Potassium channels are among the core features of life.1 They have been identified in organisms as diverse as archaebacteria and humans,2 occurring in virtually every eukaryotic and prokaryotic cell. Their ability to control the transmembrane-potential underpins elementary cellular functions such as excitability, proliferation, secretion, and volume regulation.
Recently, the three-dimensional structures of the prokaryotic potassium channels KcsA3 and MthK,4 as well as the voltage-gated channel KvAP,5 have been solved by X-ray crystallography. These crystal structures confirmed the remarkably conserved architecture of all potassium channels. As shown in Figure 1, four subunits assemble to form the central pore. Amino acid residues close to the pore line the so-called outer vestibule, a shallow depression on the extracellular surface of the channels.
Figure 1.
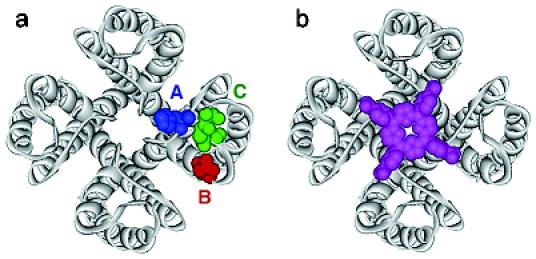
(a) X-ray structure of KcsA. The approximate location of “hot spot” residues A-C on the surface of potassium channels is indicated. (b) Overlay of KcsA with tetraphenylporphyrin 1 (magenta).
Due to the detailed structural information that is now available, potassium channels provide a unique platform with which to explore the principles of protein surface recognition.6 The interaction of ligands with channels can be determined in binding assays using radiolabeled peptide inhibitors7 or in functional assays involving radioactive 86Rb+ ions.8 Electrophysiological methods can also be used to investigate the blocking characteristics of ligands, either at the whole cell (voltage clamp) or single channel (patch clamp) level.
A large number of natural peptide inhibitors that bind to the pore region of these channels have been isolated from scorpions, snakes, spiders, and other organisms.9 In addition to this, a limited set of small-molecule blockers that bind in the same region of the channel, such as tetraethylammonium ion (TEA), have been identified. None of these inhibitors, however, make full use of the four-fold symmetry of a homotetrameric channel. Double-cycle mutant studies have identified residues in the channel's outer vestibule that are essential for high-affinity interaction of peptide inhibitors.10 Some of these “hot spot” residues A-C are highlighted in a sequence alignment of KcsA with several potassium channels of the Kv1x class. They are mapped onto the crystal structure of KcsA in Figure 1. Note that the residues shown in blue (A) and green (C) are highly variable. By contrast, the residue highlighted in red (B), aspartate or glutamate, is conserved within the Kv1x family. The conserved GYG signature sequence of potassium channels is highlighted in yellow.
We now report the development of a new class of synthetic ligands for potassium channels. These four-fold symmetrical molecules are designed to mimic the peptide toxins and to interact with all four channel subunits simultaneously, resulting in a strong polyvalency effect.11 They bind to voltage-gated potassium channels of the Kv1x class, such as Shaker and Kv1.3, with nanomolar affinities, and partially block the conductance of the channels in a reversible fashion.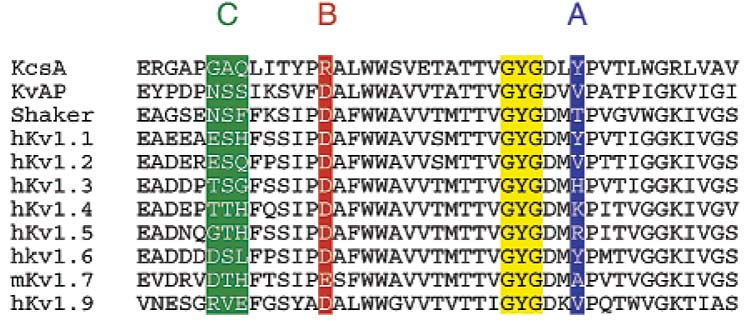
Our ligands are water-soluble tetraphenylporphyrin derivatives whose structures are shown below.12 The compounds were synthesized by coupling of tetraphenylporphyrin tetracarboxylic acid 1 or its acid chloride 2 with protected amino acids, peptides, or diamines, followed by deprotection (see Supporting Information). A similar molecular architecture has been used by Hamilton to recognize the surface of cytochrome C.13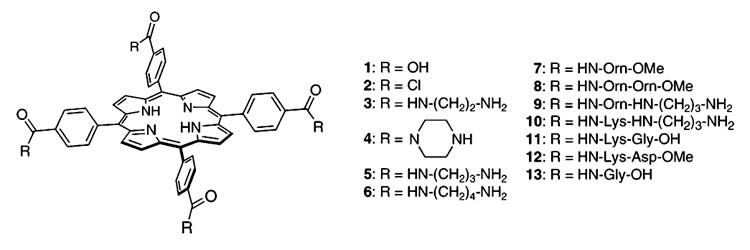
In aqueous solution near physiological pH, compounds 3-7 are, most likely, tetracations, compounds 8-10 would contain eight positively charged groups, whereas 11 and 12 are mostly neutral. Compound 13, on the other hand, contains four negatively charged side chains.
The interaction of the porphyrin ligands with potassium channels was investigated in competitive binding assays with 125I-hongotoxin1-A19Y/Y37F (125I-HgTX1A19Y/Y37F) and in electrophysiological assays using the Xenopus oocyte system. Figure 2 shows the effect of our ligands on 125I-HgTX1A19Y/Y37F binding to membranes prepared from HEK293 cells stably transfected with the human Kv1.3 channel. The voltage-gated potassium channel Kv1.3 plays a crucial role in human T-lymphocyte activation.14 125I-HgTX1A19Y/Y37F binds to the outer vestibule of Kv1.3 channels with femtomolar affinity (Kd) 0.046 pM), in a bimolecular, reversible reaction.15
Figure 2.
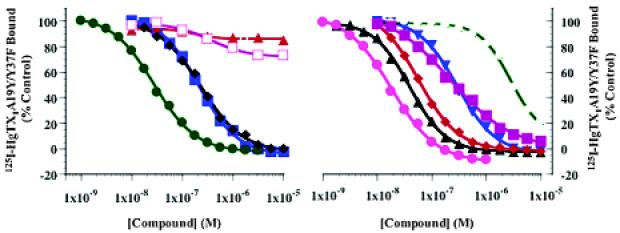
Binding of 125I-HgTX1A19Y/Y37F to Kv1.3 channels. Membranes prepared from HEK293 cells stably transfected with the Kv1.3 channel were incubated with ∼0.13 pM 125I-HgTX1A19Y/Y37F, in the absence or presence of increasing concentrations of (left panel) compound 3 (solid green circles), compound 4 (solid red triangles), compound 6 (solid black tilted squares), compound 7 (solid blue squares), compound 13 (open magenta squares); (right panel) compound 5 (solid blue triangles), compound 8 (solid black triangles), compound 9 (solid pink circles), compound 10 (solid red tilted squares), compound 11 (open green circles), compound 12 (solid magenta squares), for 20 h at room temperature. Inhibition of binding was assessed relative to an untreated control. Specific binding data can be fit to a single-site inhibition model, as described in the Supporting Information.
Ligands 3 and 5-12 inhibit 125I-HgTX1A19Y/Y37F, binding to Kv1.3 in a dose-dependent manner, whereas ligands 4 and 13 have no effect at concentrations up to 10 μM (Figure 2). The Ki values for inhibition of 125I-HgTX1A19Y/Y37F binding for these ligands are presented in Table 1.
Table 1.
Ki's for Ligands Based on Displacement Assays
| ligand | Ki[μM] | ligand | Ki[μM] | ligand | Ki[μM] |
|---|---|---|---|---|---|
| 3 | 0.020 | 7 | 0.144 | 10 | 0.038 |
| 5 | 0.138 | 8 | 0.026 | 11 | 1.918 |
| 6 | 0.157 | 9 | 0.013 | 12 | 0.156 |
Data from Figure 2 and Table 1 indicate that positively charged ligands with appropriate geometry, such as 3, 9, and 10, strongly interact with the Kv1.3 channel. According to our binding model, their cationic side chains could form salt bridges to the conserved anionic aspartate residues in position B (D381). By contrast, tetracationic ligand 4, which is a conformationally restricted and bulkier version of ligand 3, does not compete for 125I-HgTX1A19Y/Y37F binding to Kv1.3 channels. Moreover, negatively charged porphyrins such as 1 and its glycine derivative 13 do not interfere with 125I-HgTX1A19Y/Y37F binding. Note, that the overall neutral ligands 11 and 12 bind with markedly different affinities.
Further evidence for the proposed interactions between the ligands and the pore of the potassium channel was obtained from electrophysiological experiments. Ionic currents were measured in Xenopus oocytes that express Shaker, the archetypical Kv1x channel.16 The effect of two ligands on these currents is shown in Figure 3. Cationic porphyrin 3, one of the ligands to bind with the highest affinity, significantly inhibited the Shaker current, in a reversible fashion, whereas anionic porphyrins, such as 13, which lacked specific/high affinity binding, had virtually no effect. It is interesting to note that our ligands do not completely block ionic current through Shaker channels even at high concentrations. A similar observation concerning the lack of complete inhibition of currents has been observed for the peptide δ-dendrotoxin and the inward rectifier ROMK1 potassium channel.17
Figure 3.
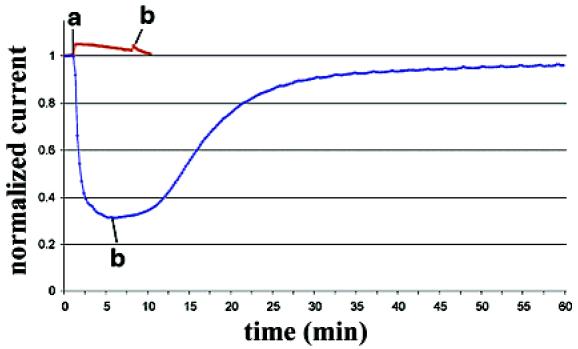
Effect of 2.5 μM ligand 3 (blue) and 100 μM ligand 13 (red) on Xenopus oocytes expressing Shaker channels. Ligand was applied at a. Perfusion with control solution began at b.
These results, taken together, provide strong evidence that our ligands bind in the outer vestibule of potassium channels. Whether they simultaneously interact with all four subunits of the tetrameric channel remains to be determined. The modular composition of our ligands allows easy modifications and should provide a large set of synthetic probes that discriminate among different potassium channels. Future directions will concern the synthesis of metalloporphyrins and fluorescent porphyrins with extended side chains. Ideally, our compounds could surpass antibodies by discriminating between closely related channels, such as the Kv1x channel subtypes, or different functional states. Ultimately, the development of this class of compounds may lead to novel therapeutic agents against various ailments such as autoimmune disorders, diabetes, epilepsy, or cardiac diseases.18
Supporting Information Available
Synthetic procedures and spectroscopic data for compounds 3-13 as well as experimental details concerning the 125I-HgTX1A19Y/Y37F binding and electrophysiological assays (PDF). This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgment
D.T. thanks NARSAD for a Young Investigator Award (Grant 16594) and the Hellman family for a Hellman Faculty Fund Award. E.Y.I. thanks the National Institutes of Health (R01NS35549) for financial support. S.N.G. thanks the Schering Research Foundation (Berlin) for a graduate fellowship. Financial support by Merck & Co is gratefully acknowledged.
References
- 1.(a) Hille B. Ion Channels of Excitable Membranes. 3rd Sinauer; Sunderland: 2001. [Google Scholar]; (b) Ashcroft FM. Ion Channels and Disease. Academic Press; San Diego: 2000. [Google Scholar]
- 2.Ruta V, Jiang Y, Lee A, Chen J, MacKinnon R. Nature. 2003;422:180. doi: 10.1038/nature01473. [DOI] [PubMed] [Google Scholar]
- 3.(a) Doyle DA, Cabral JM, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. Science. 1998;280:69. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]; (b) Zhou Y, Morals-Cabral JH, Kaufman A, MacKinnon R. Nature. 2001;414:43. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 4.Jiang YX, Lee A, Chen JY, Cadene M, Chait BT, MacKinnon R. Nature. 2002;417:515. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 5.(a) Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. Nature. 2003;423:33. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]; (b) Jiang Y, Ruta V, Chen J, Lee A, MacKinnon R. Nature. 2003;423:42. doi: 10.1038/nature01581. [DOI] [PubMed] [Google Scholar]
- 6.Peczuh MW, Hamilton AD. Chem. Rev. 2000;100:2479. doi: 10.1021/cr9900026. [DOI] [PubMed] [Google Scholar]
- 7.Garcia ML, Hanner M, Knaus H-G, Slaughter RS, Kaczorowski GJ. Methods and Tools in Biosciences and Medicine. Birkhäuser; Basel: 2000. [Google Scholar]
- 8.Felix JP, Bugianesi RM, Schmalhofer WA, Borris R, Goetz MA, Hensens OD, Bao J-M, Kayser F, Parsons WH, Rupprecht K, Garcia ML, Kaczorowski GJ, Slaughter RS. Biochemistry. 1999;38:4922. doi: 10.1021/bi982954w. [DOI] [PubMed] [Google Scholar]
- 9.(a) Garcia ML, Hanner M, Kaczorowski GJ. Toxicon. 1998;36:1641. doi: 10.1016/s0041-0101(98)00157-3. [DOI] [PubMed] [Google Scholar]; (b) Garcia ML, Gao YD, McManus OB, Kaczorowski GJ. Toxicon. 2001;39:739. doi: 10.1016/s0041-0101(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 10.(a) MacKinnon R, Cohen SL, Kuo A, Lee A, Chait BT. Science. 1998;280:106. doi: 10.1126/science.280.5360.106. [DOI] [PubMed] [Google Scholar]; (b) Imredy JP, MacKinnon R. J. Mol. Biol. 2000;296:1283. doi: 10.1006/jmbi.2000.3522. [DOI] [PubMed] [Google Scholar]
- 11.Mammen M, Choi SK, Whitesides GM. Angew. Chem., Int. Ed. 1998;37:2755. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 12.Hambright P. In: The Porphyrin Handbook. Kadish KM, Guilard R, Smith KM, editors. Vol. 3. Academic Press; San Diego: 2000. [Google Scholar]
- 13.Jain RK, Hamilton AD. Org. Lett. 2000;2:1721. doi: 10.1021/ol005871s. [DOI] [PubMed] [Google Scholar]
- 14.Kaczorowski GJ, Garcia ML. Curr. Opin. Chem. Biol. 1999;3:448. doi: 10.1016/S1367-5931(99)80066-0. [DOI] [PubMed] [Google Scholar]
- 15.Koschak A, Bugianesi RM, Mitterdorfer J, Kaczorowski GJ, Garcia ML, Knaus H-G. J. Biol. Chem. 1998;273:2639. doi: 10.1074/jbc.273.5.2639. [DOI] [PubMed] [Google Scholar]
- 16.Gross A, Abramson T, MacKinnon R. Neuron. 1994;13:961. doi: 10.1016/0896-6273(94)90261-5. [DOI] [PubMed] [Google Scholar]
- 17.Imredy JP, Chen C, MacKinnon R. Biochemistry. 1998;37:14867. doi: 10.1021/bi980929k. [DOI] [PubMed] [Google Scholar]
- 18.Coghlan MJ, Carroll WA, Gopalakrishnan M. J. Med. Chem. 2001;44:1627. doi: 10.1021/jm000484+. [DOI] [PubMed] [Google Scholar]


