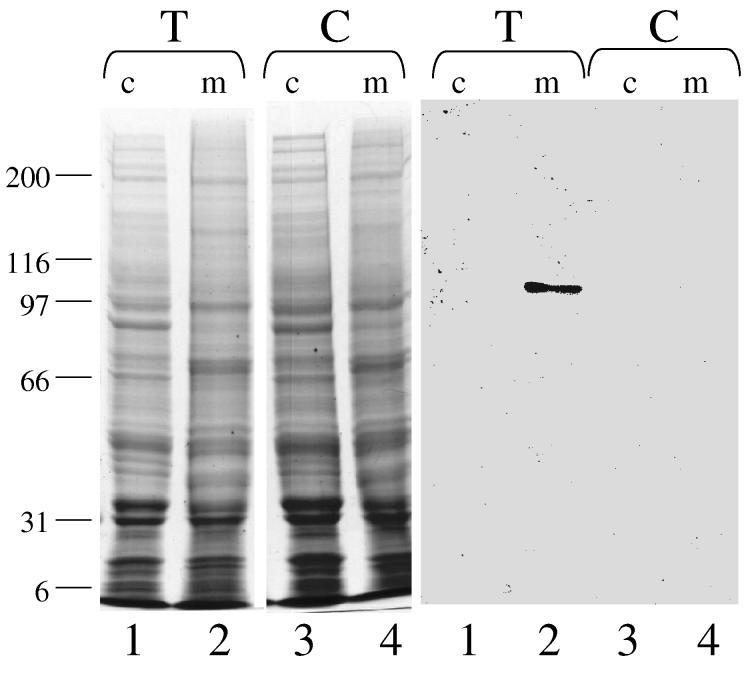
CHO Cell Expression and Subcellular Localization—In order to determine whether the protein encoded by this cDNA was, in fact, mitochondrial, we expressed the cDNA in CHO cells. The full-length cDNA was cloned into the mammalian expression vector, pcDNA3.1D/V5-His-TOPO. This construct fused the 14-amino acid V5 epitope and a 6-His tag to the C-terminus of the 2934 bp coding region. Expression of the insert in mammalian cells is driven by the CMV promoter. This plasmid, pcDNA/humito, was transfected into CHO cells and G418-resistant colonies were selected and grown. The cytosolic and mitochondrial fractions from transfected and untransfected (control) CHO cells were isolated as described in Experimental Procedures. Each fraction was assayed for the mitochondrial marker enzyme glutamate dehydrogenase and the cytoplasmic marker enzyme lactate dehydrogenase. Glutamate dehydrogenase activity ranged from 68-95 μmol/min/mg protein in the mitochondrial fractions, as compared with 2.4 - 4 μmol/min/mg protein in the cytoplasmic fractions. The lactate dehydrogenase activity of the mitochondrial fraction was only 1/7 that of the cytoplasmic fraction. These subcellular fractions were then subjected to SDS PAGE and immunoblotting using antibodies against the V5 epitope (Fig. 3). A clear signal at approximately 107 kDa was detected in the mitochondrial fraction of the transfected CHO cell line (lane 2), but not the cytoplasmic fraction (lane 1). This mobility is consistent with the expected size of the epitope-tagged construct (∼1000 amino acids). No signal was seen in either fraction of the untransfected CHO cell line (lanes 3, 4). These results confirm that this cDNA encodes a protein that localizes exclusively to mitochondria in a mammalian cell line.
Figure 3.
Subcellular localization of epitope-tagged human mitochondrial C1-THF synthase expressed in CHO cells. Coomassie blue-stained SDS PAGE (left) and immunoblot (right) of cytoplasmic (c) and mitochondrial (m) fractions from CHO cells transfected (T) with either pcDNA3/humito (lanes 1,2) or untransfected control (C) (lanes 3,4). Each lane contains 80 μg total protein.
Expression in Yeast—The full-length human mitochondrial C1-THF synthase cDNA, including the 62 codon N-terminal extension, was subcloned into a yeast expression vector (pVT103U) and transformed into an ade3 deletion strain, DAY3. Disruption of the ADE3 gene, encoding the cytoplasmic C1-THF synthase, results in yeast cells with very low 10-formyl-THF synthetase and 5,10-methylene-THF dehydrogenase activities; the residual activity is due to the mitochondrial isozyme (34). DAY3 cells transformed with pVT-humito overexpressed 10-formyl-THF synthetase activity approximately 9-fold compared to cells transformed with empty vector (64.6 vs. 7.1 milliunits/mg protein). However, we did not detect any increase in 5,10-methylene-THF dehydrogenase activity in cells carrying the pVT-humito plasmid, using either NADP+ or NAD+ as cofactor. The third activity of C1-THF synthase, 5,10-methenyl-THF cyclohydrolase, was not assayed since it is difficult to accurately measure this activity in crude extracts. These results, together with the mitochondrial localization data above, confirm that this cDNA encodes a protein with 10-formyl-THF synthetase activity, further supporting its identification as the human mitochondrial C1-THF synthase.
Gene Structure—The human gene encoding C1-THF synthase spans 236 kbp on chromosome 6 (Fig. 4). The coding sequence consists of 28 exons and is interrupted by 27 introns of length ranging from 89-55,350 bp. The start codon is present in the first exon, and the 5′ end of exon 1 extends 107 bp upstream of the ATG start codon (see Transcript Mapping, below). The stop codon is present in exon 27 and exon 28 encodes 360 nt of 3′ UTR, including a polyadenylation signal (AATAAA). Exon 1 is very GC-rich (>80% G+C), containing a CpG island and a Not1 restriction enzyme site (GCGGCCGC). The existence of this Not1 site prevented the cloning of a full-length cDNA since Not1 linkers were used in the cloning procedure (20). All of the intron-exon splice sites follow the GT/AG rule (35), except after the terminal exons (8a and 28) (Table II). A scan of the 5′ flanking sequences by the TESS web server (http://www.cbil.upenn.edu/tess) using the TRANSFAC v4.0 database predicts numerous potential transcription factor binding sites, including Sp1, RARαq and C/EBPα. The 5′ flanking sequence contains a TATAAA sequence at -985.
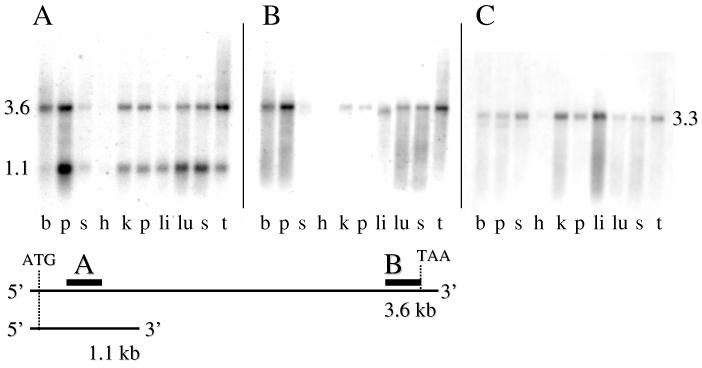
Northern Analysis— A Northern blot membrane pre-bound with human poly(A) RNA from several tissues was obtained from Ambion. A 304 base pair 5′ end probe, spanning nt 215-518 of the human mitochondrial C1-THF synthase cDNA revealed two bands: one at about 3.6 kb and the other at approximately 1.1 kb (Fig. 5A). The upper band corresponded to the expected size of the full-length transcript. To ensure that the 1.1 kb band was not an artifact, we washed the membrane for an additional 30 minutes in high stringency wash buffer at 50°C. The additional wash did not eliminate either band. The upper and lower band distributions were very similar with the highest transcript levels being in placenta, thymus, and brain. Expression was low in liver and skeletal muscle, and barely detectable in heart.
Figure 5.
Northern blot analysis of mitochondrial C1-THF synthase transcripts in adult human tissues. A multiple human tissue RNA blot was hybridized with 32P-labeled probes to the 5′ end (panel A) or 3′ end (panel B) of human mitochondrial C1-THF synthase cDNA. In panel C, the membrane was hybridized with a probe from the 3′ end of the human cytoplasmic C1-THF synthase cDNA. The lanes on each membrane contain RNA from (left to right) brain, placenta, skeletal muscle, heart, kidney, pancreas, liver, lung, spleen, and thymus. The schematic diagram below shows the relative locations of the probes used for panels A and B on the 3.6 and 1.1 kb transcripts.
To determine the relationship of the 3.6 kb and 1.1 kb transcripts, a 465 bp probe was synthesized that ended just before the stop codon. This 3′ probe detected only the 3.6 kb transcript (Fig. 5B), suggesting that the 1.1 kb transcript represented just the 5′ end of the cDNA.
We also compared the tissue distribution of the mitochondrial C1-THF synthase transcript to that of the cytoplasmic isozyme. Using a 230 bp probe from the 3′ end of the cytoplasmic C1-THF synthase cDNA (21), a 3.3 kb transcript was observed (Fig. 5C). The tissue distribution of this transcript differed from that of the mitochondrial isozyme, being highest in the liver, kidney, and skeletal muscle. Thus, the human mitochondrial and cytoplasmic C1-THF synthase isozymes are encoded by distinct transcripts that do not cross hybridize under these probe and wash conditions.
Transcript mapping—A 5′ RACE experiment was done to determine the transcriptional start site(s). 5′ RACE was performed as described under Experimental Procedures using 10 μg of human placental total RNA for first strand cDNA synthesis by reverse transcription. This was followed by a first round of PCR (outer PCR), which gave no detectable specific product. Two μl of the outer PCR product were used in a second round of PCR with nested primers (inner PCR), yielding a specific product of size <300 bp. The final PCR product was gel-purified and subcloned. Nine colonies were screened by PCR and all of them gave a product of size between 220 to 298 bp. Three of the 9 clones were sequenced and all of them exhibited the same 5′ end 107 bp upstream of the ATG start codon (Fig. 6). These results suggest that the majority of the transcripts from this gene initiate at or near –107, and it appears that both the 3.6 and 1.1 kb transcripts initiate from this site.
Figure 6.
5′ flanking sequence and exon 1 of human mitochondrial C1-THF synthase gene. Nucleotides are numbered with the A of the ATG start codon as +1. Coding nucleotides are in upper case; lower case nucleotides represent 5′ non-coding or intron sequences. The cDNA-specific outer and inner primers used in the 5′RACE are indicated by the arrows labeled GSO2 and GSI2, respectively. The arrow beginning at –107 indicates the transcriptional start site based on 5′RACE. The asterisks indicate the 5′ ends of ESTs found in the Human EST Database.
Alternative Splicing—A 3′ RACE experiment was performed to determine the 3′ end of the short 1.1 kb transcript observed on Northern blots (Fig. 5A). One μg of human placental total RNA was used for first strand cDNA synthesis. This was followed by a first round of PCR (outer PCR), which gave no detectable specific RACE product. One μl of the outer PCR product was used in a second round of PCR with nested primers (inner PCR). Four distinct PCR products of sizes 500 bp, 350 bp, 200 bp, and 100 bp were detectable on a 2% agarose gel (a smear at the top of the gel was also observed, produced from full-length transcript). Based on the 1.1 kb length of the short transcript, and the position of the inner primer, the 500 and 350 bp RACE products were gel-purified and cloned separately. Six of the clones were sequenced to determine the 3′ extent of the clones. All of these clones represented the short transcript, in which exon 7 was spliced to a previously unrecognized exon, termed exon 8a, which sits in the intron between exons 7 and 8 (Fig. 7A). Exon 8a appears to be 139 long, although in one clone the 3′ end extended 162 bp. It contains a stop codon after 45 nucleotides and a polyadenylation signal near its 3′ end. Thus the 3.6 and 1.1 kb transcripts share the first 7 exons, and then diverge at exon 8/8a. The 1.1 kb transcript would be translated into a 275 amino acid protein in which the first 260 amino acids are identical to the full-length protein, followed by 15 unrelated amino acids (Fig. 7B,C)(GenBank Accession # AY374131).
Figure 7.
Alternative splicing of human mitochondrial C1-THF synthase transcript. A. Gene structure and splicing pattern. Alternative exon 8a is in the intron between exons 7 and 8. In the 3.6 kb transcript, exon 7 is spliced to exon 8. In the 1.1 kb transcript, exon 7 is spliced to exon 8a. Exon 8a contains a stop codon (black dot) after 15 sense codons, and contains a polyadenylation signal (AATAAA) near its 3′ end. There is no homology between exons 8a and 8. B. Potential protein products. The long transcript is translated into a 978 aa protein, whereas the short transcript is translated into a 275 aa protein. The amino acid sequence of the two proteins is identical through the first 7 exons, encoding 260 aa. The short protein has 15 aa from exon 8a in place of the aa encoded by exon 8. The junction between the dehydrogenase/cyclohydrolase (D/C) and synthetase (SYN) domains is predicted to lie within aa 330-350 (Fig. 2). The asterisk (*) indicates variable 3′ splice site selection at the exon 6/7 junction (see Fig. 8). C. Nucleotide and amino acid sequence of coding sequence of exon 8a. Sequences 3′ to the stop codon not shown.
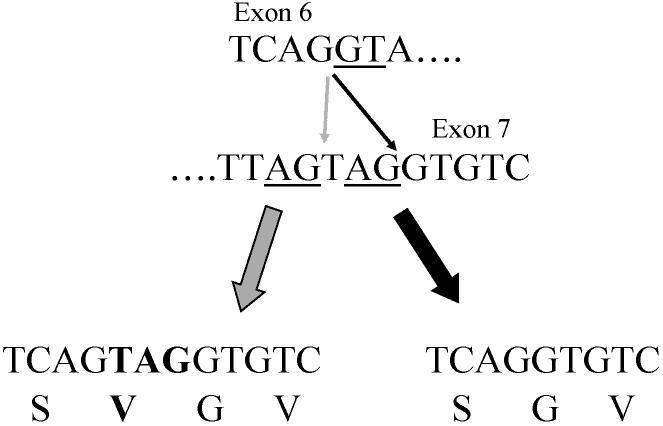
An additional variation was observed upon sequencing the 3′RACE clones. Several of the clones contained extra codon at position +643, at the junction between exons 6 and 7 (Fig. 8). This extra valine codon appears to arise from variation in the 3′ splice acceptor site during the splicing of exon 6 to exon 7. The 5′ splice site has the consensus GT as the first 2 nt of the intron. The 3′ splice site has two AG consensus dinucleotides at the 3′ end of the intron. If the first AG dinucleotide is used, exon 7 contains 3 additional nt; if the second is used, these 3 nt are not present in exon 7.
Figure 8.
Variable 3′ splice site selection at exon 6/7 junction. Top, intron/exon junctions for 3′ end of exon 6 and 5′ end of exon 7. The first two nt of the intron, and the alternative AG splice acceptors sites, are underlined. Bottom, alternative splicing products if the first (left) or second (right) AG acceptor site is used. The amino acid sequence encoded by each product is shown below the nt sequence, and the extra codon and amino acid are in bold.
Discussion
The experiments described here confirm that humans express a mitochondrial C1-THF synthase, with properties very similar to the cytoplasmic homologues previously characterized. The full-length human cDNA encodes a protein of 978 amino acids, including an N-terminal mitochondrial targeting sequence. When the full-length cDNA was expressed in CHO cells, the targeting sequence directed the protein exclusively to mitochondria (Fig. 3). Alignment of the deduced amino acid sequence with the human cytoplasmic C1-THF synthase (935 residues) revealed a 62 residue N-terminal extension in the putative mitochondrial protein (Fig. 2). PSORT II analysis (http://psort.nibb.ac.jp/form2.html) predicts a mitochondrial targeting sequence with a cleavage site between residues 31 and 32. The next 31 residues, before alignment with the cytoplasmic protein begins, include an unusual run of 9 consecutive glycines and several basic residues. A very similar N-terminal extension is predicted for the mouse protein (Fig. 2). Excluding this N-terminal extension, homology with the human cytoplasmic C1-THF synthase is quite high (61% identity), and the putative mitochondrial protein appears to possess the same domain structure. In the cytoplasmic protein, the N-terminal dehydrogenase/cyclohydrolase domain is about 300 residues and the C-terminal synthetase domain is about 700 residues (9). The two human proteins share 31% identity in the dehydrogenase/cyclohydrolase domains, and 73% identity in the synthetase domains, including conserved active site residues and the 10-formyl-THF binding site in the synthetase domain (31). However, the putative mitochondrial proteins from human and mouse lack 12 amino acids near the junction between the two domains (position 318, Fig. 2).
Expression of the full-length cDNA in yeast revealed elevated 10-formyl-THF synthetase activity, further supporting its identification as the human mitochondrial C1-THF synthase. We were unable to detect increased 5,10-methylene-THF dehydrogenase activity in these cells, using either NADP+ or NAD+ as cofactor. Is the human mitochondrial enzyme multifunctional like its yeast counterpart? Given the low identity between the human cytoplasmic and mitochondrial isozymes in the dehydrogenase/cyclohydrolase domain (31%), it is conceivable that the mitochondrial protein has lost these activities. However, other members of this family have diverged as much or more (e.g. the yeast MTD1p, ref. (30) and still retained 5,10-methylene-THF dehydrogenase activity. A second possibility is that the dehydrogenase activity of the human enzyme is simply below detection in crude yeast extracts. Depending on the species, the dehydrogenase activity of these trifunctional enzymes is only 1/2 to 1/10 that of the synthetase activity (2-9). Finally, the construct we expressed in yeast contained the entire 62 amino acid N-terminal extension. The 10-formyl-THF synthetase activity was found in the soluble, cytoplasmic fraction, but not the mitochondrial fraction (data not shown), suggesting that the presequence was not processed. If it is retained, this extension could interfere with the dehydrogenase/cyclohydrolase activities contained in the N-terminal domain of the protein, while leaving the C-terminal synthetase domain unaffected. These questions will have to await purification of the recombinant enzyme.
Expression of the gene was detected in most human tissues, but transcripts were highest in placenta, thymus, and brain. Expression was low in liver and skeletal muscle, and barely detectable in heart. A mouse cDNA has also been identified that predicts a protein with 88% identity to the human protein, including the N-terminal extension, suggesting that this mitochondrial C1-THF synthase will be found in all mammals.
The human gene encoding this enzyme has several interesting features. The gene is large, spanning 236 kbp on chromosome 6 at 6q25.2. The gene contains 29 exons (Table II), including the alternative exon 8a found in the intron between exons 7 and 8 (Fig. 4). This same intron/exon structure is observed for the mouse homolog found on mouse chromosome 10, except that the alternative exon 8a is absent from the mouse gene. Moreover, the genes for the cytoplasmic C1-THF synthase from rat, mouse, and human have all been shown to contain 28 exons, with introns in nearly identical positions (36,37). This suggests that an ancestral C1-THF synthase gene arose before the divergence of the human and rodent lineages, more than 75 million years ago (38), and genes encoding the mitochondrial and cytoplasmic isozymes are probably related by a gene duplication event.
The full-length 3.6 kb transcript is encoded in 28 exons. A shorter, 1.1 kb transcript is produced by an alternative splicing event, in which exon 7 is spliced to exon 8a instead of 8 (Fig. 7). This transcript encodes a 275 amino acid protein in which the first 260 amino acids are identical to the full-length protein, followed by 15 amino acids not found in any other C1-THF synthase. The first 11 aa of these 15 terminal amino acids are also found, with one mismatch, near the C-terminus of isoform 2 of the human α1A-adrenergic receptor, and the nucleotide sequence encoding these amino acids has high homology to an Alu repeat subfamily (39). The first 91 nt of exon 8a share 87% identity with the right half of the consensus Alu-Sc subfamily (GenBank accession # U14571), suggesting that exon 8a was derived from an Alu element that inserted, in the antisense orientation, into the intron between exons 7 and 8. This insertion is not present in the mouse homolog, since Alu elements are found only in primates (40). The exonization and alternative splicing of this Alu sequence in the human mitochondrial C1-THF synthase gene is apparently due to accumulated mutations in the Alu element that produce a functional 3′ splice site (41).
Assuming the short transcript is translated in vivo, it is unlikely that the resulting protein would retain 5,10-methylene-THF dehydrogenase or 5,10-methenyl-THF cyclohydrolase activity. Modeling the human mitochondrial protein sequence onto the X-ray structure of the dehydrogenase/cyclohydrolase domain of the human cytoplasmic C1-THF synthase (42) reveals that exons 8 and 9, which are missing in the short transcript, encode the major portion of the Rossman fold of the NADP-binding site, and a critical α-helix that forms one wall of the folate-binding site. It is likely that a truncated protein lacking these structural elements would not fold into a stable structure and would be rapidly degraded. However, without knowing how the 15 novel amino acids affect the structure, it remains possible that a stable protein with altered function could be produced. Experiments are underway to determine whether a truncated form of the protein is expressed in vivo.
Using RNA from human placenta, a single 5′ transcriptional start site at –107 was identified by 5′RACE (Fig. 6). It appears that both the 3.6 kb and 1.1 kb transcripts initiate from this site, since only a single 5′ end was identified. A Blast search of the Human EST database with the 5′ end of the human cDNA revealed more than 100 entries. Four ESTs extended beyond position –107 (Fig. 6). BG481636 (-276) and BE735249 (-268) were isolated from choriocarcinoma mRNA; BQ062382 (-119); and BQ055629 (-118) were isolated from a lymphoma cell line. Thus, it appears there may be some heterogeneity in the 5′ transcriptional start site, depending on the tissue or cell type.
One additional splicing variation was discovered. Some transcripts contained an extra codon at position +643, at the junction between exons 6 and 7 (Fig. 8). This valine codon appears to arise from alternative usage of AG splice acceptor sites separated by 1 nt. This type of variation in splice site selection has been seen in several other mammalian genes, including human prothymosin α (43) and rat TGF-β type I receptor (44). The extra codon was observed in most of the 3′RACE clones we sequenced, and can be found in numerous human ESTs that represent the full-length transcript. There is no evidence to suggest that this alternative splice site selection is a regulated process. It may simply be due to “sloppiness” in the splicing mechanism when two AG splice acceptor sites fall so closely together.
Based on the X-ray structure of the dehydrogenase/cyclohydrolase domain of the human cytoplasmic C1-THF synthase (42), the extra valine is predicted to reside on the exposed loop between α-helix D2 and β-strand e. This loop is not part of the dehydrogenase/cyclohydrolase active site or the dimerization interface for this domain. It is thus possible that an extra valine at this position could be tolerated without affecting stability or activity of the protein. On the other hand, we do not know how the dehydrogenase/cyclohydrolase and synthetase domains interact, so it will be necessary to express the protein containing the extra valine to determine its effect.
The tissue distribution of the mitochondrial C1-THF synthase is quite different from that of the cytoplasmic isozyme (Fig. 5). Whereas the cytoplasmic transcript is most abundant in liver and kidney, the transcripts for the mitochondrial isozyme are relatively low in those tissues, but highest in placenta, followed by thymus, spleen, brain and lung. The low expression of the mitochondrial isozyme in liver probably contributed to our earlier difficulties in purifying the protein from liver mitochondria. Although the ratio of the two transcripts varies somewhat from tissue to tissue, both are present in every tissue assayed, even heart (Fig. 5). The short transcript is significantly reduced in brain. Future work will be directed towards understanding the metabolic role of the mitochondrial isozyme and how that role relates to the observed tissue distribution.
The discovery of the human gene for this mitochondrial C1-THF synthase confirms our model for the compartmentation of folate-mediated one-carbon metabolism in mammalian cells (Fig. 1). Based on the well-documented existence of a mitochondrial C1-THF synthase in yeast (3,45), we proposed that mammalian mitochondria also contained this trifunctional enzyme (10). All three activities of C1-THF synthase are found in mammalian mitochondria (10). More importantly, intact rat liver mitochondria and mitochondrial extracts were shown to oxidize carbon 3 of serine to formate by the folate-dependent pathway outlined in Fig. 1 (mitochondrial reactions 1-4) (11). However, all our attempts to purify these activities from rat liver mitochondria were unsuccessful.
During this same time period, MacKenzie and coworkers characterized a mammalian bifunctional NAD-dependent 5,10-methylene-THF dehydrogenase/5,10-methenyl-THF cyclohydrolase, originally isolated from ascites tumor cells (12,13). This bifunctional enzyme lacks the large C-terminal domain catalyzing the 10-formyl-THF synthetase activity, and thus is unable to produce formate. When this enzyme was shown to be localized in mitochondria (14,15), MacKenzie proposed that mammalian mitochondria lack a trifunctional C1-THF synthase, and that this bifunctional NAD-dependent dehydrogenase/cyclohydrolase is the mammalian homolog of the mitochondrial trifunctional enzyme (18,19). There are, however, several problems with this proposal. First, the bifunctional enzyme is detectable mainly in transformed mammalian cells and embryonic or nondifferentiated tissues (12). Among adult differentiated tissues, NAD-dependent 5,10-methylene-THF dehydrogenase activity is detectable only in rat adrenal tissue, but not adult liver (16). Secondly, the 5,10-methylene-THF dehydrogenase activity we detected in rat liver mitochondria was dependent on NADP+, not NAD+ (11). Finally, adult rat liver mitochondria are capable of producing formate by the folate-dependent pathway (11), and formate production requires the 10-formyl-THF synthetase activity (Fig. 1, reaction 1) that is missing from the bifunctional enzyme. Clearly, only a trifunctional C1-THF synthase, with an NADP-dependent 5,10-methylene-THF dehydrogenase activity, is consistent with the biochemical data.
Mitochondrial C1-THF synthase probably supports several metabolic processes in mammalian mitochondria. Folate-mediated one-carbon metabolism is involved in the synthesis of formyl-methionyl-tRNA for mitochondrial protein synthesis (46,47) (Fig. 1, reaction 8), and the oxidation of choline methyl groups via dimethylglycine dehydrogenase and sarcosine dehydrogenase (48). Mitochondrial C1-THF synthase may also play an important role in homocysteine metabolism. Recent studies of patients with nonketotic hyperglycinemia (NKH) reveal a connection between the mitochondrially-localized glycine cleavage system (GCS) (Fig. 1, reaction 5) and homocysteine metabolism. NKH is an autosomal recessive brain disease caused by defects in subunits of the GCS, resulting in elevated glycine levels (49). Loss of GCS activity might be expected to cause, in addition to elevated glycine, a deficiency of mitochondrial one-carbon units. Consistent with this hypothesis, two recent studies report mild elevations of homocysteine in plasma and cerebrospinal fluid of NKH patients (50,51). Furthermore, Randak et al (51) found that the mildly elevated plasma homocysteine levels could be reduced in their three patients by treatment with the one-carbon donor 5-formyl-THF (folinic acid, leucovorin). This observation provides strong evidence that the homocysteine elevations are due to a defect in homocysteine remethylation resulting from a deficiency of one-carbon units. Examination of Fig. 1 suggests two ways in which a loss of mitochondrial GCS activity could cause a deficiency of cytoplasmic one-carbon units. First, as suggested by Van Hove et al (50), cells lacking a functional GCS might increase the transport of serine into mitochondria for metabolism by SHMT in order to compensate for the deficiency of mitochondrial 5,10-methylene-THF. This could in turn cause a deficiency of serine, and thus one-carbon units, in the cytoplasm. A second possibility is that formate production is defective in mitochondria from NKH patients. As we showed both in vitro, with rat liver mitochondria (10,11), and in vivo in yeast (28,52), mitochondrial 5,10-methylene-THF is rapidly converted to formate and transported to the cytosol where it is activated to 10-formyl-THF via cytoplasmic 10-formyl-THF synthetase (Fig. 1, mitochondrial reactions 3, 2, 1 + cytoplasmic reaction 1). The 10-formyl-THF is then reduced by cytoplasmic reactions to the 5-methyl-THF required for homocysteine remethylation. Consistent with this explanation is the observation that GCS activity is stimulated by glucagon (53), and glucagon lowers plasma homocysteine in rats (54), presumably by increasing the mitochondrial production of formate. An elegant stable isotope study in humans (55) provides further support for the role of mitochondrial one-carbon units in the remethylation of homocysteine. Gregory et al (55) showed that both cytoplasmic and mitochondrial one-carbon units end up in the methyl group of methionine following infusion of deuterated serine. This result strongly supports mitochondrial formate production as a significant contributor to cytoplasmic one-carbon units in vivo in mammals and places the mitochondrial C1-THF synthase in the center of this pathway.
Table I.
Intron/exon splice junctions of the human mitochondrial C1-THF synthase gene
| Exon number | Exon Size (bp) | Intron sequence | Exon sequence | Intron sequence | Intron Size (bp) | Intron numberb |
|---|---|---|---|---|---|---|
| 5′ junction | 3′ junction | |||||
| 1 | 356a | TCCTTC---CGTCAG | gtgagtgtc | 9860 | 1 | |
| 2 | 85 | tcaatgtag | AGAAGT---ATCCAG | gtaagccga | 1460 | 2 |
| 3 | 51 | cttcctaag | GCAGGT---GAGGAG | gtgaggact | 89 | 3 |
| 4 | 54 | ctttttcag | GCTGGT---GCCGAG | gtaataatg | 4936 | 4 |
| 5 | 125 | tctctttag | ATTATA---GGATGG | gtaagaaaa | 2748 | 5 |
| 6 | 101 | ctgattcag | AGTAAC---AATCAG | gtaggatgc | 2108 | 6 |
| 7 | 137 | tttggttag | GTGTCA---AGCAAG | gtaaatttc | 10879 | 7 |
| 8ac | 139 | ctttttcag | ACGGAG---TCCCAG | aatggtgct | 6652 | 8a |
| 8 | 112 | tccttttag | CTTCAC---TGTCAG | gtaaatgtc | 12816 | 8 |
| 9 | 92 | tttttacag | GGAAGG---ATTCAG | gtttgttca | 3537 | 9 |
| 10 | 98 | ctgctgaag | AACATG---GCCAAG | gtaacactg | 3820 | 10 |
| 11 | 174 | tcccctcag | TGACAT---TGCTGG | gtaagacac | 10509 | 11 |
| 12 | 137 | cgaactcag | GATCAC---TGAAAG | gtactgtct | 1759 | 12 |
| 13 | 47 | gttgtttag | GAGGAG---GAGGAG | gtaagacct | 5741 | 13 |
| 14 | 108 | tttctctag | TTCAAC---GATAAG | gtaagaagg | 874 | 14 |
| 15 | 75 | tgtttttag | GCTCTG---CTAAAA | gtaagtttc | 3490 | 15 |
| 16 | 103 | aatcattag | AAACTG---AGAGAG | gtgggtgct | 6862 | 16 |
| 17 | 77 | ctctttcag | TATTGG---CGGCAG | gtaggtggt | 4204 | 17 |
| 18 | 141 | ctggttcag | GCGCAG--GATTTG | gtaagtgtt | 4554 | 18 |
| 19 | 69 | ctcctgtag | GGGGTG--CTGGAA | gtaagtggt | 6910 | 19 |
| 20 | 112 | cttctctag | GGGACA--TTGTAG | gtaagttat | 37900 | 20 |
| 21 | 140 | tccccacag | TGACCG---CCAAGT | gtaagtgcc | 3821 | 21 |
| 22 | 42 | ttatttcag | GTAACG---GAGGAG | gtaagagga | 1060 | 22 |
| 23 | 101 | tttttacag | AACATC---CTTCAA | gtaagtcca | 536 | 23 |
| 24 | 178 | cttcactag | GACCGA---GTTCAG | gtaagatct | 18800 | 24 |
| 25 | 108 | tttctccag | GTTCCA---CAACAG | gtaaaagtt | 2365 | 25 |
| 26 | 153 | ttctcacag | GGTTTT---GGAACG | gtgagtgag | 55350 | 26 |
| 27 | 121 | ctcccccag | ATGAGC---ATGCAG | gtaggctga | 8939 | 27 |
| 28 | 360 | atttttcag | ACTCCT---AAAACC | aaccagcaa | ||
The 5′ end of exon one, and therefore its length, is based on the longest 5′ RACE clone isolated (see text).
Introns are numbered starting with intron 1 betweens exon 1 and 2.
In one 3′RACE clone, exon 8a extended an additional 23 bp, for a total length of 162 bp.
Footnotes
Short Title: HUMAN MITOCHONDRIAL C1-THF SYNTHASE
The cDNA nucleotide sequences reported in this paper have been submitted to the GenBank/EBI Data Bank with accession numbers AY374130 and AY374131
This work was supported by National Institutes of Health Grants DK61428 (to D.R.A.) and DK42033 (to B.S.).
- aa
- amino acid
- BSA
- bovine serum albumin
- CHO
- chinese hamster ovary
- CMV
- cytomegalovirus
- DTT
- dithiothreitol
- PCR
- polymerase chain reaction
- UTR
- untranslated region
- aa
- amino acids
- FPGS
- folylpolyglutamate synthetase
- HMS
- homogenization solution
- kbp
- kilobase pair(s)
- nt
- nucleotide(s)
- PNS
- post nuclear supernatant
- RACE
- rapid amplification of cDNA ends
- SSC
- saline/sodium citrate
- THF
- tetrahydrofolate
References
- 1.Appling DR. FASEB J. 1991;5:2645–2651. doi: 10.1096/fasebj.5.12.1916088. [DOI] [PubMed] [Google Scholar]
- 2.Paukert JL, Williams GR, Rabinowitz JC. Biochem. Biophys. Res. Comm. 1977;77:147–154. doi: 10.1016/s0006-291x(77)80176-9. [DOI] [PubMed] [Google Scholar]
- 3.Shannon KW, Rabinowitz JC. J. Biol. Chem. 1986;261:12266–12271. [PubMed] [Google Scholar]
- 4.Paukert JL, Straus LDA, Rabinowitz JC. J. Biol. Chem. 1976;251:5104–5111. [PubMed] [Google Scholar]
- 5.Tan LUL, MacKenzie RE. Can. J. Biochem. 1979;57:806–812. doi: 10.1139/o79-100. [DOI] [PubMed] [Google Scholar]
- 6.Smith GK, Mueller WT, Wasserman GF, Taylor WD, Benkovic SJ. Biochem. 1980;19:4313–4321. doi: 10.1021/bi00559a026. [DOI] [PubMed] [Google Scholar]
- 7.Villar E, Schuster B, Peterson D, Schirch V. J. Biol. Chem. 1985;260:2245–2252. [PubMed] [Google Scholar]
- 8.Cheek WD, Appling DR. Arch. Biochem. Biophys. 1989;270:504–512. doi: 10.1016/0003-9861(89)90532-8. [DOI] [PubMed] [Google Scholar]
- 9.Hum DW, MacKenzie RE. Prot. Eng. 1991;4:493–500. doi: 10.1093/protein/4.4.493. [DOI] [PubMed] [Google Scholar]
- 10.Barlowe CK, Appling DR. Biofactors. 1988;1:171–176. [PubMed] [Google Scholar]
- 11.Garcia-Martinez LF, Appling DR. Biochem. 1993;32:4671–4676. doi: 10.1021/bi00068a027. [DOI] [PubMed] [Google Scholar]
- 12.Mejia NR, MacKenzie RE. J. Biol. Chem. 1985;260:14616–14620. [PubMed] [Google Scholar]
- 13.Mejia NR, Rios-Orlandi EM, MacKenzie RE. J. Biol. Chem. 1986;261:9509–9513. [PubMed] [Google Scholar]
- 14.Mejia NR, MacKenzie RE. Biochem. Biophys. Res. Comm. 1988;155:1–6. doi: 10.1016/s0006-291x(88)81040-4. [DOI] [PubMed] [Google Scholar]
- 15.Belanger C, MacKenzie RE. J. Biol. Chem. 1989;264:4837–4843. [PubMed] [Google Scholar]
- 16.Smith GK, Banks SD, Monaco TJ, Rigual R, Duch DS, Mullin RJ, Huber BE. Arch. Biochem. Biophys. 1990;283:367–371. doi: 10.1016/0003-9861(90)90656-j. [DOI] [PubMed] [Google Scholar]
- 17.Peri KG, MacKenzie RE. Biochim. Biophys. Acta. 1993;1171:281–287. doi: 10.1016/0167-4781(93)90066-m. [DOI] [PubMed] [Google Scholar]
- 18.Yang X-M, MacKenzie RE. Biochem. 1993;32:11118–11123. doi: 10.1021/bi00092a022. [DOI] [PubMed] [Google Scholar]
- 19.Di Pietro E, Sirois J, Tremblay ML, MacKenzie RE. Mol Cell Biol. 2002;22:4158–4166. doi: 10.1128/MCB.22.12.4158-4166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiemann S, Weil B, Wellenreuther R, Gassenhuber J, Glassl S, Ansorge W, Bocher M, Blocker H, Bauersachs S, Blum H, Lauber J, Dusterhoft A, Beyer A, Kohrer K, Strack N, Mewes HW, Ottenwalder B, Obermaier B, Tampe J, Heubner D, Wambutt R, Korn B, Klein M, Poustka A. Genome Res. 2001;11:422–435. doi: 10.1101/gr.154701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hum DW, Bell AW, Rozen R, MacKenzie RE. J. Biol. Chem. 1988;263:15946–15950. [PubMed] [Google Scholar]
- 22.Horton RM, Ho SN, Pullen JK, Hunt HD, Cai Z, Pease LR. Methods Enzymol. 1993;217:270–279. doi: 10.1016/0076-6879(93)17067-f. [DOI] [PubMed] [Google Scholar]
- 23.Lin B-F, Huang R-FS, Shane B. J. Biol. Chem. 1993;268:21674–21679. [PubMed] [Google Scholar]
- 24.Schmidt E. In: Methods of Enzymatic Analysis. 2nd Bergmeyer HU, editor. Vol. 2. Academic Press; New York: 1974. pp. 650–656. [Google Scholar]
- 25.Kornberg A. Meth. Enzymol. 1955;1:441–443. [Google Scholar]
- 26.Bradford MM. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 27.Vernet T, Dignard D, Thomas DY. Gene. 1987;52:225–233. doi: 10.1016/0378-1119(87)90049-7. [DOI] [PubMed] [Google Scholar]
- 28.West MG, Horne DW, Appling DR. Biochem. 1996;35:3122–3132. doi: 10.1021/bi952713d. [DOI] [PubMed] [Google Scholar]
- 29.Ito H, Fukuda Y, Murata K, Kimura A. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.West MG, Barlowe CK, Appling DR. J. Biol. Chem. 1993;268:153–160. [PubMed] [Google Scholar]
- 31.Kirksey TJ, Appling DR. Arch. Biochem. Biophys. 1996;333:251–259. doi: 10.1006/abbi.1996.0388. [DOI] [PubMed] [Google Scholar]
- 32.Kozak M. J Mol Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- 33.Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, Adachi J, Fukuda S, Aizawa K, Izawa M, Nishi K, Kiyosawa H, Kondo S, Yamanaka I, Saito T, Okazaki Y, Gojobori T, Bono H, Kasukawa T, Saito R, Kadota K, Matsuda HA, Ashburner M, Batalov S, Casavant T, Fleischmann W, Gaasterland T, Gissi C, King B, Kochiwa H, Kuehl P, Lewis S, Matsuo Y, Nikaido I, Pesole G, Quackenbush J, Schriml LM, Staubli F, Suzuki R, Tomita M, Wagner L, Washio T, Sakai K, Okido T, Furuno M, Aono H, Baldarelli R, Barsh G, Blake J, Boffelli D, Bojunga N, Carninci P, de Bonaldo MF, Brownstein MJ, Bult C, Fletcher C, Fujita M, Gariboldi M, Gustincich S, Hill D, Hofmann M, Hume DA, Kamiya M, Lee NH, Lyons P, Marchionni L, Mashima J, Mazzarelli J, Mombaerts P, Nordone P, Ring B, Ringwald M, Rodriguez I, Sakamoto N, Sasaki H, Sato K, Schonbach C, Seya T, Shibata Y, Storch KF, Suzuki H, Toyo-oka K, Wang KH, Weitz C, Whittaker C, Wilming L, Wynshaw-Boris A, Yoshida K, Hasegawa Y, Kawaji H, Kohtsuki S, Hayashizaki Y. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- 34.Barlowe CK, Appling DR. Mol. Cell. Biol. 1990;10:5679–5687. doi: 10.1128/mcb.10.11.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Breathnach R, Chambon P. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- 36.Patel H, Christensen KE, Mejia N, MacKenzie RE. Arch Biochem Biophys. 2002;403:145–148. doi: 10.1016/S0003-9861(02)00203-5. [DOI] [PubMed] [Google Scholar]
- 37.Howard KM, Muga SJ, Zhang L, Thigpen AE, Appling DR.Gene 2003. in press [DOI] [PubMed] [Google Scholar]
- 38.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O'Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 39.Chang DJ, Chang TK, Yamanishi SS, Salazar FH, Kosaka AH, Khare R, Bhakta S, Jasper JR, Shieh IS, Lesnick JD, Ford AP, Daniels DV, Eglen RM, Clarke DE, Bach C, Chan HW. FEBS Lett. 1998;422:279–283. doi: 10.1016/s0014-5793(98)00024-6. [DOI] [PubMed] [Google Scholar]
- 40.Sorek R, Ast G, Graur D. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lev-Maor G, Sorek R, Shomron N, Ast G. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt A, Wu H, MacKenzie RE, Chen VJ, Bewly JR, Ray JE, Toth JE, Cygler M. Biochem. 2000;39:6325–6335. doi: 10.1021/bi992734y. [DOI] [PubMed] [Google Scholar]
- 43.Manrow RE, Berger SL. J Mol Biol. 1993;234:281–288. doi: 10.1006/jmbi.1993.1583. [DOI] [PubMed] [Google Scholar]
- 44.Agrotis A, Condron M, Bobik A. FEBS Lett. 2000;467:128–132. doi: 10.1016/s0014-5793(00)01132-7. [DOI] [PubMed] [Google Scholar]
- 45.Shannon KW, Rabinowitz JC. J. Biol. Chem. 1988;263:7717–7725. [PubMed] [Google Scholar]
- 46.Galper JB, Darnell JE. Biochem. Biophys. Res. Comm. 1969;34:205–214. doi: 10.1016/0006-291x(69)90633-0. [DOI] [PubMed] [Google Scholar]
- 47.Bianchetti R, Lucchini G, Crosti P, Tortora P. J. Biol. Chem. 1977;252:2519–2523. [PubMed] [Google Scholar]
- 48.Lewis KF, Randolph VM, Nemeth E, Frisell WR. Arch. Biochem. Biophys. 1978;185:443–449. doi: 10.1016/0003-9861(78)90187-x. [DOI] [PubMed] [Google Scholar]
- 49.Hamosh A, Johnston MV, Valle D.Scriver CR, Beaudet AL, Sly WS, Valle D.The Metabolic and Molecular Bases of Inherited Disease 1995I1337–1348.McGraw-Hill; New York: 7th3 vols. [Google Scholar]
- 50.Van Hove JL, Lazeyras F, Zeisel SH, Bottiglieri T, Hyland K, Charles HC, Gray L, Jaeken J, Kahler SG. J Inherit Metab Dis. 1998;21:799–811. doi: 10.1023/a:1005462400552. [DOI] [PubMed] [Google Scholar]
- 51.Randak C, Rosschinger W, Rolinski B, Hadorn HB, Applegarth DA, Roscher AA. J Inherit Metab Dis. 2000;23:520–522. doi: 10.1023/a:1005667707355. [DOI] [PubMed] [Google Scholar]
- 52.Pasternack LB, Laude DA, Jr., Appling DR. Biochem. 1994;33:74–82. doi: 10.1021/bi00167a010. [DOI] [PubMed] [Google Scholar]
- 53.Jois M, Hall B, Fewer K, Brosnan JT. J. Biol. Chem. 1989;264:3347–3351. [PubMed] [Google Scholar]
- 54.House JD, Jacobs RL, Stead LM, Brosnan ME, Brosnan JT. Adv Enzyme Regul. 1999;39:69–91. doi: 10.1016/s0065-2571(98)00008-9. [DOI] [PubMed] [Google Scholar]
- 55.Gregory JF, 3rd, Cuskelly GJ, Shane B, Toth JP, Baumgartner TG, Stacpoole PW. Am J Clin Nutr. 2000;72:1535–1541. doi: 10.1093/ajcn/72.6.1535. [DOI] [PubMed] [Google Scholar]
- 56.Corpet F. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]