Abstract
Filopodia are highly dynamic finger-like cell protrusions filled with parallel bundles of actin filaments. Previously we have shown that Diaphanous-related formin dDia2 is involved in the formation of filopodia. Another key player for the formation of filopodia across many species is vasodilator-stimulated phosphoprotein (VASP). It has been proposed that the essential role of VASP for formation of filopodia is its competition with capping proteins for filament barbed-end interaction. To better understand the function of VASP in filopodium formation, we analyzed the in vitro and in vivo properties of Dictyostelium VASP (DdVASP) and extended our findings to human VASP. Recombinant VASP from both species nucleated and bundled actin filaments, but did not compete with capping proteins or block depolymerization from barbed ends. Together with the finding that DdVASP binds to the FH2 domain of dDia2, these data indicate that the crucial role of VASP in filopodium formation is different from uncapping of actin filaments. To identify the activity of DdVASP required in this process, rescue experiments of DdVASP-null cells with mutant DdVASP constructs were performed. Only WT DdVASP, but not a mutant lacking the F-actin bundling activity, could rescue the ability of these cells to form WT-like filopodia. Our data suggest that DdVASP is complexed with dDia2 in filopodial tips and support formin-mediated filament elongation by bundling nascent actin filaments.
Keywords: actin cytoskeleton, Dictyostelium, formin
Cytoskeletal dynamics in moving fronts, lamellipodia, or filopodia require fast and highly coordinated actin polymerization, which leads to broad filamentous networks or to spiky membrane protrusions. The Arp2/3 complex is known to nucleate branching of actin filaments, thus favoring the formation of dense F-actin meshworks, whereas formins efficiently elongate linear actin filaments (1). Although formin function could be clearly correlated with the establishment of actin cables and stress fibers (2, 3), we only recently began to understand its role in filopodium formation (4–6). The Dictyostelium Diaphanous-related formin dDia2 is associated with the distal tips of growing filopodia and is required for the elongation of filopodial actin filaments (6); however, growth and stabilization of filopodia coincide with recruitment of a number of actin-associated proteins, including the actin-bundling protein fascin (7), talin (8, 9), myosin VII and X (10, 11), Scar/Wave (12), and Irsp53 (13, 14), as well as Ena/vasodilator-stimulated phosphoprotein (VASP) proteins (15–18). It still remains to be shown how these proteins stabilize the emerging bundle (19). VASP is a member of the Ena/VASP family of proteins, which are implicated in regulation of the actin cytoskeleton and control of cell motility (20–22). Ena/VASP proteins were proposed to associate with the barbed ends of microfilaments, to protect them from capping protein (CP), and to support growth of filopodial actin filaments (17, 18, 23). Consistent with a crucial function of VASP, Dictyostelium mutants lacking Dictyostelium VASP (DdVASP) are severely impaired in filopodium formation (16). Here we show that DdVASP binds directly to the FH2 domain of dDia2, and we propose a molecular mechanism of filopodium formation in which DdVASP acts downstream of dDia2 by bundling nascent actin filaments, in turn leading to a stabilization of the filopodial tip complex.
Results
dDia2 Interacts Directly with VASP.
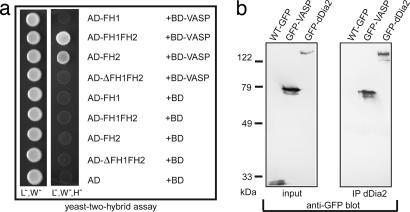
In Dictyostelium discoideum cells VASP localizes to the leading edge and the tips of filopodia and was found to play an important role in the formation of these structures (16, 24). Mouse mDia1 FH2 interacts with full-length murine VASP, and this interaction is enhanced by the presence of the FH1 domain (24). Because dDia2 is also strongly enriched in filopodial tips and is required for filopodia formation, we studied a possible interaction between dDia2 and DdVASP using the yeast two-hybrid technique. In contrast to the proline-rich FH1 domain alone and the N terminus of dDia2 (ΔFH1FH2), the FH1FH2 and the FH2 regions of dDia2 bound to VASP (Fig. 1a). This result indicated that the actin-binding FH2 domain mediates the interaction between the formin dDia2 and DdVASP. The observation that yeast cells expressing the FH1FH2 construct grew faster than those expressing FH2 alone suggested that FH1 might facilitate binding of FH2 to DdVASP. The absence of any interaction between dDia2 FH3 and DdVASP, as reported for other formins, like mDia1, is not surprising because they represent different isoforms in this protein family, are from different organisms, and exert different functions. Whereas mDia1 is a Rho effector, dDia2 more likely reflects the features of mDia2 (DRF3), having a function downstream of Cdc42 or Rif in filopodia formation.
Fig. 1.
DdVASP interacts with dDia2 in a yeast two-hybrid assay through its FH2 domain. (a) Yeast was transformed with the indicated constructs and tested for interaction by growth on selective media lacking leucine (L), tryptophane (W), or histidine (H) as indicated. AD, activation domain; BD, binding domain. (b) Coimmunoprecipitation of dDia2 and DdVASP. Cell lines expressing GFP, GFP-DdVASP, or GFP-dDia2 (Left) were used for immunoprecipitation with anti-dDia2 polyclonal antibodies. The immunoprecipitates were analyzed in Western blots with anti-GFP antibody mAb 264-449-2.
We then performed immunoprecipitations of dDia2 from cell lines expressing GFP alone, GFP-DdVASP, or GFP-dDia2 using polyclonal antibodies specific for dDia2. GFP-DdVASP was coimmunoprecipitated with dDia2, whereas GFP alone (which served as the negative control) was absent from dDia2 immunoprecipitates (Fig. 1b). These data indicate that VASP and dDia2 also interact in vivo.
VASP Nucleates Actin Assembly and Bundles but Does Not Cap Actin Filaments.
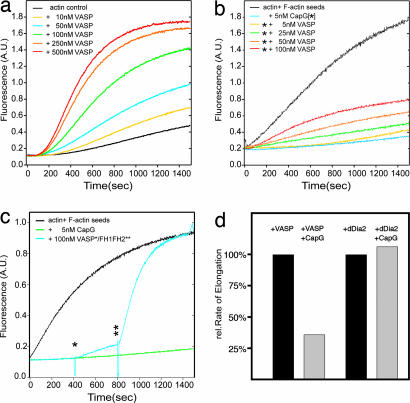
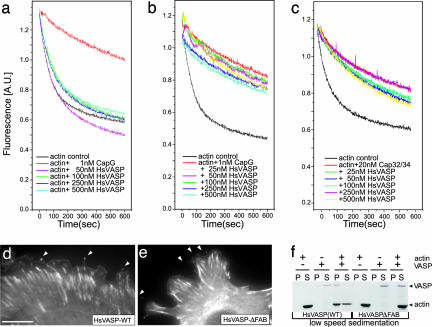
To explore the nature of the interaction between these two proteins, we characterized DdVASP on a biochemical level and compared its properties with those of dDia2 FH1FH2. DdVASP is an efficient nucleator of actin polymerization in low-ionic-strength buffers (up to 50 mM KCl) (Fig. 2a), but, in contrast to the uncapping activity of formins, DdVASP cannot remove CP such as mouse CapG or Cap32/34, the CapZ orthologue from D. discoideum, from capped filament seeds (Fig. 2b). Even a 20-fold excess of DdVASP over CP did not significantly alleviate the inhibitory effect of CP. The slight increase in polymerization is most likely the result of de novo nucleation of G-actin by DdVASP independent of the presence of capped F-actin seeds. Because of some controversy in the field, and with regard to the observation that both dDia2 and VASP localize to tips of filopodia, we compared the nucleating and uncapping activities of these two proteins in a combinatorial approach (Fig. 2c). A pyrene actin-based polymerization assay that was started in the presence of capped F-actin seeds resulted in a long lag phase, whereas the control with uncapped F-actin seeds showed immediate polymerization. The addition of 100 nM DdVASP after 400 sec to capped actin seeds caused a slow but significant actin polymerization. However, further addition of 100 nM FH1FH2 after 800 sec resulted in a steep burst of fluorescence, which indicated the uncapping of the F-actin seeds and enhanced elongation. The assumption drawn from this observation was confirmed in another assay, in which an excess of free CP was added to polymerizing actin in the presence of either DdVASP or dDia2 (Fig. 2d), showing that CP can inhibit actin polymerization in the presence of DdVASP but not in the presence of dDia2. From this result we conclude that DdVASP nucleates actin polymerization, most likely as a tetramer through its G-actin binding sites, and that efficient elongation of the de novo nuclei is inhibited by free CP. Conversely, FH1FH2 successfully competed with the CP at the barbed ends, resulting in uninterrupted elongation.
Fig. 2.
DdVASP nucleates actin assembly but does not compete with CP. (a) DdVASP promotes actin polymerization. Actin (1.1 μM) was polymerized in the presence of 0 nM (black line) to 500 nM (red line) DdVASP. (b) DdVASP does not compete with CP. Actin (1.4 μM) was polymerized in the presence of F-actin seeds to avoid de novo nucleation (black line). The presence of 5 nM CP strongly inhibited actin polymerization (light blue line). Addition of increasing amounts of DdVASP to capped filaments showed that DdVASP cannot remove the cap and merely nucleates polymerization of the remaining G-actin. (c) Direct comparison of DdVASP and formin activities. Actin was polymerized in the presence of actin seeds (black line) or capped actin seeds (green and blue lines). After 400 sec (one star) 100 nM DdVASP was added, and after 800 sec (two stars) 100 nM FH1FH2 was added to the same sample. The slope between 400 and 800 sec reflects normal DdVASP-induced de novo nucleation, whereas after addition of FH1FH2 the capped seeds were efficiently uncapped and elongated, leading to a fast increase of F-actin as indicated by the steeper slope. (d) Analysis of nucleating and uncapping activities. The intrinsic nucleation activity of 50 nM DdVASP was measured in the pyrene actin assay, and the slope of fluorescence increase was set to 100% (first bar). After addition of 30 nM CapG the fluorescence increase was strongly inhibited because the newly formed filaments were immediately capped at their barbed ends (second bar). This inhibition was not observed in the presence of FH1FH2 (third and fourth bars) because the formin competes with CP and supports elongation also in the presence of CP.
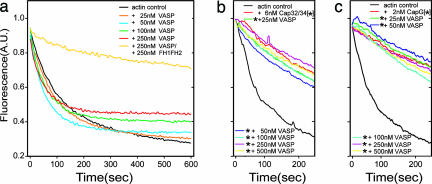
In a fourth experimental setup we used a depolymerization assay that allows the analysis of the molecular organization at the barbed end. Depolymerization occurs after dilution of F-actin solutions below the critical concentration of the barbed ends; i.e., under these conditions nucleating activity is irrelevant. The interaction of a protein of interest with the barbed end, however, should slow down the dissociation of actin monomers. Fig. 3a shows that even high concentrations of DdVASP did not inhibit depolymerization, whereas the addition of FH1FH2 from dDia2 considerably inhibited the depolymerization of actin. These findings indicate that DdVASP does not directly interact with barbed ends and thus is not able to compete with CP. Consistent with these data, actin filaments that were capped with Cap32/34 did not depolymerize from the barbed ends even at high concentrations of DdVASP (Fig. 3b). CapG, another type of barbed-end CP, could also not be removed by DdVASP under these conditions (Fig. 3c).
Fig. 3.
DdVASP does not interact with barbed filament ends. (a) Polymerized actin was diluted to 0.1 μM in polymerization buffer alone or in polymerization buffer containing DdVASP or DdVASP and FH1FH2. Note that depolymerization is not attenuated by the addition of DdVASP, indicating that it cannot alter barbed-end kinetics. (b and c) Polymerized actin with free barbed ends or with the barbed ends capped by either Cap32/34 or CapG was diluted to 0.1 μM in polymerization buffer containing 100 mM KCl and the amounts of DdVASP indicated. DdVASP removed neither Cap32/34 nor CapG from the filament ends, because this would have led to fast depolymerization. The stars in b and c indicate the presence of CPs.
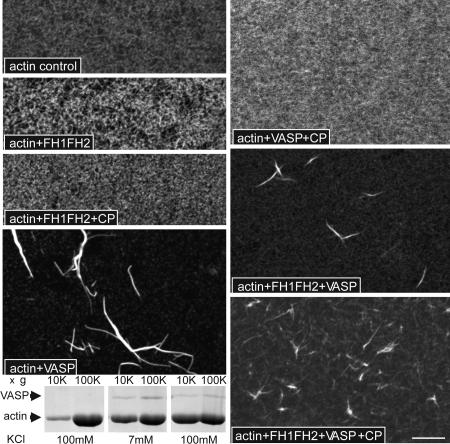
To explore the effects of DdVASP and dDia2 on the formation of actin bundles, we examined tetramethylrhodamine B isothiocyanate–phalloidin-labeled actin filaments formed in the presence of either DdVASP or FH1FH2 with or without CP (Fig. 4). In contrast to the actin control, which showed a fine filament network but no bundles, DdVASP induced the formation of prominent bundles, consistent with its expected actin-bundling activity. In the presence of CP actin filaments remained very short, and DdVASP-induced bundles were not detectable. This assay argues against an uncapping activity of DdVASP. If DdVASP were able to remove the CP and allow further elongation of filaments, then one would expect large filaments and consequently visible actin bundles.
Fig. 4.
Visualization of F-actin structures formed after incubation with DdVASP, FH1FH2, and CP. G-actin (1.1 μM) was incubated in polymerization buffer in the presence of the indicated proteins (100 nM FH1FH2, 100 nM VASP, and 5 nM CP CapG) on polyl-lysine-coated glass coverslips for 30 min. After fixation the specimens were visualized with tetramethylrhodamine B isothiocyanate–phalloidin. DdVASP induces bundling of F-actin whenever CP is absent or competed off barbed ends by FH1FH2. (Scale bar: 10 μm.) The blot shows a sedimentation assay at 10,000 × g and 100,000 × g to distinguish bundles and actin filaments. Lanes 1 and 2, actin alone plus 100 mM KCl; lanes 3 and 4, actin plus DdVASP at low salt conditions (7 mM KCl); lanes 5 and 6, actin plus DdVASP plus 100 mM KCl. The amount of actin sedimented at 10,000 × g in the presence of DdVASP and 100 mM KCl (lane 5) shows clearly that DdVASP maintains its bundling activity at higher salt concentrations.
No major difference was seen between actin samples incubated with FH1FH2 in the presence or absence of CP because, similar to the actin control, single filaments cannot be detected under the conditions used. However, the incubation of FH1FH2 together with DdVASP led to the formation of actin bundles despite the presence of CP, which demonstrated that actin filaments elongated after uncapping by FH1FH2 and formed bundles in the presence of DdVASP. Nucleation of actin polymerization by DdVASP is strongly reduced at salt concentrations above 50 mM KCl; however, its bundling activity is essentially insensitive to higher salt concentrations. The F-actin spin-down experiment (Fig. 4, blot) distinguishes sedimentation of actin bundles at 10,000 × g and actin filaments at 100,000 × g. Whereas with actin alone (lanes 1 and 2) only trace amounts of F-actin could be sedimented at 10,000 × g, the presence of DdVASP lacking its GST tag led to a substantial formation of actin bundles in the absence and presence of 100 mM KCl. The relative insensitivity of the bundling activity of VASP indicates that the bundling function is largely unaffected by these conditions and is likely to persist in vivo.
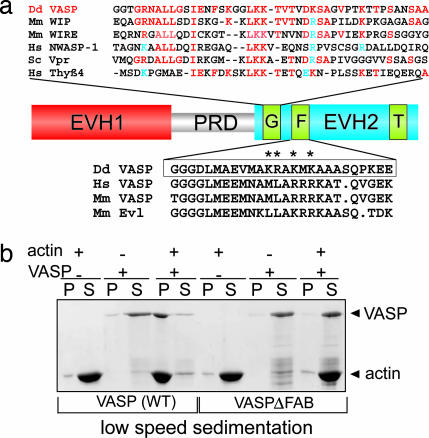
The G- and F-actin binding sites of VASP from higher eukaryotes were well characterized in the past, and Fig. 5a shows a selection of sequence alignments for both functional domains from DdVASP. In contrast to VASP from higher eukaryotes, DdVASP harbors a WH2 [WASP (Wiskott–Aldrich-syndrome protein) homology domain 2] motif as a G-actin binding site in the conserved position of the EVH2 domain (Fig. 5a Upper), also found in many regulators of the cytoskeleton, such as WIP (WASP-interacting protein), WIRE (WIP-related protein), WASP, verprolin, or thymosin β4 (reviewed in ref. 25).
Fig. 5.
Analysis of Dictyostelium VASP's bundling activity in vitro. (a) The C-terminal EVH2 domain contains three regions: a G-actin binding motif (G), an F-actin binding motif (F), and a region required for tetramerization (T). EVH, Ena/VASP homology; PRD, proline-rich domain. (Upper) Alignment of its G-actin binding site to WH2 (WASP homology domain 2) domains from other G-actin binding proteins: DdVASP, accession no. CAH05068, residues 196–239; Mm WIP, accession no. NP694778, residues 29–70; Mm WIRE, accession no. NP922922, residues 33–75; HsNWASP-1, accession no. NP003932, residues 402–442; Sc Vpr, accession no. NP013441, residues 27–68; Hs Thy-β4, accession no. NP066932, residues 1–41. Identical residues are shown in red, and conserved charged residues are shown in blue. (Lower) The alignment of the F-actin binding regions of VASP from D. discoideum and Ena/VASP-like (Evl) proteins from other species is shown: DdVASP, accession no. CAH05068, residues 264–289; HsVASP, accession no. P50552, residues 297–321; Mm VASP, accession no. P70460, residues 293–317; Mm Evl, accession no. P70429, residues 261–285. The region deleted in the DdVASPΔFAB construct is boxed, and asterisks indicate residues mutated to alanine in the point mutant. (b) Deficiency of F-actin bundling activity by DdVASPΔFAB in a low-speed sedimentation assay. Recombinant WT DdVASP and DdVASPΔFAB (1 μM) were incubated alone or with G-actin (5 μM) in polymerization buffer and spun at 15,000 × g. In contrast to WT DdVASP (left blots), DdVASPΔFAB (right blots) does not cosediment with the F-actin. P, pellet; S, supernatant.
To analyze a possible contribution of the DdVASP bundling activity for filopodia formation we generated a mutated DdVASP (DdVASPΔFAB) that lacked the F-actin binding site (residues 264–285; Fig. 5a Lower). This region is localized within the EVH2 domain and is highly conserved among other Ena/VASP family members (26). To test whether the F-actin binding site of DdVASP is indeed responsible for its bundling activity, purified GST-tagged DdVASPΔFAB was monitored for its ability to bundle actin filaments by two independent methods. In contrast to the WT protein, mutated DdVASP was not able to induce the formation of actin bundles in the coverslip assay (Fig. 8a, which is published as supporting information on the PNAS web site) or to induce bundle formation in a low-speed sedimentation assay (Fig. 5b). However, when tested for its nucleating activity in the pyrene-based actin polymerization assay, DdVASPΔFAB's behavior was virtually identical to that of WT DdVASP; i.e., it displayed actin-nucleating properties at low salt concentrations (Fig. 8b). To confirm the data obtained with the DdVASP deletion, a point-mutated DdVASP was generated in which the four basic residues (K275, R276, K278, and K280) in the F-actin binding site were replaced by alanines and analyzed accordingly. Sequential mutation of the F-actin binding site (Fig. 5a, stars) led to results similar to DdVASPΔFAB. The recombinant mutated protein was strongly reduced in its ability to bundle F-actin, as determined by microscopical and low-speed sedimentation experiments, but still was capable of nucleating actin polymerization in vitro comparable to WT DdVASP, which also suggests proper folding of the mutated protein (data not shown).
The Bundling Activity of VASP Is Required for Filopodia Formation in Vivo.
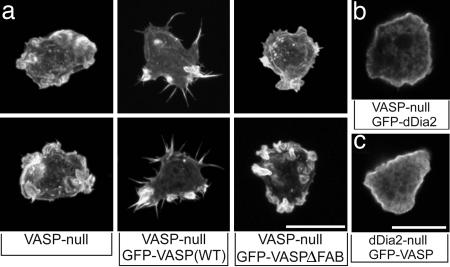
To test whether the bundling activity of DdVASP is also required for filopodium formation in vivo we disrupted the vasP gene of D. discoideum in WT cells (Fig. 9 a and b, which is published as supporting information on the PNAS web site) and used the DdVASP-null mutants for complementation with GFP-tagged WT and mutant DdVASPΔFAB. Subsequent analysis of these cells revealed that only WT DdVASP but not DdVASPΔFAB could rescue their ability to form filopodia (Fig. 6a). In line with these findings, reexpression of a mutated GFP-DdVASP with four alanine exchanges in the F-actin binding site (see above) could barely trigger the formation of filopodia in the DdVASP-null mutant (data not shown). Taken together, these findings clearly demonstrate the importance of the actin bundling activity of DdVASP for filopodium formation in vivo.
Fig. 6.
The F-actin bundling activity of DdVASP is required for filopodium formation in vivo. (a) 3D reconstructions of tetramethylrhodamine B isothiocyanate–phalloidin-labeled knockout and reconstituted cells as indicated. (Scale bar: 5 μm.) Only cells reexpressing WT DdVASP display normal filopodia. (b and c) Cellular distribution of GFP-tagged dDia2 or DdVASP in mutants lacking VASP or dDia2. Confocal sections are shown. (Scale bars: 5 μm.)
Finally, to test for a mutual recruitment of DdVASP and dDia2, we examined the localization of GFP-dDia2 in DdVASP-null cells as well as the localization of GFP-DdVASP (WT) in dDia2-null cells. As shown in Fig. 6b both cell lines largely lacked filopodia, and the GFP signals in both cell lines were predominantly confined to the cortical region. Interestingly, analysis of the few filopodia in these mutants revealed that both GFP-DdVASP and GFP-dDia2 in the opposite null strains still localized to filopodial tips. This finding strongly suggests that the targeting of these two proteins to filopodial tips is not mutually dependent, whereas the formation of filopodia requires both.
Human VASP Displays Virtually Identical Properties as Dictyostelium VASP.
To test whether the functions of DdVASP identified in this study are specific to Dictyostelium or whether they can be extended to higher organisms, we analyzed some of the key features of human VASP (HsVASP). Similar to DdVASP, recombinant HsVASP did not inhibit actin depolymerization or compete with CP (Fig. 7a–c). In analogy to DdVASP, HsVASP-ΔFAB also nucleated actin polymerization (Fig. 10a, which is published as supporting information on the PNAS web site) but failed to bundle actin filaments, as demonstrated in coverslip and low-speed sedimentation assays (Figs. 7fand 10b). We compared the subcellular localizations of both GFP-tagged WT and HsVASP-ΔFAB in MVD7 mouse fibroblasts lacking endogenous VASP and Mena but expressing low amounts of Evl (27). Both ectopically expressed proteins were targeted to focal adhesions, to lamellipodia tips, and, most importantly, to filopodia tips, demonstrating that the F-actin binding site of HsVASP is not required for recruitment to these sites (Fig. 7 d–e). Localization of DdVASP and DdVASPΔFAB in transfected mammalian cells was similar to that of the mammalian counterparts, indicating conserved functions for VASP family proteins in these evolutionarily distant systems (Fig. 11, which is published as supporting information on the PNAS web site).
Fig. 7.
In vitro and in vivo analysis of human VASP. (a) HsVASP does not interact with barbed filament ends. Polymerized pyrene-labeled actin was diluted to 0.1 μM in polymerization buffer alone or in polymerization buffer containing increasing concentrations of HsVASP. Whereas the addition of CapG inhibits depolymerization (red line), even high concentrations of HsVASP do not slow down depolymerization. (b and c) Polymerized actin with either free barbed ends (black lines) or with the barbed ends capped by CapG (b) or Cap32/34 (c) was diluted to 0.1 μM in polymerization buffer containing 100 mM KCl and the amounts of HsVASP indicated. Neither CapG nor Cap32/34 was removed by HsVASP from the filament ends. (d and e) The F-actin binding site is not required for proper cellular localization of vertebrate VASP. Shown are mouse embryonic Mena/VASP double knockout fibroblasts stably expressing either GFP-tagged human WT VASP or a mutant lacking the F-actin binding motif. White arrows mark tips of filopodia. (Scale bar: 20 μm.) (f) Deficiency of F-actin bundling activity by HsVASPΔFAB in a low-speed sedimentation assay. Recombinant WT HsVASP and HsVASPΔFAB (1 μM) were incubated alone or with G-actin (5 μM) in polymerization buffer and spun at 15,000 × g. In contrast to WT HsVASP (left blots), HsVASPΔFAB (right blots) does not cosediment with the F-actin. (P, pellet; S, supernatant). Lanes 1, 2, 7, and 8, actin alone; lanes 3, 4, 9, and 10, VASP alone; lanes 5, 6, 11, and 12, actin plus VASP.
Discussion
VASP is a key player in actin cytoskeleton rearrangements and has been localized to distal ends of lamellipodia and filopodia, where it controls dynamic actin turnover at the barbed ends of actin filaments. Previous biochemical studies with recombinant mammalian VASP revealed that this protein binds to G- and F-actin, bundles actin filaments, and is a nucleator of actin polymerization in vitro. However, conflicting results have been reported for the ability of VASP to compete with CP at filament barbed ends (23, 28). We show here that DdVASP and HsVASP nucleate and bundle actin filaments in vitro but are not capable of uncapping barbed ends. During elongation the dDia2/DdVASP complex remains at the filopodium tip, whereas other actin bundling proteins, such as fascin or fimbrin, stabilize the mature filopodial actin bundle.
Our data do not indicate a nucleating activity of VASP in filopodia formation. Moreover, the evidence for an in vivo nucleating activity of VASP is still missing. For instance, a Listeria mutant incapable of binding and activating the Arp2/3 complex can massively accumulate VASP at the bacterial surface without promoting any detectable actin assembly at this site (29). In lamellipodia actin is assembled into branched networks by the Arp2/3 complex, which nucleates daughter filaments either along mother filaments or in close vicinity to their barbed ends (30, 31). Ena/VASP proteins appear to be used for stabilization of these single actin filaments. Lamellipodia lacking Ena/VASP proteins contain actin networks with shorter filaments that display a higher branching frequency than WT, whereas lamellipodia with excess Ena/VASP proteins contain longer and less-branched actin filaments (23).
In a recent study Barzik et al. (32) investigated how mammalian VASP influenced actin polymerization in the presence of CPs. They proposed that VASP associated “at or near” the barbed ends of actin filaments and thus could restrict access of barbed-end CP. For reasons that we cannot explain, our data are different from those obtained by Barzik et al. (32). DdVASP and HsVASP do not affect the behavior of barbed ends during dilution-induced depolymerization of filaments in the absence or presence of either CapG or Cap32/34. A direct interaction of VASP with the plus ends of filaments leading to competition with CP should, however, alter the depolymerization kinetics. Furthermore, VASP as an in vitro bundling protein must bind along the length of the entire actin filament and not only “near” the barbed end. In the cellular context of a filopodium, however, its bundling activity is probably restricted to the tip region.
Possible explanations for the contradictory results could be slightly different experimental setups. Whereas Barzik et al. (32) mixed F-actin, VASP, and CP and then diluted the mixture to induce depolymerization, we tried to avoid the presence of VASP in the undiluted F-actin. We realized that the addition of VASP to F-actin before dilution caused immediate bundling and thus a slower depolymerization thereafter, presumably because of sterical hindrance or inhibition of monomer dissociation as soon as a crosslinking bridge in the bundle was reached. We therefore precapped the actin filaments and subsequently induced depolymerization by addition of buffer and VASP.
Our data demonstrate that the FH1FH2 region of the formin dDia2 interacts with DdVASP during filopodia formation and that this interaction is crucial for the efficient elongation of actin filaments at the tips of protruding filopodia. Notably, in vivo VASP remains confined to the tips of protruding filopodia despite its F-actin binding and bundling activities. From this result we conclude that the entire complex moves on with the filopodium tip, as has been already shown for dDia2 (6). In this scenario, filament bundling by VASP applies only to nascent filopodial filaments in the vicinity of the filopodial tip complex before other actin-bundling proteins, like fascin, stabilize the emerging bundles in the filopodial shaft.
Experimental measurements suggest that tens of pN are required to allow growth of a filopodium against the resistance of the cell membrane (33). Recently, formins have been identified to act as processive motors (34, 35) that can generate at least 1.3 pN per actin filament (35). Provided that the actin filaments are firmly anchored in the cortical cytoskeleton, as suggested by Svitkina et al. (17), it is presumably the combination of both protein activities, i.e., filament elongation by dDia2 and rapid bundling of the actin filaments by VASP, that ensure the appropriate cytoskeletal architecture and force required to push the membrane outward, ultimately leading to the formation of a new filopodium. Our model differs from the postulated model of Mejillano et al. (36), which suggests that the nucleating activities of the Arp2/3 complex and VASP together with CP are sufficient for this process to occur. Our results provide conclusive evidence for the requirement of both the nucleating activity of a formin and the bundling activity of VASP for the formation of filopodia. An intriguing possibility to reconcile both models could be a putative activation of the Arp2/3 complex as a first trigger to form F-actin for filopodium initiation and anchorage in the cortex, with the formin–VASP complex then being responsible for the generation and growth of the filopodium acting at its tip. Furthermore, one cannot exclude that the chronological order of the interaction between actin and actin-binding proteins may be different in distinct regions of the cell, thus regulating the sequence of activities through specific off-rates (37). This mechanism allows different cytoskeletal dynamics even with the same set of proteins.
The involvement of formins in filopodia formation was reported recently not only for Dictyostelium (6), but also for vertebrate cells. An mDia2-mediated but Cdc42-independent pathway leading to the formation of filopodia has been described for the Rho GTPase Rif (5). Interestingly, elimination of mDia1 in mice resulted in an up-regulation of mDia2 and revealed its potential role as an effector of Cdc42, which in turn is involved in filopodia formation (4). Furthermore, overexpression of an EGFP-tagged constitutively active (i.e., truncated) mDia1 in fibroblasts led to a massive formation of actin fibers as well as to a large number of filopodia-like structures that were labeled at their tips with the EGFP fusion protein (38). Finally, mDia1 was previously shown to coimmunoprecipitate with VASP (24). In the light of the finding that Dictyostelium and vertebrate VASP behave virtually identically, it is tempting to speculate that the disclosed formin/VASP pathway of this study provides a conserved mechanism of filopodia formation.
Materials and Methods
cDNA Cloning and Generation of Transformation Vectors.
For the expression of GFP-DdVASP, the entire coding region of the vasP gene was amplified from AX2 WT cDNA by PCR as a 1.2-kb BamHI/SalI fragment, cloned into pDGFP-MCS-Neo (39), and sequence-verified. For construction of the vasP targeting vector, a 510-bp 5′ BamHI/PstI fragment and a 540-bp 3′ HindIII/SalI fragment were amplified from genomic AX2 WT DNA by PCR. Both fragments cloned into the corresponding sites of pLPBLP containing the blasticidin S resistance cassette (40). The resulting vector was cleaved with BamHI and SalI and used to disrupt the vasP gene in WT cells. The mutant construct DdVASPΔFAB lacks residues 264–285 comprising the F-actin binding site.
Protein Expression and Purification.
For the expression of the dDia2 FH1FH2 domain and WT and mutated DdVASP constructs, the corresponding DNA fragments were cloned into the BamHI and SalI sites of pGEX-5x-1 or pGEX-6P-1. Human WT (amino acids 1–380) and ΔFAB (amino acids Δ259–277) VASP were amplified by using the GFP fusion vectors as templates and cloned into pGEX-6P-1. Expression and purification of GST- or His-tagged dDia2 and VASP constructs were carried out essentially as described (41). The GST tag was removed by incubating the purified proteins with PreScission protease in PBS (pH 7.4) supplemented with 1 mM DTT and 1 mM EDTA overnight at 4°C. His-tagged CapG and heterodimeric His-tagged CP Cap32/34 were purified as described (6).
Yeast Two-Hybrid Assay.
Yeast two-hybrid interactions were analyzed with Matchmaker Two-Hybrid System 3 (CLONTECH). The activation domain of pGADT7 was fused to FH1 (amino acids 548–642), FH1FH2 (amino acids 548-1087), FH2 (amino acids 616-1087), and ΔFH1FH2 (amino acids 1–547). Complete VASP (amino acids 1–380) was expressed as full-length fusion with the Gal4-binding domain in plasmid pGBKT7.
Miscellaneous.
Cells of D. discoideum AX2 WT strain and of derived transformants were cultivated and transformed by electroporation as described (41). MVD7 cells and MVD7 cells stably expressing GFP-tagged human VASP variants were described previously (42). DdVASP was cloned into a pEGFP-C vector (CLONTECH) and expressed in mouse embryonic fibroblasts immortalized by using a temperature-sensitive variant of the large T antigen and growing at 32°C as described previously (43). Images were obtained by video microscopy. Immunoprecipitations were performed with anti-dDia2 polyclonal antibodies as described (6). Immunoblotting was performed by standard procedures using GFP-specific mAb 264-449-2 (44). Microscopy was performed essentially as described (44). F-actin was labeled with tetramethylrhodamine B isothiocyanate-conjugated phalloidin (Sigma), and microscopic analysis of F-actin structures was performed as described previously (26). Protein concentration was determined according to the method of Bradford (45). Actin polymerization and depolymerization were measured by fluorescence spectroscopy with pyrene-labeled actin as described (46). Actin polymerization or depolymerization experiments were performed in a buffer containing 10 mM imidazole, 2 mM MgCl2, 0.2 mM CaCl2, 1 mM Na-ATP, and either 50 mM or 100 mM KCl (pH 7.2). For preparation of F-actin seeds 50 μM G-actin was polymerized for 1 h at room temperature. The F-actin was vortexed vigorously for 1 min to mechanically shear the filaments. Routinely we used 0.1 μM actin as seeds for each assay. Actin bundling assays were performed according to Faix et al. (47).
Supplementary Material
Acknowledgments
We thank D. Rieger and M. Borath for excellent technical assistance and Dr. J. Wehland (German Research Centre for Biotechnology) for providing stably transfected MVD7 cell lines and HsVASP constructs. This work was supported by grants from the Deutsche Forschungsgemeinschaft (to M.S. and T.E.B.S.) and a grant from the Friedrich-Baur-Stiftung (to J.F.).
Abbreviations
- VASP
vasodilator-stimulated phosphoprotein
- DdVASP
Dictyostelium VASP
- HsVASP
human VASP
- CP
capping protein.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The Dictyostelium vasP cDNA sequence has been deposited in the European Molecular Biology Laboratory database (accession no. AJ786025).
References
- 1.Zigmond S. H. Curr. Opin. Cell Biol. 2004;16:99–105. doi: 10.1016/j.ceb.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 2.Watanabe N., Kato T., Fujita A., Ishizaki T., Narumiya S. Nat. Cell Biol. 1999;1:136–143. doi: 10.1038/11056. [DOI] [PubMed] [Google Scholar]
- 3.Evangelista M., Pruyne D., Amberg D. C., Boone C., Bretscher A. Nat. Cell Biol. 2002;4:260–269. doi: 10.1038/ncb770. [DOI] [PubMed] [Google Scholar]
- 4.Peng J., Wallar B. J., Flanders A., Swiatek P. J., Alberts A. S. Curr. Biol. 2003;13:534–545. doi: 10.1016/s0960-9822(03)00170-2. [DOI] [PubMed] [Google Scholar]
- 5.Pellegrin S., Mellor H. Curr. Biol. 2005;15:129–133. doi: 10.1016/j.cub.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 6.Schirenbeck A., Bretschneider T., Arasada R., Schleicher M., Faix J. Nat. Cell Biol. 2005;7:619–625. doi: 10.1038/ncb1266. [DOI] [PubMed] [Google Scholar]
- 7.Kureishy N., Sapountzi V., Prag S., Anilkumar N., Adams J. C. BioEssays. 2002;24:350–361. doi: 10.1002/bies.10070. [DOI] [PubMed] [Google Scholar]
- 8.Kreitmeier M., Gerisch G., Heizer C., Muller-Taubenberger A. J. Cell Biol. 1995;129:179–188. doi: 10.1083/jcb.129.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sydor A. M., Su A. L., Wang F. S., Xu A., Jay D. G. J. Cell Biol. 1996;134:1197–1207. doi: 10.1083/jcb.134.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuxworth R. I., Weber I., Wessels D., Addicks G. C., Soll D. R., Gerisch G., Titus M. A. Curr. Biol. 2001;11:318–329. doi: 10.1016/s0960-9822(01)00097-5. [DOI] [PubMed] [Google Scholar]
- 11.Berg J. S., Cheney R. E. Nat. Cell Biol. 2002;4:246–250. doi: 10.1038/ncb762. [DOI] [PubMed] [Google Scholar]
- 12.Biyasheva A., Svitkina T., Kunda P., Baum B., Borisy G. J. Cell Sci. 2004;117:837–848. doi: 10.1242/jcs.00921. [DOI] [PubMed] [Google Scholar]
- 13.Krugmann S., Jordens I., Gevaert K., Driessens M., Vandekerckhove J., Hall A. Curr. Biol. 2001;11:1645–1655. doi: 10.1016/s0960-9822(01)00506-1. [DOI] [PubMed] [Google Scholar]
- 14.Nakagawa H., Miki H., Nozumi M., Takenawa T., Miyamoto S., Wehland J., Small J. V. J. Cell Sci. 2003;116:2577–2583. doi: 10.1242/jcs.00462. [DOI] [PubMed] [Google Scholar]
- 15.Rottner K., Behrendt B., Small J. V., Wehland J. Nat. Cell Biol. 1999;1:321–322. doi: 10.1038/13040. [DOI] [PubMed] [Google Scholar]
- 16.Han Y. H., Chung C. Y., Wessels D., Stephens S., Titus M. A., Soll D. R., Firtel R. A. J. Biol. Chem. 2002;277:49877–49887. doi: 10.1074/jbc.M209107200. [DOI] [PubMed] [Google Scholar]
- 17.Svitkina T. M., Bulanova E. A., Chaga O. Y., Vignjevic D. M., Kojima S., Vasiliev J. M., Borisy G. G. J. Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lebrand C., Dent E. W., Strasser G. A., Lanier L. M., Krause M., Svitkina T. M., Borisy G. G., Gertler F. B. Neuron. 2004;42:37–49. doi: 10.1016/s0896-6273(04)00108-4. [DOI] [PubMed] [Google Scholar]
- 19.Jay D. G. J. Neurobiol. 2000;44:114–125. doi: 10.1002/1097-4695(200008)44:2<114::aid-neu3>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 20.Bear J. E., Loureiro J. J., Libova I., Fassler R., Wehland J., Gertler F. B. Cell. 2000;101:717–728. doi: 10.1016/s0092-8674(00)80884-3. [DOI] [PubMed] [Google Scholar]
- 21.Reinhard M., Giehl K., Abel K., Haffner C., Jarchau T., Hoppe V., Jockusch B. M., Walter U. EMBO J. 1995;14:1583–1589. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwiatkowski A. V., Gertler F. B., Loureiro J. J. Trends Cell Biol. 2003;13:386–392. doi: 10.1016/s0962-8924(03)00130-2. [DOI] [PubMed] [Google Scholar]
- 23.Bear J. E., Svitkina T. M., Krause M., Schafer D. A., Loureiro J. J., Strasser G. A., Maly I. V., Chaga O. Y., Cooper J. A., Borisy G. G., Gertler F. B. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- 24.Grosse R., Copeland J. W., Newsome T. P., Way M., Treisman R. EMBO J. 2003;22:3050–3061. doi: 10.1093/emboj/cdg287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paunola E., Mattila P. K., Lappalainen P. FEBS Lett. 2002;513:92–97. doi: 10.1016/s0014-5793(01)03242-2. [DOI] [PubMed] [Google Scholar]
- 26.Bachmann C., Fischer L., Walter U., Reinhard M. J. Biol. Chem. 1999;274:23549–23557. doi: 10.1074/jbc.274.33.23549. [DOI] [PubMed] [Google Scholar]
- 27.Auerbuch V., Loureiro J. J., Gertler F. B., Theriot J. A., Portnoy D. A. Mol. Microbiol. 2003;49:1361–1375. doi: 10.1046/j.1365-2958.2003.03639.x. [DOI] [PubMed] [Google Scholar]
- 28.Samarin S., Romero S., Kocks C., Didry D., Pantaloni D., Carlier M. F. J. Cell Biol. 2003;163:131–142. doi: 10.1083/jcb.200303191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skoble J., Auerbuch V., Goley E. D., Welch M. D., Portnoy D. A. J. Cell Biol. 2001;155:89–100. doi: 10.1083/jcb.200106061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pollard T. D., Borisy G. G. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 31.Pantaloni D., Boujemaa R., Didry D., Gounon P., Carlier M. F. Nat. Cell Biol. 2000;2:385–391. doi: 10.1038/35017011. [DOI] [PubMed] [Google Scholar]
- 32.Barzik M., Kotova T. I., Higgs H. N., Hazelwood L., Hanein D., Gertler F. B., Schafer D. A. J. Biol. Chem. 2005;280:28653–28662. doi: 10.1074/jbc.M503957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai J., Sheetz M. P. Biophys. J. 1999;77:3363–3370. doi: 10.1016/S0006-3495(99)77168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romero S., Le Clainche C., Didry D., Egile C., Pantaloni D., Carlier M. F. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 35.Kovar D. R., Pollard T. D. Proc. Natl. Acad. Sci. USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mejillano M. R., Kojima S., Applewhite D. A., Gertler F. B., Svitkina T. M., Borisy G. G. Cell. 2004;118:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 37.Kovar D. R., Wu J. Q., Pollard T. D. Mol. Biol. Cell. 2005;16:2313–2324. doi: 10.1091/mbc.E04-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Higashida C., Miyoshi T., Fujita A., Oceguera-Yanez F., Monypenny J., Andou Y., Narumiya S., Watanabe N. Science. 2004;303:2007–2010. doi: 10.1126/science.1093923. [DOI] [PubMed] [Google Scholar]
- 39.Dumontier M., Hocht P., Mintert U., Faix J. J. Cell Sci. 2000;113:2253–2265. doi: 10.1242/jcs.113.12.2253. [DOI] [PubMed] [Google Scholar]
- 40.Sutoh K. Plasmid. 1993;30:150–154. doi: 10.1006/plas.1993.1042. [DOI] [PubMed] [Google Scholar]
- 41.Faix J., Weber I., Mintert U., Kohler J., Lottspeich F., Marriott G. EMBO J. 2001;20:3705–3715. doi: 10.1093/emboj/20.14.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geese M., Loureiro J. J., Bear J. E., Wehland J., Gertler F. B., Sechi A. S. Mol. Biol. Cell. 2002;13:2383–2396. doi: 10.1091/mbc.E02-01-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lommel S., Benesch S., Rottner K., Franz T., Wehland J., Kuhn R. EMBO Rep. 2001;2:850–857. doi: 10.1093/embo-reports/kve197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weber I., Gerisch G., Heizer C., Murphy J., Badelt K., Stock A., Schwartz J. M., Faix J. EMBO J. 1999;18:586–594. doi: 10.1093/emboj/18.3.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bradford M. M. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 46.Eichinger L., Schleicher M. Biochemistry. 1992;31:4779–4787. doi: 10.1021/bi00135a006. [DOI] [PubMed] [Google Scholar]
- 47.Faix J., Steinmetz M., Boves H., Kammerer R. A., Lottspeich F., Mintert U., Murphy J., Stock A., Aebi U., Gerisch G. Cell. 1996;86:631–642. doi: 10.1016/s0092-8674(00)80136-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.