Abstract
Approximately one-third of patients with porphyria cutanea tarda (PCT), the most common porphyria in humans, inherit a single mutant allele of the uroporphyrinogen decarboxylase (URO-D) gene. PCT associated with URO-D mutations is designated familial PCT. The phenotype is characterized by a photosensitive dermatosis with hepatic accumulation and urinary excretion of uroporphyrin and hepta-carboxylic porphyrins. Most heterozygotes for URO-D mutations do not express a porphyric phenotype unless hepatic siderosis is present. Hemochromatosis gene (HFE) mutations are frequently found when the phenotype is expressed. We used homologous recombination to disrupt one allele of murine URO-D. URO-D+/− mice had half-wild type (wt) URO-D protein and enzymatic activity in all tissues but did not accumulate hepatic porphyrins, indicating that half-normal URO-D activity is not rate limiting. When URO-D+/− mice were injected with iron-dextran and given drinking water containing δ-aminolevulinic acid for 21 days, hepatic porphyrins accumulated, and hepatic URO-D activity was reduced to 20% of wt. We bred mice homozygous for an HFE gene disruption (HFE−/−) to URO-D+/− mice, generating mice with the URO-D+/−/HFE−/− genotype. These animals developed a porphyric phenotype by 14 weeks of age without ALA supplementation, and URO-D activity was reduced to 14% of wt. These data indicate that iron overload alone is sufficient to reduce URO-D activity to rate-limiting levels in URO-D+/− mice. The URO-D+/− mouse serves as an excellent model of familial PCT and affords the opportunity to define the mechanism by which iron influences URO-D activity.
Porphyria cutanea tarda (PCT), the most common porphyric disorder in humans, is estimated to occur with a prevalence of one in 5,000 (1). Clinically, PCT is characterized by cutaneous photosensitivity associated with skin fragility and blistering. Biochemically, the disorder is characterized by the accumulation of uroporphyrin and hepta-carboxyl porphyrin in the liver. These compounds circulate in plasma, mediate the cutaneous photosensitivity, and are excreted in the urine. Diminished activity of uroporphyrinogen decarboxylase (URO-D) in the liver is present in all cases, and hepatic iron overload is a nearly constant finding (2, 3).
Heterozygosity for mutant URO-D alleles is present in approximately one-third of patients with PCT (1). The activity of URO-D and the concentration of URO-D protein are half-normal in all tissues, but porphyrins accumulate predominantly in the liver. This type of PCT is designated familial-PCT (F-PCT). Most carriers of mutant URO-D alleles do not express a porphyric phenotype unless hepatic siderosis develops, often in association with homozygosity for the cysteine 282 tyrosine (C282Y) mutation of the hemochromatosis gene (HFE; refs. 1, 4, and 5). Environmental factors such as alcohol abuse, hepatitis C virus, and estrogen use also play a role in clinical expression (1, 5). Numerous mutations of the URO-D gene have been identified in patients with F-PCT (6–15) but the most common is a splice-site mutation resulting in the deletion of exon 6 (16).
Approximately two-thirds of cases of PCT occur in individuals with wild-type (wt) URO-D alleles. This type of PCT has been designated sporadic-PCT. URO-D protein concentration and catalytic activity are normal in all tissues but in the liver; protein levels are normal, and enzymatic activity is severely reduced. As with F-PCT, both environmental factors (alcohol, hepatitis C, and estrogens) and genetic factors (HFE mutations) play a role in clinical expression (1, 4, 5), and depletion of liver iron stores is central to successful therapy.
URO-D catalyzes the decarboxylation of the four acetate side chains of uroporphyrinogen to yield coproporphyrinogen (Fig. 1). URO-D is a homodimer with a molecular mass of 82 kDa (17) and is a member of the TIM barrel protein family, with one active site per monomer (18). The URO-D gene has been cloned and characterized from many species, and there is strong amino acid sequence homology among species (2, 18). A murine URO-D cDNA has been cloned (19), but the mouse URO-D genomic locus has not yet been characterized.
Figure 1.
Enzymatic conversion of uroporphyrinogen III to coproporphyrinogen III. At physiological substrate concentrations, the four acetate groups of uroporphyrinogen are decarboxylated in a clockwise fashion, starting with the acetate group on the asymmetric D ring (37). The enzyme will use any of the uroporphyrinogen isomers as substrates, but only the III isomer is used for heme production. Under conditions of substrate excess, decarboxylations occur in a random fashion (38). URO-D, a cytosolic 80-kDa homodimer, functions without the need for a cofactor (17, 18).
An outbreak of PCT occurred in Turkey in 1960 after seed wheat treated with the fungicide hexachlorobenzene was ingested rather than planted (20). Subsequently, many murine models of PCT induced by polyhalogenated aromatic compounds have been reported, but no animal models of F-PCT exist just as no URO-D mutations have been detected in mammals other than humans. To generate a mouse model of F-PCT, we cloned and characterized the URO-D gene from a mouse genomic library and used established gene-targeting techniques to generate mice that were heterozygous for a URO-D–null allele (URO-D+/−). The URO-D+/− mouse, like most humans heterozygous for URO-D mutations, did not develop a porphyric phenotype unless additional factors were present. Injection of iron-dextran coupled with drinking water supplemented with δ-aminolevulinic acid (ALA) produced a porphyric phenotype in 3 weeks. When URO-D+/− animals were crossbred to HFE−/− animals, a porphyric phenotype developed after 14 weeks without ALA supplementation or any other manipulation. Expression of the porphyric phenotype in both iron-injected URO-D+/− mice and unperturbed URO-D+/−/HFE−/− animals was associated with reduction of hepatic URO-D activity to levels well below the half-wt value in untreated URO-D+/− animals, even though URO-D protein levels remained constant. These findings strongly suggest that an inhibitor of URO-D was generated. This mouse model of F-PCT affords the opportunity to define the factors required to generate an inhibitor of URO-D activity without the need for any exogenous perturbing factors.
Materials and Methods
Cloning of URO-D from a Mouse Genomic Library.
A total of 8.5 × 105 recombinants from a 129SvJ mouse genomic library (Stratagene) were screened as described (21) with a radiolabeled full-length human URO-D cDNA (22). Positive plaques after the third screen were further confirmed by PCR using published primer pairs (23). Bacteriophage lambda DNA was prepared by purification of DNA from small-scale liquid cultures as described (24).
Targeted Disruption of Mouse URO-D.
A DNA vector containing the disrupted allele of URO-D was introduced into murine embryonic stem (ES) cells by the method of Thomas and Capecchi (25). The linearized vector, pTKNEOURO-D (1.4 μg), was transfected by electroporation into 1.4 × 107 R1 ES cells (26) suspended in 1.2 ml of transfer buffer (20 mM Hepes, pH 7.0/6 mM dextrose/137 mM NaCl/5 mM KCl/0.7 mM Na2HPO4/2 mM 2-mercaptoethanol). Immediately after electroporation, 2 × 106 ES cells were plated in 10 ml DMEM supplemented with 15% FCS onto eight 10-cm dishes, each seeded with 2.5 × 106 radiation-inactivated embryonic fibroblasts (27). After 24 h of growth, the media were replaced and supplemented with 280 μg/ml neomycin (G418) and 2 × 106 M ganciclovir (28). After 7 days of growth, individual colonies were picked, and each was transferred to one well of a four-well culture plate (Nunc), also seeded with embryonic fibroblasts. When cells covered 80% of the well, they were removed by trypsinization and transferred to one well of a 12-well culture plate (Corning).
Screening of ES Cell Clones.
The 140 ES cells clones, which were resistant to G418 and ganciclovir, were analyzed for integration of NEOr at the correct genomic locus. Genomic DNA from each clone was purified (25) and digested with EcoRI, XbaI, or HindIII; electrophoresed through a 0.7% agarose gel; and transferred for Southern blot analysis. Membranes were probed with either a BamHI to EcoRI fragment of the “F” fragment (Fig. 2A) or a DNA fragment containing the NEOr gene. Mouse Cot-1 DNA (GIBCO/BRL) was added with the probe and hybridized for 2.5 h at 65°C in Rapid-Hyb buffer (Amersham Life Science, Arlington Heights, IL) and washed in 0.1 × SSC (0.15 M sodium chloride/0.015 M sodium citrate, pH 7.0) and 0.1% SDS. Blots were exposed to x-ray film (Kodak) for 24–72 h. Five clones with a correctly targeted integration were identified. The 1C3 clone was injected into C57BL/6J-derived blastocysts that were implanted into foster mothers who were allowed to come to term. Chimeric animals were identified by coat color, and males were mated with C57BL/6J mice to produce mice heterozygous at the URO-D locus.
Figure 2.
Characterization of the null allele. (A1) A 12-kb EcoRI fragment from the genomic clone was subcloned into pBSIISK+. A 1-kb BglII to XhoI fragment (F) was removed. This fragment was subcloned and used as an external probe for Southern blot analysis. (A2) A fragment of DNA containing the neomycin resistance gene (NEOr; ref. 27) was inserted between the BamHI and BglII sites that flank exon 6, deleting exon 6 and portions of the flanking introns. The resulting genomic fragment was purified and ligated into the vector containing two copies of the herpes simplex virus thymidine kinase gene (27). The final construct pTKNEOURO-D was linearized with XhoI and transferred by electroporation into ES cells. (B) A restriction map of the wt allele (1) and the NEOr disrupted allele (2) are shown. Approximate band size expected from digestion with EcoRI are shown as solid lines below each allele. Horizontal arrows represent primers (WF, wt forward; R, reverse; NF, NEO forward) used to generate PCR products shown in D. (C) Southern blot of wt DNA and DNA from ES cell lines carrying the targeted disruption. The DNA was digested to completion with EcoRI and probed with a BamHI, EcoRI fragment of the F probe or a portion of the NEO gene. Molecular size markers are indicated on the right. (D) Genotyping by PCR. Genomic DNA was prepared from tails of mice and used for genotypic assignment (27). The three primers shown in B1 and B2 were used to amplify genomic DNA. The bands at 196 bp and 298 bp represent the disrupted (Null) and wt alleles, respectively. Restriction enzymes: BamHI (B), BglII (Bg), EcoRI (E), HindIII (H), NotI (N), XhoI (X), and XbaI (Xb).
URO-D Assays and Porphyrin Determinations.
Activity of URO-D and tissue and urine porphyrin concentrations were determined as described (29). Porphyrin standards for HPLC determinations were purchased from Porphyrin Products (Logan, UT).
Iron Analysis in Liver Samples.
Approximately 100 mg of liver tissue was added to 400 μl of a nitric/perchloric acid mix [5:2 (vol/vol)] and incubated at 100°C for 4–6 h. Samples were centrifuged at 100,000 × g; the supernatant was diluted 1:2 (vol/vol) with water and analyzed on a Perkin–Elmer Optima 3100 XL instrument. Iron levels were normalized to original tissue wet weight.
PCR Genotyping at the URO-D Locus.
Genotyping at the URO-D locus used DNA purified from tail biopsies or embryo sacs (30). The primers used to amplify wt URO-D (5′ WF: CTGGAGAAAGCAGGGATGTTACT with 3′ primer R: CTGAGGAAGGGCTAAGAATCTAC) amplify a 298-bp product. The mutant locus was amplified by using the 5′ primer NF: CCTGTTACCCTTTTGGTTTGGATA and the common R, listed above, as the 3′ primer, yielding a 196-bp product. Reactions contained 5 ng/μl genomic DNA in a standard PCR buffer mix as described by the manufacturer (Life Technologies, Rockville, MD; ref. 28). Amplification conditions were as follows: 95°C for 30 sec, 60°C for 30 sec, and 72°C for 60 sec for 30 cycles.
Hemochromatosis Gene (HFE) Knockouts.
A generation of HFE−/− mice was accomplished as described (31).
Results
Characterization of the Mouse URO-D Gene.
The genomic architecture of the mouse URO-D gene proved to be identical to the human gene (22) with 10 exons and nine introns. The exons of murine URO-D were identical in length to the corresponding exons in the human gene. The exonic sequence corresponded to the reported sequence of a mouse URO-D cDNA isolated from a B6/CBF1J mouse liver cDNA library (19) with two exceptions. We determined amino acid 96 to be methionine (encoded by ATG) rather than isoleucine and amino acid 333 to be tyrosine (encoded by TAC) rather than serine. The sequence we determined was consistent with the deduced amino acid sequences of human, rat, and yeast URO-D (22, 32, 33). Intron lengths were similar to the human gene with the exceptions of intron 6 (799 nt in mouse vs. 362 nt in human) and intron 9 (777 nt in mouse vs. 321 nt in human). We sequenced 600 bp of the 5′ untranslated region and found the basic transcription elements of a housekeeping gene.
Disruption of the URO-D Gene in Mice.
We used established gene-targeting techniques to delete exon 6 of the murine URO-D gene in ES cells (Fig. 2 A and B; ref. 26). This deletion results in a null allele (Fig. 2A; ref. 16). The deletion was confirmed by Southern blotting (Fig. 2C). An ES cell line (1C3) containing the null allele was injected into blastocysts derived from C57BL/6J mice, and recombinant animals were generated and genotyped (Fig. 2D).
When URO-D+/− animals were intercrossed, the ratio of the URO-D+/+:URO-D+/−:URO-D−/− genotypes in offspring was 140:253:0 with approximately equal numbers of males and females of each genotype. To determine the gestational time of lethality for URO-D−/− embryos, we examined 66 embryos between day 7.5 and 9.5 after fertilization; 20 were URO-D+/+, 46 were URO-D+/−, and none were URO-D−/−, indicating that the URO-D−/− genotype is an early embryonic lethal.
Exogenous Factors Are Required to Produce a Porphyric Phenotype in URO-D+/− Mice.
The hepatic porphyrin content in URO-D+/− mice did not differ from that in URO-D+/+ littermates, but URO-D protein quantified by Western immunoblotting and URO-D enzymatic activity was half-wt in liver, lung, kidney, and muscle (data not shown). Hepatic iron concentrations in URO-D+/− mice did not differ from URO-D+/+ animals followed up to 1 year of age, indicating that URO-D deficiency does not lead to hepatic iron accumulation.
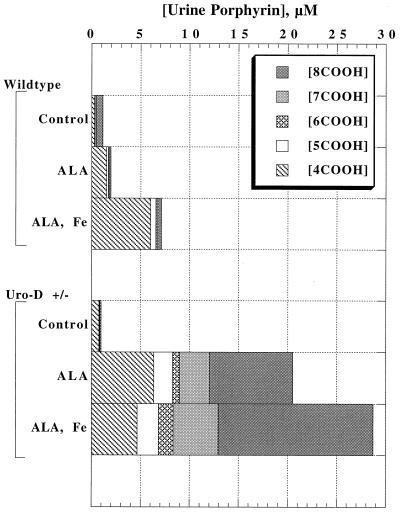
Both URO-D+/+ and URO-D+/− adult animals (age range 3 mo to 1 yr) were iron loaded by i.p. injection with iron-dextran (10 mg of iron) and were given drinking water supplemented with ALA (2 mg/ml) to bypass the rate-limiting step in hepatic porphyrin synthesis [ALA-synthase 1 (2, 3)]. Urinary porphyrin excretion before treatment was similar in animals of either genotype (Fig. 3). Animals were killed after 3 weeks of treatment when an increase in urinary porphyrin excretion was noted. In URO-D+/+ mice given iron and ALA, urinary porphyrin excretion rose 6-fold, and the predominant urinary porphyrin was coproporphyrin (Fig. 3). In contrast, urinary porphyrin excretion rose 29-fold in URO-D+/− animals. Uroporphyrin and heptacarboxyl porphyrin predominated, but coproporphyrin excretion paralleled the increase seen in URO-D+/+ animals (Fig. 3).
Figure 3.
Quantification of urinary porphyrins in wt and URO-D+/− mice 3 weeks after treatment with ALA or ALA plus iron. Animals were given drinking water containing ALA (2 mg/ml), and some received an i.p. injection of iron-dextran (Fe) containing 10 mg of iron. Urinary porphyrins were separated and quantified by HPLC. Each group represents the mean value from three animals. Within a group, neither total urinary porphyrin excretion nor the proportion of uroporphyrin or partially decarboxylated porphyrins varied by more than 10%. 8COOH, uroporphyrin; 7COOH, heptacarboxyl porphyrin; 6COOH, hexacarboxyl porphyrin; 5COOH, pentacarboxyl porphyrin; 4COOH, coproporphyrin.
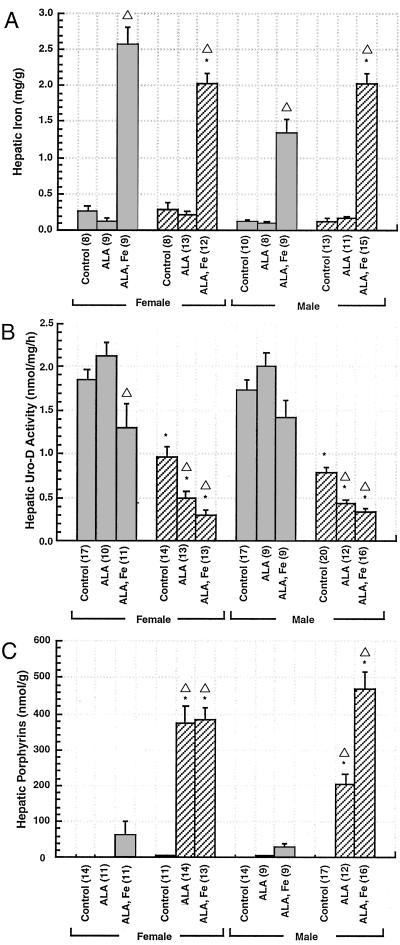
Hepatic iron concentrations in animals injected with iron-dextran increased approximately 10-fold (Fig. 4A). There was no significant effect of ALA alone on porphyrin accumulation or URO-D activity in URO-D+/+ animals of either sex (Fig. 4 B and C), but hepatic porphyrin accumulation (mainly coproporphyrin) increased slightly, and URO-D activity decreased slightly (Fig. 4 B and C). This effect was more marked in females. In contrast, ALA or ALA plus iron caused a further reduction in URO-D activity in URO-D+/− mice (Fig. 4B) without altering the levels of URO-D protein as determined by Western blotting (data not shown). Reduction of URO-D activity was accentuated by iron (Fig. 4B). Hepatic porphyrins accumulated in URO-D+/− animals given ALA or ALA plus iron (Fig. 4C) and were predominantly uroporphyrin and heptacarboxylic porphyrin. Hepatic porphyrin accumulation was accentuated by iron in URO-D+/− males but did not alter the high accumulation seen with ALA alone in females (Fig. 4C).
Figure 4.
Hepatic iron, URO-D activity, and porphyrin content in wt and URO-D+/− mice 3 weeks after treatment with ALA or ALA plus iron (A) Hepatic iron concentration, determined by atomic absorption spectroscopy. (B) Hepatic URO-D activity determined on cytosolic extracts. (C) Total hepatic porphyrin concentration. The numbers in parentheses in A–C represent the number of animals in each group. ■, wt; ▨, URO-D+/−. * indicates a value significantly different from wt. ▵ indicates a value significantly different from control animals in the same group. Error bars represent the standard error of the mean.
Genetic Hemochromatosis Produces a Porphyric Phenotype in URO-D+/− Mice.
Mice homozygous for a deletion of the HFE gene (HFE−/−) develop hepatic iron overload by 10 weeks of age with liver iron concentrations approximately 8-fold higher than wt littermates (31, 34). Animals with the HFE+/− genotype accumulate much less iron. We generated mice with the URO-D+/−/HFE−/− genotype on a C57BL/6J background through crossbreeding of the two mutant lines. Other genotypes resulting from these matings were URO-D+/−/HFE+/− and URO-D+/+/HFE+/−. Urine porphyrin excretion was monitored weekly beginning at 8 weeks of age. By 17 weeks of age, total urinary porphyrin excretion had increased 40-fold in URO-D+/−/HFE−/− mice with uroporphyrin and heptacarboxyl porphyrin predominating (data not shown). No change in urinary porphyrin excretion occurred in animals with other genotypes. Animals were killed between 14 and 17 weeks of age, and liver iron content, liver porphyrin content, and hepatic URO-D activities were measured. Hepatic porphyrins accumulated only in animals with the URO-D+/−/HFE−/− genotype, and in these animals, URO-D activity was reduced to approximately 16% of control (URO-D+/+/HFE−/−) values (Table 1). The reduction in URO-D activity was more profound than observed in URO-D+/− animals injected with iron-dextran (data not shown) even though hepatic iron levels were higher than in URO-D+/−/HFE−/− animals. Injected animals were killed 3 weeks after iron loading, whereas hepatic iron content in URO-D+/−/HFE−/− animals increased steadily from birth to the time of death, suggesting that the length of exposure to iron is important in generating the porphyric phenotype.
Table 1.
Effects of URO-D and HFE genotypes on hepatic iron and porphyrin concentration and URO-D
| Genotype
|
N | Iron, mg/g | Porphyrins, nmol/g | URO-D, nmol/h/mg | |
|---|---|---|---|---|---|
| URO-D | HFE | ||||
| +/+ | +/+ | 4 | 0.13 ± 0.04 | 0.75 ± 0.41 | 1.68 ± 0.09 |
| +/+ | +/− | 5 | 0.15 ± 0.04 | 1.26 ± 0.56 | 1.53 ± 0.14 |
| +/+ | −/− | 3 | 0.73 ± 0.14* | 1.77 ± 1.42 | 1.36 ± 0.06 |
| +/− | +/+ | 8 | 0.11 ± 0.02 | 8.46 ± 4.81 | 0.79 ± 0.07† |
| +/− | +/− | 4 | 0.22 ± 0.05 | 2.13 ± 0.58 | 0.64 ± 0.07† |
| +/− | −/− | 5 | 0.89 ± 0.22* | 259.32 ± 43.15*† | 0.22 ± 0.07*† |
Significantly different (P < 0.05) from HFE+/+ and identical URO-D genotype.
Significantly different (P < 0.05) from URO-D+/+ and identical HFE genotype.
Discussion
The URO-D+/− mouse, like most humans heterozygous for URO-D mutations, does not develop a porphyric phenotype unless additional factors are present, indicating that half-wt activity of URO-D does not create a rate-limiting step in the heme biosynthetic pathway. Supplementation of the drinking water with ALA to bypass the rate-limiting step caused an increase in urinary porphyrin excretion and hepatic accumulation in URO-D+/− mice over a 3-week period. This effect was markedly accentuated by the injection of iron-dextran. URO-D activity fell well below the half-wt value, even though URO-D protein levels remained constant, suggesting that an inhibitor of URO-D is generated. Smith and Francis (35) reported that iron overload, produced by injection of iron-dextran, can cause hepatic porphyrin accumulation and diminished URO-D activity in mice with wt URO-D alleles, but these effects occurred 25 weeks after iron injection and varied widely between strains. Elder et al. (36) reported diminished URO-D activity, but no reduction in URO-D protein, in liver biopsy specimens from humans with sporadic-PCT, and iron depletion led to normalization of URO-D activity. Collectively, these data suggest that a common mechanism is responsible for expression of both F-PCT and sporadic-PCT, namely generation of an inhibitor of URO-D by an iron-dependent mechanism.
When URO-D+/− animals were crossbred to HFE−/− animals a porphyric phenotype appeared after 14 weeks without ALA supplementation. Thus, we have constructed a murine model of PCT in which no exogenous factors are required. We and others have reported that HFE mutations, hepatitis C virus infection, alcohol abuse, and estrogen use are factors contributing to the pathogenesis of PCT (1, 4, 5). In our study of 108 patients with PCT, multiple risk factors were identified in most cases (1). In three patients with F-PCT, however, homozygosity for the C282Y HFE mutation was the only risk factor identified. All three of these patients were children. Our murine model of F-PCT appears to reflect the clinical findings in humans. Unperturbed URO-D+/− mice and URO-D+/− humans appear normal, but the URO-D+/−/HFE−/− genotype is sufficient to generate the porphyric phenotype.
The importance of the genetic background on which iron overload occurs has been established in mice and is emphasized by the low incidence of PCT in humans with hemochromatosis. In a recent study of 184 hemochromatosis pedigrees, we detected PCT in 19 probands (10.3%; ref. 39). This value likely is higher than the true incidence of PCT in individuals with hemochromatosis, as some patients were ascertained for study because of signs and symptoms of PCT.
The URO-D+/− genotype appears to both accelerate and accentuate the porphyric phenotype in the presence of iron overload. Our mouse model of F-PCT affords the opportunity to define the iron-dependent mechanism leading to the generation of an inhibitor of URO-D activity without the confounding effects of exogenous perturbing factors.
Acknowledgments
We acknowledge Hector A. Bergonia for HPLC analysis of porphyrins and enzyme assays and the contributions of the Transgenic and Recombinant Mouse Core of the Center of Excellence in Molecular Hematology. This work was supported in part by National Institutes of Health Grants DK20503, DK49219, RR00064, CA42014, and KO8-HL03503 (to J.E.L.). N.C.A. is an Associate Investigator of the Howard Hughes Medical Institute.
Abbreviations
- URO-D
uroporphyrinogen decarboxylase
- HFE
hemochromatosis gene
- PCT
porphyria cutanea tarda
- F-PCT
familial PCT
- ALA
δ-aminolevulinic acid
- wt
wild-type
- ES
embryonic stem
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011481398.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011481398
References
- 1.Bulaj Z J, Phillips J D, Ajioka R S, Franklin M R, Griffen L M, Guinee D J, Edwards C Q, Kushner J P. Blood. 2000;95:1565–1571. [PubMed] [Google Scholar]
- 2.Wyckoff E E, Kushner J P. In: The Liver: Biology and Pathobiology. Arias I M, Boyer J L, Fausto N, Jakoby W B, Schachter D A, Shafritz D A, editors. New York: Raven; 1994. pp. 505–527. [Google Scholar]
- 3.Kappas A, Sassa S, Galbraith R A, Nordmann Y. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. Vol. 2. New York: McGraw–Hill; 1995. pp. 2103–2160. [Google Scholar]
- 4.Elder G H. Semin Liver Dis. 1998;18:67–75. doi: 10.1055/s-2007-1007142. [DOI] [PubMed] [Google Scholar]
- 5.Bonkovsky H L, Poh-Fitzpatrick M, Pimstone N, Obando J, Di Bisceglie A, Tattrie C, Tortorelli K, LeClair P, Mercurio M G, Lambrecht R W. Hepatology. 1998;27:1661–1669. doi: 10.1002/hep.510270627. [DOI] [PubMed] [Google Scholar]
- 6.Garey J R, Hansen J L, Harrison L M, Kennedy J B, Kushner J P. Blood. 1989;73:892–895. [PubMed] [Google Scholar]
- 7.de Verneuil H, Sassa S, Kappas A. J Biol Chem. 1983;258:2454–2560. [PubMed] [Google Scholar]
- 8.de Verneuil H, Bourgeois F, de Rooij F, Siersema P D, Wilson J H, Grandchamp B, Nordmann Y. Hum Genet. 1992;89:548–552. doi: 10.1007/BF00219182. [DOI] [PubMed] [Google Scholar]
- 9.Romana M, Grandchamp B, Dubart A, Amselem S, Chabret C, Nordmann Y, Goossens M, Romeo P H. Eur J Clin Invest. 1991;21:225–229. doi: 10.1111/j.1365-2362.1991.tb01814.x. [DOI] [PubMed] [Google Scholar]
- 10.de Verneuil H, Grandchamp B, Beaumont C, Picat C, Nordmann Y. Science. 1986;234:732–734. doi: 10.1126/science.3775362. [DOI] [PubMed] [Google Scholar]
- 11.Meguro K, Fujita H, Ishida N, Akagi R, Kurihara T, Galbraith R A, Kappas A, Zabriskie J B, Toback A C, Harber L C, Sassa S. J Invest Dermatol. 1994;102:681–685. doi: 10.1111/1523-1747.ep12374134. [DOI] [PubMed] [Google Scholar]
- 12.Moran-Jimenez M J, Ged C, Romana M, Enriquez-De-Salamanca R, Taieb A, Topi G, D'Alessandro L, de-Verneuil H. Am J Hum Genet. 1996;58:712–721. [PMC free article] [PubMed] [Google Scholar]
- 13.Mendez M, Sorkin L, Rossetti M V, Astrin K H, del C. Batlle A M, Parera V E, Aizencang G, Desnick R J. Am J Hum Genet. 1998;63:1363–1375. doi: 10.1086/302119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McManus J F, Begley C G, Sassa S, Ratnaike S. Hum Mutat. 1999;13:412. doi: 10.1002/(SICI)1098-1004(1999)13:5<412::AID-HUMU13>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 15.Christiansen L, Ged C, Hombrados I, Brons-Poulsen J, Fontanellas A, de Verneuil H, Horder M, Petersen N E. Hum Mutat. 1999;14:222–232. doi: 10.1002/(SICI)1098-1004(1999)14:3<222::AID-HUMU5>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 16.Garey J R, Harrison L M, Franklin K F, Metcalf K M, Radisky E S, Kushner J P. J Clin Invest. 1990;86:1416–1422. doi: 10.1172/JCI114856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips J D, Whitby F G, Kushner J P, Hill C P. Protein Sci. 1997;6:1343–1346. doi: 10.1002/pro.5560060624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitby F G, Phillips J D, Kushner J P, Hill C P. EMBO J. 1998;17:2463–2471. doi: 10.1093/emboj/17.9.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu C, Xu W, Kozak C A, Desnick R J. Mamm Genome. 1996;7:349–352. doi: 10.1007/s003359900101. [DOI] [PubMed] [Google Scholar]
- 20.Schmid R. N Engl J Med. 1960;263:397–398. doi: 10.1056/NEJM196008252630807. [DOI] [PubMed] [Google Scholar]
- 21.Strauss W M. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Vol. 1. New York: Wiley; 1995. pp. 6.3.1–6.3.4. [Google Scholar]
- 22.Romeo P-H, Raich N, Dubart A, Beaupain D, Pryor M, Kushner J, Cohen-Solal M, Goossens M. J Biol Chem. 1986;261:9825–9831. [PubMed] [Google Scholar]
- 23.Garey J R, Franklin K F, Brown D A, Harrison L M, Metcalf K M, Kushner J P. Gastroenterology. 1993;105:165–169. doi: 10.1016/0016-5085(93)90022-5. [DOI] [PubMed] [Google Scholar]
- 24.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 25.Thomas K R, Capecchi M R. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 26.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hogan B, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 28.Mansour S L, Thomas K R, Capecchi M R. Nature (London) 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 29.Franklin M R, Phillips J D, Kushner J P. Toxicol Appl Pharmacol. 1997;147:289–299. doi: 10.1006/taap.1997.8282. [DOI] [PubMed] [Google Scholar]
- 30.Thomas K R, Deng C, Capecchi M R. Mol Cell Biol. 1992;12:2919–2923. doi: 10.1128/mcb.12.7.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy J E, Montross L K, Cohen D E, Fleming M D, Andrews N C. Blood. 1999;94:9–11. [PubMed] [Google Scholar]
- 32.Romeo P H, Dubart A, Grandchamp B, de Verneuil H, Rosa J, Nordmann Y, Goossens M. Proc Natl Acad Sci USA. 1984;81:3346–3350. doi: 10.1073/pnas.81.11.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garey J R, Labbe-Bois R, Chelstowska A, Rytka J, Harrison L, Kushner J, Labbe P. Eur J Biochem. 1992;205:1011–1016. doi: 10.1111/j.1432-1033.1992.tb16868.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhou X Y, Tomatsu S, Fleming R E, Parkkila S, Waheed A, Jiang J, Fei Y, Brunt E M, Ruddy D A, Prass C E, et al. Proc Natl Acad Sci USA. 1998;95:2492–2497. doi: 10.1073/pnas.95.5.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith A G, Francis J E. Biochem J. 1993;291:29–35. doi: 10.1042/bj2910029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elder G H, Urquhart A J, De Salamanca R E, Munzo J J, Bonkovsky H L. Lancet. 1985;II:229–232. doi: 10.1016/s0140-6736(85)90287-9. [DOI] [PubMed] [Google Scholar]
- 37.Jackson A H, Sancovich H A, Ferrmola A M, Evans N, Games D E, Matlin S A, Elder G H, Smith S G. Philos Trans R Soc London. 1976;273:191–206. doi: 10.1098/rstb.1976.0009. [DOI] [PubMed] [Google Scholar]
- 38.Luo J, Lim C K. Biochem J. 1993;289:529–532. doi: 10.1042/bj2890529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bulaj Z J, Ajioka R S, Phillips J D, LaSalle B S, Jorde L B, Griffin L M, Edwards C Q, Kushner J P. N Engl J Med. 2000;343:1529–1535. doi: 10.1056/NEJM200011233432104. [DOI] [PubMed] [Google Scholar]