Abstract
Protein kinase D (PKD) regulates the fission of vesicles from the trans-Golgi-network1,2. We show that phosphatidylinositol-4-kinase III beta (PI4KIIIβ), a key player in Golgi complex structure and function3, is a physiological substrate of PKD. Of the three PKD isoforms, only PKD1 and PKD2 phosphorylated PI4KIIIβ at a motif highly conserved from yeast to man. PKD mediated phosphorylation stimulated lipid kinase activity of PI4KIIIβ and enhanced VSV-G transport to plasma membrane. The identification of PI4KIIIβ as one of the PKD substrates should help to reveal the molecular events leading to transport carrier formation.
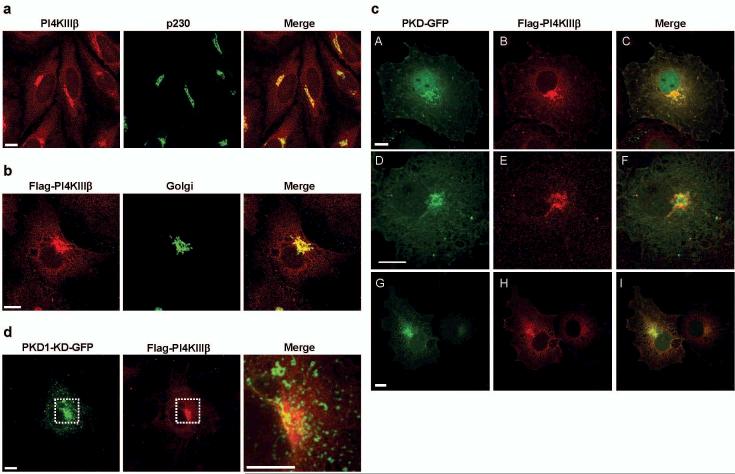
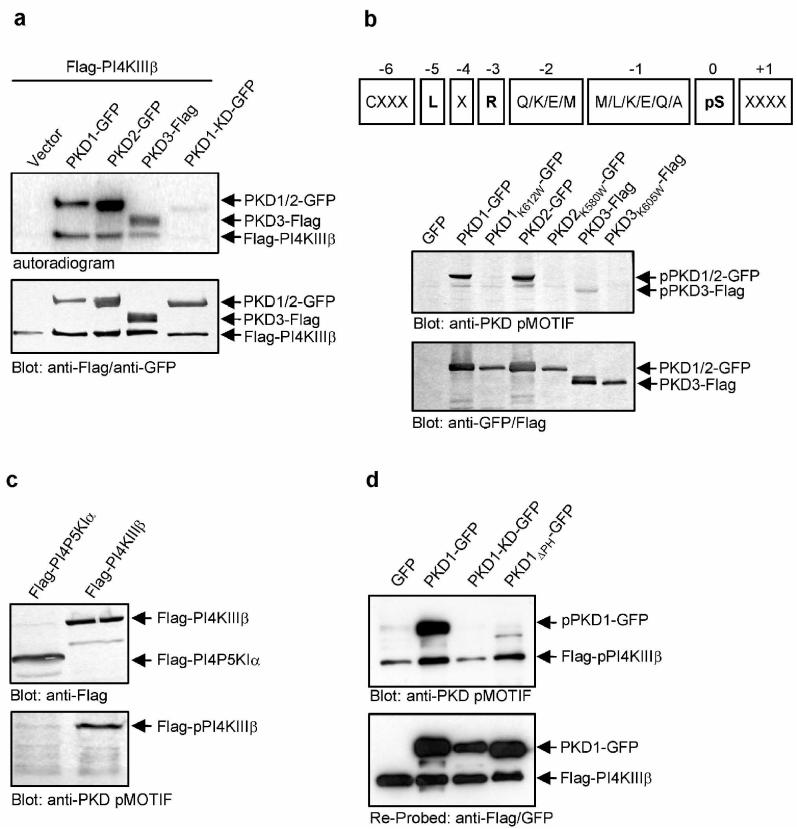
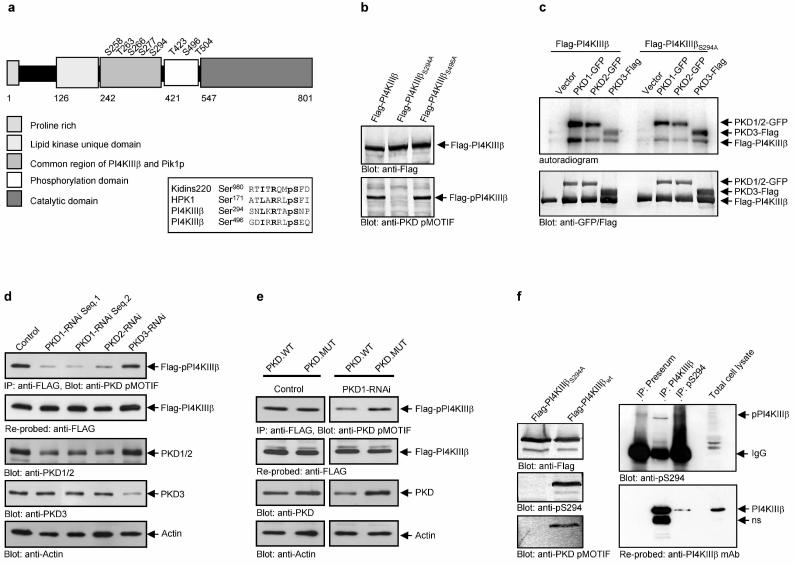
Protein kinase D (PKD) is a family of serine/threonine specific protein kinases composed thus far of 3 members, PKD1/PKCμ, PKD2 and PKD3/PKCν. Depending on the cell type and external stimulus, PKDs are reported to localize to the cytosol, nucleus, Golgi complex and plasma membrane4. In cells of epithelial origin, all three PKD isoforms localize predominantly at the trans-Golgi network (TGN)1,2,5 (Fig. 1). PKD1 has been shown to bind to Golgi membranes through its aminoterminal domain in a diacyl glycerol (DAG) dependent manner6,7. Activation of PKD1 at the Golgi complex involves heterotrimeric G protein subunits ß1γ28,9 and phosphorylation of the activation loop by nPKCη9. Typically, for established cell lines under tissue culture growth conditions a significant level of basal PKD kinase activity is observed, which is upregulated by extrinsic signals through nPKCs9,10,11. PKD is involved in fission of transport carriers from the TGN to the cell surface1,2. The mechanism, by which PKD regulates vesicle fission, however remains unknown. PI(4)P is regarded to be an important lipid mediator in vesicular trafficking. PI(4)P is generated from PI by PI4-Kinases. In yeast, the PI4-Kinase Pik1p is required for Golgi to cell surface transport12,13. Mammalian cells express at least three Golgi membrane-localized PtIns-4-kinases, PI4KIIα14, PI4KIIIα15 and PI4KIIIβ3 of which PI4KIIα14 and PI4KIIIβ16 have been implicated in Golgi complex to cell surface transport based on knock-down by siRNA and overexpression of a kinase-dead mutant, respectively. We tested for a potential connection between PKD and PI4-Kinases in Golgi complex to cell surface transport. Anti-PI4KIIIβ antibodies stain perinuclear reticular structures that are also stained by antibodies directed against trans-Golgi membrane associated proteins (Fig. 1a). When expressed as a Flag-tagged protein, PI4KIIIβ also localized to the Golgi complex (Fig. 1b). We visualized the intracellular distribution of Flag-PI4KIIIβ and PKD with GFP tagged wild-type PKD1, PKD2 or PKD3 proteins (Fig. 1c). All three PKD-GFP wild-type isoforms colocalized with Flag-tagged PI4KIIIβ at the Golgi complex. Overexpression of the kinase-dead (KD) mutant of PKD1-GFP revealed partial segregation from Flag-PI4KIIIβ, with PKD1-KD-GFP detectable in extended post TGN tubular structures and vesicles, a morphological phenotype of PKD dysfunction at the TGN1,10. In contrast, Flag-PI4KIIIβ remained exclusively localized at the Golgi complex, costaining with PKD1-KD-GFP at vesicles and tubules was not detectable in these cells (Fig. 1d). Using in vitro kinase assays, we examined whether Flag-PI4KIIIβ is a substrate for any of the three PKD isoforms. HEK293 cells were cotransfected with cDNAs encoding Flag-PI4KIIIβ and PKD1-GFP, PKD2-GFP and Flag-tagged PKD3, respectively. Flag-PI4KIIIβ and PKD1-GFP, PKD2-GFP or PKD3-Flag were immunoprecipitated using Flag and GFP specific antibodies, respectively, and were subjected to in vitro kinase assays using [γ-32P]-ATP (Fig. 2a). All three PKD isoforms phosphorylated Flag-PI4KIIIβ, with PKD1 being the strongest and PKD3 the weakest compared with the level of PKD autophosphorylation. Lack of Flag-PI4KIIIβ phosphorylation in the presence of a kinase-dead mutant, PKD1-KD-GFP, indicates that phosphorylation is directly mediated by PKD (Fig. 2a). We could not detect a direct interaction between PKD and Flag-PI4KIIIβ by co-immunoprecipitation in cells co-expressing the two proteins. However, this interaction may be transient and stimulated by signals that promote trafficking, which were not reconstituted under our experimental conditions. Is Flag-PI4KIIIβ also phosphorylated by PKD in intact cells? To address this question we used an antibody that specifically recognizes a phosphorylated PKD consensus motif (anti-PKD pMOTIF17). PKD preferentially phosphorylates serines or threonines with a leucine or isoleucine in the −5 position and an arginine in the −3 position18,19. PKD contains this motif and we used PKD autophosphorylation to control the specificity and functionality of this antibody. In HEK293 cells PKD-GFP fusion protein migrates in a SDS-PAGE as a double band, representing differentially phosphorylated PKD species, whereas the kinase-dead PKD isoforms, due to lack of autophosphorylation, appear as a single protein band10. The PKD pMOTIF antibody recognizes all three wild-type PKDs, but not their respective kinase-dead mutants, demonstrating the antibody specificity for PKD phosphorylation motifs (Fig. 2b). The antibody also recognizes this PKD pMOTIF in PI4KIIIβ , ectopically expressed in HEK293 cells (Fig. 2c). PKD1-KD is dominant-negative for the endogenous, wild-type PKD function2,20. In vivo phosphorylation of PI4KIIIβ by PKD should therefore be inhibited by overexpression of the dominant negative PKD variant. We transiently expressed wild-type PKD1, PKD1ΔPH, a variant lacking the negative regulatory PH-domain and displaying enhanced enzymatic activity10, or PKD1-KD, cultured the cells for 24 hours and subsequently transfected the cells with Flag-PI4KIIIβ followed by additional 24 hour incubation. PKD-GFP and Flag-PI4KIIIβ expression and phosphorylation levels were analyzed by western blotting the total cell lysates. Overexpression of PKD1-KD-GFP fusion protein reduced the recognition of Flag-PI4KIIIβ with the PKD pMOTIF specific antibody, whereas expression of wild-type PKD1-GFP or PKD1ΔPH-GFP markedly increased its detection compared with the vector control (Fig. 2d). This finding provides strong evidence that PI4KIIIβ is a physiological substrate of PKD. Of note, the pMOTIF antibody does not detect PKD1ΔPH which is in accordance with the localization of the recognized phosphorylation site within the PH-domain (P.Storz, unpublished data). Fig. 3a is a schematic depiction of the eight known phosphorylation sites in PI4KIIIβ21. Alignment with the PKD consensus motif in Kidins220 or HPK1, known PKD substrates22,23, revealed that two of these phosphorylation residues, Ser294 and Ser496, match the potential PKD consensus sequence. Ser294 is present in the common region between PI4KIIIβ and its yeast homolog Pik1p and is highly conserved from yeast to man. Both serines were replaced with alanine and expressed in HEK293 cells as Flag-PI4KIIIβS294A and Flag-PI4KIIIβS496A, respectively. Only the S294A mutation abolished the detection of Flag-PI4KIIIβ by the PKD pMOTIF antibody (Fig. 3b). Moreover, this mutation strongly reduced the ability of PKD1-GFP and PKD2-GFP fusion proteins to phosphorylate Flag-PI4KIIIβ in an in vitro kinase assay (Fig. 3c). Interestingly, in vitro phosphorylation of Flag-PI4KIIIβ by PKD3-Flag was not influenced by this mutation. This suggests that other phosphorylation sites in PI4KIIIβ are recognized by PKD3, which are not detected by the PKD pMOTIF specific antibody. In addition this implicates a major role for PKD1 and, to a lesser extent, PKD2, in phosphorylation of PI4KIIIβ at Ser294. We depleted PKD1, PKD2 and PKD3 expression by RNAi in order to assess their specific roles in the phosphorylation of PI4KIIIβ. HEK293 cells were transfected with pSuper-PKD RNAi vectors to knock-down expression of PKD1, PKD2 and PKD3, respectively. Because of the long in vivo half life of PKD (t1/2 = >25 hrs., data not shown), cells were cultured for 24h before transfection with the plasmid encoding Flag-PI4KIIIβ. PKD expression level and phosphorylation of immunoprecipitated Flag-PI4KIIIβ were evaluated 24h after Flag-PI4KIIIβ transfection. Loss of PKD1 and PKD2 expression was accompanied by a significant reduction in Flag-PI4KIIIβ phosphorylation, whereas loss of PKD3 did not effect phosphorylation of Flag-PI4KIIIβ at Ser294 (Fig. 3d). Cotransfection of a PKD mutant that is not targeted by the RNAi vectors rescues phosphorylation of Flag-PI4KIIIβ and confirms the specificity of the PKD mediated phosphorylation (Fig. 3e and supplementary figure S1). Interestingly, knock-down of one of the two PI4KIIIβ phosphorylating PKD isoforms was sufficient in inhibiting Ser294 phosphorylation. With the readily detectable levels of endogeneous PKD, this indicates an at least partially non-redundant function of PKD1 and PKD2 and suggests that the two isoforms may target distinct pools of PI4KIIIβ at the TGN. In support of this reasoning is the recent proposal that PI(4)P is located in functionally distinct TGN domains that can be discriminated by their specific lipid and protein content16. PKD1 and PKD2 could be recruited to these separate domains at the TGN, where different cargo is sorted into distinct carriers. The latter is in accordance with the proposal that PKD isoforms serve non-redundant functions in the transport of distinct cargo proteins2.
Figure 1.
PKD-GFP and Flag-PI4KIIIβ colocalize at the Golgi compartment. (a) PI4KIIIβ localizes to the Golgi complex. Hela cells were double-labelled with antibodies against PI4KIIIβ (red) and a trans-Golgi protein p230 (green). (b) Flag-PI4KIIIβ localizes to the Golgi complex. COS7 cells expressing Flag-PI4KIIIβ and a Golgi-specific YFP tagged protein (green) were stained with antibodies against PI4KIIIβ (red). (c) COS7 cells expressing wildtype PKD-GFP fusion proteins (green, A: PKD1-GFP; D: PKD2-GFP; G: PKD3-GFP) and Flag-PI4KIIIβ (B, E, H) were labelled with antibodies against PI4KIIIβ (red). (d) COS7 cells expressing a kinase-dead PKD1-GFP fusion protein (PKD1-KD-GFP, green) and Flag-PI4KIIIβ (red). Scale bar represents 10μm. Shown are parallel projections of stacks taken at 0.5 μm distance in z-direction.
Figure 2.
PKD phosphorylates Flag-PI4KIIIβ in vitro and in whole cells.(a) PKD-GFP phosphorylates Flag-PI4KIIIβ in vitro. HEK293 cells were transfected with plasmids encoding the indicated proteins, lysed and PKD-GFP and Flag-PI4KIIIβ were precipitated using anti-GFP and anti-Flag antibodies. Samples were further processed as described in Methods. (b) A PKD pMOTIF antibody is specific for PKD consensus phosphorylation sites. Upper panel: The sequence of the peptide used for immunization of rabbits, derived from the PKD consensus motif is shown. X represents any amino acid, pS represents phospho-serine. Lower panel: HEK293 transfected with the indicated plasmids were lysed and samples were subjected to SDS-PAGE. Proteins were detected using anti-Flag/anti-GFP or anti-PKD pMOTIF antibodies. (c) The PKD pMOTIF antibody specifically detects PI4KIIIβ. HEK293 transfected with the indicated plasmids were lysed and samples were subjected to SDS-PAGE. Proteins were detected using anti-Flag or anti-PKD pMOTIF antibodies. (d) Expression of PKD1-KD-GFP abrogates detection of Flag-PI4KIIIβ with the anti-PKD pMOTIF antibody. HEK293 cells were transfected with the indicated plasmids. Expression of PKD1-GFP and Flag-PI4KIIIβ proteins was controlled by Western Blot using anti-GFP, anti-Flag and anti-PKD pMOTIF antibodies. Results shown are representative of at least three independent experiments.
Figure 3.
PKD phosphorylates serine 294 in PI4KIIIβ. (a) Schematic view of the domain structure of PI4KIIIβ and the eight known phosphorylation sites and alignment of the putative PKD phosphorylation sites in PI4KIIIβ, serine 294 and 496 with the PKD substrate Kidins220 (bold: critical amino acids: pS: phospho-serine; R: arginine at −3; L/I: critical leucine or isoleucine at −5) (b) the anti-PKD pMOTIF antibody detects the phosphorylated serine 294. HEK293 cells expressing the indicated proteins were lysed and Flag-PI4KIIIβ was detected by Western Blot using anti-Flag and anti-PKD pMOTIF antibodies. (c) PKD phosphorylates serine 294 in Flag-PI4KIIIβ in vitro. HEK293 cells were transfected with plasmids encoding the indicated proteins, lysed and PKD-GFP and Flag-PI4KIIIβ were precipitated using anti-GFP and anti-Flag antibodies. Samples were further processed as described in Methods. (d) Knock-down of PKD1 or PKD2 isoenzymes inhibits Flag-PI4KIIIβ phosphorylation. HEK293E cells were transfected with pSUPER-PKD1-RNAi (2 different sequences; Seq.1 or Seq.2), pSUPER-PKD2-RNAi or pSUPER-PKD3-RNAi. Flag-PI4KIIIβ was precipitated with anti-Flag M2 antibodies and analyzed for phosphorylation at Ser294 (anti-PKD pMOTIF). The blot was re-probed against Flag-PI4KIIIβ. Silencing of endogenous PKD1/2 or PKD3 was monitored by immunoblot analysis with anti-PKD antibodies. Detection of Actin served as a loading control. (e) A PKD mutant with silent mutations in the RNAi-targeted sequence rescues PKD-mediated PI4KIIIβ phosphorylation. HEK293E cells were transfected with pSUPER (control) or pSUPER-PKD1-RNAi. After 24hr cells were transfected with Flag-PI4KIIIβ, combined with PKD1.WT (wild-type) or a PKD1 mutant with two silent mutations in the PKD1-RNAi-targeted sequence (PKD1.MUT). Flag-PI4KIIIβ was precipitated with anti-Flag M2 antibodies and analyzed for phosphorylation at Ser294 (anti-PKD pMOTIF). The blot was re-probed against Flag-PI4KIIIβ. Silencing of endogenous PKD1 was monitored by immunoblot analysis with anti-PKD antibodies. Detection of Actin served as a loading control. (f) A phospho-antibody specific for pS294 in PI4KIIIβ detects endogenous PI4KIIIβ. Endogenous PI4KIIIβ was precipitated from HEK293 cells either with the polyclonal PI4KIIIβ antibody or the polyclonal pS294 serum and detected with the polyclonal pS294 serum. The blot was reprobed with a monoclonal PI4KIIIβ antibody. The preserum served as a control.
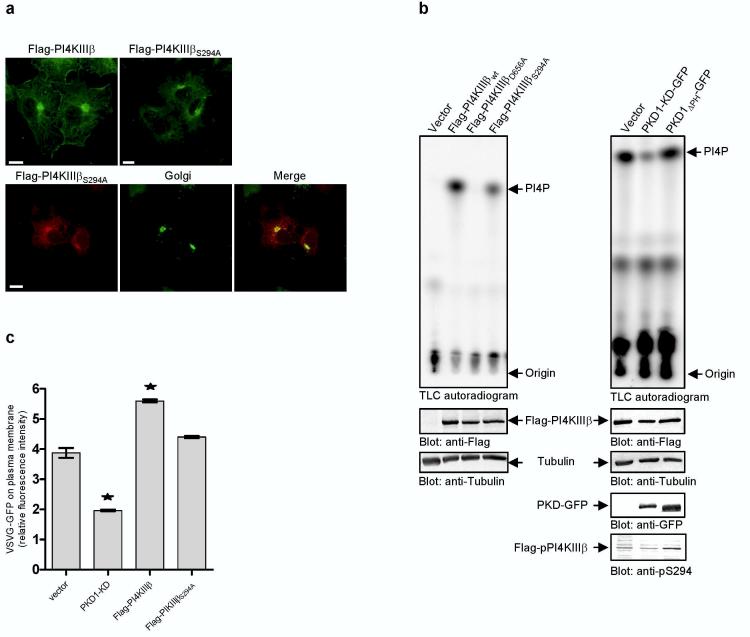
To further investigate phosphorylation of PI4KIIIβ at Ser294 in intact cells, we developed a phospho-specific antibody against the peptide epitope surrounding this residue in PI4KIIIß. Transfected Flag-PI4KIIIβwt was detected with the pS294 phospho-antibody, but not Flag-PI4KIIIβS294A (Fig. 3f). The phospho-antibody pS294 also detects immunprecipitated, endogenously phosphorylated PI4KIIIβ. Likewise endogenous p-PI4KIIIβ can be precipitated with this antibody, although less efficiently compared with a polyclonal immune serum developed against different epitopes (Figure 3f). What is the functional consequence of Ser294 phosphorylation of PI4KIIIβ by PKD1 and PKD2? To address this question we first investigated the localization of the mutated lipid kinase in COS7 cells. The S294A mutated Flag-PI4KIIIβ also localized to the Golgi compartment like its wild-type counterpart (Fig. 4a). Of note, neither overexpression of PI4KIIIβ nor PI4KIIIβS294A caused apparent changes in the distribution of Mannose-6-phosphate-receptor (data not shown). To investigate whether the phosphorylation of Ser294 in PI4KIIIβ is relevant for its activity, we performed lipid kinase assays. HEK293 cells transfected with Flag-tagged PI4KIIIβ constructs were used. Flag-PI4KIIIβ proteins were precipitated with anti-Flag M2 antibodies and analyzed for lipid kinase activity using [γ-32P]-ATP and phosphatidylinositol (PI) as a substrate. Wild-type Flag-PI4KIIIβ and a kinase-dead mutant, Flag-PI4KIIIβD656A3 served as positive and negative controls, respectively. The lipid kinase activity of PI4KIIIβS294A was 60% less than its wild-type counterpart (Fig 4b, left panel). To further analyze the functional consequences of PKD mediated phosphorylation on lipid kinase activity, we investigated the effect of overexpression of PKD1-GFP variants on Flag-PI4KIIIβ wild-type lipid kinase activity. As expected from basal kinase activity of endogeneously expressed PKD10, phosphorylated species of PI4KIIIβ and lipid kinase activity were readily deteced in the vector control (Fig.4b, right panel). Both, PI4KIIIβ phosphorylation and lipid kinase activity were further enhanced upon expression of PKD1ΔPH (Fig. 4b, right panel). In contrast, overexpression of PKD1-KD decreased PI4KIIIβ phosphorylation and lipid kinase activity below vector control (Fig. 4b, right panel), in accordance with the known dominant negative action of kinase inactive PKD mutants. These data, together with those shown in Fig. 2d, provide compelling evidence that PKD phosphorylation of PI4KIIIβ at Ser294regulates lipid kinase activity.
Figure 4.
Phosphorylation of PI4KIIIβ at serine 294 is essential for lipid kinase activity and important for transport of VSVG-GFP. (a) Mutation at serine 294 does not affect Golgi localisation. Upper panel: COS7 cells expressing either Flag-PI4KIIIβ or Flag-PI4KIIIβS294A were labelled with antibodies against PI4KIIIβ (green). Lower panel: COS7 cells expressing Flag-PI4KIIIβS294A variants and an YFP-Golgi marker (green) were labelled with antibodies against PI4KIIIβ (red) and analyzed by confocal microscopy. Shown are parallel projections of stacks taken with 0.5 μm width in z-direction. Scale bar represents 10 μm. (b) Phosphorylation at serine 294 affects lipid kinase activity. HEK293 cells were transfected with Flag-PI4KIIIβ variants (left panel) or PKD1-GFP variants and Flag-PI4KIIIβ wild-type (right panel). Flag-PI4KIIIβ variants were precipitated with Flag-antibodies and subjected to lipid kinase assay using phosphatidylinositol (PI) as a substrate. Shown is one representative experiment (n=4). To control the expression of the proteins Western Blots of total cell lysates were performed using anti-Flag, anti-GFP and anti-Tubulin antibodies. The amount of radioactive labelled PI(4)P was normalized to Flag-PI4KIIIβ protein levels. (c) Expression of Flag-PI4KIIIβS294A fails to enhance secretory transport of VSVGGFP protein. HEK293 cells were cotransfected with VSVG-GFP and the indicated plasmids. The amount of VSVG-GFP protein on the plasma membrane is shown as the ratio of red fluorescence (phycoerythrin, staining of plasma membrane localized VSVG protein by indirect immunofluorescence with the 8G5F11 antibody) to green fluorescence (GFP, total VSVG protein expressed). Results are representative (± SEM) of two independent experiments performed. Significance of changes were analysed by student's t-test and indicated by an asterisk (*p < 0.05).
It has been reported that kinase-dead PI4KIIIβD656A partially blocked transport of VSVG-GFP protein to the plasma membrane16. Likewise, through dominant negative action, PKD1-KD and PKD2-KD inhibit transport of basolateral cargo from the TGN to the plasma membrane2. Our findings reveal that PKD1 and PKD2 regulate lipid kinase activity of PI4KIIIβ by phosphorylation. But is there a functional link between phosphorylation of PI4KIIIβ by PKD and Golgi to cell surface transport? We compared the effect of overexpression of PI4KIIIβ and of PI4KIIIβS294A on transport of VSVG-GFP protein. HEK293 cells were transfected with plasmids encoding Flag-PI4KIIIβ or Flag-PI4KIIIβS294A. PKD1-KD was used as a positive control. After 24h incubation at 37°C cells were transfected with a plasmid for VSVG-GFP and incubated for 20h at 39.5°C. Cells were then shifted to 20°C for 2h in the presence of cycloheximide to accumulate VSVG-GFP in the TGN and subsequently shifted to 31°C to permit its transport from the TGN. The amount of VSVG-GFP protein on the cell surface was quantified by immunofluorescence flow cytometry and normalised to the amount of total cellular VSVG-GFP protein. As expected, the expression of PKD1-KD significantly reduced transport of VSVG-GFP protein to the plasma membrane (Fig. 4c, upper panel). Expression of wild-type Flag-PI4KIIIβ led to an increase in TGN-to-plasma membrane export of VSVG-GFP (145% of vector control), whereas the phosphorylation site deficient mutant, PI4KIIIβS294A, did not affect basal VSVG-GFP transport. This indicates that, unlike PKD1-KD, the PI4KIIIβS294A mutant does not act in a dominant-negative manner. In support of this, the expression of kinase-dead Flag-PI4KIIαK152A alone or together with the phosphorylation site deficient mutant, Flag-PI4KIIIβS294A, also did not inhibit secretory transport of VSVGGFP (Supplementary Figure S2). This is not surprising since mammalian cells contain at least three Golgi-complex localized PI4-Kinases and all three PI4-Kinases have to be depleted to significantly reduce PI(4)P levels at the Golgi24. The lack of dominant negative action of overexpressed lipid kinase mutants has already been reported. In yeast, Pik1p function is highly allele specific12. In mammalian cells, a catalytically inactive variant of PI4KIIα is not dominant negative14. Moreover, PKD1-KD is likely to have additional substrates involved in vesicle trafficking from the Golgi complex. Malhotra and coworkers have recently proposed a model where DAG kinase is suggested to be a PKD target9. In summary, PI4KIIIβ is involved in Golgi to cell surface transport. PI4KIIIβ is phosphorylated at Ser294 by PKD1 and PKD2, but not PKD3. This phosphorylation increases the lipid kinase activity of the PI4KIIIβ. We propose that PKD acts as a “bottleneck” at the TGN, regulating secretory transport processes by phosphorylating a set of proteins involved in the generation and/or fission of transport vesicles. PI4KIIIβ is one of the PKD substrates at the Golgi apparatus. Our findings reveal that the trafficking from the TGN requires the combined action of a protein and a lipid kinase. How these two kinases promote membrane fission during vesicle formation is the obvious next challenge.
METHODS
Antibodies and reagents
Anti-PI4KIIIβ antibodies were purchased from Becton Dickinson and Upstate Biotechnology. Alexa488 or Alexa546 labelled anti-mouse or anti-rabbit IgG for immunofluorescence microscopy were obtained from Molecular Probes. The anti-GFP antibody was from Roche, the anti-Flag M2 and the anti-Actin antibodies from Sigma-Aldrich, the anti-HA antibody from NEB, the monoclonal anti-Tubulin antibody was from Neomarkers, the anti-p230 monoclonal antibody was from Becton-Dickinson. The antibody anti-PKD C-20 recognizing the C-terminal domain in PKD1 and PKD2 was from Santa Cruz. The anti-PKD3 antibody was a gift from Sharon Matthews and Andrew Scharenberg (University of Washington, Seattle, USA). The monoclonal antibody I1 (8G5F11) recognizing the extracellular domain of VSVG was kindly provided by Doug Lyles (Wake Forest University School of Medicine, North Carolina, USA). The anti-PKD pMOTIF antibody was generated as described in detail in17. The polyclonal anti-pS294 antibody was generated by immunizing rabbits with the peptide NH2-CLKRTApSNPL-CONH2 (amino acids 289-297 in human PI4KIIIβ).
DNA constructs
Flag-tagged PI4KIIIβ was generated by amplification of full-length human PI4KIIIβ from HEK293 cDNA by polymerase chain reaction (PCR), using a 5′sense primer containing an EcoRV site and a 3′antisense primer containing an XhoI site. The fragment was cloned into EcoRV and XhoI digested pCR3V62-Met-Flag (kindly provided by J. Tschopp, University of Lausanne, Switzerland) to generate pCR3V62-Met-Flag-PI4KIIIβ. The point mutations in Flag-PI4KIIIβ were generated from this construct by site-directed mutagenesis (Stratagene) following the manufacturers instructions. The following oligonucleotides were used: Flag-PI4KIIIβD656A: 5′-ctgcaagcaaggccagacacaatggg-3′ and 5′-cccattgtgtctggccttgacttgcag-3′, Flag-PI4KIIIβS294A: 5′-aaacgaacagccgccaaccctaaagtg-3′ and 5′-cactttagggttggcggctgttcgttt-3′, Flag-PI4KIIIβS496A: 5′-cgccggcgccttgcggaacagctggct-3′ and 5′-agccagctgttccgcaaggcgccggcg-3′. The plasmids pEGFP-N1-PKD1, pEGFPN1-PKD1ΔPH and pEGFP-N1-PKD1K612W (PKD1-KD) were described previously10. The plasmids pEGFP-N1-PKD2, pEGFP-N1-PKD3 and pCR3.V62-PKD3-Flag were generated by amplification of full length human PKD2 and PKD3 from HEK293 cDNA by polymerase chain reaction, using a 5′sense primer containing an EcoRI site (PKD2) or a SacI site (PKD3) and a 3′antisense primer containing a BamHI site (PKD2) or a XmaI site (PKD3). Mutations at position lysine 580 (PKD2) and lysine K605 (PKD3) to generate kinase-dead versions of PKD2 and PKD3 were introduced as described for PKD1-KD. The integrity of all constructs was verified by DNA sequencing. To silence the expression of PKD1 and PKD2, pSuper-PKD1 RNAi vectors (Seq.1) and pSuper-PKD2 RNAi were used as detailed in25. PKD1-RNAi Seq.2: 5′-gatccccgttccctgaatgtggtttcttcaagagagaaaccacattcagggaactttttggaaa-3′ and 5′-agcttttccaaaaagttccctgaatgtggtttctctcttgaagaaaccacattcagggaacggg-3′. PKD3-RNAi: 5′-gatccccgtcctaagacgggactctcttcaagagagagagtcccgtcttaggactttttggaaa-3′ and 5′-agcttttccaaaaagtcctaagacgggactctctctcttgaagagagtcccgtcttaggacggg-3′. The RNAi Sequences for PKD1-RNAi (Seq.1) and PKD2-RNAi have been described before25. The PKD1 mutant with silent mutations in the RNAi-targeted sequence has also been described before26. Temperature-sensitive folding mutant of Vesicular Stomatitis Virus G protein VSVG-ts045-GFP in pEGFP-N1 vector (Clontech) was kindly provided by Kai Simons (MPI Dresden, Germany). The pEYFP-Golgi plasmid contains the sequence for the N-terminal 81 amino acids of the human beta 1, 4-galactosyltransferase fused to the N-terminus of EYFP (Clontech).
Cell culture and transfection
HEK293 and COS7 cells were grown in RPMI supplemented with 5% or 10% foetal calf serum (FCS), respectively, in a humidified atmosphere containing 5% CO2. For immunofluorescence, COS7 cells were grown on glass coverslips for 24h and transfected with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturers instructions. HEK293 cells grown on petri dishes were transfected using TransIT293 reagent (Mirus).
Immunofluorescence microscopy
Transfected COS7 cells were grown on coverslips, washed with PBS, fixed in 4% paraformaldehyde at room temperature for 20min, washed, blocked and permeabilized with blocking buffer (5% normal goat serum and 0.05% Tween 20 in PBS) for 30min. The coverslips were incubated with the primary antibodies diluted in blocking buffer (1:300) for 2h, washed, incubated with secondary antibodies diluted in blocking buffer for 1h, washed, mounted in Fluormount G (Southern Biotechnology) and analyzed on a confocal Laser scanning microscope (TCS SP2, Leica) using 488-and 568 nm excitation and a 100x HL Apo objective. Images were processed with Adobe Photoshop.
Vesicular Stomatitis Virus-G Protein-Green Fluorescent Protein (VSVG-GFP) transport assay
HEK293 cells grown on Petri dishes were transfected with vectors encoding Flag-tagged PI4KIIIβ constructs or PKD1-GFP vectors. After 24h cultivation cells were again transfected with the plasmid VSVG-ts045-GFP, cultured at 39.5°C in a humidified atmosphere for 20h and received 100μg/ml cycloheximide prior to incubation at 20°C for 2h. The cells were then transferred to 31°C for 1h. Labelling of surface VSVG-GFP was performed with the anti-VSVG mAb I1 (8G5F11), which is specific for the extracellular domain of VSVG-GFP: In brief, cells were harvested on ice and washed once with PBS containing 2% serum and 0,02% sodium acide (PSA) to avoid internalisation. Incubation with the primary antibody was performed in PSA for 1h on ice. The cells were then incubated with the secondary phycoerythrin (PE) labelled anti-mouse IgG antibody (Sigma-Aldrich) for 45 min on ice. After washing with PSA 5000 cells were analysed by flow cytometry (EPICS XL-MCL, Beckmann-Coulter). The relative amount of cell surface (PE, red fluorescence) and total (GFP, green fluorescence) VSVG-GFP protein of gated viable cells was recorded as mean fluorescence intensity. PE values were normalised with GFP values and the respective ratios are shown in the figures.
Lipid kinase assay
The activity of Flag-tagged PI4KIIIβ was measured as incorporation of radioactivity from [γ-32P]-ATP into organic solvent-extractable material. HEK293 cells grown on Petri dishes were transfected with vectors encoding Flag-tagged PI4KIIIβ constructs. The cells were cultured for 48h, harvested, lysed and Flag-tagged PI4KIIIβ was immunoprecipitated using anti-Flag M2 antibody. The standard reaction mixture for PI4KIIIβ (100μl volume) contained 100mM MgCl2, 10mM HEPES pH 7, 4μg Phosphatidylinositol (Biomol) and 10μCi [γ-32P]-ATP. Reactions were started by addition of [γ-32P]-ATP and terminated after 10min by the addition of 25μl 5M HCL. Lipids were extracted by adding 160 μl Chloroform/Methanol followed by vigorously mixing and high-speed centrifugation. The bottom phase containing the lipids was transferred to a new tube, 20μl at a time were spotted onto a potassium oxalate coated TLC plate, and lipids were separated using 0.7 M acetic acid in N-Propanol as solvent. TLC was exposed to phosphoimager (Molecular Dynamics) over night. Density of spots representing PI(4)P were quantified using ImageQuant software (Molecular Dynamics).
Immunoprecipitation and Western Blot
HEK293 cells were harvested, lysed in lysis buffer (20mM Tris pH 7,4; 150mM NaCl, 5mM MgCl2, 1% Triton X-100, plus protease and phosphatase inhibitors) and the proteins precipitated using anti-GFP antibodies, anti-Flag M2 antibodies or anti-PI4Kβ antibodies. Immunocomplexes were pelleted using Protein G-Sepharose. After washing samples were subjected to 12.5% SDS-PAGE, blotted onto Nitrocellulose membrane and blocked with 3% milk. Incubation with the primary antibodies was performed in PBS at 4°C over night. After washing with PBS incubation with secondary alkaline phosphatase (AP)-labelled or horseradish peroxidase (HRP)-labelled anti-mouse or anti-rabbit IgG antibodies was performed in PBS for 1h at RT. Detection was done either with NBT/BCIP as substrate or ECL.
PKD in vitro protein kinase assay
After immunoprecipitation (with anti-GFP or anti-Flag antibodies for transfected PKD), the kinase reaction was carried out for 15min at 37°C in 30μl kinase buffer (50mM Tris pH 7.4, 10mM MgCl2 and 2mM DTT). Reaction was started by addition of 10μl of a kinase buffer mixture containing 2μCi [γ-32P]-ATP. To terminate reaction, 10μl of 5x SDS-sample buffer was added and the samples were resolved by SDS-PAGE, blotted onto nitrocellulose and analyzed on a phosphoimager (Molecular Dynamics). Quantification was done with ImageQuant software (Molecular Dynamics).
RNAi
Hela cells and HEK293 cells were transfected with pSuper or pSuper-PKD-RNAi vectors with the Hela Monster or the TransIT293 transfection reagent (Mirus). In all experiments the cells were transfected at 30% confluency. Transfection efficiencies were ≥ 90%. After 24hr cells were transfected with Flag-PI4KIIIβ. Experiments were performed 48 h after initial transfection. Western Blot was used to evaluate the expression of target proteins.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Lucia Rameh for help with the lipid kinase assay and E. Behrle and O. Selchow for help with confocal microscopy; this work was supported by grants of the Deutsche Forschungsgemeinschaft (SFB 495 to K.P.), the Landesstiftung Baden-Württemberg (to A.H.) and by the National Institutes of Health (CA075134 to A. T.).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
References
- 1.Liljedahl M, et al. Protein kinase D regulates the fission of cell surface destined transport carriers from the trans-Golgi network. Cell. 2001;104:409–420. doi: 10.1016/s0092-8674(01)00228-8. [DOI] [PubMed] [Google Scholar]
- 2.Yeaman C, et al. Protein kinase D regulates basolateral membrane protein exit from trans-Golgi network. Nat. Cell Biol. 2004;6:106–112. doi: 10.1038/ncb1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Godi A, et al. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulatessynthesis of PtdIns(4,5)P2 on the Golgi complex. Nat. Cell Biol. 1999;1:280–287. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- 4.Rykx A, et al. Protein kinase D: a family affair. FEBS Lett. 2003;546:81–86. doi: 10.1016/s0014-5793(03)00487-3. [DOI] [PubMed] [Google Scholar]
- 5.Prestle J, Pfizenmaier K, Brenner J, Johannes FJ. Protein kinase C mu is located at the Golgi compartment. J. Cell Biol. 1996;134:1401–1410. doi: 10.1083/jcb.134.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maeda Y, Beznoussenko GV, Van Lint J, Mironov AA, Malhotra V. Recruitment of protein kinase D to the trans-Golgi network via the first cysteine-rich domain. EMBO J. 2001;20:5982–5990. doi: 10.1093/emboj/20.21.5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baron CL, Malhotra V. Role of diacylglycerol in PKD recruitment to the TGN and protein transport to the plasma membrane. Science. 2002;295:325–328. doi: 10.1126/science.1066759. [DOI] [PubMed] [Google Scholar]
- 8.Jamora C, et al. Gbetagamma-mediated regulation of Golgi organization is through the direct activation of protein kinase D. Cell. 1999;98:59–68. doi: 10.1016/S0092-8674(00)80606-6. [DOI] [PubMed] [Google Scholar]
- 9.Anel AM, Malhotra V. PKC{eta} is required for {beta}1{gamma}2/{beta}3{gamma}2-and PKD-mediated transport to the cell surface and the organization of the Golgi apparatus. J. Cell Biol. 2005;169:83–91. doi: 10.1083/jcb.200412089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hausser A, et al. Structural requirements for localization and activation of protein kinase C mu (PKC mu) at the Golgi compartment. J. Cell Biol. 2002;156:65–74. doi: 10.1083/jcb.200110047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waldron RT, et al. Activation loop Ser744 and Ser748 in protein kinase D are transphosphorylated in vivo. J. Biol. Chem. 2001;276:32606–32615. doi: 10.1074/jbc.M101648200. [DOI] [PubMed] [Google Scholar]
- 12.Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat. Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- 13.Sciorra VA, et al. Synthetic Genetic Array Analysis of the PtdIns 4-kinase Pik1p Identifies Components in a Golgi-specific Ypt31/rab-GTPase Signaling Pathway. Mol. Biol. Cell. 2004 doi: 10.1091/mbc.E04-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang YJ, et al. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 2003;114:299–310. doi: 10.1016/s0092-8674(03)00603-2. [DOI] [PubMed] [Google Scholar]
- 15.Nakagawa T, Goto K, Kondo H. Cloning, expression, and localization of 230-kDa phosphatidylinositol 4-kinase. J. Biol. Chem. 1996;271:12088–12094. doi: 10.1074/jbc.271.20.12088. [DOI] [PubMed] [Google Scholar]
- 16.Godi A, et al. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- 17.Doppler H, Storz P, Li J, Comb MJ, Toker A. A phosphorylation state-specific antibody recognizes HSP27, a novel substrate of protein kinase D. J. Biol. Chem. 2005 doi: 10.1074/jbc.C400575200. [DOI] [PubMed] [Google Scholar]
- 18.Nishikawa K, Toker A, Johannes FJ, Songyang Z, Cantley LC. Determination of the specific substrate sequence motifs of protein kinase C isozymes. J. Biol. Chem. 1997;272:952–960. doi: 10.1074/jbc.272.2.952. [DOI] [PubMed] [Google Scholar]
- 19.Hutti JE, et al. A rapid method for determining protein kinase phosphorylation specificity. Nat. Methods. 2004;1:27–29. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- 20.Prigozhina NL, Waterman-Storer CM. Protein kinase D-mediated anterograde membrane trafficking is required for fibroblast motility. Curr. Biol. 2004;14:88–98. doi: 10.1016/j.cub.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Suer S, Sickmann A, Meyer HE, Herberg FW, Heilmeyer LM., Jr. Human phosphatidylinositol 4-kinase isoform PI4K92. Expression of the recombinant enzyme and determination of multiple phosphorylation sites. Eur. J. Biochem. 2001;268:2099–2106. doi: 10.1046/j.1432-1327.2001.02089.x. [DOI] [PubMed] [Google Scholar]
- 22.Iglesias T, et al. Identification and cloning of Kidins220, a novel neuronal substrate of protein kinase D. J. Biol. Chem. 2000;275:40048–40056. doi: 10.1074/jbc.M005261200. [DOI] [PubMed] [Google Scholar]
- 23.Arnold R, et al. Activation of hematopoietic progenitor kinase 1 involves relocation, autophosphorylation, and transphosphorylation by protein kinase d1. Mol. Cell Biol. 2005;25:2364–2383. doi: 10.1128/MCB.25.6.2364-2383.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balla A, Tuymetova G, Tsiomenko A, Varnai P, Balla T. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol. Biol. Cell. 2005;16:1282–1295. doi: 10.1091/mbc.E04-07-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storz P, Doppler H, Toker A. Protein kinase Cdelta selectively regulates protein kinase D-dependent activation of NF-kappaB in oxidative stress signaling. Mol. Cell Biol. 2004;24:2614–2626. doi: 10.1128/MCB.24.7.2614-2626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Storz P, Toker A. Protein kinase D mediates a stress-induced NF-kappaB activation and survival pathway. EMBO J. 2003;22:109–120. doi: 10.1093/emboj/cdg009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.