Abstract
In Gram-negative bacteria, TEM-1 β-lactamase provides the major mechanism of plasmid-mediated β-lactam resistance. Natural variants of TEM-1 with increased antibiotic resistance have appeared in response to the use of extended-spectrum β-lactam antibiotics (e.g., ceftazidime) and β-lactamase inhibitors (e.g., clavulanic acid). Some of the variant enzymes are more efficient at catalyzing β-lactam hydrolysis, whereas others are more resistant to inhibitors. M182T is a substitution observed in both types of variant TEM-1 β-lactamases. This mutation is found only in combination with other amino acid substitutions, suggesting that it may correct defects introduced by other mutations that alter the specificity. An engineered core mutation, L76N, which diminishes the periplasmic β-lactamase activity by 100-fold, was used as a model to understand the mechanism of suppression of the M182T mutation. Biochemical studies of the L76N enzyme alone and in combination with the M182T mutation indicate that the M182T substitution acts at the level of folding but does not affect the thermodynamic stability of TEM-1 β-lactamase. Thus, the M182T substitution is an example of a naturally occurring mutation that has evolved to alter the folding pathway of a protein and confer a selective advantage during the evolution of drug resistance.
In bacteria and viruses where the spontaneous mutation rate is often high and the generation time is short, the rapid emergence of drug-resistant organisms has created a problem for the use and development of antibiotics. In response to the selective pressure of drug exposure, amino acid substitutions that alter the specificity of the drug target accumulate and confer a selective advantage when the antibiotic is encountered (1). A puzzling phenomenon has been the accumulation of additional mutations that do not appear to confer a selective advantage by altering substrate specificity (2, 3). A plausible hypothesis is that these “secondary” mutations compensate for defects in catalytic activity or enzymatic stability arising from specificity-altering primary mutations (4).
Bacterial resistance toward β-lactam antibiotics such as penicillins and cephalosporins is most often mediated by the bacterial synthesis of β-lactamases, which inactivate these compounds by catalyzing their hydrolysis. One of the most common plasmid-mediated β-lactamase enzymes is TEM-1. This enzyme utilizes a catalytic serine residue (S70) to cleave the lactam ring (5, 6). In response to the clinical use of extended-spectrum β-lactam antibiotics, naturally occurring variants of TEM-1 β-lactamase have been isolated that contain amino acid substitutions that alter the enzyme's substrate specificity (7). Some of the new variants exhibit increased activity (kcat/Km) against the extended-spectrum antibiotics without losing their activity against penicillins (7). Other mutations can render the enzyme resistant to β-lactamase inhibitors such as clavulanic acid (8).
Among the 90 TEM-1 natural isolates found to date, substitutions at residues that directly affect substrate binding and catalysis (R164, G238, and E104) or inhibitor binding (M69 and R244) are repeatedly found (9–15). The substitution of threonine for methionine at residue 182 of TEM-1 β-lactamase is also a frequent mutation in natural isolates (16–20). M182T does not have a direct role in catalysis or substrate/inhibitor binding (17) and is never found alone, but always in combination with other mutations (Table 1). Interestingly, M182T accompanies mutations that affect inhibitor binding (as in TEM-32) as well as mutations that extend the substrate specificity of the enzyme (TEM-20 and TEM-52). In the three-dimensional structure, Met-182 is far away from the sites of the coupled primary mutation so that any suppression mechanism must operate at a distance. The nature of the evolutionary advantage conferred by M182T to mutant β-lactamase strains has yet to be systematically explored although M182T increases the thermal stability of β-lactamase mutant M69I when measured in crude periplasmic extracts (21).
Table 1.
Naturally occurring extended-spectrum and inhibitor-resistant β-lactamases*
| β-Lactamase | Substitutions |
|---|---|
| TEM-1 | Wild type |
| TEM-20 | M182T, G238S |
| TEM-32 | M182T, M69I |
| TEM-43 | M182T, E104K, R164H |
| TEM-52 | M182T, E104K, G238S |
| TEM-63/TEM-64 | M182T, L21F, E104K, R164S |
| TEM-72 | M182T, G238S, E240K |
Taken from the table of TEM-1 variants at http://www.lahey.hitchcock.org/pages/lhc/studies/webt.htm.
In natural isolates, a very small selective advantage because of a mutation may lead to enhanced survival over many generations. Consequently, it is not surprising that relatively small defects may accompany the primary mutations that are associated with M182T in the wild (21, 22). For this reason, we used an engineered substitution at an invariant, buried residue of TEM-1 β-lactamase, Leu-76, as a probe to select for second-site mutations that suppressed its deleterious effects (21). Leu-76 was chosen as the site of a potentially suppressible “primary” mutation because it resides in the hydrophobic core of the enzyme within the important helix H2 in the α-helical domain of TEM-1 β-lactamase (23). Moreover, it is one of the few amino acids within the sequence that were intolerant of substitutions in a functional mutagenesis study (24). We therefore expected that a mutation at this site would have a pronounced effect on the structure or function of the enzyme. Indeed, the L76N substitution results in a dramatically reduced resistance toward ampicillin in Escherichia coli (21). More importantly, genetic reversion analysis identified M182T as the only secondary mutation that could restore the loss of β-lactamase function (21).
In the present work, we explore the mechanisms by which M182T corrects deleterious effects stemming from the L76N mutation. The results show that M182T restores the periplasmic β-lactamase activity that is lost because of the L76N mutation by correcting a folding defect introduced by L76N. By itself, the M182T mutation has only a small effect on the enzyme activity and stability, but its presence suppresses pathways that lead the L76N mutant protein toward misfolding, aggregation, and proteolysis.
Materials and Methods
Strains, Plasmids, Construction of Mutants, and Protein Purification.
Mutants were constructed by oligonucleotide-directed mutagenesis as described previously (21). E. coli strains XL1-Blue and SB646 (21) harboring the pJ1522 plasmid carrying the blaTEM-1 gene under control of a constitutive promoter were used for the analysis of in vivo expression of β-lactamase. TEM-1 β-lactamase and its mutants were purified to >90% homogeneity from E. coli XL1-Blue cells containing the pBG66 plasmid under the control of the natural β-lactamase promoter. These strains were grown in culture for 9 h to late log phase. Periplasmic proteins were isolated by osmotic shock (25). The solution was adjusted to a final concentration of 100 mM NaAc (pH 7.5) and 800 mM NaCl (Buffer A) and concentrated with an Amicon Centriprep-10 concentrator. The protein solution was then applied to a 1-ml HiTrap zinc chelating column (Pharmacia) equilibrated in Buffer A. After washing with Buffer A, β-lactamase was eluted by using a linear gradient of Buffer B (100 mM NaAc (pH 4.0)/800 mM NaCl). Fractions containing β-lactamase activity were identified by nitrocefin hydrolysis and SDS/PAGE and were pooled and concentrated with a Centriprep-10 concentrator. The β-lactamase was further purified by Sephadex G-75 gel filtration chromatography using a 25 mM sodium phosphate buffer (pH 7.0). The purity was verified by SDS/PAGE.
Enzyme Assays.
β-lactamase activity was determined by observing the rate of nitrocefin (10 μM) hydrolysis at 486 nm at 25°C in 100 mM sodium phosphate buffer (pH 7.0) (26). Protein concentrations were determined by the method of Bradford (27). The kinetics of ampicillin hydrolysis were determined at 235 nm (Δɛ = 900 M−1⋅cm−1) (12) at 30°C in 0.05 M phosphate buffer (pH 7.0). Spectrophotometric measurements were made on a Beckman DU640 or DU70 spectrophotometer using a 0.1- or 1.0-cm pathlength cuvette as needed. Kinetic parameters Vmax and Km were determined by fitting to the Michaelis-Menten equation using unweighted nonlinear least squares.
Western Blots.
Proteins were separated by SDS/PAGE (MiniProtean II, Bio-Rad) using 12% Bio-Rad Ready Gels. Proteins were transferred to Hybond ECL nitrocellulose membrane (Amersham Life Science), incubated with a rabbit anti-β-lactamase antibody (28) (1:12,000 dilution) followed by a donkey anti-rabbit horseradish peroxidase conjugate (1:3,000 dilution). Binding was detected by using the Amersham ECL Detection kit according to the manufacturer's instructions. Quantitation was performed by densitometry using authentic β-lactamase standards in the linear range of the response.
Equilibrium Denaturation.
For equilibrium denaturation, purified β-lactamase was incubated overnight at room temperature in 50 mM sodium phosphate and 50 mM NaCl (pH 7.0) containing various concentrations of guanidine hydrochloride (29). Fluorescence spectroscopy was performed on a Perkin–Elmer LS-5 fluorescence spectrometer. The protein concentration was 0.6 μM, and emission was measured at 340 nm (290 nm excitation). Circular dichroism spectroscopy was performed on a Jasco (Easton, MD) Spectropolarimeter J-710 instrument at 25°C. The far UV CD signal of the samples was monitored at 220 nm at 25°C. The protein concentration was 15 μM. The concentration of guanidine hydrochloride was determined by using the refractive index (30).
Equilibrium denaturation transitions followed by fluorescence were analyzed by using a two-state (Eq. 1) model, which includes parameters for a linear pre- and/or posttransition dependence on denaturant concentration (31):
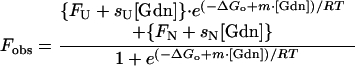 |
1 |
where Fobs is the observed fluorescence signal [Gdn] is the concentration of Gdn⋅HCl, FU is the fluorescence of the unfolded protein, sU is the slope of the posttransition baseline because of the effect of Gdn⋅HCl on the fluorescence of the unfolded protein, FN is the fluorescence signal of the native protein, sN is the slope of the pretransition baseline because of the effect of Gdn⋅HCl on the fluorescence of the native protein, ΔGo is the free energy change from protein unfolding, m is the slope of a plot of observed ΔG (−RT ln (U)/(N)) against [Gdn⋅HCl], R is the gas constant, and T is the temperature in °K. The error limits shown are the standard deviations of the parameter estimates as determined by non-linear least squares fitting.
The CD denaturation data were fit to a three-state model, including an intermediate species, I, according to Eq. 2:
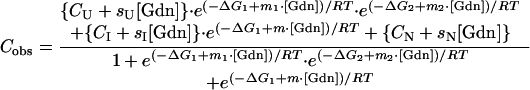 |
2 |
where the symbols have essentially the same definition as given for Eq. 1, except for the inclusion of an intermediate species. ΔG1 corresponds to the free energy of the N to I transition extrapolated to zero denaturant, and ΔG2 corresponds to the free energy of the I to U transition. To simplify the fitting, the ΔG1 and m1 for the first denaturation transition were held at the values determined by fitting to the simpler fluorescence data. This fitting procedure assumes that there is only one intermediate and that it is observed by both fluorescence and CD (29, 32).
Periplasmic Aggregation Experiments.
Crude periplasmic extracts were denatured in 6 M Gdn⋅HCl, and aliquots were diluted directly into 100 mM sodium phosphate buffer (pH 7.0). The rate of aggregation was recorded as an increase in the absorbance at 350 nm because of light scattering by increased turbidity at 25°C by using a Beckman DU70 spectrophotometer (33). When renaturation was complete, the amount of recovered β-lactamase activity was determined by measuring the hydrolysis of ampicillin (300 μM) as described above.
Inclusion Body Isolation.
Overnight cultures of E. coli XL1-Blue carrying β-lactamase plasmids (plasmid pJ1522) were grown to an A600 of 1.0 at 37°C in LB medium in the presence of 12.5 μg/ml chloramphenicol. The cells were harvested by centrifugation at 8,000 × g for 10 min. The soluble periplasmic fraction was released by osmotic shock by resuspending the pellet in 1 ml of 0.03 M Tris, 1 mM EDTA, 20% sucrose buffer (pH 8.0) for 10 min (0°C), centrifuging in a microfuge at 15,000 rpm for 5 min, and resuspending the pellet in 2 ml of cold water for 10 min. After centrifugation at 15,000 rpm for 5 min, the supernatant, containing soluble periplasmic proteins, was set aside. The pellet was resuspended in 1 ml of lysis buffer and extracted repeatedly with detergent to isolate inclusion bodies (34).
Results
Effects of Mutations on Ampicillin Resistance and β-Lactamase Activity.
The substitution of L76 in the hydrophobic core of TEM-1 β-lactamase to a polar asparagine (L76N) seriously compromises the ampicillin resistance of a wild-type E. coli strain carrying a plasmid with the mutant blaTEM gene (21). The L76N mutation increases the sensitivity to ampicillin (as measured by the minimum inhibitory concentration value), and causes a dramatic, 100-fold reduction in the periplasmic β-lactamase specific activity (Table 2 and ref. 21). The M182T substitution is a common clinical isolate that was also found in a suppressor screen for second-site mutations that restore ampicillin resistance when combined with L76N (21). By itself, M182T causes a slight (20–30%) increase in sensitivity to ampicillin and a comparable decrease in the β-lactamase activity (Table 2). However, in combination with L76N, the M182T suppressor restores ampicillin resistance to near wild-type values and increases the β-lactamase activity to 40% of the wild type. Changing Leu to Asn within the hydrophobic core of the protein clearly has a detrimental effect on the amount of β-lactamase activity in vivo, which is largely suppressed by the second-site mutation, M182T.
Table 2.
Values for minimal inhibitory concentration of ampicillin and wild-type and mutant β-lactamase-specific activities in a wild-type E. coli strain (XL1-Blue)
| Sample | MIC* | Specific activity† | Relative activity, % |
|---|---|---|---|
| Wild-type | 3,000 | 38 ± 8 | 100 |
| L76N | 100 | 0.4 ± 0.2 | 1.0 |
| M182T | 2,000 | 31 ± 13 | 81.6 |
| L76N/M182T | 2,000 | 15 ± 4 | 39.5 |
MIC (minimal inhibitory concentration) measured in μg/ml of ampicillin. Data from ref. 21.
Specific activity in mM ampicillin per mg of total protein per min at 25 °C.
Kinetic Parameters of Mutant and Wild-Type β-Lactamases.
To determine whether M182T suppresses the effects of the L76N mutation by altering the catalytic parameters of the enzyme, the two single mutants (L76N and M182T) and the double mutant (L76N/M182T) were purified to homogeneity, and the kinetic parameters were measured by using ampicillin as the substrate (Table 3). The catalytic efficiency of L76N is 30% of wild type, but M182T by itself has no effect on kcat or Km values. When M182T is combined with L76N, it causes a slight increase in activity against ampicillin, resulting in a double mutant with 53% the catalytic efficiency of the wild type.
Table 3.
Kinetic parameters of wild-type and mutant β-lactamase catalysis of ampicillin hydrolysis*
| Sample | kcat (s−1) | Km, μM | 10−7 × kcat/Km, M−1·s−1 | Ratio† |
|---|---|---|---|---|
| Wild type | 830 ± 30 | 58 ± 1 | 1.4 | 1.00 |
| L76N | 660 ± 13 | 150 ± 5 | 0.43 | 0.30 |
| M182T | 820 ± 50 | 59 ± 7 | 1.4 | 0.96 |
| L76N/M182T | 820 ± 40 | 110 ± 10 | 0.8 | 0.53 |
The kinetic parameters were measured using ampicillin as the substrate as described in Materials and Methods.
Ratio of kcat/Km of mutant relative to wild-type β-lactamase.
None of the three mutant enzymes shows appreciable deviations from wild-type structure as judged by far UV and near UV circular dichroism spectroscopy (data not shown). The small effects of these mutations on the structure and intrinsic catalytic properties of β-lactamase cannot account for the overall effect of these mutations on the ampicillin resistance in vivo.
Expression Levels of Mutant and Wild-Type β-Lactamases.
The effects of the L76N and M182T mutations on the amounts of β-lactamase protein expression were examined by Western blots (n = 7) using a polyclonal antibody to TEM-1 β-lactamase (28). The L76N mutation decreases the steady-state levels of the protein in whole-cell extracts to 20 ± 6% of the wild type (Fig. 1, lanes 2 and 4). When periplasmic fractions are isolated, only half of the total L76N is periplasmic and soluble (Fig. 1, lane 5). The other half fractionates with the insoluble, cellular fraction. Because the β-lactamase signal sequence is completely removed, the insoluble fraction represents periplasmic inclusion bodies and membrane-associated material. Unlike L76N, the M182T mutant is present at wild-type levels (107 ± 15%), and the majority is soluble (Fig. 1, lane 7). The combination of both mutations in L76N/M182T restores the total amount of β-lactamase protein to near wild-type levels (70%) (Fig. 1, lane 8). The majority of the double mutant is also soluble and periplasmic (lane 9). Thus, when combined with L76N, the suppressor mutation M182T largely restores the steady-state amount of TEM-1 β-lactamase protein to wild-type levels.
Figure 1.
Steady-state levels of wild-type and mutant β-lactamases from a wild-type strain of E. coli (XL1-Blue). Lane 1, control of purified wild-type β-lactamase; lanes 2, 4, 6, and 8, whole cell extracts; lanes 3, 5, 7, and 9, periplasmic extracts; lanes 2 and 3, wild-type; lanes 4 and 5, L76N; lanes 6 and 7, M182T; and lanes 8 and 9, L76N/M182T. Total β-lactamase was determined from whole-cell extracts boiled in sample buffer; periplasmic extracts were prepared as described in Materials and Methods.
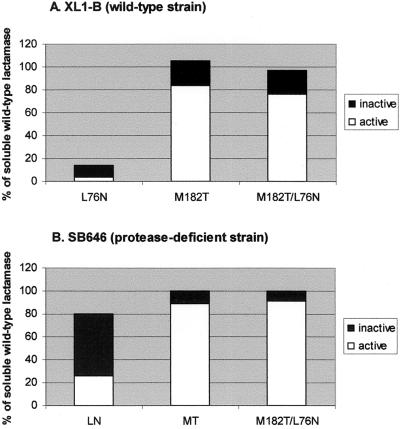
If the low expression of L76N in wild-type E. coli is due to increased proteolysis, expressing it in a protease-deficient strain should enhance its periplasmic expression levels. In accord with this prediction, L76N was present at virtually wild-type levels when expressed in an E. coli strain (SB646) lacking an outer membrane and four periplasmic proteases (21) (Fig. 2). Despite the near wild-type amount of β-lactamase protein, the periplasmic extracts from the protease-deficient strain expressing L76N have only 8% of the wild-type activity (21). When the actual β-lactamase activity measured in the periplasmic extracts is compared with the activity expected from the same amount of purified enzyme (kinetic constants from Table 3), the majority (67%) of L76N is in a catalytically inactive form that is likely misfolded (Fig. 2B). Whereas misfolded, inactive L76N persists in a protease-deficient strain like SB646, it is almost entirely proteolyzed in a wild-type E. coli strain (Fig. 2A). By contrast, the M182T mutation alone results in wild-type levels of mostly active β-lactamase protein (Fig. 2 A and B). When combined with L76N, the suppressor mutation M182T restores β-lactamase expression to the correctly folded, soluble, and enzymatically active form (Fig. 2).
Figure 2.
Effects of expression in a protease-deficient strain. (A) XL1-Blue (wild-type strain); (B) SB646 (protease-deficient strain). Active and inactive β-lactamases within the periplasmic space of a protease-deficient strain. The amount of active β-lactamase was calculated from the activity of periplasmic extracts and the specific activity of the purified protein. The total amount of β-lactamase protein was determined from densitometry of Western blots (see text).
Thermodynamic Properties of Mutant β-Lactamases.
The enhanced proteolysis of L76N in wild-type cells could result from the reduced thermodynamic stability caused by the substitution of a hydrophobic leucine by a polar asparagine residue in the hydrophobic core of the enzyme. M182T might then restore global stability simply by stabilizing other regions of the protein.
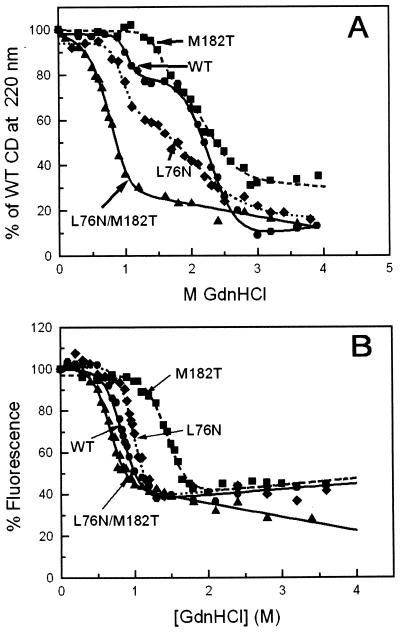
To test this possibility, the effects of the L76N and M182T substitutions on TEM-1 stability separately or in combination were examined by reversible denaturation of β-lactamase in Gdn⋅HCl. The equilibrium unfolding of wild-type β-lactamase monitored by the loss of CD signal at 220 nm proceeds through two transitions involving the equilibrium formation of a “molten-globule” intermediate (Fig. 3), consistent with previous reports (29, 32, 35). For the wild-type protein, the first transition (N → I) has a mid-point at 0.8 M Gdn⋅HCl and the second (I → U) at 2.3 M Gdn⋅HCl (Fig. 3A and Table 4). Because there is little difference in the fluorescence properties of I and U, fluorescence (Fig. 3B) reveals only one transition, which approximately corresponds to the first transition observed by CD (Fig. 3A).
Figure 3.
Equilibrium denaturation of wild-type and mutant TEM-1 β-lactamase at 25°C. (A) Gdn⋅HCl denaturation monitored by CD spectroscopy. The final protein concentration was 15 μM. Points represent averages of four experiments. The curves are drawn according to a three-state model (Eq. 2; Table 4). (B) Gdn⋅HCl denaturation monitored by fluorescence spectroscopy. The β-lactamase samples (0.6 μM final concentration) were excited at 290 nm, and their fluorescence emission was monitored at 340 nm. The theoretical curves are drawn by using a two-state model (Eq. 1; Table 4).
Table 4.
Thermodynamic parameters for the equilibrium transitions observed with wild-type and mutant β-lactamase
| Sample | ΔGNI*, kcal/mol | mNI*, kcal/liter | FI†, % N | ΔGIU‡, kcal/mol | mIU‡, kcal/liter | ΔGNU§, kcal/mol |
|---|---|---|---|---|---|---|
| Wild type | 4.1 ± 0.2 | 4.9 ± 0.3 | 82 ± 1 | 5.9 ± 0.5 | 2.6 ± 0.3 | 10.0 ± 0.7 |
| M182T | 6.3 ± 0.7 | 6.4 ± 0.7 | 67 ± 4 | 3.0 ± 0.6 | 1.5 ± 0.2 | 9.3 ± 1.3 |
| L76N | 6.2 ± 0.6 | 4.3 ± 0.4 | 90 ± 7 | 4 ± 1 | 1.9 ± 0.4 | 10.2 ± 1.6 |
| L76N:M182T | 3.4 ± 0.2 | 5.1 ± 0.3 | 56 ± 8 | 2.7 ± 1.6 | 1.2 ± 0.8 | 6.1 ± 1.8 |
ΔGo for the transition from N to I as measured by fluorescence. As a single transition, this first denaturation process is more accurately determined by fluorescence. The second transition by CD spectroscopy was determined by forcing the first transition to correspond to that monitored by fluorescence. The m values are slopes of plots of ΔG against the Gdn·HCl concentration. Pre- and post-transition changes in the signal were treated as linear functions as described (31).
The CD signal (at 220 nm) of the intermediate as a percentage of the CD signal of the native protein.
ΔGo and m for the transition from I to U.
ΔGo for the transition between N and U.
Both single mutations L76N and M182T significantly destabilize the folding intermediate relative to the unfolded state (Table 4, ΔGIU). By contrast, the native protein is destabilized only in the double mutant. Individually, the L76N and M182T substitutions have small effects on the stability of the native structure relative to the unfolded state (ΔGNU, Table 4). However, the native structure of the double mutant is destabilized by approximately 4 kcal/mol relative to the unfolded state, implying that, in the native protein, the two mutations synergistically destabilize the folded state. The effects of these mutations on the thermodynamic stability of the native and intermediate states are counter to their effects on the amount of correctly folded β-lactamase in the periplasm. Surprisingly, for these β-lactamase variants, periplasmic stability is not dependent on the thermodynamic stability of the protein or its folding intermediate.
Periplasmic Misfolding/Aggregation and Inclusion Body Formation.
Because the effects of these mutations cannot be attributed to changes in stability or intrinsic catalytic activity, the remaining possibility is an effect on protein folding and/or aggregation. To directly address whether L76N promotes the accumulation of aggregated or misfolded forms within the periplasm, a phenotype that is subsequently suppressed by M182T, periplasmic extracts from cells harboring wild-type or mutant β-lactamase were denatured in Gdn⋅HCl and then diluted to allow refolding. Under these conditions, only the extract containing L76N showed a low efficiency of refolding (7 ± 10% activity regain) (Table 5). M182T, L76N/M182T, and the wild-type enzyme refolded to a significantly higher extent (62–82%). Light scattering (turbidity) measurements at 350 nm also showed that extracts containing the L76N mutant were the only ones that exhibited pronounced aggregate formation. The rate of aggregation depended on total protein concentration (not shown). None of the other extracts formed light-scattering aggregates on refolding (Table 5). Whereas aggregation is a complex kinetic process, particularly in a mixture of denatured periplasmic proteins, it is significant that only the L76N mutant showed an increased tendency to form aggregates under these conditions.
Table 5.
Aggregation properties of wild-type and mutant β-lactamases
| Wild type | L76N | M182T | L76N:M182T | |
|---|---|---|---|---|
| % inclusion bodies* | 21 ± 7 | 31 ± 2 | 13 ± 5 | 19 ± 5 |
| Light scattering at 350 nm† | None | + | None | None |
| % activity recovered‡ | 82 ± 11 | 7 ± 12 | 65 ± 10 | 62 ± 4 |
The percentage of β-lactamase protein found in periplasmic inclusion body preparations as a percentage of the β-lactamase in whole-cell extracts. The amount of β-lactamase protein was determined by densitometric quantitation of Western blots from multiple (n = 4–6) independent experiments.
Aggregation was recorded as light scattering at 350 nm after diluting periplasmic extracts from a GdnHCl concentration of 6M into 100 mM sodium phosphate buffer, pH 7.0. The final GdnHCl was 0.88 M.
Activity recovered after completion of refolding was measured using ampicillin as the substrate. Averages are from multiple, independent experiments (n = 5–7). Per cent recovered activity was based on the activity of non-denatured extracts, which had the same final concentration of GdnHCl.
The L76N substitution also increases the formation of β-lactamase-containing periplasmic inclusion bodies. Densitometry of multiple, independent experiments shows a 1.5-fold increase (P < 0.05) in the amount of insoluble L76N compared with wild type. By contrast, M182T forms few inclusion bodies, and the majority is present in the soluble fraction (Table 5). Collectively, these results suggest that the L76N mutation promotes the accumulation of misfolded and aggregated β-lactamase within the periplasmic space. Because the majority of the double mutant (L76N/M182T) is soluble and active within the periplasm, the M182T mutation suppresses this enhanced aggregation and misfolding caused by the L76N mutation. M182T may destroy an aggregation-prone site created by L76N or alter the folding mechanism to kinetically disfavor aggregation and misfolding.
Discussion
While never isolated alone in naturally occurring variants of TEM-1 β-lactamase, a threonine in place of methionine at position 182 of the TEM-1 sequence has been found in combination with mutations that render the enzyme resistant to inhibitors (M69I) or extend its substrate specificity (G238S, R164H/S, E104K, and E240K, Table 1). In addition, M182T restores functionality to the L76N or L76S substitution engineered into the TEM-1 sequence, as well as to a I47Y:E48C double mutant isolated from a library of random substitutions (21). Strikingly, the M182T change was also found to restore the ampicillin resistance in hybrid fusion constructs between the TEM-1 and Proteus vulgaris β-lactamases (36). Clearly, whether in natural or engineered variants, this mutation is beneficial to the structure/function of the enzyme although by itself it does not affect substrate binding or catalysis (17). Because of its pleiotropic effects on the function of a number of mutations throughout the molecule, M182T has been proposed to be a global suppressor of defects introduced into TEM-1 β-lactamase by primary mutations (21, 37).
Our results show that one mechanism by which M182T complements the primary mutations in drug-resistant β-lactamase is through suppression of defects in the protein folding pathway. The L76N mutant is, by itself, highly susceptible to proteolysis, as shown by the high expression levels for this protein in a protease-deficient strain (21). This increased susceptibility is not a consequence of the mutant's lowered thermodynamic stability, but arises because of the propensity of L76N to form insoluble aggregates and inactive, misfolded forms. M182T corrects the low expression levels of L76N, increasing soluble β-lactamase levels 10-fold and decreasing the amount of inactive β-lactamase that accumulates in a protease-deficient strain. M182T also slightly improves catalysis, probably from a subtle rearrangement of the geometry of the active site, resulting in a double mutant roughly half as efficient as the wild-type enzyme. It should be noted that Met-182 lies quite far (17 Å) from Leu-76 within the structure of the TEM-1 β-lactamase. Leu-76 is located in the hydrophobic core of the enzyme, buried within helix H2 in the α-helical domain (23). Helix H2 also contains residues Ser-70 and Lys-73, which are crucial for catalysis. Met-182 lies within a small stretch of residues (180) found before helix H8 and after the omega loop (residues 161–179) of the protein (23). It is therefore surprising that a Thr at this position can address defects associated not only with mutations in the remotely located Leu-76, but also mutations around the substrate or inhibitor binding site (e.g., G238S).
Whereas an engineered core mutation such as L76N causes a drastic impairment in in vivo β-lactamase folding in the wild, the deleterious effects on expression of substitutions such as M69I and G238S must be relatively small. During the natural selection of resistant bacteria, a specificity-altering mutation like G238S has to retain sufficient function to provide a competitive advantage to the bacterial host. Nevertheless, correction of subtle folding defects by M182T could provide a competitive advantage after many generations of selection among the large populations of bacteria found in infections. Furthermore, the complex and changing environment of a bacterial infection (e.g., the body temperature of a patient with a fever) may also favor the selection of subtle changes that are difficult to measure in the laboratory.
The subtle character of defects introduced into β-lactamase by naturally occurring specificity-altering mutations is evident in the case of M69 and G238 mutants. Both M69I and G238S have been isolated as single TEM mutants, suggesting that these mutations alone can produce viable β-lactamases (15, 38). Expression levels for these mutants are near wild type, unlike the low levels observed with L76N (refs. 21 and 39, and unpublished results). Whereas G238S is a destabilizing mutation to the enzyme (40), it is not known what effect M69I has on the thermodynamic stability of the protein. However, M182T increases the thermal stability of extracts containing M69I (21). Molecular modeling provides scant insight into how a substitution at residue 182 specifically addresses defects associated with changes at Met-69, other than that a Thr at position 182 may form a new hydrogen bond anchoring the two domains of the protein and potentially stabilizing its active site (41). Mutations like G238S and M69I may destabilize the protein, yet leave β-lactamase expression levels essentially unchanged. Other mutations associated in natural isolates with M182T such as E104K, have, in fact, a stabilizing effect on the structure of β-lactamase (40), providing another argument that the effects of M182T are due to alterations in folding rather than stability.
Phenotypes associated with drug-resistant β-lactamase variants have usually been attributed to the cumulative effects on specificity and catalysis arising from multiple mutations. Obviously, the increased resistance of bacteria to a range of antibiotics requires the re-engineering of β-lactamase specificity and catalysis through evolution. Suppressor mutations such as M182T could play an important role in the evolution of new activities or altered specificity. M182T, by complementing folding defects introduced by other mutations in the molecule, would permit β-lactamase to sample a much larger repertoire of sequence space by further expanding the number and types of positions that may tolerate substitution. Our results suggest that the effects of drug-resistance mutations on protein folding and aggregation should be considered as a significant contributor to the mechanisms by which drug resistance evolves.
Acknowledgments
This work was supported by National Institutes of Health Grants GM-40379 (to H.G.) and AI-32956 (to T.P.).
Abbreviation
- Gdn⋅HCl
guanidine hydrochloride
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011454198.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011454198
References
- 1.Davies J. Science. 1994;264:375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- 2.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, et al. Nature (London) 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 3.Lin Y, Lin X, Hong L, Foundling S, Heinrikson R L, Thaisrivongs S, Leelamanit W, Raterman D, Shah M, Dunn B M, Tang J. Biochemistry. 1995;34:1143–1152. doi: 10.1021/bi00004a007. [DOI] [PubMed] [Google Scholar]
- 4.Richman D D. Nature (London) 1995;374:494. doi: 10.1038/374494a0. [DOI] [PubMed] [Google Scholar]
- 5.Jacoby G A. Trends Microbiol. 1994;2:357–360. doi: 10.1016/0966-842x(94)90611-4. [DOI] [PubMed] [Google Scholar]
- 6.Strynadka N C J, Adachi H, Jensen S E, Johns K, Sielecki A, Betzel C, Sutoh K, James M N G. Nature (London) 1992;359:700–705. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- 7.Jacoby G A, Medeiros A A. Antimicrob Agents Chemother. 1991;35:1697–1704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaibi E B, Sirot D, Paul G, Labia R. Antimicrob Agents Chemother. 1999;43:447–458. doi: 10.1093/jac/43.4.447. [DOI] [PubMed] [Google Scholar]
- 9.Arlet G, Brami G, Decre D, Flippo A, Gaillot O, Lagrange P H, Philippon A. FEMS Microbiol Lett. 1995;134:203–208. doi: 10.1111/j.1574-6968.1995.tb07938.x. [DOI] [PubMed] [Google Scholar]
- 10.Collatz E, Tran Van Nhieu G, Billot-Klein D, Williamson R, Gutmann L. Gene. 1989;78:349–354. doi: 10.1016/0378-1119(89)90237-0. [DOI] [PubMed] [Google Scholar]
- 11.Venkatachalam K V, Huang W, LaRocco M, Palzkill T. J Biol Chem. 1994;269:23444–23450. [PubMed] [Google Scholar]
- 12.Cantu III C, Huang W, Palzkill T. J Biol Chem. 1996;271:22538–22545. doi: 10.1074/jbc.271.37.22538. [DOI] [PubMed] [Google Scholar]
- 13.Cantu III C, Palzkill T. J Biol Chem. 1998;273:26603–26609. doi: 10.1074/jbc.273.41.26603. [DOI] [PubMed] [Google Scholar]
- 14.Vladiu H, Osuna J, Fink A L, Soberon X. J Biol Chem. 1995;270:781–787. [PubMed] [Google Scholar]
- 15.Stapleton P, Wu P J, King A, Shannon K, French G, Phillips I. Antimicrob Agents Chemother. 1995;39:2478–2483. doi: 10.1128/aac.39.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arlet G, Goussard S, Courvalin P, Philippon A. Antimicrob Agents Chemother. 1999;43:969–971. doi: 10.1128/aac.43.4.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blazquez J, Baquero M-R, Canton R, Alos I, Baquero F. Antimicrob Agents Chemother. 1993;37:2059–2063. doi: 10.1128/aac.37.10.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y, Bhachech N, Bradford P A, Jett B D, Sahm D F, Bush K. Antimicrob Agents Chemother. 1998;42:1671–1676. doi: 10.1128/aac.42.7.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poyart C, Mugnier P, Quesne G, Berche P, Trieu-Cuot P. Antimicrob Agents Chemother. 1998;42:108–113. doi: 10.1128/aac.42.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perilli M, Segatore B, De Massis R M, Riccio M L, Bianchi C, Zol Rossolini G M, Amicosante G. Antimicrob Agents Chemother. 2000;44:2537–2539. doi: 10.1128/aac.44.9.2537-2539.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang W, Palzkill T. Proc Natl Acad Sci USA. 1997;94:8801–8806. doi: 10.1073/pnas.94.16.8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dean A M. Genetics. 1989;110:441–454. doi: 10.1093/genetics/123.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jelsch C, Mourey L, Masson J-M, Samama J-P. Proteins Struct Funct Genet. 1993;16:364–383. doi: 10.1002/prot.340160406. [DOI] [PubMed] [Google Scholar]
- 24.Huang W, Petrosino J, Hirsch M, Shenkin P S, Palzkill T. J Mol Biol. 1996;258:688–703. doi: 10.1006/jmbi.1996.0279. [DOI] [PubMed] [Google Scholar]
- 25.Neu H C, Heppel L A. J Biol Chem. 1965;240:3685–3692. [PubMed] [Google Scholar]
- 26.O'Callaghan C H, Morris A, Kirby S M, Shingler A H. Antimicrob Agents Chemother. 1972;1:283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bradford M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 28.Palzkill T, Le Q Q, Venkatachalan K V, LaRocco M, Ocera H. Mol Microbiol. 1994;12:217–229. doi: 10.1111/j.1365-2958.1994.tb01011.x. [DOI] [PubMed] [Google Scholar]
- 29.Vanhove M, Raquet X, Frère J-M. Proteins. 1995;22:110–118. doi: 10.1002/prot.340220204. [DOI] [PubMed] [Google Scholar]
- 30.Pace C N, Shirley B A, Thomson J A. In: Protein Structure. Creighton T E, editor. Oxford: Oxford Univ. Press; 1990. pp. 311–329. [Google Scholar]
- 31.Senear D F, Bolen D W. Methods Enzymol. 1992;210:463–481. doi: 10.1016/0076-6879(92)10024-8. [DOI] [PubMed] [Google Scholar]
- 32.Frech C, Wunderlich M, Glockshuber R, Schmid F X. Biochemistry. 1996;35:11386–11395. doi: 10.1021/bi9608525. [DOI] [PubMed] [Google Scholar]
- 33.Primm T P, Walker K W, Gilbert H F. J Biol Chem. 1996;271:33664–33669. doi: 10.1074/jbc.271.52.33664. [DOI] [PubMed] [Google Scholar]
- 34.Georgiou G, Valax P, Ostermeier M, Horowitz P M. Protein Sci. 1994;3:1953–1960. doi: 10.1002/pro.5560031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zahn R, Axmann S E, Rücknagel K-P, Jaeger E, Laminet A A, Plückthun A. J Mol Biol. 1994;242:150–164. doi: 10.1006/jmbi.1994.1566. [DOI] [PubMed] [Google Scholar]
- 36.Hosseini-Mazinani S M, Nakajima E, Ihara Y, Kameyama K, Sugimoto K. Antimicrob Agents Chemother. 1996;40:2152–2158. doi: 10.1128/aac.40.9.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrosino J, Cantu III C, Palzkill T. Trends Microbiol. 1998;8:323–327. doi: 10.1016/s0966-842x(98)01317-1. [DOI] [PubMed] [Google Scholar]
- 38.Mabilat C, Courvalin P. Antimicrob Agents Chemother. 1990;34:2210–2216. doi: 10.1128/aac.34.11.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaccolo M, Gherardi E. J Mol Biol. 1999;285:775–783. doi: 10.1006/jmbi.1998.2262. [DOI] [PubMed] [Google Scholar]
- 40.Raquet X, Vanhove M, Lamotte-Brasseur J, Goussard S, Courvalin P, Frère J-M. Proteins. 1995;23:63–72. doi: 10.1002/prot.340230108. [DOI] [PubMed] [Google Scholar]
- 41.Farzaneh S, Chaibi E B, Peduzzi J, Barthelemy M, Labia R, Blazquez J, Baquero F. Antimicrob Agents Chemother. 1996;40:2434–2436. doi: 10.1128/aac.40.10.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]