Abstract
Retinoid X receptor α (RXRα) is involved in multiple signaling pathways, as a heterodimeric partner of several nuclear receptors. To investigate its function in energy homeostasis, we have selectively ablated the RXRα gene in adipocytes of 4-week-old transgenic mice by using the tamoxifen-inducible Cre-ERT2 recombination system. Mice lacking RXRα in adipocytes were resistant to dietary and chemically induced obesity and impaired in fasting-induced lipolysis. Our results also indicate that RXRα is involved in adipocyte differentiation. Thus, our data demonstrate the feasibility of adipocyte-selective temporally controlled gene engineering and reveal a central role of RXRα in adipogenesis, probably as a heterodimeric partner for peroxisome proliferator-activated receptor γ.
Keywords: peroxisome proliferator-activated receptor γ, conditional mutagenesis, obesity, fasting, adipocyte fatty acid binding protein
In vertebrates, the adipose tissue is critical for energy homeostasis (1, 2). Whereas the brown adipose tissue can dissipate energy through thermogenesis, the white adipose tissue (WAT) stores excess energy in the form of triglycerides, when caloric intake exceeds expenditure, and releases free fatty acids (FFAs) when expenditure exceeds intake. The size of the adipose tissue can be modulated by the formation of new adipocytes from precursor cells (adipocyte differentiation) and/or increase in adipocyte size (adipocyte hypertrophy). Adipocyte differentiation has been studied mainly with preadipocyte culture systems (1). Three classes of transcription factors have been identified that influence fat cell development. These include CCAAT/enhancer-binding proteins (C/EBPs), adipocyte differentiation determinant-dependent factor 1 (ADD1)/sterol response element-binding protein (SREBP1), and the peroxisome proliferator-activated receptor (PPAR) family proteins (3–7). PPARγ, one of the three PPAR isotypes, is abundantly expressed in adipose tissue and has been recently shown to play a critical role in both adipocyte differentiation and hypertrophy in vivo (8–10). As other members of the nuclear hormone receptor superfamily, such as the retinoic acid, vitamin D3, and thyroid hormone receptors, it forms heterodimers with the three retinoid X receptor (RXR) isotypes (α, β, or γ) (6, 11, 12).
That RXR–PPAR heterodimers could mediate adipocyte differentiation has been demonstrated in vitro by using cultured cells and RXR- and PPARγ-specific ligands (13). RXRα is expressed in a number of mouse tissues, including the adipose tissue, in which high levels of transcripts were detected (14). However, its role in adipocyte differentiation and hypertrophy could not be investigated in vivo in RXRα knockout (KO) mice, because RXRα−/− fetuses die in utero (15, 16).
We have used the Cre–lox system to generate mice in which RXRα could be specifically ablated in adipocytes in a temporally controlled manner. To this end, we have generated a transgenic mouse line that specifically expresses the tamoxifen (Tam)-inducible fusion protein between the Cre recombinase and a mutated ligand-binding domain of the human estrogen receptor (Cre-ERT2) (17, 18) in adipocytes, under the control of the promoter of the adipocyte fatty acid binding protein (aP2) (19). After treatment with Tam, transgenic mice harboring the aP2-Cre-ERT2 transgene and a LoxP site containing (floxed) RXRα gene, yielded animals in which RXRα was specifically ablated in adipocytes. We show that such mice have impaired adipogenesis and lipolysis.
Materials and Methods
Generation of Transgenic Mice.
The aP2-Cre-ERT2 transgene was constructed as follows: a 5.4-kb blunt-ended NotI fragment containing the aP2 promoter, amplified from mouse genomic DNA by PCR with the LA-PCR kit (Perkin–Elmer) using the oligonucleotides 5′-ATACGCGGCCGCGAATTCCAGCAGGAATCAGGTAGCT-3′ and 5′-ATAGCGCCGGCGCTGCAGCACAGGAGGGTGCTATGAG-3′. The product was cloned into the blunt-ended SalI site of pGS-Cre-ERT2 (17), resulting in paP2-Cre-ERT2. The aP2-L-EGFP-L transgene was constructed as follows: the 5.4-kb blunt-ended NotI fragment containing the aP2 promoter was cloned into the blunt-ended SalI site of pL-EGFP-L, resulting in paP2-L-EGFP-L. pL-EGFP-L was obtained by cloning the 0.7-kb blunt-ended EcoRI fragment isolated from pCX-EGFP (20) into the blunt-ended BamHI site of pLox2 (21). The 8.3-kb and 7-kb NotI DNA fragments were purified from paP2-Cre-ERT2 and paP2-L-EGFP-L, respectively, and injected into (C57BL/6 × SJL) F1 zygotes at a concentration of 4 ng/ml, as described (22, 23). Four aP2-Cre-ERT2(tg/0) transgenic lines were obtained from 54 founder animals. RXRα+/−, RXRα+/af2(I), and RXRαL2/+ mice were as described (15, 18, 24).
Animal Treatments.
Under normal conditions, mice were fed a standard laboratory chow (regular diet, RD; 2,800 kcal/kg; 1 cal = 4.18 J; Usine d'Alimentation Rationnelle, Villemoisson-sur-Orge, France). The high-fat and high-sucrose diet (HFD) study was carried out with a chow containing 4,056 kcal/kg (fat = 1,600 kcal/kg and sucrose = 1,600 kcal/kg; Research Diets, New Brunswick, NJ). HFD was given to animals at weaning.
Monosodium glutamate (MSG) dissolved in saline solution was injected at 0.1 g/ml s.c. [2 mg per g (body weight) per day] from postnatal days 1 to 7 (19, 25, 26).
Tam (1 mg in 100 μl of sunflower oil) was injected i.p. into mice for 5 consecutive days, as described (18, 27).
Reverse Transcription–PCR (RT-PCR) and Northern and Southern Blot Analyses.
RNA was prepared from mouse organs by guanidine thiocyanate extraction (28). Cre-ERT2 expression was analyzed by RT-PCR. cDNA was synthesized for 20 min at 50°C from 1 μg of RNA, 50 units of Moloney murine leukemia virus reverse transcriptase and the 3′-primer 5′-GGCGATCCCTGAACATGTCC-3′ and was amplified by 30 cycles of PCR using the same 3′-primer and the 5′-primer 5′-TTGACCTCCATAGAAGACA-3′. The 254-bp Cre-ERT2 cDNA fragment was identified by Southern blotting using the 32P-radiolabeled oligonucleotide 5′-ATCCGAAAAGAAAACGTTGA-3′. The primers used to amplify the 310-bp Pref-1 cDNA fragment were 5′-CAACAAGGAGGCTGGTGATG-3′ and 5′-CGAGATGAAATTCTTATTAT-3′. The probe corresponds to the 32P-radiolabeled oligonucleotide 5′-GAGTTTGCTCTATTGTGAGG-3′. Hypoxanthine phosphoribosyltransferase (HPRT) was used as an internal control, as described (23). For Northern blotting, 10 μg of total RNA was electrophoresed on a 0.8% agarose gel, transferred to a nylon membrane, hybridized with radiolabeled RXRβ (29); RXRγ (30); PPARγ, aP2, and lipoprotein lipase (LPL) (31); and 36B4 (32) probes, as described (33).
aP2-Cre-ERT2-mediated DNA excision was determined by Southern blot analysis, performed on BamHI-digested genomic DNA isolated from organs of aP2-Cre-ERT2(tg/0)/RXRα+/af2(I) or aP2-Cre-ERT2(tg/0)/RXRαL2/− double-transgenic mice, or from RXRα+/af2(II) mouse tail (18, 23). Purification of adipocytes by collagenase II treatment of adipose tissue and centrifugation was as described (34, 35).
Histological Analysis.
Organs were taken from mice perfused with paraformaldehyde (4% in PBS), fixed in formaldehyde (20% in PBS), and frozen in tissue-Tek OCT compound (SAKURA, Zoeterwoude, The Netherlands). Ten-micrometer cryosections were stained with hematoxylin and eosin or Oil red O and examined by light microscopy. 4′,6-Diamidino-2-phenylindole dihydrochloride and green fluorescence was analyzed on cryosections by confocal microscopy as described (36).
Plasma-Free Fatty Acids Analysis.
Plasma-free fatty acids were determined by an enzymatic assay, adapted to microtiter plates, and commercially available reagents (Boehringer Mannheim) (37).
Statistical Analysis.
Values are reported as mean ± SEM. Statistical significance (P < 0.05) was determined by unpaired Student's t test (statview, Abacus Concepts, Berkeley, CA).
Results and Discussion
Cre-ERT2-Mediated Targeted Mutagenesis in Mouse Adipocytes.
Transgenic mice expressing the conditional Cre-ERT2 recombinase selectively in adipocytes were obtained by using the aP2 promoter (19). Cre-ERT2 transcripts were found to be expressed specifically in WATs and brown adipose tissues in three of four transgenic lines (Fig. 1A and data not shown). The line expressing the highest level of Cre-ERT2 (referred to as the aP2-Cre-ERT2(tg/0) line in the hemizygous state) was then crossed with an aP2-L-EGFP-L(tg/0) reporter line that expresses a floxed enhanced green fluorescent protein (EGFP) gene under the control of the aP2 promoter. A green fluorescence was detected in ≈30% of WAT adipocytes of aP2-Cre-ERT2(tg/0)/aP2-L-EGFP-L(tg/0) bigenic mice, but not in other cell types of adipose tissue nor in cells of other organs (Fig. 1Bb and data not shown). Interestingly, the floxed EGFP cassette was efficiently excised upon Tam treatment, as judged from the total disappearance of the green fluorescence (Fig. 1Bc). In contrast, no aP2-Cre-ERT2-mediated excision occurred in other cell types (e.g., pancreas, skeletal muscle, and myocardium cells), as no 5-bromo-4-chloro-3-indolyl β-D-galactoside staining was observed upon Tam treatment of aP2-Cre-ERT2(tg/0)/ACZL bigenic mice, under conditions where Cre-mediated recombination in the ACZL reporter transgenic mouse is known to result in β-galactosidase expression in these cells (refs. 38 and 39 and unpublished results).
Figure 1.
Characterization of aP2-Cre-ERT2 transgenic mice. (A) Cre-ERT2 mRNA is selectively expressed in adipose tissue of aP2-Cre-ERT2 transgenic mice. Cre-ERT2 expression was analyzed by RT-PCR on RNA extracted from WAT of a 3-month-old WT mouse and from the indicated tissues of a 3-month-old aP2-Cre-ERT2(tg/0) transgenic mouse. The PCR products corresponding to Cre-ERT2 and HPRT mRNA are indicated. (B) After Tam treatment, aP2-Cre-ERT2 transgenic mice efficiently excise a floxed EGFP cassette in adipocytes. Cryosections of WAT isolated from a 3-month-old Tam-treated aP2-Cre-ERT2(tg/0) mouse (a), vehicle-treated aP2-Cre-ERT2(tg/0)/aP2-L-EGFP-L(tg/0) bigenic mouse (b), and Tam-treated aP2-Cre-ERT2(tg/0)/aP2-L-EGFP-L(tg/0) bigenic mouse (c), 30 days after the injection, were analyzed by confocal microscopy. The blue color corresponds to 4′,6-diamidino-2-phenylindole dihydrochloride-stained nuclei, and the green color corresponds to GFP fluorescence. (Scale bar, 20 μm.) (C) Cre-ERT2 mice selectively excise floxed DNA in adipocytes after Tam treatment. Cre-ERT2-mediated DNA excision was determined by Southern blot analysis, performed on genomic DNA isolated from organs of 6-month-old aP2-Cre-ERT2(tg/0)/RXRα+/af2(I) bigenic mice, 7 days after the last injection of vehicle (lanes 2–12) or Tam (lanes 13–23) or from RXRα+/af2(II) mouse tail (lane 1). The position of the WT (+), floxed [af2(I)], and recombined [af2(II)] RXRα alleles are indicated. BAT, brown adipose tissue.
Tissue-specific Cre-ERT2-mediated excision upon Tam treatment also was observed in aP2-Cre-ERT2(tg/0)/RXRα+/af2(I) mice in which a floxed neomycin-resistance cassette is present within the RXRα locus (24). Excision of the cassette was restricted to brown adipose tissue and WAT, as shown by the presence of the “recombined” RXRα af2(II) allele 7 days after Tam treatment (Fig. 1C, compare lanes 22 and 23 with lanes 11 and 12; similar blots were obtained 30 and 90 days after Tam treatment, data not shown).
Temporally Controlled RXRα Ablation in Adipocytes.
To selectively disrupt the RXRα gene in adipocytes, we generated aP2-Cre-ERT2(tg/0)/RXRαL2/L2 mice, in which the exon encoding the RXRα DNA binding domain is floxed on both alleles (18). These mice then were crossed with RXRα+/− mice (15) to generate aP2-Cre-ERT2(tg/0)/RXRαL2/− mice which, after Tam-induced Cre-mediated excision, should yield mice in which adipocytes have a disrupted RXRαL−/− genotype (hereafter called RXRαadL−/− mice), whereas other cell types have a RXRαL2/− genotype. Their littermates, particularly, aP2-Cre-ERT2(tg/0)/ RXRα+/− mice, were indistinguishable from wild-type (WT) mice and used as control (CT) animals. Four-week-old aP2-Cre-ERT2(tg/0)/RXRαL2/− mice were treated with Tam. Three months later, the selective presence of the L− allele in the adipose tissue was found to depend on Tam treatment (Fig. 2A, compare lanes 1 and 4, and data not shown). Importantly, WAT fractionation into purified adipocytes [supernatant (S) fraction] and stromal-vascular cells [pellet (P) fraction] indicated that at least 80% of the purified adipocytes had a RXRαL−/− genotype, whereas no Cre-mediated recombination had occurred in stromal-vascular cells (Fig. 2A, compare lanes 5 and 6).
Figure 2.
Adipocyte-selective RXRα ablation in mice. (A) Efficiency of RXRα inactivation in adipocytes. Cre-ERT2-mediated RXRα disruption was analyzed by Southern blotting DNA extracted from WAT (T) isolated from 4-month-old aP2-Cre-ERT2(tg/0)/RXRαL2/− mice, 3 months after vehicle (−) and Tam (+) treatments, and from the supernatant (S) and pellet (P) fractions after centrifugation of collagenase-treated WAT. Positions of RXRα L2, L−, and (−) alleles are indicated. (B) Expression of nuclear receptors, PPARγ target genes, and Pref-1 in RXRα-deficient adipose tissue. RXRβ, RXRγ, PPARγ, aP2, and LPL mRNA levels were analyzed by Northern blotting RNA isolated from WAT of 6-month-old CT (lanes 1, 3, and 5) and RXRαadL−/− (KO, lanes 2, 4, and 6) mice, under RD (lanes 1 and 2), after MSG treatment (lanes 3 and 4) and under HFD (lanes 5 and 6). 36B4 was used as an internal control. Pref-1 expression was analyzed by RT-PCR performed on the same RNA samples, and HPRT was used as an internal control. (C) Altered RXRγ and aP2 expression in the adipose tissue of RXRαadL−/− mice 2 weeks after Tam injection. RXRγ and aP2 mRNA levels were analyzed by Northern blotting RNA isolated from adipose tissue of two 6-week-old CT (lanes 1 and 2) and RXRαadL−/− (KO, lanes 3 and 4) mice, 2 weeks after Tam treatment. 36B4 was used as an internal control.
The above results demonstrate that RXRα was selectively and efficiently disrupted in adipocytes upon Tam treatment of aP2-Cre-ERT2(tg/0)/RXRαL2/− mice. Although this disruption did not affect RXRβ levels in the adipose tissue of RXRαadL−/− mice, RXRγ levels were increased (Fig. 2B, compare lanes 1 and 2), indicating a possible compensatory up-regulation of RXRγ expression. Interestingly, the transcripts of the aP2 and LPL genes, whose expression is known to be under PPARγ control (14, 31), were drastically reduced in WAT of RXRαadL−/− mice, even though PPARγ levels were not modified (Fig. 2B). These observations suggest that aP2 and LPL expressions are controlled by RXRα–PPARγ heterodimers. Note that RXRγ and aP2 expressions were already up- and down-regulated, respectively, 2 weeks after Tam treatment (Fig. 2C), which indicates that RXRα must be functionally ablated in RXRαadL−/− mice shortly after Tam-induced Cre-mediated disruption of the RXRα gene.
RXRα Ablation in Adipocytes Results in Alteration of Preadipocyte Differentiation and Resistance to Obesity.
The body weight of RXRαadL−/− mice generated by Tam treatment at the age of 4 weeks and fed with a RD (named RD-KO mice) was followed for 28 weeks and compared with that of CT mice, named RD-CT (Fig. 3Aa). The weight of RD-KO mice was significantly lower by approximately 10% during the first 4 weeks after Tam treatment and progressively reached that of adult RD-CT mice. From the age of 10 weeks, the weight of RD-KO mice was indistinguishable to that of RD-CT animals, suggesting that RXRα ablation may delay the formation of fat deposits. No differences in s.c. (inguinal and interscapular) and retroperitoneal WAT was observed between 6-month-old RD-KO and RD-CT mice (Fig. 3B), and their adipocytes exhibited a similar size distribution (Fig. 3C a and b).
Figure 3.
RXRαadL−/− mice are resistant to obesity and are impaired in preadipocyte differentiation. (A) RXRαadL−/− mice are resistant to HFD- and MSG-induced obesity. Total body weight of CT and RXRαadL−/− (KO) mice under RD (a and b), under HFD (a) and after MSG treatment (b) was measured weekly. The number of animals monitored in each group was 10–15 males. Values are expressed as the mean ± SEM. (B) Reduced adipose tissue weight in RXRαadL−/− mice under HFD or after MSG treatment compared with CT mice. Inguinal (ING), interscapular (INT), and retroperitoneal (RET) WAT weight was determined in 6-month-old CT and RXRαadL−/− (KO) mice, fed with RD or HFD and after MSG treatment. Each group was composed of 5–7 males. Values are expressed as the mean ± SEM. (C) Impaired adipogenesis in RXRαadL−/− mice. Cryosections of s.c. inguinal WAT from 6-month-old CT (a, c, and e) and RXRαadL−/− (b, d, and f) mice, under RD (a and b), HFD (c and d), and after MSG treatment (e and f) were stained with hematoxylin and eosin. (Scale bar, 160 μm.) The areas of at least 400 cells per sample were determined with the NSURFX software (J. L. Vonesch, Institut de Génétique et de Biologie Moléculaire et Cellulaire Illkirch). The distribution of the cell size is shown in the corresponding graphs to the right. Arrows point to small adipocytes and preadipocytes or poorly differentiated adipocytes in e and f, respectively.
We next investigated whether RXRα ablation in adipocytes may affect obesity induced in mice fed with a HFD. As expected, CT animals became obese under HFD (HFD-CT in Fig. 3 Aa and B). In contrast, no obesity was induced by HFD in HFD-KO mice (Fig. 3Aa), and their WAT weight remained similar to that of RD-CT and RD-KO animals (Fig. 3B). As would be predicted, the adipocytes of HFD-CT mice were much larger than those of RD-CT mice, whereas those of HFD-KO mice exhibited a size distribution similar to those of RD-CT and RD-KO animals. Clearly, RXRγ, which is increased upon RXRα ablation in adipose tissue (Fig. 2B), cannot replace RXRα to mediate the HFD-induced (and chemically induced, see below) hypertrophy of adipocytes, which leads to obesity. Interestingly, it has been reported that heterozygous PPARγ+/− mice are partially resistant to HFD-induced obesity and maintain a smaller size of their adipocytes relative to WT adipocytes under HFD conditions (10). Thus, these observations suggest that HFD-induced adipocyte hypertrophy is mediated by RXRα-PPARγ heterodimers. However, we note that, in contrast to PPARγ+/− heterozygous mice, heterozygous RXRα+/− mice developed HFD-induced obesity and adipocyte hypertrophy that were similar to those of WT animals (data not shown).
MSG administration to newborn mice induces hypothalamic lesions that result in a marked obesity in the adult (refs. 19, 25, and 26 and see Fig. 3Ab). To investigate whether ablation of RXRα in adipocytes also will prevent against the development of MSG-induced obesity, MSG was administered to aP2-Cre-ERT2(tg/0)/RXRαL2/− newborn mice that were treated with Tam at the age of 4 weeks. As expected, MSG-treated CT animals (MSG-CT) developed obesity from the age of 10 weeks onward, compared with CT mice fed with a RD (RD-CT) (Fig. 3 Ab and B). Interestingly, the adipocyte population in WAT of obese MSG-CT animals included not only hypertrophic adipocytes but also a markedly increased population of small adipocytes that was not found in HFD-induced obesity [Fig. 3C, compare e (MSG-CT, arrows) with a (RD-CT) and c (HFD-CT)]. These smaller adipocytes most probably represent maturing adipocytes derived from an increased population of preadipocytes in MSG-treated animals. There is indeed in MSG-CT animals an increase in RNA transcripts encoding Pref-1, a protein abundant in preadipocytes but not expressed in mature adipocytes (refs. 40 and 41; see Fig. 2B, lanes 1 and 3). Note that, in contrast, Pref-1 expression was not increased by a high-fat diet (Fig. 2B, lane 5) that results in adipocyte hypertrophy only (Fig. 3Ce). Thus, MSG treatment appears to cause both generation of preadipocytes and adipocyte hypertrophy in adult mice.
In contrast, the weights of the body and WAT deposits of adult MSG-treated RXRαadL−/− (MSG-KO) mice were similar to those of adult RD-CT animals (Fig. 3 Ab and B). Most of the adipocytes present in MSG-KO WAT were reduced in size compared with MSG-CT adipocytes and were only slightly larger than adipocytes present in mice fed a RD (RD-CT; in Fig. 3C, compare f with e and a). Interestingly, much smaller cells, which were not or only poorly stained with Oil red O, were seen in MSG-KO WAT deposits (Fig. 3Cf, arrows, and data not shown). It is likely that these cells correspond to preadipocytes or poorly differentiated adipocytes, because their presence was accompanied by a further increase in Pref-1 transcripts compared with MSG-CT mice (Fig. 2B, compare lanes 3 and 4). Thus, in MSG-treated mice, RXRα appears to be required for adipogenesis during both the differentiation of preadipocytes to adipocytes and the formation of hypertrophic adipocytes.
RXRα Ablation in Adipocytes Impairs Lipolysis During Fasting.
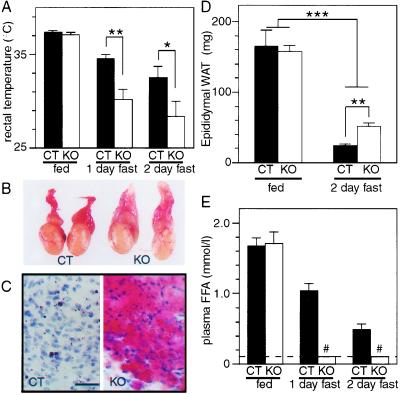
The liberation of fatty acids from the adipose tissue (i.e., lipolysis) is a critical event in the fasting response aimed at maintaining whole-body energy homeostasis in the absence of an external energy supply. That RXRαadL−/− mice, in which RXRα is specifically ablated in adipocytes, could suffer from decreased lipolysis and, therefore, from a fuel shortage of FFAs was first suggested by the observations that three of nine RXRαadL−/− mice, but none of the 10 littermate controls, died after 2 days of fasting. Furthermore, a more pronounced hypothermia was observed upon fasting in RXRαadL−/− mice than in CT littermates (Fig. 4A). After 2 days of fasting, WAT deposits completely disappeared in CT mice but not in RXRαadL−/− (KO) mice, as illustrated by the epididymal fat pads in Fig. 4 B and D. There was an almost total lack of Oil red O neutral lipid staining in a section of the epididymal fat pad of a CT mouse, whereas a corresponding section from a RXRαadL−/− mutant mouse was intensely stained (Fig. 4C). Defective lipolysis in mutant mice was confirmed by the analysis of the FFA levels in the blood of fasting animals (Fig. 4E). No FFA could be detected above background level in plasma of RXRαadL−/− mice after 1 day of fasting. Under similar conditions FFA levels were readily detectable in CT animals. Thus, the above data indicate that the presence of RXRα in adipocytes is required for the efficient release of FFA from white adipose fat.
Figure 4.
Impaired lipolysis in RXRαadL−/− mice during fasting. (A) Fasted RXRαadL−/− mice suffer from hypothermia. Rectal temperature was determined on 10-week-old CT and RXRαadL−/− (KO) mice at the beginning of the light cycle (fed state) and after a 1- or 2-day fast that was started at the beginning of the light cycle. (Error bars, SEM.) *, P < 0.05; **, P < 0.001. (B) Gross appearance of epididymal fat pad after fasting. Testes and epididymal fat pads from 3-month-old CT and RXRαadL−/− (KO) mice after a 2-day fast are presented. (C) Histological analysis of epididymal fat pad. Sections of the epididymal fat pads shown in B were stained with Oil red O. (Scale bar, 20 μm.) (D) Weight of epididymal fat pad. Epididymal fat pad weight of fed and fasted (2 days) 10-week-old CT and KO mice was measured. Values are expressed as the mean ± SEM (n = 9–10). **, P < 0.001; ***, P < 0.0001. (E) Plasma FFA concentrations. FFA was determined in 10-week-old fed and fasted (1 and 2 days) CT and KO mice. Values are expressed as the mean ± SEM (n = 9–10). # indicates values below background levels (dashed line, 0.1 mmol/liter).
Conclusion
We show herein the effectiveness of the Tam-inducible Cre-ERT2 chimeric recombinase expressed under the control of the aP2 promoter to generate adipocyte-selective temporally controlled targeted mutations in transgenic mice. These mice will be useful to analyze the function of many genes that are involved in energy homeostasis obesity and diabetes and often are expressed in several tissues where they exert pleiotropic effects (refs. 4, 6, 7, and 42 and references therein). In the present study, we have used the aP2-Cre-ERT2 transgenic line to investigate the functions of the RXRα gene in adipocytes.
We found that mice lacking RXRα in their adipocytes are resistant to obesity induced by a HFD or a treatment with MSG. It has been reported (10) that, under a HFD, the size of adipocytes from PPARγ+/− mice is significantly smaller than that of adipocytes from WT mice. Thus, the formation of hypertrophic adipocytes appears to be mediated by RXRα–PPARγ heterodimers. Furthermore, our results indicate that RXRα not only is involved in adipocyte hypertrophy, as seen after HFD or MSG treatment, but also is probably involved in de novo differentiation of preadipocytes. Because PPARγ has been shown to be required for adipocyte differentiation in vitro and probably in vivo (refs. 8–10 and references therein), RXRα–PPARγ heterodimers may mediate fat accretion during both adipocyte differentiation and hypertrophy.
Finally, our results also reveal that RXRα is required during fasting for efficient lipolysis, demonstrating that RXRα plays an important role in both fat storage and mobilization. The molecular mechanisms involved in FFA release from the adipocytes and controlled by RXRα are at present unknown. Whether PPARγ also is involved in the control of lipolysis as an heterodimeric partner for RXRα will require its temporally controlled adipocyte-specific ablation.
Acknowledgments
We thank J. Auwerx, B. Desvergne, W. Wahli, and G. Richards for probes, helpful discussions, and critical reading of the manuscript; S. Bronner, M. F. Champy, C. Gérard, R. Lorentz, and J. L. Vonesch for excellent technical help; M. LeMeur and the animal facility staff for animal care; and the secretarial staff for preparation of the manuscript. This work was supported by funds from the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, the Collège de France, the Hôpital Universitaire de Strasbourg, the Association pour la Recherche sur le Cancer, the Fondation pour la Recherche Médicale, the Human Frontier Science Program, European Economic Community Contract FAIR-CT97–3220, and the Ministère de l'Éducation Nationale de la Recherche et de la Technologie Déc.97.C.0115. T.I. was supported by postdoctoral fellowships from the Centre National de la Recherche Scientifique, the Fondation de la Recherche Médicale, and the Toyobo Science Foundation.
Abbreviations
- aP2
adipocyte fatty acid binding protein
- Cre-ERT2
fusion protein between the Cre recombinase and a mutated ligand-binding domain of the human estrogen receptor
- EGFP
enhanced green fluorescent protein
- FFA
free fatty acid
- HFD
high-fat and high-sucrose diet
- KO
knockout
- LPL
lipoprotein lipase
- MSG
monosodium glutamate
- PPAR
peroxisome proliferator-activated receptor
- RD
regular diet
- RXR
retinoid X receptor
- Tam
tamoxifen
- WAT
white adipose tissue
- RT-PCR
reverse transcription–PCR
- WT
wild type
- CT
control
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011528898.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011528898
References
- 1.Spiegelman B M, Flier J S. Cell. 1996;87:377–389. doi: 10.1016/s0092-8674(00)81359-8. [DOI] [PubMed] [Google Scholar]
- 2.Mandrup S, Lane M D. J Biol Chem. 1997;272:5367–5370. doi: 10.1074/jbc.272.9.5367. [DOI] [PubMed] [Google Scholar]
- 3.Fajas L, Fruchart J C, Auwerx J. Curr Opin Cell Biol. 1998;10:165–173. doi: 10.1016/s0955-0674(98)80138-5. [DOI] [PubMed] [Google Scholar]
- 4.Loftus T M, Lane M D. Curr Opin Genet Dev. 1997;7:603–608. doi: 10.1016/s0959-437x(97)80006-8. [DOI] [PubMed] [Google Scholar]
- 5.Lowell B B. Cell. 1999;99:239–242. doi: 10.1016/s0092-8674(00)81654-2. [DOI] [PubMed] [Google Scholar]
- 6.Desvergne B, Wahli W. Endocr Rev. 1999;20:649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 7.Rosen E D, Walkey C J, Puigserver P, Spiegelman B M. Genes Dev. 2000;14:1293–1307. [PubMed] [Google Scholar]
- 8.Barak Y, Nelson M C, Ong E S, Jones Y Z, Ruiz-Lozano P, Chien K R, Koder A, Evans R. Mol Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 9.Rosen E D, Sarraf P, Troy A E, Bradwin G, Moore K, Milstone D S, Spiegelman B M, Mortensen R M. Mol Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- 10.Kubota N, Terauchi Y, Miki H, Tamemoto H, Yamauchi T, Komeda K, Satoh S, Nakano R, Ishii C, Sugiyama T, et al. Mol Cell. 1999;4:597–609. doi: 10.1016/s1097-2765(00)80210-5. [DOI] [PubMed] [Google Scholar]
- 11.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Um K, Blumberg B, Kastner P, Mark M, Chambon P, et al. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambon P. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 13.Tontonoz P, Singer S, Forman B M, Sarraf P, Fletcher J A, Fletcher C D M, Brun R P, Mueller E, Altiok S, Oppenheimer H, et al. Proc Natl Acad Sci USA. 1997;94:237–241. doi: 10.1073/pnas.94.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tontonoz P, Hu E, Spiegelman B M. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 15.Kastner P, Grondona J M, Mark M, Gansmuller A, LeMeur M, Decimo D, Vonesch J-L, Dollé P, Chambon P. Cell. 1994;78:987–1003. doi: 10.1016/0092-8674(94)90274-7. [DOI] [PubMed] [Google Scholar]
- 16.Sucov H M, Dyson E, Gumeringer C L, Price J, Chien K R, Evans R M, et al. Gene Dev. 1994;8:1007–1018. doi: 10.1101/gad.8.9.1007. [DOI] [PubMed] [Google Scholar]
- 17.Indra A K, Warot X, Brocard J, Bornert J-M, Xiao J-H, Chambon P, Metzger D. Nucleic Acid Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Indra A K, Warot X, Brocard J, Messaddeq N, Kato S, Metzger D, Chambon P. Nature (London) 2000;407:633–636. doi: 10.1038/35036595. [DOI] [PubMed] [Google Scholar]
- 19.Ross S R, Graves R A, Spiegelman B M. Genes Dev. 1993;7:1318–1324. doi: 10.1101/gad.7.7b.1318. [DOI] [PubMed] [Google Scholar]
- 20.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. FEBS Lett. 1997;407:311–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 21.Sumi-Ichinose C, Ichinose H, Metzger D, Chambon P. Mol Cell Biol. 1997;17:5976–5986. doi: 10.1128/mcb.17.10.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Proc Natl Acad Sci USA. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imai T, Chambon P, Metzger D. Genesis. (Dev Genet) 2000;26:147–148. doi: 10.1002/(sici)1526-968x(200002)26:2<147::aid-gene15>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 24.Mascrez B, Mark M, Dierich A, Ghyselinck N B, Kastner P, Chambon P. Development (Cambridge, UK) 1998;125:4691–4707. doi: 10.1242/dev.125.23.4691. [DOI] [PubMed] [Google Scholar]
- 25.Olney J W. Science. 1969;164:719–721. doi: 10.1126/science.164.3880.719. [DOI] [PubMed] [Google Scholar]
- 26.Pizzi W J, Barnhart J E. Pharmacol Biochem Behav. 1976;5:551–557. doi: 10.1016/0091-3057(76)90268-9. [DOI] [PubMed] [Google Scholar]
- 27.Metzger, D. & Chambon, P. (2001) Methods, in press. [DOI] [PubMed]
- 28.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:159–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 29.Kastner P, Mark M, Leid M, Gansmuller A, Grondona J M, Décimo D, Krezel W, Dierich A, Chambon P. Genes Dev. 1996;10:80–92. doi: 10.1101/gad.10.1.80. [DOI] [PubMed] [Google Scholar]
- 30.Krezel W, Dupé V, Mark M, Dierich A, Kastner P, Chambon P. Proc Natl Acad Sci USA. 1996;93:9010–9014. doi: 10.1073/pnas.93.17.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoonjans K, Peinado-Onsurbe A M, Heyman R A, Briggs M, Deeb S, Staels B, Auwerx J. EMBO J. 1996;15:5336–5348. [PMC free article] [PubMed] [Google Scholar]
- 32.Bouillet P, Oulad-Abdelghini M, Vicaire S, Garnier J M, Schuhbaur B, Dollé P, Chambon P. Dev Biol. 1995;170:420–433. doi: 10.1006/dbio.1995.1226. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 34.Rodbell M. J Biol Chem. 1964;239:375–380. [PubMed] [Google Scholar]
- 35.Hotamisligil G S, Shargill N S, Spiegelman B M. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 36.Brocard J, Warot X, Wendling O, Messaddeq N, Vonesch J-L, Chambon P, Metzger D. Proc Natl Acad Sci USA. 1997;94:14559–14563. doi: 10.1073/pnas.94.26.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peters J M, Hennuyer N, Staels B, Fruchart J C, Fievet C, Gonzalez F J, Auwerx J. J Biol Chem. 1997;272:27307–27312. doi: 10.1074/jbc.272.43.27307. [DOI] [PubMed] [Google Scholar]
- 38.Akagi K, Sandig V, Vooijs M, Van der Valk M, Giovannini M, Strauss M, Berns A. Nucleic Acids Res. 1997;25:1766–1773. doi: 10.1093/nar/25.9.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Postic C, Shiota M, Niswender K D, Jetton T L, Chen Y, Moates J M, Shelton K D, Lindner J, Cherrington A D, Magnuson M A. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 40.Smas C M, Sul H S. Cell. 1993;73:725–734. doi: 10.1016/0092-8674(93)90252-l. [DOI] [PubMed] [Google Scholar]
- 41.Smas C M, Chen L, Sul H S. Mol Cell Biol. 1997;17:977–988. doi: 10.1128/mcb.17.2.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kadowaki T. J Clin Invest. 2000;106:459–465. doi: 10.1172/JCI10830. [DOI] [PMC free article] [PubMed] [Google Scholar]