Abstract
Platelet production is induced by the cytokine thrombopoietin (TPO). It is physiologically critical that TPO expression is tightly regulated, because lack of TPO causes life-threatening thrombocytopenia while an excess of TPO results in thrombocytosis. The plasma concentration of TPO is controlled by a negative feedback loop involving receptor-mediated uptake of TPO by platelets. Furthermore, TPO biosynthesis is limited by upstream open reading frames (uORFs) that curtail the translation of the TPO mRNA. uORFs are suggested to activate RNA degradation by nonsense-mediated decay (NMD) in a number of physiological transcripts. Here, we determine whether NMD affects TPO expression. We show that reporter mRNAs bearing the seventh TPO uORF escape NMD. Importantly, endogenously expressed TPO mRNA from HuH7 cells is unaffected by abrogation of NMD by RNAi. Thus, regulation of TPO expression is independent of NMD, implying that mRNAs bearing uORFs cannot generally be considered to represent NMD targets.
INTRODUCTION
Physiological platelet production by megakaryocytes depends on the cytokine thrombopoietin (TPO). The circulating serum levels of TPO are very low (<100 pg/ml) and tightly regulated. Lack of TPO signaling results in life-threatening thrombocytopenia (1) and an excess of TPO causes dangerous thrombocytosis (2,3). The regulation involves an important negative feedback loop of TPO uptake and degradation by platelets. The plasma TPO level reciprocally correlates to the mass of megakaryocytes and platelets, which degrade this cytokine following its uptake by specific membrane receptors (2). Furthermore, TPO synthesis is tightly regulated in hepatocytes and bone marrow stromal cells. Translation of TPO is physiologically inhibited by seven upstream open reading frames (uORFs) in the 5′-untranslated region (5′-UTR) of the TPO mRNA (4). The seventh uORF has been shown to be of particular importance, because it overlaps with the start codon of the TPO-coding open reading frame (ORF) and strongly inhibits initiation of TPO translation (5). Mutations in the 5′-UTR of the TPO gene, which cause hereditary thrombocytosis (HT), inactivate the inhibitory function of uORF 7 and abolish this translational control (4–7). In these cases, pathologically high TPO levels are observed, leading to an increased number of platelets in the peripheral blood. Thus, the tight regulation of TPO expression at multiple levels appears to be critical for its physiological function.
The specific architecture of the TPO mRNA with its uORFs suggests that this mRNA might be targeted not only by translational regulation but also by a mRNA surveillance pathway called nonsense-mediated mRNA decay (NMD) (8,9). NMD represents a splicing- and translation-dependent pathway of RNA degradation which limits the expression of transcripts bearing premature translation termination codons (10–12). In the TPO mRNA, the physiological uORFs introduce several upstream translation termination codons that would, according to current models of NMD, be expected to induce down-modulation of the transcript by NMD (10–14).
The splicing dependence of NMD is mediated by a multi-protein complex that is deposited ∼20 nts upstream of the exon–exon junction (15). This exon-junction complex (EJC) provides a potential binding platform for factors involved in mRNA export and NMD (15). Some components of the EJC accompany the mRNA to the cytoplasm and remain bound until they are removed by translating ribosomes (16). NMD-activating stop codons have at least one downstream EJC that is proposed to tag aberrant mRNAs for rapid decay (10,17). In contrast, normal stop codons are usually not followed by an exon–exon junction (18). In the case of TPO mRNA, the stop codons at the end of the uORFs are followed by several introns. For the recognition of the spatial relationship between the termination codon and the EJC, active translation of the respective mRNA is required. Interference with any step of translation abrogates NMD and thus stabilizes PTC-containing transcripts (11,19).
NMD activation by uORFs has mostly been studied in Saccharomyces cerevisiae. In yeast, the effects of both natural as well as artificially introduced uORFs were analyzed. In the presence of artificial uORFs, mRNA levels of the yeast genes CAT (20) and CYC1 (21) are reduced. The genes GCN4 and YAP1 naturally harbor several uORFs. Interestingly, they were shown to contain stabilizing elements (STE) that actively prevent degradation of their mRNAs via NMD (22–24). These STEs are located between the uORFs and the start codon. For the yeast gene YAP2, a natural uORF overlapping with the ORF strongly destabilizes the mRNA (25). However, this destabilization occurs independently of the central NMD factor Upf1p (26). Thus, the canonical NMD pathway seems not to be involved.
Genome-wide microarray analyses revealed that uORF-containing transcripts in mammals as well as in yeast represent a major class of natural NMD targets (13,14). In yeast, a large number of uORF-containing mRNAs were up-modulated when either of the essential NMD factors (Upf1p, Upf2p or Upf3p) were deleted (14). Likewise, depletion of hUpf1 (also called Rent1) increases the expression levels of a large number of mammalian mRNAs containing natural uORFs (13).
Since the TPO mRNA harbors several uORFs that are followed by downstream exon–exon junctions, it would also be expected to represent a natural NMD target. NMD might thus add to levels of regulation of TPO synthesis.
Surprisingly, our analysis of normal and mutant TPO shows that the TPO mRNA escapes NMD and naturally occurring uORFs in mammalian mRNAs do not necessarily trigger NMD. We suggest that the NMD resistance of TPO may be physiologically required to maintain normal levels of TPO synthesis and avoid thrombocytopenia.
MATERIALS AND METHODS
Plasmid constructs
The TPO-minigene was generated by PCR amplification of the TPO exons 3, 4 and 5 with the translational initiation codon and part of the 5′-UTR from human genomic DNA (sense primer, 5′-TTTTCTCGAGCCTCACCCTTGGCCCGCC-3′; antisense primer, 5′-TTTTTTTTGCGGCCGCTTATCACTTGTCGTCATCGTCCTTGTAGTCGGTTTTCCATTCTCCC-3′containing the sequence of a FLAG epitope and a stop codon). The PCR product was gel purified, digested with XhoI and NotI and inserted into the multiple cloning site of the pCI-neo vector (Promega).
The intron of the TPO ΔIntron 4 construct was deleted by overlapping PCR using the primer 5′-CTGGGCACTGGCTCAGTCTGCTGTGAAG-3′ together with the TPO sense and antisense primers.
The mutants ΔG and ΔATG were constructed by site-directed mutagenesis using overlapping PCR (primer sequences available upon request). TPO ΔG [for position of the ΔG mutation see (5)] was used as parental vector for the NS 27, NS 29, NS 40 and NS 40 ΔIntron 4 constructs. Construct TPO NS 40 ΔATG was derived from TPO ΔATG construct using the same mutagenesis primer as for the NS 40 construct. The β-globin+300+eIII transfection control and β-globin wild-type (wt) and NS39 were described previously (11,27).
The identity of all constructs was confirmed by DNA sequencing on an ABI Prism 3100 Sequencer (Applied Biosystems) using the BigDye Terminator Sequencing Kit 3.1 (Applied Biosystems).
Cell culture and transfection
HeLa (cervix carcinoma) and HuH7 cells (hepatoma) were grown in DMEM supplemented with 10% fetal calf serum (FCS) and 1% penicillin/streptomycin at 37°C and 5% CO2. Cells were transiently transfected by calcium phosphate precipitation (28) in 10 cm plates using 30 µg of the test construct DNA, 1 µg of a green fluorescent protein (GFP) expression vector and 5 µg β-globin+300 DNA as a control for transfection efficiency. Cells were washed after 20 h and harvested 24 h later.
Transient transfection of siRNA
Transient transfection of siRNAs was carried out using Oligofectamine reagent (Invitrogen) according to the manufacturer's recommendations as described (29). The siRNA oligonucleotides used for transfections were purchased as annealed, ready-to-use duplexes from Dharmacon. The siRNA target sequences were: Luciferase (AA-CGUACGCGGAAUACUUCGA), hUpf1 (AA-GAUGCAGUUCCGCUCCAUU).
Immunoblot analysis
An aliquot of 10 µg of protein lysate was separated on an polyacrylamide gel according to standard protocols and transferred to PVDF membranes. Monoclonal anti-FLAG antibody (Sigma) was used at 1:2000, monoclonal anti-tubulin at 1:7500, polyclonal anti-β-globin at 1:2000 and polyclonal anti-hUpf1 at 1:500 dilutions. Horseradish peroxidase (HRP)-conjugated anti-mouse IgG or anti-rabbit IgG antiserums (Sigma) were used at 1:5000 dilutions. Signals were detected using ECL or ECL-plus reagent (Amersham).
RNA analysis
Total cytoplasmic RNA was isolated as described (29,30). Northern blot analysis was performed with 3 µg of total cytoplasmic RNA. Probes were prepared by in vitro transcription in the presence of [α-32P]GTP (Perkin Elmer). The TPO probe was transcribed with T3 polymerase from an EcoRI-linearized pCI-neo TPO ΔIntron 4 plasmid. The β-globin probe was transcribed with SP6 polymerase from a BamHI-linearized plasmid containing exon III of the β-globin gene.
Hybridization was carried out overnight in Church buffer [0.5 M Na2HPO4, 1 mM EDTA, 7% SDS (pH 7,2)] at 65°C.
Band intensities were quantified by imaging in a FLA-3000 PhosphorImager (Fujifilm). Percentages correspond to mean values that were calculated from at least three independent experiments. A representative blot is shown.
Quantitative real-time PCR (LightCycler)
First strand cDNA was synthesized using MuMLV RNase H- RT (MBI Fermentas) according to the manufacturer's protocol using 1 µg of RNA. We carried out real-time PCR, using the LightCycler™ system (Roche Diagnostics, Mannheim, Germany). Expression analyses was performed with single-stranded cDNA and gene-specific primers (primer sequences are available on request) using the FastStart DNA Master SYBR Green kit (Roche Diagnostics). The expression levels are the means of three independent experiments. Melting curves of the PCR products were performed for quality control.
RESULTS
Transfected TPO reporter mRNAs are NMD-resistant
We generated a series of mutant TPO minigenes consisting of exons 3 to 5 including the important uORF7 (4–7). The minigenes contain a C-terminal FLAG-tag to facilitate identification of the protein product (Figure 1A).
Figure 1.
The TPO wt mRNA is not subjected to NMD. (A) Schematic representation of the TPO minigene used in this study. Boxes and lines represent exons and introns, respectively. The bold arrow (8) marks the position of the translation initiation codon for TPO. Numbers and arrows indicate the position of the seven uORFs of TPO. The minigene contains uORF 7 and the indicated parts of exons 3, 4 and 5. A FLAG-tag for immunoblot detection is present at the 3′ end of the minigene. The positions of the ΔG and ΔATG 7 mutations are indicated. The arrows below the different construct schemes represent the major translation products of the respective mRNAs. (B) Northern blot of cytoplasmic RNA from HeLa cells transfected with the TPO minigene wt (lane 2) and mutants (ΔG, lane 3; ΔATG 7, lane 4; ΔIntron 4, lane 5). Messenger RNA levels were calculated after normalization to the level of the control mRNA (β-globin+300+eIII) to control for variations in transfection efficiencies and gel loading. Signals were quantified as described in Materials and Methods. Percentages represent mean values that were calculated from at least three independent experiments. (C) Immunoblot analysis of TPO protein expression. The TPO protein was detected with an anti-FLAG antibody. The asterisk indicates a nonspecific cross-reactive band. To control for equal transfection efficiency and gel loading, the blot was reprobed with a β-globin-specific antibody that detects the β-globin control protein (β-globin+300+eIII). (D) Northern blot analysis of total cytoplasmic RNA from cells that were transfected with siRNAs directed against hUpf1 to specifically deplete endogenous hUpf1 protein (lanes 4–6, 9, 10). Cells transfected with siRNAs directed against luciferase (lanes 1–3, 7, 8) served as controls. 20 h after siRNA treatment, cells were cotransfected with TPO minigene constructs wt (lanes 1 and 4), ΔIntron 4 (lanes 2 and 5), ΔG (lanes 3 and 6) and β-globin wt (lanes 7 and 9) and NS 39 (lanes 8 and 10) together with the control plasmid. (E) Immunoblot analysis of HeLa cell extracts transfected with hUpf1 siRNA shown in (D). Immunoblotting was performed with a hUpf1-specific antibody (lower panel). The membrane was reprobed with a tubulin-specific antibody (upper panel) to control for equal loading. (F) Northern blot of cytoplasmic RNA from HeLa cells transfected with the TPO minigene wt (lanes 1 and 4) and mutants (ΔIntron 4, lanes 2 and 5; ΔG, lanes 3 and 6) together with the control plasmid. Cycloheximide (CHX, 12 µg/ml) was added to the cell culture medium 5 h prior to harvest. CHX-treated (lanes 1–3) or untreated (lanes 4–6) cells were transfected, washed and harvested simultaneously. Expression of the mRNAs was analyzed as described in Figure 1B.
The minigene series was designed to specifically address two central requirements for NMD in mammals: translation and splicing. Interference with either translation or splicing has previously been shown to abolish NMD (10–12,17,19,27,31). The ΔG and ΔATG variants of our TPO minigene address the translation dependence, and the ΔIntron 4 construct the splicing dependence.
A single G-nucleotide deletion in the 5′-UTR of the TPO gene has been shown to cause HT in a Japanese family (5). This deletion, referred to as ΔG (Figure 1A), dramatically increases TPO synthesis, because it bypasses the translation inhibition of uORF7 by shifting it into the same reading frame as TPO, thus allowing the synthesis of an N-terminally extended TPO polypeptide with full biological activity. Consequently, the ΔG mRNA does not contain a PTC and is thus not expected to be targeted by NMD. Another mutant, called ΔATG7, was constructed by removing the start codon of uORF 7. In this mRNA, translation would be predicted to initiate exclusively at the ATG of the TPO-coding ORF (ATG8). Analogous to the ΔG mRNA, no degradation by the NMD pathway is predicted. TPO intron 4 is located in the minigene at a NMD-supporting position 127 nt downstream of the termination codon of the uORF7, i.e. more than the required 55 nt downstream of the stop codon (10,11). The splicing of this intron is therefore expected to be essential to direct the corresponding reporter mRNA to the NMD pathway, if translation is initiated at uORF7 (15,17). This intron was removed in the construct referred to as ΔIntron 4. This manipulation should render the resulting mRNA NMD insensitive, because no EJC can be deposited at a position downstream of the termination codon of uORF7. If NMD contributes to the control of the TPO expression, we expect to observe an increase of mRNA levels of all three mutant constructs in comparison to wt.
Expression of wt and mutant mRNAs were analyzed in HeLa cells that were transiently transfected with different minigene constructs together with a control for transfection efficiency. Efficient splicing of intron 3 and intron 4 in the wt, ΔG, ΔATG 7 and ΔIntron 4 (only intron 3 splicing assessed) mRNAs was confirmend by RT–PCR (data not shown) and by the identical mobility of the TPO wt, ΔG, ΔATG 7 and ΔIntron 4 mRNAs during RNA-gel electrophoresis (Figure 1B). With this experimental approach, we observed slightly, but reproducibly higher expression levels for the ΔG and ΔATG 7 mRNAs when compared to the wt mRNA (Figure 1B, lanes 1–3). In contrast, the TPO ΔIntron 4 mRNA was expressed at wt levels (Figure 1B, lane 4). Notably, the deletion of intron 4 removes the only exon–exon junction downstream of the uORF 7 termination codon. Therefore, the small increase of steady-state mRNA levels observed for ΔG and ΔATG 7 is likely caused independent of NMD. In conclusion, the steady state-levels of the TPO reporter mRNAs were not strongly affected by manipulations that typically inhibit NMD.
Expression of the TPO-FLAG protein was only observed when either the ΔG or the ΔATG 7 constructs were transfected (Figure 1C). This indicates that translation is efficiently initiated at AUG 7 (ΔG), but can also be initiated at AUG 8 (ΔATG 7), if AUG 7 is lacking. In the wt and ΔIntron 4 mRNAs, translation initiates at AUG 7 and terminates downstream of AUG 8 (Figure 1A). In this arrangement, the translation of uORF7 is known to interfere with the initiation at the TPO ORF (5), which explains the absence of TPO-FLAG proteins encoded by these constructs (Figure 1C). In the ΔG mRNA, translation starts from AUG 7 and terminates at the physiological stop codon. The resulting protein also contains amino acids encoded by uORF7 and is therefore 23 amino acids longer than the protein translated from the TPO ΔATG 7 mRNA.
A specific way to address the contribution of NMD to the expression levels of TPO wt is to deplete endogenous hUpf1 (29,32,33). This was accomplished by transfecting HeLa cells with siRNAs targeting hUpf1. Efficient hUpf1 depletion was confirmed by immunoblot anylysis with a hUpf1-specfic antibody (Figure 1E). We compared TPO wt expression to TPO ΔIntron 4 and TPO ΔG as NMD specificity controls, because these mRNAs are not targeted by NMD. β-globin wt and NS 39 served as validated positive NMD controls (11,29). hUpf1 depletion up-modulated the positive control, NS 39 mRNA, about 4-fold (Figure 1D lanes 7 and 8 versus lanes 9 and 10), which is similar to levels observed in previous studies (29) and proves the functional effect of the hUpf1 depletion on NMD. In contrast, depletion of hUpf1 led to a comparable up-modulation of TPO wt, TPO ΔIntron 4 and TPO ΔG (negative controls) mRNAs. As TPO ΔIntron 4 and TPO ΔG do not represent NMD targets, the equal up-modulation of TPO wt, ΔIntron 4 and ΔG mRNA is likely not a consequence of NMD inhibition (Figure 1D compare lanes 1–3 versus lanes 4–6). A slight up-modulation was also observed for β-globin wt (Figure 1D, compare lanes 7 and 9). Thus, the observed increase of TPO wt mRNA is probably not caused by inhibition of NMD, but rather represents an NMD-unrelated response that enhances mRNA expression.
An important characteristic of NMD is its translation dependence. This hallmark can be used to discriminate NMD from translation-independent degradation pathways. We analyzed the mRNA expression levels of TPO wt, ΔIntron 4 and ΔG in the presence of cycloheximide, which stabilizes PTC-containing mRNAs (19). Steady state levels of all tested TPO mRNAs are slightly increased in cells treated with cycloheximide when compared to untreated cells (Figure 1F). We conclude that the equal up-modulation of TPO wt, ΔIntron 4 and ΔG mRNA by cycloheximide treatment is not caused by inhibition of NMD, but rather represents another, translation-dependent effect.
These data further corroborate that the TPO reporter mRNA is not degraded by NMD despite the presence of upstream termination codons in a NMD-competent position.
Endogenous TPO mRNA is NMD-resistant
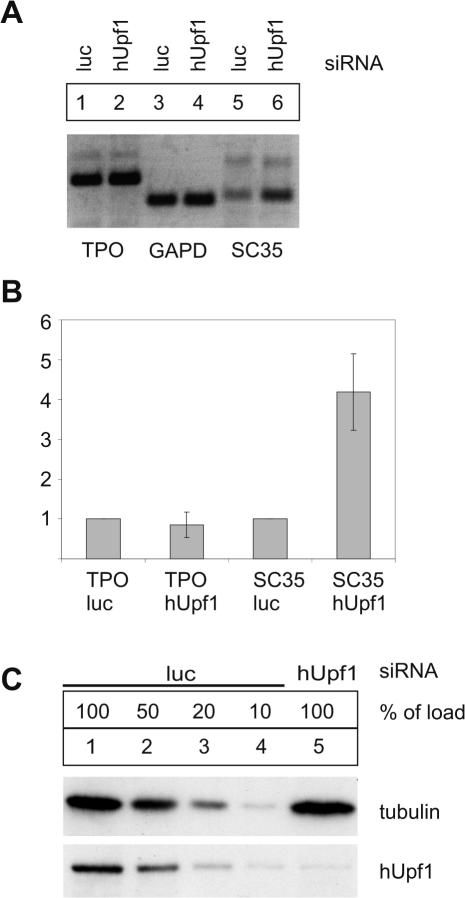
Because the experiments described above were performed with transfected minigenes that lack some features of the physiological architecture of the TPO mRNA, we wanted to challenge the conclusions of these data by an analysis of endogenous full length TPO mRNA. We established by RT–PCR that the hepatoma cell line HuH7 endogenously expresses TPO mRNA. The expression levels of TPO mRNA in HuH7 cells were comparable to that in HepG2 cells (data not shown). HuH7 cells were transfected with siRNA against hUpf1 and mRNA levels of TPO, GAPD and SC35 were measured by RT–PCR and quantitative real-time PCR (LightCycler™). Immunoblot analysis of protein lysates from HuH7 cells transfected with siRNAs against luciferase or hUpf1 showed that hUpf1 was depleted to less than 10% (Figure 2C). GAPD mRNA steady-state levels served as an internal standard for normalization that is not affected by hUpf1 depletion. The splicing factor SC35 (also referred to as SFRS2) regulates its own expression by an autoregulatory feedback loop that involves activation of splice-sites in the 3′-UTR of the SC35 mRNA (40). The transcripts that undergo splicing in the 3′-UTR are degraded by NMD and thus serve as a positive control for effective NMD-inhibition by hUpf1 depletion (33). Accordingly, the expression of NMD-sensitive SC35 was increased after hUpf1 depletion (Figure 2B), confirming the efficient inhibition of NMD by the hUpf1 depletion in HuH7 cells. In contrast to SC35, the expression levels of TPO mRNA were not increased, showing directly that the endogenously expressed TPO transcripts in HuH7 cells do not represent NMD targets (Figure 2B).
Figure 2.
Endogenous full-length TPO mRNA is NMD resistant. (A) RT–PCR analysis of endogenous HuH7 TPO after hUpf1 depletion. Cells were transfected with siRNAs specific for hUpf1 to deplete endogenous hUpf1 protein (cDNA used in lanes 2, 4, 6). Cells transfected with siRNAs directed against luciferase (cDNA used in lanes 1, 3, 5) served as negative controls. Cells were harvested 72 h after siRNA transfection. After first strand cDNA synthesis, PCR with gene specific primers for TPO (lanes 1 and 2), GAPD (lanes 3 and 4) and SC35 (lanes 5 and 6) were performed using cDNA from luciferase- (lanes 1, 3, 5) or hUpf1-siRNA transfected cells (lanes 2, 4, 6). (B) Quantitative RT–PCR analysis of endogenous HuH7 TPO after hUpf1 depletion. LightCycler™ analysis was performed to determine the expression levels of TPO, GAPD and SC35 in cells transfected with hUpf1 siRNAs compared to luciferase-treated cells. cDNAs from the first strand synthesis described in (A) was used for LightCycler analysis. All expression levels were normalized to GAPD expression levels. NMD-sensitive SC35 served as positive control for the inhibition of NMD. SC 35 primers specifically amplify the spliced 3′-UTR with two splice variants (1,6 and 1,7 kb) as described previously (33). The level of up-modulation and standard deviations are indicated. Values and standard errors were calculated from five independent experiments. (C) Immunoblot analysis of protein lysates from HuH7 cells transfected with siRNAs against luciferase as a negative control (luc, lanes 1–4) or hUpf1 (lane 5) using a hUpf1 specific antibody. Dilutions corresponding to 50, 20 or 10% (lanes 2–4) of the initial protein amount (lane 1) from luciferase-siRNA transfected cells were loaded to assess the efficiency of the hUpf1 depletion. Reprobing with a tubulin specific antibody was performed as a loading control.
Elongation of uORF 7 leads to a hUpf1-independent reduction of TPO mRNA expression
Early PTCs in the β-globin and BRCA1 mRNAs do not activate NMD suggesting that transcripts with short ORFs may not be efficient NMD substrates (30,34,35). It is therefore conceivable that the uORF7 of the TPO mRNA is too short to elicit NMD. We thus increased the length of the short uORF 7 by introducing a termination codon into the ΔG construct at codon 40 (NS 40 ΔG) (Figure 3A). Here, the ΔG construct was used to prevent translation termination at the normal termination codon of uORF 7. The length of this ORF is thus similar to that of β-globin NS 39, which is known to efficiently induce NMD. The mRNA expression of NS 40 ΔG was reduced to 40% of the wt mRNA and to 29% of the ΔG mRNA (Figure 3B) suggesting that this longer ORF is sufficient to activate NMD.
Figure 3.
An elongated uORF decreases TPO mRNA expression. (A) Schematic representation of mutated TPO minigenes. The positions of the respective termination codons (Ter) are indicated. Arrows are indicating the length of the respective mRNAs. Nonsense codon positions are counted from the (Met) at position 1. (B) Northern blot of cytoplasmic RNA from HeLa cells cotransfected with either the TPO minigene wt (lane 3) or mutants (ΔG, lane 4; ΔATG 7, lane5; ΔIntron 4, lane 6; NS40 ΔG, lane 7; NS40 ΔG ΔIntron 4, lane 8; ΔG ΔIntron 4, lane 9; NS40 ΔATG7, lane 10; NS26 ΔG, lane 11; NS28 ΔG, lane 12) together with the control plasmid. Cytoplasmic RNA from untransfected HeLa-cells (lane 1) or cells transfected with only the control plasmid (lane 2) is shown. Expression of the mRNAs was analyzed as described in Figure 1B. Percentages and standard deviations (SD) are based on three independent experiments.
We analyzed the splicing dependence of the decreased expression of NS40 ΔG by deleting intron 4. Indeed, deletion of intron 4 increases the abundance of this mRNA 1.5-fold (Figure 3B, lane 8), but this level of expression is still significantly lower than the ΔG mRNA expression. Deletion of intron 4 in the ΔG mRNA without the NS 40 mutation decreased mRNA expression ∼1.8-fold (Figure 3B, lane 9). These results are consistent with the possibility that the TPO NS40 mRNA could be subjected to NMD.
If the short length of the uORF determines the weak NMD-efficiency of the TPO wt mRNA, decreasing the length of the uORF in TPO NS40 ΔG is expected to lower the NMD efficiency. This was tested using the construct NS 40 ΔATG 7, which lacks the start codon of uORF 7 (Figure 3A). Thus, translation initiates exclusively at AUG 8 and terminates at the 17th codon, a position corresponding to NS 40 in the ΔG mRNA (NS40 ΔG). This preserves the position of the termination codon, but shortens the length of the translated sequence. The mRNA expression levels of this mutant were slightly lower than wt mRNA levels, markedly lower than ΔATG 7 mRNA levels (i.e. the parental vector), but higher than the TPO NS40 ΔG expression levels. This suggests that the length of the translated uORF modulates NMD-efficiency, but additional determinants also play a role.
Could the position of the termination codon affect NMD-efficiency of the TPO wt mRNA and hence represent such an additional determinant? The termination codon of uORF7 is created by splicing, i.e. 2 nt of the codon are located in the third exon and 1 nt in the fourth. It has been demonstrated previously that nonsense codons which surround the splice site are efficiently recognized by NMD (36). To recapitulate this important aspect in the context of the TPO mRNA, we created constructs NS 26 ΔG and NS28 ΔG that harbor termination codons either in exon 3 (NS26) or in exon 4 (NS28) at positions adjacent to the physiological termination codon of uORF 7 at position 27 (Figure 3A). These mutations were constructed in the ΔG backbone in order to prevent translation termination at the physiological termination codon of uORF 7. None of these manipulations significantly altered mRNA expression when compared to wt, although in all cases translation of the uORF terminates more than 50 nt upstream of the last exon junction. This indicates that the splitting of the termination codon by an intron is not causing the NMD-insensitivity of the TPO mRNA, confirming earlier findings with the TCR-beta mRNA (36).
As only elongation of the uORF from 27 to 40 amino acids (NS 40 ΔG) significantly decreased mRNA steady-state levels, the short length of the uORF of the transcript rather than the position of its termination codon could be responsible for the NMD-insensitivity.
However, when hUpf1 was depleted to a level that resulted in a more than 3.5-fold up-modulation of the β-globin NS 39 control mRNA (29), TPO NS40 ΔG increased less than 1.8-fold when compared to TPO ΔG (Figure 4). This remarkable difference between β-globin NS 39 and TPO NS40 ΔG suggests that the TPO NS40 ΔG mRNA is not efficiently regulated by hUpf1. As in Figure 1D and E, efficient hUpf1 depletion was verified by immunoblot analysis with a hUpf1-specific antibody (Figure 4B).
Figure 4.
Increasing the length of the TPO uORF results in a hUpf1-independent decline of mRNA steady-state levels. (A) Northern blot analysis of total cytoplasmic RNA from HeLa cells that were transfected with siRNAs directed against hUpf1 to specifically deplete endogenous hUpf1 protein (lanes 5–8, 11, 12). Cells transfected with siRNAs directed against luciferase (lanes 1–4, 9, 10) served as negative controls. After 20 h the siRNA treatment, cells were cotransfected with TPO minigene constructs wt (lanes 1 and 5), ΔIntron 4 (lanes 2 and 6), ΔG (lanes 3 and 7), NS40 ΔG (lanes 4 and 8), β-globin wt (lanes 9 and 11) and NS 39 (lanes 10 and 12) together with the control plasmid. Percentages are means that were calculated from three independent experiments. (B) Immunoblot analysis of HeLa cell extracts transfected with hUpf1 siRNA. Immunoblotting was performed with a hUpf1-specific antibody (lower panel). The membrane was reprobed with a tubulin-specific antibody (upper panel) to control for equal loading.
In summary, our data indicate that the down-modulation of the TPO NS 40 ΔG transcript in comparison to TPO wt and TPO ΔG is only partially hUpf1-dependent and not predominantly caused by canonical hUpf1-dependent NMD.
DISCUSSION
A significant proportion of mammalian mRNAs contain short uORFs that precede the ORF encoding the actual gene product (37,38). These uORFs are located in the 5′ part of the transcript and thus terminate upstream of one or more downstream introns. Such arrangements of termination codons located more than 50 nt upstream of the final exon–exon junction commonly activate an mRNA surveillance pathway, referred to as nonsense-mediated mRNA-decay (10–12). In yeast as well as in mammals, mRNAs with uORFs represent a major class of natural NMD targets (13,14). The human TPO mRNA contains seven uORFs, all of them terminating more than 50 nt 5′ of at least six exon–exon junctions. Therefore we expected that the TPO mRNA represents one of the natural targets of the NMD pathway and that NMD represented an additional level to limit physiological TPO expression (9).
We tested this hypothesis by using a minigene that contains the region harboring all previously described mutations causing HT and also includes uORF7, AUG 8 (start-codon for TPO) and intron 4 that is located in a NMD-competent position >50 nt downstream of the termination codon of uORF7. Our analysis of different TPO mutants revealed that TPO mRNA steady-state levels are slightly elevated when produced from a construct without a translated uORF (ΔATG 7 and ΔG), as compared to wt levels. In contrast, deletion of intron 4 did not increase TPO mRNA expression. These data indicate that NMD does not play a major role in the regulation of the TPO mRNA expression.
Our data employing the reporter constructs were validated by hUpf1 depletion in transfected HeLa cells as well as in HuH7 cells that express endogenous TPO. RNAi against hUpf1 has been demonstrated previously to specifically inhibit NMD of nonsense-mutated β-globin or T-cell receptor mRNAs (29,32). Despite efficient depletion of hUpf1 that significantly stabilized known NMD-targets (β-globin NS 39, SC35), expression levels of the TPO wt mRNA did not display a stronger increase than the negative controls TPO ΔG and TPO ΔIntron 4. A similar up-modulation was also observed for β-globin wt mRNA after hUpf1 depletion when expressed from the same expression vector as TPO and is therefore not necessarily related to NMD inhibition. These data demonstrate that both the endogenous full-length TPO mRNA and the mRNA expressed from the TPO reporter are - at most - only marginally regulated by a canonical hUpf1-dependent NMD. However, our data cannot rule out the possibility that the TPO wt mRNA is targeted by another, hUpf1 independent degradation mechanism.
The uORF 7 is relatively short and spans only 27 codons. Human β-globin mRNAs with such short ORFs (i.e. early termination codons) do not undergo NMD (30). We therefore investigated if the potential NMD insensitivity is due to the short length of the uORF. We show that increasing the length of the TPO uORF does indeed lead to a decrease of TPO mRNA expression. This decrease was only partly reverted by a hUpf1 knockdown. Thus, hUpf1-dependent NMD does not represent the main cause of the decreased TPO NS40 ΔG expression, but may be partially involved to modulate TPO NS40 ΔG mRNA expression.
What prevents the TPO wt mRNA from being degraded via the NMD pathway although it bears all the necessary hallmarks of a NMD-sensitive transcript? The presence of a stabilizer element, as has been reported for the yeast GCN4 and PGK1 mRNAs (22,39) might offer a potential explanation. The mechanism of action of stabilizer elements in yeast is unknown, but a typical feature seems to be their position between the uORF Ter and the ORF AUG (39). In contrast, TPO uORF7 overlaps with the coding ORF (AUG 8) suggesting that any putative TPO stabilizer element would have to act in a different manner than the described yeast stabilizer elements. Such a difference may thus explain the lack of any apparent sequence homologies between the 5′-UTR of the TPO gene with the yeast stabilizer elements.
It is well conceivable that the NMD-insensitivity of the TPO mRNA is the sum of a combination of NMD-efficiency decreasing features. These include the short length of the uORF7, that seems to represent a weak suppressor of TPO wt NMD. Addtional modulators could be (A) an unfavorable sequence context at the exon–exon junctions that do not allow for the efficient recruitment of NMD-activating EJC components, or (B) the presence of an unusual stabilizing element (as outlined above). Such an arrangement of NMD-limiting characteristics could be the reason, why a solitary basis for the NMD-insensitivity of the TPO-mRNA could not be determined.
In conclusion, the data presented here demonstrate that the TPO mRNA does not represent an endogenous NMD target and that the tight control of physiological TPO expression is independent of NMD-mediated regulation of mRNA stability. On the contrary, NMD resistance of TPO mRNA might have evolved to maintain adequate TPO mRNA levels and avoid thrombocytopenia.
Our data also demonstrate that naturally occurring mammalian uORFs do not necessarily trigger NMD. The presence of uORFs in the 5′-UTRs of many genes involved in cellular growth and differentiation (37,38) therefore does not qualify them as endogenous NMD targets per se.
Supplementary Material
Acknowledgments
The authors would like to thank Sven Danckwardt, Jill Holbrook and Joachim Kunz for helpful discussions. The authors would also like to thank Marcelo Viegas for designing the SC35 LightCycler primer pairs. C.S. was supported by the young medical investigator award program of the medical faculty of the Ruprecht-Karls Universität Heidelberg. This work was supported by grants KU 563/7-2 and KU 563/8-2 from the Deutsche Forschungsgemeinschaft. Funding to pay the Open Access publication charges for this article was provided by the medical faculty of the Ruprecht-Karls Universität Heidelberg.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ballmaier M., Schulze H., Strauss G., Cherkaoui K., Wittner N., Lynen S., Wolters S., Bogenberger J., Welte K. Thrombopoietin in patients with congenital thrombocytopenia and absent radii: elevated serum levels, normal receptor expression, but defective reactivity to thrombopoietin. Blood. 1997;90:612–619. [PubMed] [Google Scholar]
- 2.Wolber E.M., Jelkmann W. Thrombopoietin: the novel hepatic hormone. News Physiol. Sci. 2002;17:6–10. doi: 10.1152/physiologyonline.2002.17.1.6. [DOI] [PubMed] [Google Scholar]
- 3.Kaushansky K. Thrombopoietin. N. Engl. J. Med. 1998;339:746–754. doi: 10.1056/NEJM199809103391107. [DOI] [PubMed] [Google Scholar]
- 4.Cazzola M., Skoda R.C. Translational pathophysiology: a novel molecular mechanism of human disease. Blood. 2000;95:3280–3288. [PubMed] [Google Scholar]
- 5.Ghilardi N., Skoda R.C. A single-base deletion in the thrombopoietin (TPO) gene causes familial essential thrombocythemia through a mechanism of more efficient translation of TPO mRNA. Blood. 1999;94:1480–1482. [PubMed] [Google Scholar]
- 6.Wiestner A., Schlemper R.J., van der Maas A.P., Skoda R.C. An activating splice donor mutation in the thrombopoietin gene causes hereditary thrombocythaemia. Nature Genet. 1998;18:49–52. doi: 10.1038/ng0198-49. [DOI] [PubMed] [Google Scholar]
- 7.Ghilardi N., Wiestner A., Kikuchi M., Ohsaka A., Skoda R.C. Hereditary thrombocythaemia in a Japanese family is caused by a novel point mutation in the thrombopoietin gene. Br. J. Haematol. 1999;107:310–316. doi: 10.1046/j.1365-2141.1999.01710.x. [DOI] [PubMed] [Google Scholar]
- 8.Maquat L.E., Carmichael G.G. Quality control of mRNA function. Cell. 2001;104:173–176. doi: 10.1016/s0092-8674(01)00202-1. [DOI] [PubMed] [Google Scholar]
- 9.Mendell J.T., Dietz H.C. When the message goes awry: disease-producing mutations that influence mRNA content and performance. Cell. 2001;107:411–414. doi: 10.1016/s0092-8674(01)00583-9. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J., Sun X., Qian Y., LaDuca J.P., Maquat L.E. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol. Cell. Biol. 1998;18:5272–5283. doi: 10.1128/mcb.18.9.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thermann R., Neu-Yilik G., Deters A., Frede U., Wehr K., Hagemeier C., Hentze M.W., Kulozik A.E. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–3494. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hentze M.W., Kulozik A.E. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 13.Mendell J.T., Sharifi N.A., Meyers J.L., Martinez-Murillo F., Dietz H.C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nature Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 14.He F., Li X., Spatrick P., Casillo R., Dong S., Jacobson A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell. 2003;12:1439–1452. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- 15.Le Hir H., Izaurralde E., Maquat L.E., Moore M.J. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dostie J., Dreyfuss G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr. Biol. 2002;12:1060–1067. doi: 10.1016/s0960-9822(02)00902-8. [DOI] [PubMed] [Google Scholar]
- 17.Schell T., Kulozik A.E., Hentze M.W. Integration of splicing, transport and translation to achieve mRNA quality control by the nonsense-mediated decay pathway. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-reviews1006. reviews1006.1–reviews1006.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagy E., Maquat L.E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 19.Carter M.S., Doskow J., Morris P., Li S., Nhim R.P., Sandstedt S., Wilkinson M.F. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 20.Oliveira C.C., McCarthy J.E. The relationship between eukaryotic translation and mRNA stability. A short upstream open reading frame strongly inhibits translational initiation and greatly accelerates mRNA degradation in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:8936–8943. doi: 10.1074/jbc.270.15.8936. [DOI] [PubMed] [Google Scholar]
- 21.Pinto I., Na J.G., Sherman F., Hampsey M. cis- and trans-acting suppressors of a translation initiation defect at the cyc1 locus of Saccharomyces cerevisiae. Genetics. 1992;132:97–112. doi: 10.1093/genetics/132.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruiz-Echevarria M.J., Peltz S.W. The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell. 2000;101:741–751. doi: 10.1016/s0092-8674(00)80886-7. [DOI] [PubMed] [Google Scholar]
- 23.Ruiz-Echevarria M.J., Gonzalez C.I., Peltz S.W. Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 1998;17:575–589. doi: 10.1093/emboj/17.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruiz-Echevarria M.J., Peltz S.W. Utilizing the GCN4 leader region to investigate the role of the sequence determinants in nonsense-mediated mRNA decay. EMBO J. 1996;15:2810–2819. [PMC free article] [PubMed] [Google Scholar]
- 25.Vilela C., Linz B., Rodrigues-Pousada C., McCarthy J.E. The yeast transcription factor genes YAP1 and YAP2 are subject to differential control at the levels of both translation and mRNA stability. Nucleic Acids Res. 1998;26:1150–1159. doi: 10.1093/nar/26.5.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vilela C., Ramirez C.V., Linz B., Rodrigues-Pousada C., McCarthy J.E. Post-termination ribosome interactions with the 5′-UTR modulate yeast mRNA stability. EMBO J. 1999;18:3139–3152. doi: 10.1093/emboj/18.11.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neu-Yilik G., Gehring N.H., Thermann R., Frede U., Hentze M.W., Kulozik A.E. Splicing and 3′ end formation in the definition of nonsense-mediated decay-competent human beta-globin mRNPs. EMBO J. 2001;20:532–540. doi: 10.1093/emboj/20.3.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J., Russell D.W. Molecular cloning: a laboratory manual. 3rd edn. Cold Spring Harbour, NY: Cold Spring Harbour Laboratory Press; 2001. [Google Scholar]
- 29.Gehring N.H., Neu-Yilik G., Schell T., Hentze M.W., Kulozik A.E. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell. 2003;11:939–949. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 30.Danckwardt S., Neu-Yilik G., Thermann R., Frede U., Hentze M.W., Kulozik A.E. Abnormally spliced beta-globin mRNAs: a single point mutation generates transcripts sensitive and insensitive to nonsense-mediated mRNA decay. Blood. 2002;99:1811–1816. doi: 10.1182/blood.v99.5.1811. [DOI] [PubMed] [Google Scholar]
- 31.Brocke K.S., Neu-Yilik G., Gehring N.H., Hentze M.W., Kulozik A.E. The human intronless melanocortin 4-receptor gene is NMD insensitive. Hum. Mol. Genet. 2002;11:331–335. doi: 10.1093/hmg/11.3.331. [DOI] [PubMed] [Google Scholar]
- 32.Mendell J.T., ap Rhys C.M., Dietz H.C. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science. 2002;298:419–422. doi: 10.1126/science.1074428. [DOI] [PubMed] [Google Scholar]
- 33.Gehring N.H., Kunz J.B., Neu-Yilik G., Breit S., Viegas M.H., Hentze M.W., Kulozik A.E. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol. Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 34.Inacio A., Silva A.L., Pinto J., Ji X., Morgado A., Almeida F., Faustino P., Lavinha J., Liebhaber S.A., Romao L. Nonsense mutations in close proximity to the initiation codon fail to trigger full nonsense-mediated mRNA decay. J. Biol. Chem. 2004 doi: 10.1074/jbc.M405024200. [DOI] [PubMed] [Google Scholar]
- 35.Perrin-Vidoz L., Sinilnikova O.M., Stoppa-Lyonnet D., Lenoir G.M., Mazoyer S. The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Hum. Mol. Genet. 2002;11:2805–2814. doi: 10.1093/hmg/11.23.2805. [DOI] [PubMed] [Google Scholar]
- 36.Carter M.S., Li S., Wilkinson M.F. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 1996;15:5965–5975. [PMC free article] [PubMed] [Google Scholar]
- 37.Morris D.R., Geballe A.P. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruiz-Echevarria M.J., Munshi R., Tomback J., Kinzy T.G., Peltz S.W. Characterization of a general stabilizer element that blocks deadenylation-dependent mRNA decay. J. Biol. Chem. 2001;276:30995–31003. doi: 10.1074/jbc.M010833200. [DOI] [PubMed] [Google Scholar]
- 40.Sureau A., Gattoni R., Dooghe Y., Stevenin J., Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.