Abstract
The cleavage model for signal transduction by receptors of the LIN-12/Notch family posits that ligand binding leads to cleavage within the transmembrane domain, so that the intracellular domain is released to translocate to the nucleus and activate target gene expression. The familial Alzheimer's disease-associated protein Presenilin is required for LIN-12/Notch signaling, and several lines of evidence suggest that Presenilin mediates the transmembrane cleavage event that releases the LIN-12/Notch intracellular domain. However, doubt was cast on this possibility by a report that Presenilin is not required for the transducing activity of NECN, a constitutively active transmembrane form of Notch, in Drosophila. Here, we have reassessed this finding and show instead that Presenilin is required for activity of NECN for all cell fate decisions examined. Our results indicate that transmembrane cleavage and signal transduction are strictly correlated, supporting the cleavage model for signal transduction by LIN-12/Notch and a role for Presenilin in mediating the ligand-induced transmembrane cleavage.
Receptors of the LIN-12/Notch family mediate cell–cell interactions during normal development, and aberrations of LIN-12/Notch signaling have been linked to many different diseases (reviewed in refs. 1 and 2). LIN-12/Notch proteins are single-pass membrane proteins that undergo at least three proteolytic processing events during maturation and signal transduction. First, the protease Furin cleaves at a site in the extracellular domain (site 1) during transport of the receptor to the cell surface; the resulting amino-terminal domain remains associated with a carboxy-terminal transmembrane domain (3, 4). Second, ligand binding triggers an additional cleavage of the extracellular region of the carboxy terminal domain (site 2), probably by a protease of the ADAM family; this second cleavage thereby shortens the extracellular region to 12 amino acids (5, 6). Third, proteolytic cleavage within the transmembrane domain (site 3) releases the intracellular domain so that it can translocate to the nucleus and activate transcription of target genes (see refs. 7–9).
This paper is concerned with the role of Presenilin in the proteolytic cleavage event that releases the LIN-12/Notch intracellular domain, and the requirement for that proteolytic cleavage event for signal transduction. Presenilin was identified in studies of familial forms of Alzheimer's disease (reviewed in ref. 10), a devastating neurological disease that is associated with deposits in the brain of Aβ peptide processing products produced from a transmembrane precursor protein, β-APP. Two cleavage events release Aβ from β-APP: one cleavage occurs at a site in the extracellular domain, called β, and one cleavage occurs within the transmembrane domain, at a site called γ (reviewed in 10). The γ “site” is not a specific sequence, and the cleavage within the transmembrane domain may occur after a few different amino acids. Processing at the γ site requires Presenilin activity (11) and recent studies with putative aspartyl protease active site inhibitors have provided evidence that Presenilin may itself be γ-secretase, the long-elusive enzyme that performs the cleavage event (12, 13).
Genetic studies in Caenorhabditis elegans, Drosophila, and mice have shown that Presenilin is essential for LIN-12/Notch signaling (14–19), and several lines of evidence have suggested that Presenilin mediates the transmembrane cleavage event that releases the LIN-12/Notch intracellular domain (18, 20–23). However, doubt was cast on this view (e.g., 24, 25) because two contemporaneous studies in Drosophila reached conflicting conclusions about the role of Presenilin in Notch processing and signal transduction.
One study, performed by us, examined the effect of PS null mutations on nuclear access by the Notch intracellular domain by using a sensitive, in vivo assay (18). This assay utilizes functional Notch proteins in which the chimeric transcription factor Gal4-VP16 (GV) is inserted in frame into Notch (N) just after the transmembrane domain, so that nuclear access of the intracellular domain results in expression of a UAS-lacZ reporter gene under GV control (9). Biochemical evidence has validated this assay as a measurement of transmembrane cleavage (26). We found that nuclear access of N+-GV3, a wild-type protein, depended on Presenilin activity. Similarly, nuclear access of NECN-GV3, a constitutively active transmembrane form that mimics a site 2 processing product (see Fig. 1), also depended on Presenilin activity. However, Nintra-GV3, a constitutively active form that consists of just the intracellular domain and mimics a site 3 processing product (Fig. 1), gains nuclear access even in the absence of Presenilin activity. From these results, we concluded that Presenilin is required for the site 3 transmembrane cleavage and release of the intracellular domain of Notch from the plasma membrane.
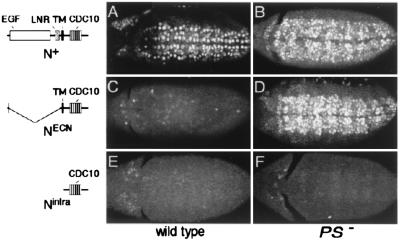
Figure 1.
Presenilin-dependent activity of Notch in the embryonic central nervous system. Neuroblasts marked by the expression of Hb protein are shown in wild-type (A, C, and E) and PS− (B, D, and F) embryos that ubiquitously express either N+ (A and B), NECN (C and D), or Nintra (E and F) under Gal4/UAS control. (A) Ubiquitous expression of N+ in otherwise wild-type embryos has little or no effect on neuroblast segregation. (C and E) In contrast, ubiquitous expression of either NECN or Nintra suppresses neuroblast segregations, indicating constitutive transducing activity. In PS− embryos, most or all ventral ectodermal cells segregate as neuroblasts, even when N+ or NECN are ubiquitously expressed (B and D), indicating that the transducing activity of these transmembrane forms requires Presenilin. In contrast, neuroblast segregation is suppressed in PS− embryos expressing Nintra (F), indicating that this form of Notch bypasses the requirement for Presenilin. All embryos are shown in ventral aspect just after completion of germ-band elongation (anterior to the left). The structure of the Notch proteins are shown schematically: EGF, epidermal growth factor motifs; LNR, LIN-12/Notch repeat motifs; TM, transmembrane domain; CDC10, CDC10/SWI6 motifs.
The other study, conducted by Ye et al. (19), examined the requirement for Presenilin in Notch signal transduction by using a phenotypic assay. The key genetic experiment was to express NECN or Nintra under the control of a heat shock promoter, in wild-type and PS− wing imaginal disks, and then to examine the effect on neurogenesis: when Notch is constitutively active, neurogenesis is suppressed. Ye et al. reported that both NECN and Nintra were able to suppress neurogenesis in PS− wing disks, as they do in wild-type disks. From this observation, they concluded that Presenilin is not required for transmembrane cleavage per se, but rather for an upstream event necessary to generate the activated, transmembrane form of the receptor.
If the cleavage model for LIN-12/Notch signal transduction is correct, then transmembrane cleavage and Notch activity should be strictly correlated. However, the findings of Ye et al. suggest that Notch activity does not correlate with transmembrane cleavage for the case of NECN protein. Therefore, we reassessed whether Presenilin is required for the constitutive transducing activities of NECN and Nintra in several different developmental contexts, including neurogenesis in the wing disk, the same context assayed by Ye et al. (19). In all cases, and in contrast to Ye et al., we find that Presenilin is strictly required for the constitutive transducing activity of NECN, but not for that of Nintra. Thus, our results strongly favor the transmembrane cleavage model for Notch signal transduction as well as a specific role for Presenilin in mediating the ligand-induced transmembrane cleavage.
Materials and Methods
Generating Ubiquitous Expression of N+, NECN, and Nintra in Wild-Type Embryos.
A modified version of the Gal4/UAS method (9, 27) was used to drive expression of UAS-N+, UAS-NECN, and UAS-Nintra transgenes in wild-type embryos. Wild-type females were crossed to males of the following genotypes: (i) Tubulinβ1-flp/Y; UAS-GFPnls Tubulinα1>CD2, y+>Gal4-VP16/+; UAS-N+ PSC2 FRT2A ftz-lacZ/TM2; (ii) Tubulinβ1-flp/Y; UAS-GFPnls Tubulinα1>CD2, y+>Gal4-VP16/ftz-lacZ; UAS-NECN PSC2 FRT2A/TM3, ftz-lacZ; and (iii) Tubulinβ1-flp/Y; UAS-GFPnls Tubulinα1>CD2, y+>Gal4-VP16/ftz-lacZ; UAS-Nintra PSC2 FRT2A/TM3, ftz-lacZ. The Tubulinβ1-flp transgene expresses the yeast recombinase Flp specifically during spermatogenesis and catalyzes excision of the >CD2, y+> Flp-out cassette from the Tubulinα1>CD2, y+>Gal4-VP16 transgene in most of the mature sperm [zygotes carrying the resulting Tubulinα1>Gal4-VP16 survive during embryogenesis, but die just before or during larval development; hence, the need to generate the Tubulinα1>Gal4-VP16 transgene during spermatogenesis (see refs. 9 and 28)]. Progeny of the correct genotype, UAS-GFPnls Tubulinα1>Gal4-VP16/+; UAS-N/+ were identified by the expression of the UAS-GFPnls gene (which encodes a nuclear-localized form of Green Fluorescent Protein) and the presence (UAS-N+) or absence (UAS-NECN and UAS-Nintra) of ftz-lacZ expression.
Generating Ubiquitous Expression of N+, NECN, and Nintra in PS− Embryos.
Females that lay eggs derived solely from PS− germ cells (18) were fertilized by males of the same three genotypes listed above, and progeny of the correct genotype identified in the same fashion.
Generating Clones of PS− Cells That Express the UAS-NECN or UAS-Nintra Transgenes Marked by the Coincident Expression of the UAS-GFPnls and UAS-y+ Transgenes.
The MARCM system (29), employing the use of a Tubulinα1-Gal80 transgene in conjunction with the Gal4/UAS method was used to activate UAS-NECN and UAS-Nintra expression in marked clones of PS− cells. Females of the genotype y w hsp70-flp Tubulinα1-Gal4 UAS-GFPnls; UAS-y+; Tubulinα1-Gal80 FRT2A/TM6B were crossed to males of the following three genotypes: (i) y w hsp70-flp; smc-Z; UAS-NECN PSC2 FRT2A/TM6B; (ii) y w hsp70-flp; smc-Z; UAS-Nintra PSC2 FRT2A/TM6B; and (iii) y w hsp70-flp Tubulinα1-PS+, y+; smc-Z; UAS-NECN PSC2 FRT2A/TM6B. The progeny were heat shocked during the first or second instar to induce clones by Flp/FRT mediated somatic recombination (30). Wing imaginal disks were then dissected from late third instar larvae of the correct genotype [non-TM6B for all three crosses and, in the case of (iii), also female and y+] and fixed for immunofluorescent staining.
Immunofluorescence Microscopy.
Embryos were stained by standard procedures and βGal and Hunchback (Hb) protein expression detected by using rabbit anti-βGal and rat anti-Hb antisera. Imaginal disks were similarly fixed and stained by using standard procedures and βGal and Cut expression detected by using rabbit anti-βGal and mouse anti-Cut antisera. All images shown were obtained by using confocal microscopy.
Results
Presenilin Is Required for the Constitutive Transducing Activity of NECN in the Developing Embryonic Nervous System.
The classic Notch-mediated neurogenic interaction occurs during embryonic development, so that some cells in the “proneural” portion of the ventral ectoderm segregate as neuroblasts, while the others remain in the ectoderm and eventually differentiate into the ventral epidermis. The absence of Notch activity results in neural hyperplasia at the expense of the epidermis, whereas constitutive Notch activity suppresses neuroblast segregation so that all ectodermal cells differentiate as epidermis (see ref. 28).
Early neuroblast segregation can be readily visualized by the expression of the transcription factor Hb. During wild type development, the initial rounds of neuroblast segregations generate a stereotyped pattern of three anteroposterior columns of Hb-expressing neuroblasts on each side of the ventral midline (see ref. 28). Early neural segregations also appear normal in embryos in which N+ is ubiquitously expressed from a transgene (Fig. 1A). In contrast, embryos lacking Notch activity form a broad swath of Hb-expressing neuroblasts in place of the normal pattern of three columns, whereas embryos in which constitutively activated forms of Notch (NECN or Nintra) are ubiquitously expressed lack Hb expression (ref. 28; Fig. 1 C and E).
Embryos lacking maternal and zygotic Presenilin activity, referred to as PS− embryos, resemble Notch− embryos (refs. 18 and 19; Fig. 1B). This phenotype results from the absence of Notch signal transducing activity rather than from a marked decrease in Notch protein levels at the plasma membrane (18). Here, we have examined the ability of N+, NECN, and Nintra to suppress neuroblast formation in PS− embryos (Fig. 1 B, D, and F). We find that ubiquitous expression of N+ or NECN fails to suppress neuroblast segregations (Fig. 1 B and D), so that such embryos appear indistinguishable from PS− embryos. In contrast, ubiquitous expression of Nintra in PS− embryos efficiently suppresses neuroblast segregations (Fig. 1F), as it does in otherwise wild-type embryos (Fig. 1E).
We previously showed that the intracellular domains of N+-GV3 and NECN-GV3 do not gain access to the nucleus in PS− embryos, in contrast to Nintra-GV3, which appears to have ready access (18). Thus, Notch nuclear access in PS− embryos appears to correlate with Notch transducing activity: Nintra has access and retains constitutive transducing activity, whereas NECN and N+ lack access and show no evidence of transducing activity.
Presenilin Is Required for the Constitutive Transducing Activity of NECN in the Developing Wing Imaginal Disk.
Notch activity is required in several distinct processes during the development of the wing imaginal disk. The eponymous Notch phenotype is a notched wing, a consequence of reduced Notch-mediated signaling across the dorsoventral compartment boundary. Notch-mediated signaling also regulates classic neural/ectodermal decisions that control the pattern of mechanosensory bristles on the mesonotum (the dorsal portion of the fuselage of the adult thorax). Finally, Notch signaling is required to resolve thin stripes of wing vein cells from initially broader stripes of “prevein” tissue, a process essential for normal vein development. Here we analyze the consequences of expressing NECN and Nintra in genetically marked clones of PS− cells for each of these processes. In all cases, we have found that in the absence of Presenilin, Nintra retains constitutive transducing activity, whereas NECN shows no evidence of transducing activity.
Notch Signaling Across the Dorsoventral Compartment Boundary.
Activation of Notch signaling across the dorsoventral compartment boundary in wing imaginal disks induces a thin stripe of “edge cells” that straddle the boundary to express the target genes Cut and Wingless (Wg; refs. 31 and 32). Cut is a transcription factor that is required for differentiation of the edge cells (31, 32) and Wg is a morphogen that controls growth and patterning of the wing, including specification of the mechanosensory bristles that decorate the wing margin (see ref. 33). Clones of cells that lack either Notch or Presenilin activity fail to express either Cut or Wg along the presumptive wing margin. The loss of Cut expression can be visualized in disks by antibody staining; furthermore, in adults, the loss of Wg signaling can be readily assayed morphologically by the presence of large wing notches. Conversely, clones of cells that express constitutively active forms of Notch, such as Nintra or NECN, ectopically express both Cut and Wg wherever they arise within the wing blade primordium. Ectopic expression of Wg in turn induces the formation of ectopic sensory mother cells (SMCs) in neighboring wing tissue and also causes ectopic wing outgrowths.
Clones of PS− cells that express NECN or Nintra, as well as a nuclearly localized form of Green Fluorescent Protein (GFPnls) and the Yellow (Y) protein (which allows adult structures to be genetically marked), were generated early during wing disk development by using the MARCM technique (29) and their effects on Cut expression and growth in the wing blade were assayed (see Materials and Methods). The MARCM technique (29) is a modification of the Gal4/UAS (27) and Flp/FRT (30) methods in which the yeast Gal80 protein, a dominant antagonist of transcriptional activation by yeast Gal4 protein, is expressed constitutively from a Tubulinα1-G80 transgene positioned in trans to a mutation of interest, in this case PS−. Mitotic recombination is induced by using the Flp/FRT method (30), to generate homozygous PS−/PS− cells that lack the Tubulinα1-G80 transgene. Within these PS− clones, therefore, UAS-target genes such as UAS-NECN or UAS-Nintra, together with UAS-GFPnls and UAS-yellow, can be expressed under the control of Gal4.
PS− clones expressing NECN that straddle the dorsoventral compartment boundary fail to express Cut (Fig. 2B). In addition, they are associated with severe notching of the adult wing (Fig. 3A), consistent with loss of Wg signaling. These phenotypes indicate that the constitutive activity of NECN in the developing wing depends on Presenilin activity.
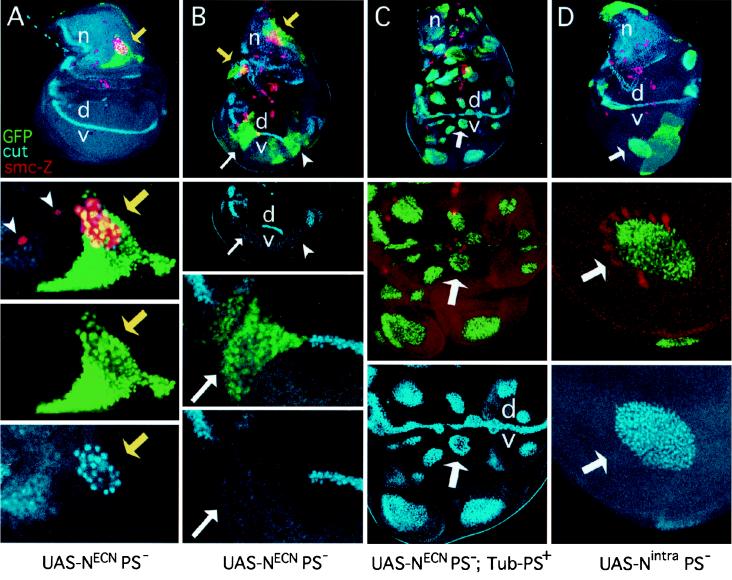
Figure 2.
Presenilin is required for the transducing activity of transmembrane but not intracellular forms of Notch during wing development. Each column contains views of a single mosaic imaginal wing disk. Clones that lack PS activity are marked by GFP expression (green). These clones also express either NECN or Nintra, which are constitutively active in a wild-type genetic background. smc-Z expression (red) labels SMCs that will form mechanosensory bristles in the presumptive notum (n) as well as sense organs in other portions of the disk. Cut expression (blue) marks subsets of SMCs. Cut is also expressed in a thin stripe of “edge cell” straddling the dorsoventral compartment boundary (vd) in the wing blade, and in adepithelial cells associated with the notum. The disk shown in A contains only a single small clone and hence serves as a reference for the normal disk. Disks are shown anterior to the left and ventral down. (A) Series of views of a disk carrying one small clone of PS− UAS-NECN-expressing cells (yellow arrow). Cells within the clone form a cluster of SMCs (marked by smc-Z and Cut expression) instead of the single SMC that would normally form at this position, indicating the absence of Notch transducing activity. Single SMCs (marked by smc-Z expression) in neighboring wild-type tissue are indicated by arrowheads. (B) This disk carries multiple PS− clones that express NECN and display phenotypes associated with the absence of Notch transducing activity. Two clones in the notum (yellow arrows) show the formation of SMC clusters in place of single SMCs; two clones in the wing blade span the dorsoventral compartment boundary (white arrow and arrowhead) and show a cell autonomous loss of Cut expression. (C) This disk serves as a control. Clones of PS− UAS-NECN-expressing cells were generated in the presence of a rescuing Tub-PS+ transgene. Restoration of Presenilin activity from the rescuing transgene reveals the constitutive activity of NECN within clones. In the notum, SMCs do not form, indicating that neurogenesis is suppressed. There is no overlap between the clone (green) and smc-Z expression (red); the few apparent cases are due to superimposition of the nuclear GFP and cytoplasmic smc-Z signals in different focal planes. In the wing blade, clones autonomously express Cut, indicating constitutive activity of Notch (white arrow marks a representative clone). These clones are also associated with overgrowth of neighboring, wild-type tissue, an indication that the mutant cells are ectopically expressing Wg as well as Cut. (D) This disk carries several clones of PS− UAS-Nintra-expressing cells. Clones in the wing blade autonomously express Cut. The wing blade clone marked by the white arrow has also induced neighboring wild-type cells to express the smc-Z reporter, an indication that these cells are developing as wing margin bristles in response to Wg ectopically expressed by the mutant cells within the clone (see also Fig. 3C).
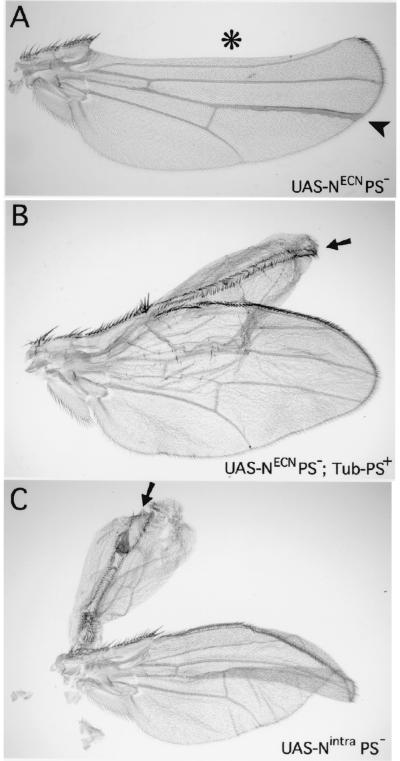
Figure 3.
Adult wing phenotypes associated with PS− clones that express transmembrane or nuclear forms of Notch. Clones are marked by expression of Yellow under UAS/Gal4 control, which darkens cuticular structures, including wing hairs and margin bristles (not visible in the figure). (A) PS− clones that express NECN display phenotypes associated with the absence of Notch signal transduction. One clone (*) crosses the wing margin (which coincides with the dorsoventral compartment boundary) and displays extensive wing notching. A second clone (arrowhead) within the dorsal compartment of the blade forms an abnormally thick vein. (B) A clone of PS− cells that express NECN in the presence of the rescuing Tub-PS+ transgene displays constitutive Notch activity (arrow). The clone is associated with a double dorsal wing outgrowth which is flanked by adjacent rows of dorsal wing margin bristles (here, as in C, an arrow points down the plane of mirror symmetry). Note that in this experiment, the presence of the clone could not be scored on the margin bristles because the Tub-PS+ transgene used in this experiment includes a yellow+ gene which rescues normal (dark) pigmentation in the bristles. However, the contribution of the clone to the rest of the wing blade could still be scored because this yellow+ gene does not rescue pigmentation of the wing hairs. (C) A clone of Nintra-expressing PS− cells (arrow) associated with a double dorsal wing outgrowth which coincides with a thin strip of mutant cells flanked by adjacent rows of dorsal wing margin bristles formed by neighboring wild-type cells.
In contrast, the constitutive activity of Nintra does not require Presenilin activity. Clones of PS− cells that express Nintra autonomously express Cut (Fig. 2D). In addition, they are associated with two phenotypes that indicate that they ectopically express Wg. First, they induce ectopic wing margin bristles in neighboring wild-type cells (Fig. 2D). Second, they are associated with bulges in the disk epithelium suggesting excessive wing growth, a possibility confirmed by the behavior of the clones in the adult wing where they are associated with large outgrowths of wing tissue and ectopic rows of margin bristles formed by wild-type cells adjacent to the clone (Fig. 3C).
As an additional control for the NECN experiment, we also generated NECN-expressing PS− clones by using the same genetic configuration described above, except for the additional presence of a Tubulinα1-PS+ (Tub-PS+) transgene, which restores Presenilin activity (see Materials and Methods). We found that restoring Presenilin activity restored the constitutive activity of NECN (Figs. 2C and 3B), confirming that the absence of Notch transducing activity in NECN-expressing PS− cells is specifically due to the absence of Presenilin activity.
Notch Signaling and the Control of Neural/Ectodermal Decisions in the Mesonotum.
During the development of the mesonotum, small “proneural clusters” of ectodermal cells undergo Notch-mediated interactions so that one cell within the cluster becomes an SMC, whereas the others remain ectodermal. In the absence of Notch or Presenilin function, all cells of the cluster choose the SMC fate, so that a cluster of neurons is produced at the expense of the epidermis. Conversely, the constitutive activity of NECN or Nintra prevents any cell from choosing the SMC fate, thereby suppressing bristle formation. All of the SMCs can be marked by the expression of the smc-Z reporter gene (34), and a subset of these also expresses Cut.
We find that Presenilin activity is essential for the constitutive transducing activity of NECN during SMC specification. Clones of PS− cells expressing NECN that arise within the mesonotum primordium cause clusters of SMCs to form in place of a single SMC (Fig. 2 A and B). In contrast, no SMCs appear to segregate within clones of PS− cells expressing Nintra or clones of PS− cells expressing NECN which also carry the rescuing Tubulinα1-PS+ transgene (Fig. 2C and data not shown). Thus, the constitutive transducing activity of NECN in this context also depends on Presenilin. These results directly contradict the findings reported by Ye et al. (19) and will be considered further in the Discussion.
Notch Signaling During Wing Vein Development.
Cells of initially broad “provein” regions undergo Notch-mediated cell–cell interactions so that some cells become vein cells whereas the others become intervein cells (35–37). In the absence of Notch or Presenilin function, most or all provein cells become vein cells, so that the wing veins are abnormally thick; conversely, constitutive activation of the Notch pathway suppresses vein cell formation (28, 35). As shown in Fig. 3A, clones of PS− cells that express NECN can contribute to the adult wing blade, provided that they do not cross the wing margin where Notch signal transduction is essential for activating Wg. Such clones cause a thickened vein phenotype indicating a failure of Notch signal transduction in the provein cells. Because Nintra-expressing PS− cells as well as Tubulinα1-PS+ NECN-expressing PS− cells strongly activate Wg expression and cause outgrowths composed primarily of surrounding, wild-type wing cells, we cannot readily assess whether they have the ability to differentiate as vein. Nevertheless, our finding that NECN-expressing PS− cells form abnormally thickened veins indicates that Presenilin is essential for NECN transducing activity in this context as well.
Discussion
Previously, we showed that Presenilin activity is necessary for nuclear access of the Notch intracellular domain, for both the wild-type receptor, N+, as well as a constitutively active transmembrane form, NECN, but not for nuclear access of a constitutively active cytosolic form, Nintra (18). Here, we have examined the requirement for Presenilin for the constitutive transducing activities of both NECN and Nintra in several different cell fate decisions during Drosophila development. Transduction was assayed by phenotypic outputs, using well established criteria for absent or constitutive Notch transducing activity. In all cases examined, we find that Presenilin is required for the transducing activity of NECN, but not for that of Nintra. These observations, in conjunction with our previous experiments showing that Presenilin activity is required for nuclear access for NECN but not Nintra (18), indicate that transmembrane cleavage and Notch transduction are coupled in these decisions.
One of the developmental decisions we examined was a neural/epidermal decision in the developing notum that singles out SMCs from proneural clusters. For this cell fate decision, we found that NECN requires Presenilin activity for its constitutive activity, to suppress SMC segregation. Our results directly conflict with those of Ye et al. (19), who reported that Presenilin is not required for NECN activity in this decision. Ye et al. also scored suppression of SMC segregation to assess Notch signal transduction activity. However, their experimental methodology differed from ours. Ye et al. expressed NECN under the control of a heat shock promoter in wild-type and PS− wing imaginal disks. They then subjected the disks to multiple heat shocks to activate the promoter, and stained for neural markers to assess SMC differentiation. The accuracy of this approach depends critically on developmental stage because SMCs normally form only in mid-late third instar wing disks. In the absence of an independent marker for the developmental stage, it is possible that the disks they selected were too young or were developmentally delayed by the multiple heat shock regime. In contrast, the method we used was internally controlled: we could see that SMC specification was normal in nonmutant areas of the same disk where mutant clones were scored.
Ye et al. (19) also reported biochemical evidence that the cleavage event mediated by Presenilin occurs in the extracellular domain, to produce a carboxy-terminal transmembrane fragment analogous to the site 2 processing product. This finding is in contrast to our inference from the N-GV assay that the cleavage site is in or near the transmembrane domain (18). We have not addressed this discrepancy directly in this study. However, we have verified biochemically in Drosophila that our N-GV assay reflects a transmembrane cleavage event that depends on Presenilin (26). In addition, we have observed that any single-pass transmembrane protein can serve as a substrate for Presenilin-dependent cleavage, provided that the extracellular domain is relatively small (26). Indeed, effective substrates include transmembrane proteins that consist only of an extracellular Myc tag, a heterologous transmembrane domain, and an intracellular GV domain, an observation that is difficult to reconcile with a specific requirement for Presenilin upstream of the Notch transmembrane cleavage.
In support of our view, two biochemical studies in mammalian cells (20, 21) have also provided evidence that Presenilin is required for the transmembrane cleavage event, rather than in an upstream processing or trafficking event as suggested by Ye et al. (19). Both mammalian studies provided evidence that proteolytic processing of a form equivalent to NECN and release of the intracellular domain was impaired in cells lacking PS1 activity. In addition, one study showed that transcription of the Notch target gene HES1 was reduced in PS1-deficient cells (21), which is consistent with our finding that signal transduction and transmembrane cleavage are linked. However, another study (38) reached a different conclusion, as they still saw substantial HES1 activation despite the reduced cleavage in PS1-deficient cells.
One potential complication in the original mammalian cell culture experiments is the redundant activity of PS2. This issue was addressed recently by performing similar experiments in cells derived from mice lacking both PS1 and PS2 activity (17, 39). In the double-null situation, studies by Herreman et al. (22) and Zhang et al. (23) have shown that transmembrane proteolysis of Notch is abolished. Furthermore, Herreman et al. have shown that transcription of HES1 is not seen after transfection of NECN, again demonstrating that transmembrane cleavage and signal transduction are linked in this assay.
In sum, for several canonical Notch-mediated decisions in Drosophila examined here, Presenilin-dependent transmembrane cleavage appears to be essential for Notch signal transduction. We believe that this correlation will be generally true, given that in all systems examined, the phenotype resulting from the absence of Presenilin activity is essentially the same as mutants lacking LIN-12/Notch activity (14, 17–19, 39). Furthermore, if the cleavage product—the intracellular domain—were not generally relevant to normal signal transduction, it would be difficult to account for the many different experimental systems in which expression of the intracellular domain mimics the effects of activating LIN-12/Notch. Indeed, the cleavage model was first proposed as a possible way to account for the phenotypic consequences of expressing LIN-12/Notch intracellular domains (28).
We note that our results do not rule out the possibility that there may be some situations where LIN-12/Notch signal transduction involves an alternative mechanism that does not depend on cleavage of the transmembrane domain and nuclear translocation of the intracellular domain. However, at this point there is no evidence that such an alternative mechanism is used in Drosophila. The main body of evidence in mammalian cell culture systems also supports the centrality of transmembrane cleavage to LIN-12/Notch signal transduction.
Acknowledgments
We thank Xiao-Jing Qui for technical assistance, Liqun Luo for the MARCM system, Joaquim Culi for smc-Z, and Paul Macdonald for the anti-Hb antiserum. We also thank Sophie Jarriault, Laura Johnston, and Hui-Min Chung for comments on the manuscript, and Chiann-mun Chen for help with imaging. G.S. and I.G. are Investigators of the Howard Hughes Medical Institute.
Abbreviations
- GV
Gal14-Vp16
- N
Notch
- SMC
sensory mother cell
- Wg
wingless
- Hb
Hunchback
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011530298.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011530298
References
- 1.Greenwald I. Genes Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- 2.Joutel A, Tournier-Lasserve E. Semin Cell Dev Biol. 1998;9:619–625. doi: 10.1006/scdb.1998.0261. [DOI] [PubMed] [Google Scholar]
- 3.Blaumueller C M, Qui H, Zagouras P, Artavanis-Tsakonas S. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 4.Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah N G, Israel A. Proc Natl Acad Sci USA. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens J R, Cumano A, Roux P, Black R A, Israel A. Molec Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 6.Mumm J S, Schroeter E H, Saxena M T, Griesemer A, Tian X, Pan D J, Ray W J, Kopan R. Molec Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 7.Lecourtois M, Schweisguth F. Curr Biol. 1998;8:771–774. doi: 10.1016/s0960-9822(98)70300-8. [DOI] [PubMed] [Google Scholar]
- 8.Schroeter E H, Kisslinger J A, Kopan R. Nature (London) 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 9.Struhl G, Adachi A. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 10.Selkoe D J. Trends Cell Biol. 1998;8:447–453. doi: 10.1016/s0962-8924(98)01363-4. [DOI] [PubMed] [Google Scholar]
- 11.De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. Nature (London) 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 12.Esler W P, Kimberly W T, Ostaszewski B L, Diehl T S, Moore C L, Tsai J Y, Rahmati T, Xia W, Selkoe D J, Wolfe M S. Nat Cell Biol. 2000;2:428–434. doi: 10.1038/35017062. [DOI] [PubMed] [Google Scholar]
- 13.Li Y M, Xu M, Lai M T, Huang Q, Castro J L, DiMuzio-Mower J, Harrison T, Lellis C, Nadin A, Neduvelil J G. Nature (London) 2000;405:689–694. doi: 10.1038/35015085. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Greenwald I. Proc Natl Acad Sci USA. 1997;94:12204–12209. doi: 10.1073/pnas.94.22.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen J, Bronson R T, Chen D F, Xia W, Selkoe D J, Tonegawa S. Cell. 1997;89:629–639. doi: 10.1016/s0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 16.Wong P C, Zheng H, Chen H, Becher M W, Sirinathsinghji D J S, Trumbauer M W, Chen H Y, Price D L, Van der Ploeg L H T, Sisodia S S. Nature (London) 1997;387:288–292. doi: 10.1038/387288a0. [DOI] [PubMed] [Google Scholar]
- 17.Donoviel D B, Hadjantonakis A K, Ikeda M, Zheng H, Hyslop P S, Bernstein A. Genes Dev. 1999;13:2801–2810. doi: 10.1101/gad.13.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Struhl G, Greenwald I. Nature (London) 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 19.Ye Y, Lukinova N, Fortini M E. Nature (London) 1999;398:525–529. doi: 10.1038/19096. [DOI] [PubMed] [Google Scholar]
- 20.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm J S, Schroeter E H, Schrijvers V, Wolfe M S, Ray W J. Nature (London) 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 21.Song W, Nadeau P, Yuan M, Yang X, Shen J, Yankner B A. Proc Natl Acad Sci USA. 1999;96:6959–6963. doi: 10.1073/pnas.96.12.6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herreman A, Serneels L, Annaert W, Collen D, Schoonjans L, De Strooper B. Nat Cell Biol. 2000;2:461–462. doi: 10.1038/35017105. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, Nadeau P, Song W, Donoviel D, Yuan M, Bernstein A, Yankner B A. Nat Cell Biol. 2000;2:463–464. doi: 10.1038/35017108. [DOI] [PubMed] [Google Scholar]
- 24.Hardy J, Israel A. Nature (London) 1999;398:466–467. doi: 10.1038/18979. [DOI] [PubMed] [Google Scholar]
- 25.Chan Y M, Jan Y N. Neuron. 1999;23:201–204. doi: 10.1016/s0896-6273(00)80771-0. [DOI] [PubMed] [Google Scholar]
- 26.Struhl G, Adachi A. Mol Cell. 2000;6:625–636. doi: 10.1016/s1097-2765(00)00061-7. [DOI] [PubMed] [Google Scholar]
- 27.Brand A H, Perrimon N. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 28.Struhl G, Fitzgerald K, Greenwald I. Cell. 1993;74:331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- 29.Lee T, Luo L. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 30.Golic K G. Science. 1991;252:958–961. doi: 10.1126/science.2035025. [DOI] [PubMed] [Google Scholar]
- 31.de Celis J F, Garcia-Bellido A, Bray S J. Development. 1996;122:359–369. doi: 10.1242/dev.122.1.359. [DOI] [PubMed] [Google Scholar]
- 32.Micchelli C A, Rulifson E J, Blair S S. Development. 1997;124:1485–1495. doi: 10.1242/dev.124.8.1485. [DOI] [PubMed] [Google Scholar]
- 33.Zecca M, Basler K, Struhl G. Cell. 1996;87:833–844. doi: 10.1016/s0092-8674(00)81991-1. [DOI] [PubMed] [Google Scholar]
- 34.Culi J, Modolell J. Genes Dev. 1998;12:2036–2047. doi: 10.1101/gad.12.13.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Celis J F, Garcia-Bellido A. Mech Dev. 1994;46:109–122. doi: 10.1016/0925-4773(94)90080-9. [DOI] [PubMed] [Google Scholar]
- 36.de Celis J F, Bray S, Garcia-Bellido A. Development. 1997;124:1919–1928. doi: 10.1242/dev.124.10.1919. [DOI] [PubMed] [Google Scholar]
- 37.Huppert S S, Jacobsen T L, Muskavitch M A. Development. 1997;124:3283–3291. doi: 10.1242/dev.124.17.3283. [DOI] [PubMed] [Google Scholar]
- 38.Berechid B E, Thinakaran G, Wong P C, Sisodia S S, Nye J S. Curr Biol. 1999;9:1493–1496. doi: 10.1016/s0960-9822(00)80121-9. [DOI] [PubMed] [Google Scholar]
- 39.Herreman A, Hartmann D, Annaert W, Saftig P, Craessaerts K, Serneels L, Umans L, Schrijvers V, Checler F, Vanderstichele H, et al. Proc Natl Acad Sci USA. 1999;96:11872–11877. doi: 10.1073/pnas.96.21.11872. [DOI] [PMC free article] [PubMed] [Google Scholar]